Abstract
Many denitrifying organisms contain the norEF gene cluster, which codes for two proteins that are thought to be involved in denitrification because they are expressed during the reduction of nitrite and nitric oxide. The products of both genes are predicted to be membrane associated, and the norE product is a member of the cytochrome c oxidase subunit III family. However, the specific role of norEF is unknown. The denitrification phenotypes of Rhodobacter sphaeroides strains with and without norEF genes were studied, and it was found that loss of norEF lowered the rate of denitrification from nitrate and resulted in accumulation of micromolar concentrations of nitric oxide during denitrification from nitrite. norEF appears to have no direct role in the reduction of nitric oxide; however, since deletion of norEF in the wild-type 2.4.3 strain had essentially no influence on the kinetics of potential nitric oxide reduction (Vmax and Ks), as measured by monitoring the depletion of a bolus of nitric oxide injected into anoxic cultures without any other electron acceptors. However, norEF-deficient cells that had undergone a more chronic exposure to micromolar concentrations of nitric oxide showed an ∼50% reduction in Vmax but no change in apparent Ks. These results can explain the occurrence of norEF in the 2.4.3 strain of R. sphaeroides, which can reduce nitrate to nitrous oxide, and their absence from strains such as 2.4.1, which likely use nitric oxide reductase to mitigate stress due to episodic exposure to nitric oxide from exogenous sources.
INTRODUCTION
Heme-copper oxygen reductases (HCO) are a large family of proteins that serve as terminal enzymes in electron transport chains. The aa3-type cytochrome oxidases of bacteria and mitochondria are the most well-characterized HCO. Recently, it was proposed that the HCO family be subdivided into types A, B, and C, along with nitric oxide reductase (Nor) as a fourth type (1). The A to C types all reduce oxygen (O2) to water, while Nor reduces nitric oxide (NO) to nitrous oxide. All of these proteins share an active-site binuclear center consisting of a high-spin heme and a nonheme metal which is contained within a protein designated subunit I (2). In the aa3-type HCO, subunit I and two other subunits form the conserved enzymatic core (3). Subunit II contains a copper center, while subunit III lacks cofactors. While they are essential for aa3 activity, subunit II and III family proteins are not defining subunits of the HCO family, since some members lack orthologs of these proteins. For example, the bacterial cbb3-type oxidase complex consists of subunit I and two additional subunits that contain c-type hemes (4).
While these additional subunits are not orthologous across HCO families, most are required to transfer electrons from extramembranous sources to the binuclear center (3). However, the absence of cofactors from members of the subunit III family makes their role in the active HCO complex harder to establish. Experiments measuring the activity of subunit III-depleted aa3-type oxidase have found that the enzyme is rapidly inactivated through a process termed suicide inactivation (5). The mechanism underlying suicide inactivation is unclear, but its occurrence does provide evidence that subunit III, like subunit II, is critical for HCO function.
Nor is the only type of HCO for which O2 is not the primary substrate, and it is also distinct in having a nonheme iron at the active site (6, 7). There are two Nor subtypes, with one, cNor, oxidizing cytochrome c and the other, qNor, oxidizing quinol. Purified cNor contains two subunits; a subunit I and a second c-heme-containing subunit that is used to transfer electrons to the active site (8). There is evidence suggesting subunit III orthologs also play a role in cNor function. Analysis of the cNor gene cluster in many denitrifiers has identified a gene, frequently designated norE, encoding a protein that is a member of the subunit III family (10). In most denitrifiers, norE is immediately upstream of a second gene that has been designated norF (11, 12). norF encodes a protein that is also likely membrane associated. Inactivation of the norEF cluster has been shown to slow nitrogen oxide reduction in both Paracoccus denitrificans and Rhodobacter sphaeroides 2.4.3 (11, 12). Complementation experiments have shown that the phenotype of a norEF deletion cannot be restored by norE alone but can be restored with norEF together, indicating both NorE and NorF are involved in nitrogen oxide reduction (12). Orthologs of norEF are not typically found in denitrifiers with qNor.
While most bacteria with cNor have norEF, there are examples of denitrifiers lacking this gene pair. For example, there are currently five R. sphaeroides strains with sequenced genomes, and all five have the norCBQD cluster. However, strain 2.4.3 is the only one of these five that contains the norEF gene pair (12). Strain 2.4.3 also has two periplasmic nitrate reductases (Nap) and a copper-containing nitrite reductase (Nir) (13). One of the two Nap proteins, referred to as Nap-β, is expressed under hypoxic conditions, and this activity, coupled with Nir and Nor, allows 2.4.3 to reduce nitrate to nitrous oxide. This strain also contains the genes for nitrous oxide reductase (Nos), but an apparent frameshift in nosR prevents expression of an active Nos (12). Since it has been demonstrated that loss of NorE and NorF reduces the rate of denitrification, it is surprising there is no positive selection for norEF in these other strains. This indicates that, despite previous observations, loss of the norEF products does not actually lead to a significant change in the cells' ability to reduce nitrogen oxides, and that NorEF are required for some function indirectly related to N-oxide reduction. It is also possible the norEF products play a critical role during nitrogen oxide reduction, and the strains lacking these genes have adapted to their loss. To better define the role of NorEF specifically and the subunit III family more generally, experiments were undertaken to directly establish how inactivation of norEF impacts N-oxide reduction in a strain having these genes. Experiments were also carried out to determine how addition of norEF affected nitrogen oxide reduction in a strain that naturally lacks these genes.
MATERIALS AND METHODS
Strains and media.
The denitrifying strain 2.4.3 (ATCC 17025) of Rhodobacter sphaeroides was used in this study along with the partial denitrifier strain 2.4.1 (ATCC 17023). When necessary, the following antibiotics and concentrations were used: tetracycline, 1.0 μg/ml; streptomycin, 50 μg/ml; and gentamicin, 20 μg/ml. The 2.4.3 ΔnorEF, 2.4.1 pAK1 (2.4.1 supplemented with nirK from 2.4.3), 2.4.1 pAK1 + norEF, and 2.4.3 Nap-β-deficient strains have been previously described (12, 13). Taxis assays were carried out essentially as described previously (12). In short, cells were resuspended in 0.4% agar, and a 2% agar plug containing the nitrite source was inserted into the center of the plate. Incubations of resuspended cells were done under an N2 atmosphere.
Gas measurements.
Detailed profiles of denitrification phenotypes were obtained by headspace gas measurements of batch cultures during the transition from oxic to anoxic respiration. All incubations were run at 30°C in Sistrom's medium (14), and liquid cultures were continuously stirred at 600 rpm to maximize transport rates of gases between headspace and liquid.
Cultures were raised on plates from frozen glycerol stocks before they were transferred to liquid medium and allowed to grow aerobically to an optical density at 600 nm of ∼0.5 to 1.5. Depending on cell density, 0.5 to 1.5 ml of culture was transferred to experimental vials to reach an initial optical density (OD) of ∼0.02. These 120-ml serum vials contained 50 ml Sistrom's medium and 70 ml headspace, and they were supplemented with various concentrations of KNO2 or KNO3 in the low-mM range (specific concentrations are reported in Results). Vials containing sterile medium were sealed with rubber septa and aluminum caps and made anoxic (residual O2, <150 μl liter−1) through repeated cycles of evacuation and He filling. Pure O2 was injected into some of the vials for experiments where the transitions from oxic to anoxic respiration by O2 depletion were to be measured. Remaining overpressure was released by piercing the septum with a needle coupled to a syringe with no piston but containing water to monitor gas release. The washing and injection of O2 were done prior to inoculation of the vials.
Transitions from oxic to anoxic respiration were monitored using a semiautomatic system consisting of a thermostatic water bath holding 15 stirred vials and an autosampler connected to a Varian CP 4700 micro-gas chromatograph with a 10 m poraPLOT U and a 20 m molesieve 5 A column (in parallel), each equipped with a thermal conductivity detector (TCD), and a chemiluminescence NOx analyzer (model 200A; Advanced Pollution Instrumentation, USA). This system allows frequent measurements of O2, CO2, NO, N2O, N2, and CH4 in the headspace of soil samples, slurries, or cultures. The system is described in detail by Molstad et al. (15), who also describe the routines for calculating net production/consumption of gases, which necessitates a correction for sampling loss. Routines have also been developed for calculating O2 concentration in the liquid based on measured transport rates, since O2 in the liquid is not in equilibrium with headspace concentrations. This is essential when analyzing kinetics of respiration as a function of O2 concentrations in the liquid and for NO reduction as a function of NO concentrations in the liquid. The system has been used for similar analyses of the transition from oxic to anoxic respiration in other bacteria (16–18). The measured rates of O2 consumption were routinely converted to electron flow rates (4 mol electrons per mol O2 consumed) to be compared with the anoxic electron flow rates (calculated from the rates of NO2− reduction to NO and NO reduction to N2O (1 mol electron per mol N). In an experiment with NO3− in the medium, nitrite accumulation was assumed to be negligible (19); hence, 3 electrons per mol NO produced was assumed.
To test every strain's ability to denitrify, aerobically grown cells of 2.4.3 wild-type, ΔnorEF, Δ-napβ, 2.4.1 pAK1, and 2.4.1 pAK1 + ΔnorEF strains were transferred to vials with 0 or 10 ml liter−1 O2 in the headspace and 1 mM nitrate or nitrite. The rates of oxic and anoxic respiration were monitored by frequent sampling of the headspace for determination of O2, NO, N2O, and N2. Every treatment was run in duplicate or triplicate for each strain.
Reduction of NO added to the headspace was measured in wild-type 2.4.3 and the ΔnorEF mutant by initially growing cultures with 1% O2 and 0.1 mM KNO3. Headspace gases were monitored while the cultures were allowed to deplete the O2 and subsequently deplete nitrate by anoxic respiration. When nitrate had been depleted, as seen by 100% recovery of nitrate-N as N2O, NO was injected into the headspace to reach ∼300 to 4,500 nM in the liquid, and the rate of NO reduction was monitored by frequent sampling (every 9 to 17 min). The analytical approach is explained in more detail in Fig. S1 in the supplemental material. The observed rates were used to estimate the maximum respiration rates (Vmax; mol NO cell−1 min−1) and apparent half-saturation constant (Ks; nM NO) for the 2.4.3 wild-type and ΔnorEF strains.
Nitrite sensitivity of O2 respiration and denitrification in 2.4.3 wild-type and ΔnorEF strains used cultures containing 10 ml liter−1 O2 and 0.5, 1, 2, 5, and 10 mM nitrite, and gas levels were monitored during the transition to anoxia. An additional experiment with 2% initial O2 and 0.5, 1, 2, and 4 mM nitrite was conducted using the wild type and the norEF-deficient strain. To test if 1 mM nitrite would cause permanent damage to the cells' ability to respire nitrite, exposed cells were harvested by centrifugation (10,000 × g, 10 min, 4°C) and then washed twice in nitrate-free Sistrom's medium to remove residual nitrite. These cells were tested for oxic respiration and also for the ability to reduce NO, as described above. The initial OD600 of the cells in these vials was ∼0.005, which equaled 0.09 mg cell dry weight (dw) per flask.
RESULTS
Oxic respiration comparison.
Inspection of the O2 depletion curves and estimations of Vmax and apparent Ks for oxic respiration showed that the oxic respiration rates were practically identical for wild-type 2.4.3, ΔnorEF, and 2.4.1 pAK1 strains (see Fig. S2 and S3 in the supplemental material). The estimated maximum respiration rate (Vmax; mmol O2 g−1 cell dry weight h−1) was 30 (standard error [SE], 5.9) and 27 (SE, 3.4) for 2.4.3 and ΔnorEF strains, respectively. The apparent half-saturation constant, Ks (μM O2), was 18 (SE, 5.2) and 14 (SE, 2.7) for 2.4.3 and ΔnorEF strains, respectively. Very similar results were obtained for the pAK1 strain, although somewhat more variable results precluded parameter estimations.
Denitrification in 2.4.3 and 2.4.3 ΔnorEF.
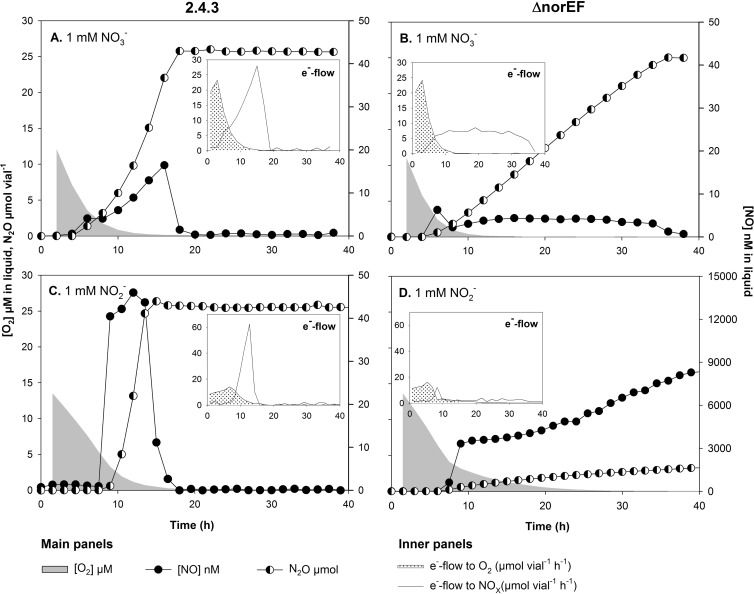
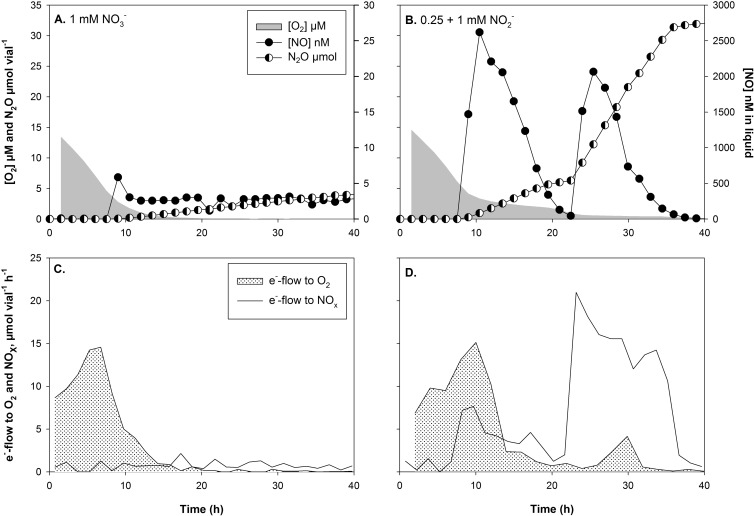
To evaluate the importance of NorEF in N-oxide respiration, the denitrification phenotype of wild-type 2.4.3 was compared to that of 2.4.3 ΔnorEF by a series of incubation experiments with 0 or 1% initial O2 and with either nitrate or nitrite in the medium. As can be seen in Fig. 1, inactivation of norEF in 2.4.3 dramatically changed the denitrification phenotype. In the treatments with 1 mM nitrate, 2.4.3 ΔnorEF showed a much lower rate of N2O production than the wild type, and the rate was almost constant until nitrate depletion, whereas the wild type showed an exponentially increasing rate of N2O production. As expected, N2O is the final product of denitrification in 2.4.3 (12). Nitrite was not measured in these experiments, but previous work has shown that nitrite is undetectable by the standard assay when 2.4.3 is denitrifying from nitrate (19).
FIG 1.
Respiration profiles during the switch from oxic to anoxic conditions in batch cultures of 2.4.3 with norEF intact (A and C) and 2.4.3 ΔnorEF (B and D) with 1% initial O2 and 1 mM nitrate (A and B) or nitrite (C and D). The main graphs show O2 reduction (μM in liquid), NO (nM in liquid), and N2O (μmol vial−1) accumulation. The insets show electron flow (μmol e− vial−1 h−1) to O2 or NOx assuming no or negligible nitrite accumulation in nitrate-treated cultures.
In the presence of 1 mM nitrite, there was an even more dramatic effect of the deletion of norEF. Three repeats of this experiment were conducted, and in two of these incubations all of the replicate vials with the ΔnorEF mutant and 1 mM nitrite showed a sudden accumulation of micromolar concentrations of NO (Fig. 1). To ease the interpretation of the gas data and facilitate comparison of respiratory metabolism through the transition from oxic to anoxic respiration, electron flow (μmol e− vial−1 h−1) to the terminal electron acceptors O2, nitrite, and NO, assuming no or negligible transient nitrite accumulation in nitrate-supplemented cultures, were estimated (Fig. 1, insets). These graphs show that the 2.4.3 ΔnorEF strain experiencing runaway NO production had severely restricted anoxic respiration rates compared to the 2.4.3 wild type, and the rates did not increase with time.
A closer inspection of the constant versus exponentially increasing rates of N2O production is shown in Fig. S4 in the supplemental material. This figure demonstrates that the rate of N2O production by the 2.4.3 wild type is clearly exponential, with an r2 of >0.99 for exponential functions fitted to data and apparent specific growth rates (μ) ranging from 0.09 to 0.21 h−1. In contrast, the rate of N2O production in the ΔnorEF strain was more or less constant and in some cases decreased gradually, long before depletion of nitrate. The one exception was the ΔnorEF strain grown in vials with 0 vol% initial O2 (with a µ of ~0.028 and an r2 of 0.97 to 0.98). The experiment illustrated in Fig. 1 was run for all of the strains, and these results are summarized in Table 1.
TABLE 1.
Phenotypic characteristics of R. sphaeroides cultures in Sistrom's medium supplied with 1% initial O2 and 1 mM nitrite or nitrate during 40 h of incubation at 30°Ca
| Strain | Added NOx | Initial [O2] (%) | [NO]max (nM) | Δtstart-N depl (h) | μNOx (h−1) | End FN2O |
|---|---|---|---|---|---|---|
| 2.4.3 | NO3− | 0 | 276 ± 44 | 27 ± 1 | 0.09 ± 0.03 | 1 |
| 1 | 16 ± 1 | 18 ± 2 | 0.16 ± 0.05 | 1 | ||
| NO2− | 0 | 75 ± 3 | 13 ± 2 | >0d | 1 | |
| 1 | 38 ± 1 | 15 ± 2 | >0d | 1 | ||
| 2.4.3 ΔnorEF | NO3− | 0 | 823 ± 240 | ∼40 | 0.027 ± 0.002 | 1 |
| 1 | 5 ± 1 | 35 ± 2 | ≤0 | 1 | ||
| NO2− | 0 | 5,846 ± 4,211 | >40 | ≤0 | 0.057 ± 0.065 | |
| 1.5 | 4,179 ± 3,821 | >40 | ≤0 | 0.192 ± 0.139 | ||
| 2.4.1 pAK1b | NO3− | 0 | 245 ± 9 | >40 | ≤0 | 0.158 ± 0.008 |
| 1 | 2 ± 1 | >40 | ≤0 | 0.523 ± 0.069 | ||
| NO2− | 0 | 14,745 ± 1,011 | >40 | ≤0 | 0.004 ± 0.001 | |
| 1 | 3,230 ± 1,604 | >40 | ≤0 | 0.012 ± 0.002 | ||
| 2.4.1 pAK1 + norEFb | NO3−c | 1 | 37 ± 44 | >40 | ≤0 | 0.157 ± 0.013 |
| NO2−c | 1 | 236 ± 85 | 27 ± 1 | 0.06 ± 0.02 | 1 |
[NO]max is the maximum measured concentration of either transiently accumulated NO or NO still increasing at the end of an incubation. Δtstart-N depl is the time from inoculation to nitrate or nitrite depletion (100% recovery as N2O). μNOx is apparent anoxic growth rates based on electron flow kinetics. R. sphaeroides usually initiates denitrification long before O2 is depleted; thus, μNOx as estimated represents a combination of anoxic growth and recruitment of cells to denitrification while still respiring O2. μNOx of ≤0 indicates nearly constant or slightly declining rates of denitrification after O2 depletion. End FN2O is the fraction of added NOx recovered as N2O at t = 40 h. The values given are averages from 2 or 3 replicates ± standard deviations.
norEF deficient.
Two mM initial nitrate or nitrite.
Depletion was too fast to estimate growth rate (Fig. 1).
N oxide reduction in 2.4.1.
Since the 2.4.1 strain naturally lacks norEF, it is of interest to determine if their absence affects N oxide respiration, as seen in the ΔnorEF mutant of 2.4.3. To permit direct comparison to 2.4.3 activity, the nitrite reductase deficiency in 2.4.1 was overcome by introducing nirK from strain 2.4.3 in trans. The 2.4.1 strain with nirK, designated 2.4.1 pAK1, showed a phenotype comparable to that of 2.4.3 ΔnorEF. Cultures treated with nitrate accumulated nM concentrations of NO transiently spiking to >100 nM in 0% O2 treatments, followed by a slow, linear accumulation of N2O (Table 1). Unlike in the corresponding 2.4.3 ΔnorEF cultures, not all of the available N oxide had been reduced to N2O by the end of the incubation.
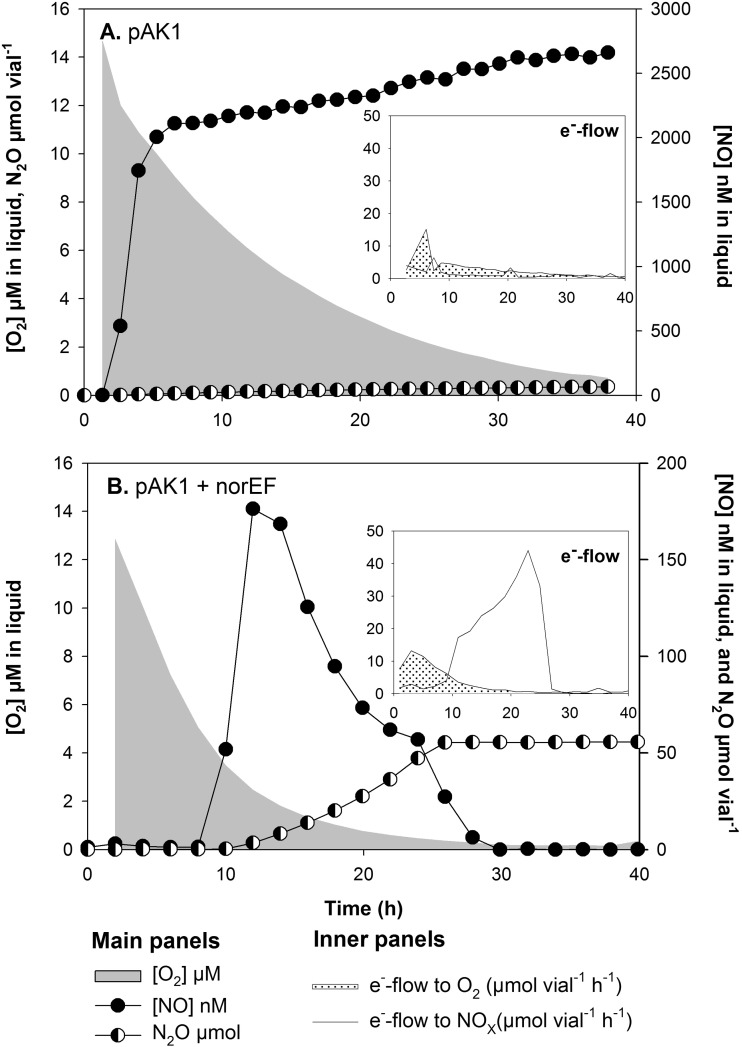
When treated with nitrite, 2.4.1 pAK1 showed a more pronounced respiratory inhibition than that seen in 2.4.3ΔnorEF. In cultures with 1 mM nitrite and 1% initial O2, this was reflected by a slowing of the aerobic respiration rate due to excessive NO accumulation and minimal recovery of nitrite-N as N2O within 40 h of incubation (Fig. 2A and Table 1). The 2.4.1 pAK1 culture with 0% initial O2 accumulated the highest levels of NO of any strain while having the lowest rate of denitrification.
FIG 2.
O2 depletion, NO and N2O accumulation, and electron flow to O2 and NOx in 2.4.1 pAK1 and 2.4.1 pAK1 + norEF when treated with 1% initial O2 and 1 or 2 mM nitrite, respectively. (A) Arrest in denitrification and accumulation of μM concentrations of NO in 2.4.1 pAK1. (B) Restoration of Nor function (as seen by the 100% recovery of added nitrite as N2O) in 2.4.1 pAK1 upon the inclusion of 2.4.3-derived norEF.
2.4.1 pAK1 + norEF.
To confirm that the limited denitrification capacity of 2.4.1 pAK1 is attributable to the absence of norEF, a plasmid containing the genes from 2.4.3 was mobilized into 2.4.1 pAK1. A treatment starting with 1% O2 and 2 mM nitrite was chosen for this experiment, since this level of nitrite was readily reduced by 2.4.3, while in both 2.4.3 ΔnorEF and 2.4.1 pAK1 similar or slightly lower levels of nitrite led to limited N2O production and μM levels of NO accumulation (Table 1 and Fig. 1C and D and 2A). A striking increase in nitrite reduction capacity was observed in 2.4.1 pAK1 + norEF to the point that this strain's respiration profile in 2 mM initial nitrite and 1% O2 was comparable to that of the 2.4.3 wild type in cultures. During nitrite respiration, NO was kept within the nM range, and anoxic growth was evident both by an exponential N2O accumulation rate and a corresponding exponential increase in electron flow to NOx as O2 approached zero (Fig. 2B).
Nitrite tolerance of 2.4.3 and 2.4.3 ΔnorEF.
In previous taxis experiments with 2.4.3 ΔnorEF, it was observed that the mutant was still able to form a ring similar to that of the wild type, but the distance from the ring to the nitrite source was greater (12). This suggests that the respiratory disruption caused by nitrite is not absolute but proportional to its concentration. To test this hypothesis, gas-phase analyses were run with a range of initial nitrite concentrations. In this case, initial O2 levels were 1% in all treatments but the nitrite concentrations were varied (Table 2). Wild-type 2.4.3 rapidly reduced 0.5, 1, and 2 mM nitrite to equivalent amounts of N2O while keeping NO in the low-nM range. Respiration of higher concentrations of nitrite was problematic. At 5 mM nitrite, only 50% of the cultures made a balanced transition to denitrification (indicated by +− in Table 2), while 10 mM nitrite resulted in a general respiratory inhibition of all cultures. Balanced transition refers to a situation where a batch culture switches from aerobic respiration to denitrification without runaway NO causing arrest and with 100% recovery of available NOx as N2O.
TABLE 2.
Nitrite sensitivity test in 2.4.3 wild-type and ΔnorEF mutant strainsa
| Initial [NO2−] (mM) | Nitrite sensitivity of strainb: |
|
|---|---|---|
| 2.4.3 | 2.4.3 ΔnorEF | |
| 0.5 | ++ | ++ |
| 1 | ++ | −(+) |
| 2 | ++ | −− |
| 5 | +− | −− |
| 10 | −− | −− |
Cultures (n = 2 to 7) were treated with ∼1% initial O2 and 0.5, 1, 2, 5, or 10 mM nitrite.
++, Balanced transition to denitrification, all of the available nitrite reduced to N2O. −−, nitrite toxicity and arrest reflected either by the accumulation of μM concentrations of NO or a more general inhibition of oxic respiration and denitrification. +−, 1 out of 2 tested cultures reduced the available nitrite to N2O. −(+), 3 out of 7 tested cultures reduced the available nitrite to N2O.
As expected, 2.4.3 ΔnorEF exhibited respiratory sensitivity to nitrite different from that of wild-type 2.4.3. The ΔnorEF mutant had an apparent critical threshold at 1 mM nitrite, where 4 out of 7 cultures accumulated μM concentrations of NO, followed by an arrest in subsequent denitrification. Higher concentrations of nitrite inhibited respiration in all cultures (Table 2). 2.4.1 pAK1 was even more sensitive to nitrite and was unable to perform a balanced transition to nitrite respiration at nitrite concentrations as low as 0.25 mM (data not shown).
To confirm that loss of norEF did not impact the nitrite sensitivity of oxic respiration, O2 respiration was compared in strains with and without these genes. In this experiment, the initial O2 level was 2% in all treatments while the nitrite concentrations were varied. The respiration rates of the wild type and the ΔnorEF mutant showed no statistically significant difference. In both strains the respiration rate declined linearly with increasing nitrite concentration, with a 16 to 20% reduction per mM nitrite. The respiration kinetics are shown in more detail in Fig. S5 and S6 in the supplemental material.
Taxis experiments.
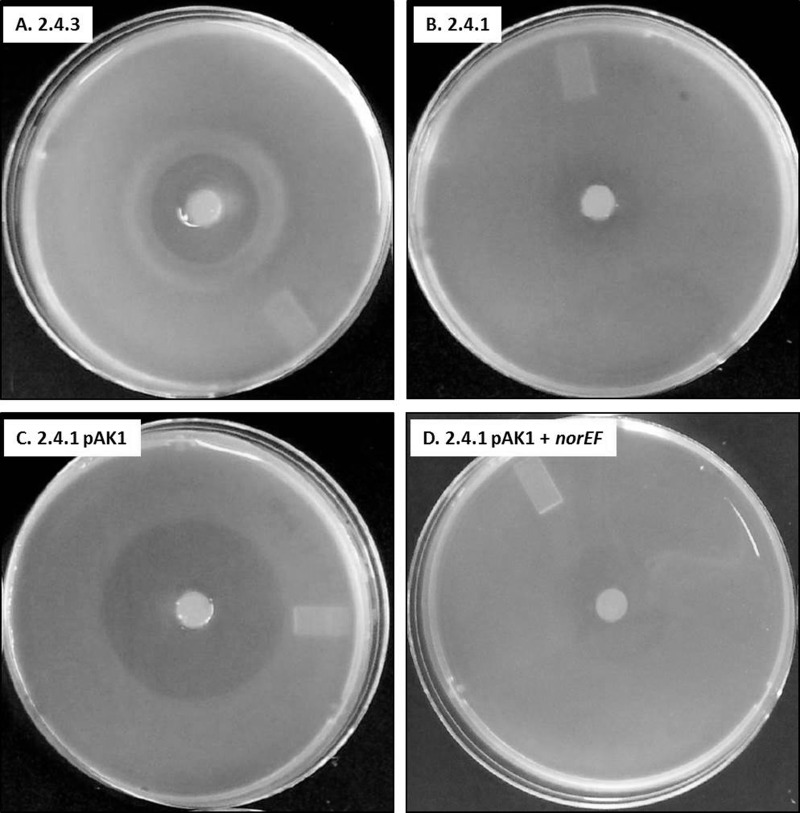
Gas measurements suggest that the observed differences in the taxis of 2.4.3 and 2.4.3 ΔnorEF to nitrite are due to uncontrolled NO production (compare Fig. 1C to D). 2.4.1 pAK1 also showed runaway NO production in the presence of nitrite (Fig. 2A). The latter result is inconsistent with previous results which found that 2.4.1 pAK1 showed a taxis response to nitrite similar to that of wild-type 2.4.3 (20). Therefore, taxis experiments were rerun using the 2.4.1 pAK1 strain which had been constructed for this work. This 2.4.1 pAK1 strain showed a large area of clearing around the nitrite but did not form the tight ring of cells seen in 2.4.3 (Fig. 3). Wild-type 2.4.1 showed only a slight clearing around the nitrite source, which is consistent with its lack of nirK. Therefore, it seems likely that the previous results with 2.4.1 containing nirK were actually obtained using a 2.4.3 strain. As expected from the gas measurements, introduction of norEF to 2.4.1 pAK1 greatly reduced the zone of clearing around the nitrite source, but there was no ring of cells like that seen in 2.4.3 (Fig. 3).
FIG 3.
Taxis response of wild-type 2.4.3, wild-type 2.4.1, 2.4.1 with nirK (pAK1), or 2.4.1 with nirK and norEF (pAK1 + norEF) to a source of nitrite. The nitrite plug is visible in the center of the plate. Plates had been incubated under hypoxic conditions for 18 h. The thick band in the 2.4.3 plate is an accumulation of cells which does not occur in the 2.4.1 strains.
In vivo assay of NO reduction.
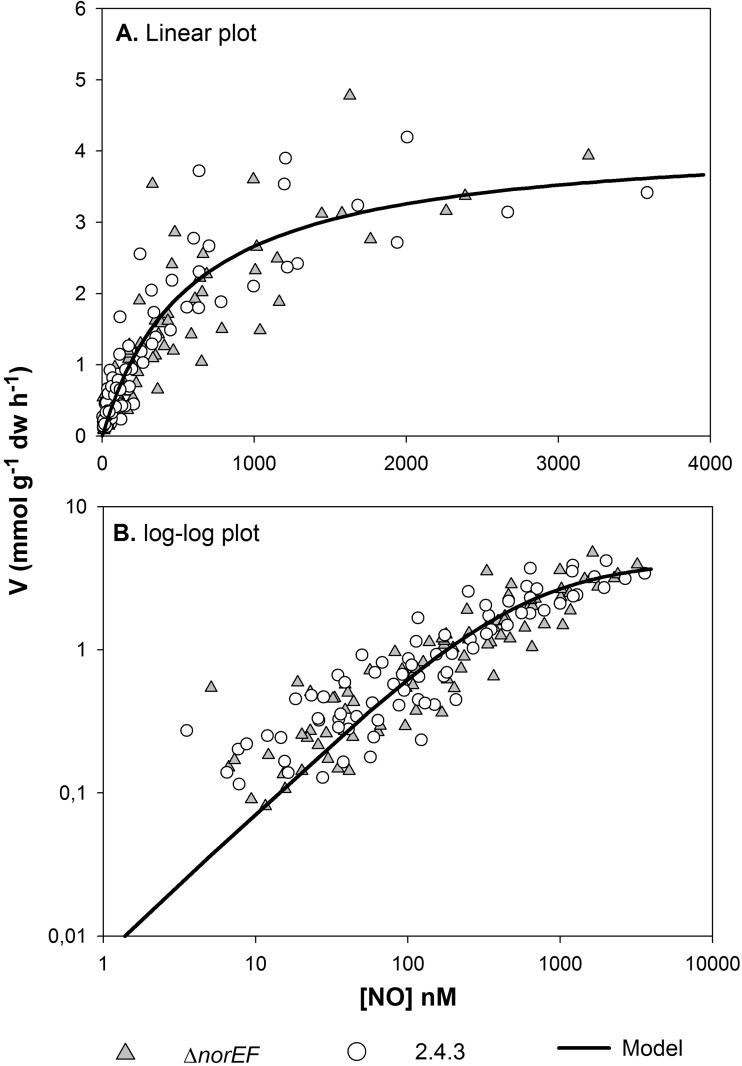
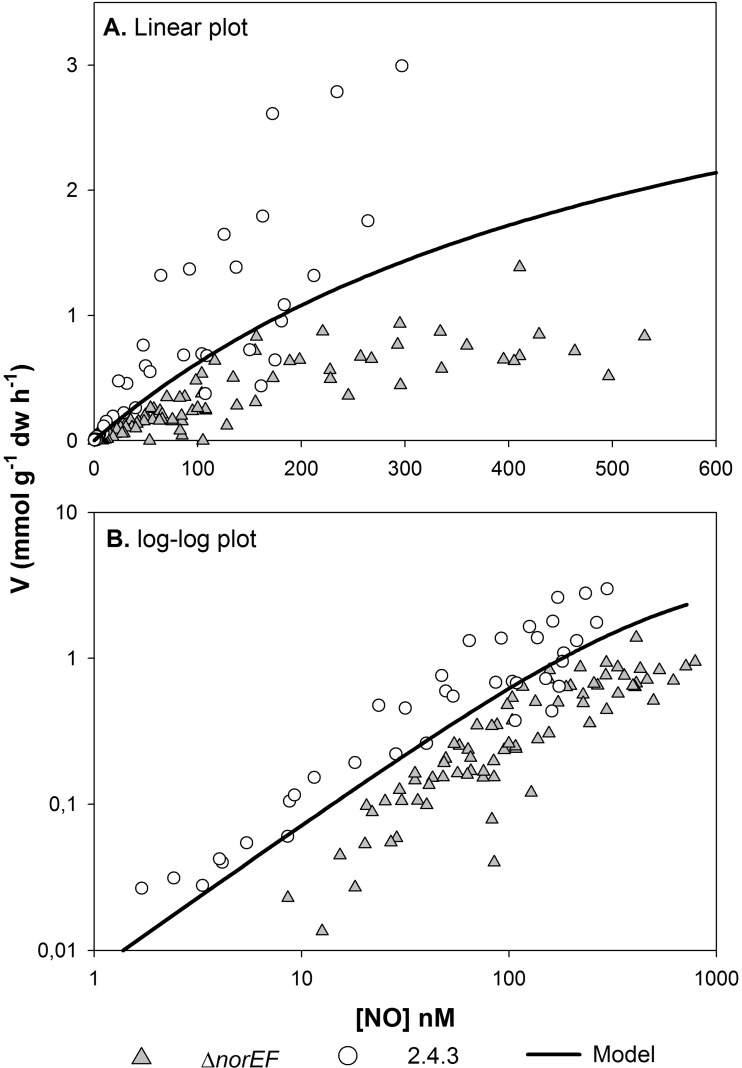
The most obvious difference in nitrogen oxide respiration between wild-type 2.4.3 and the norEF-negative strains is the excessive accumulation of NO in the presence of nitrite by the latter. This suggests that norEF is necessary to sustain high Nor activity. To test this more directly, the ability of wild-type 2.4.3 and 2.4.3 ΔnorEF to reduce NO was compared. Initially, cells of wild-type and ΔnorEF 2.4.3 were grown under hypoxic conditions in the presence of 0.1 mM nitrate to induce expression of Nor. After depletion of nitrite, several rounds of NO, ranging from ∼300 to 4,500 nM, were injected, and the rates of NO reduction were monitored by frequent analyses of the headspace concentration (every 9 to 17 min) until NO was depleted. A typical result and the analytic approach for analyzing the data are provided in Fig. S1 in the supplemental material. The rates were clearly concentration dependent and could be described by standard enzyme kinetics, i.e., V = Vmax × [NO]/([NO] + Ks), where V is the rate of NO reduction, Vmax is the estimated maximum rate, and Ks is the apparent half saturation concentration (nM NO in the liquid). Figure 4 shows measured rates plotted against NO concentrations in the liquid for the 2.4.3 wild type and ΔnorEF mutant (combining data for 5 replicate vials for each strain). The kinetic parameters for each strain were individually estimated by nonlinear regression. For wild-type 2.4.3, the estimates were a Vmax of 4.1 mmol g−1 dw h−1 (3.6 to 4.7) and Ks of 509 nM (369 to 707) (95% confidence limits are in parentheses; n = 68). For the ΔnorEF mutant, the estimates were a Vmax of 4.3 mmol g−1 dw h−1 (3.6 to 5.1) and Ks of 644 nM (445 to 938). The differences between the two strains were not statistically significant. Thus, when grown in nitrate medium, norEF has no obvious effect on the cell's capacity to reduce NO regarding either the amount of active NO reductase or the apparent substrate affinity (Ks).
FIG 4.
Kinetics of NO reduction in the 2.4.3 wild type versus the ΔnorEF mutant. (A) Rates (mmol g−1 cell dry weight h−1) plotted against the NO concentration in liquid (nM), together with the model fitted to the entire data set (Vmax = 4.2 mmol g−1 cell dw h−1, Ks = 577 nM). (B) Log-log plot of the same data to visualize the relationships between the strains and model at low NO concentrations.
It is possible that the amounts of NO used were insufficient to permit significant levels of Nor inactivation, as seen during cultivation with nitrite in the medium. To determine if this was the case, experiments were done using cells that had been exposed to levels of nitrite that lead to accumulation of NO or general respiratory inhibition. For this comparison, cells were first grown with 1% O2 and 1 mM nitrite. As expected, the ΔnorEF strain accumulated NO to μM concentrations in the liquid while the wild type reduced 1 mM nitrite rapidly (see Fig. S7 in the supplemental material), as observed previously (Fig. 1). The cells were then washed by repeated centrifugation and resuspension in nitrite-free medium and reinoculated to new vials with or without O2. In response to NO injection to the anoxic vials, the wild-type cells rapidly reduced NO, and the estimated specific rates were somewhat higher than that predicted by the model based on cells raised in a medium with 0.1 mM NO3− (Fig. 5). The ΔnorEF strain, which had been exposed to high NO, was able to reduce NO, but the rates were only 55% of that predicted by the model. The apparent Ks for the ΔnorEF strain, estimated by nonlinear regression of the rates against concentration, was not significantly different from that of the wild type. The estimated Ks of the mutant was 359 nM (SE, 67; 95% confidential interval, 213 to 647 nM; the Ks of the wild type was 577 nM). Oxic respiration in the vials with initial O2 at 1% showed normal rates for the 2.4.3 cells, with a Vmax of 28.5 mmol O2 g−1 cell dry weight h−1 and an apparent Ks of 16 μM O2. Unexpectedly, the 2.4.3 ΔnorEF cells were unable to respire O2 efficiently, and the respiration rate remained about 1 to 2 mmol O2 g−1 cell dry weight h−1 throughout the incubation of 20 h (see Fig. S8 in the supplemental material).
FIG 5.
Kinetics of NO reduction in norEF-deficient cells after exposure to high NO concentrations. Wild-type 2.4.3 and the ΔnorEF mutant were first cultured in medium with 1 mM NO2−, leading to accumulation of μM concentrations of NO in the ΔnorEF mutant (as shown in Fig. 1D; also see Fig. S7 in the supplemental material) but not in wild-type 2.4.3. The cells were then harvested, washed once in sterile Sistrom's medium, and transferred to anoxic vials with fresh medium without NO2− and NO3− for measuring rates of NO reduction. (A) Rates (mmol g−1 cell dry weight h−1) plotted against NO concentration in the liquid. The line shows the predictions by the Michaelis-Menten model (Vmax = 4.2 mmol g−1 h−1, Ks = 577 nM) (Fig. 4). (B) Log-log plot for inspection of data at low concentrations.
Strains lacking Nap-β.
Comparisons of denitrification phenotypes of the various strains during nitrate respiration revealed a significant decrease in the rate of N2O accumulation by the 2.4.1 pAK1 strain compared to both wild-type 2.4.3 and 2.4.3 ΔnorEF. Comparing the time at which 100% recovery of nitrate-N as N2O occurred in 2.4.3 to those of both 2.4.3 ΔnorEF and 2.4.1 pAK1 cultures showed that while denitrification rates of both of the norEF-deficient strains were slower than that of the wild type, 2.4.1 pAK1 was notably slower than 2.4.3 ΔnorEF (Table 1). This indicates that the loss of norEF alone does not account for the differences. Another notable difference in the 2.4.1 pAK1 strain not observed in either 2.4.3 strain was a slight decrease in N2O production rate as the experiment progressed (μNOx, <0) (see Fig. S4 in the supplemental material).
This trend of decreasing nitrate reduction under denitrifying conditions has also been observed in 2.4.3 strains reducing nitrate under denitrifying conditions with Nap-α, which is expressed under oxic conditions but downregulated under hypoxic conditions, due to inactivation of Nap-β (13). The two gene clusters can be readily distinguished, since the Nap-β cluster contains narGH while Nap-α does not. The observation in the Nap-β mutant of 2.4.3 could explain the slower nitrate reduction in 2.4.1, since its Nap is orthologous to the Nap-α cluster of 2.4.3. To test the hypothesis that the presence of Nap-β enhances N2O production in 2.4.3 relative to 2.4.1, N2O production was measured in a strain of 2.4.3 in which the genes encoding nap-β were insertionally inactivated (13). When treated with 1% initial O2 and 1 mM nitrate, nitrate respiration of the Nap-β mutant of 2.4.3 was considerably slower than that of the wild type (Fig. 6A and C). Electron flow to NOx was low throughout the 40 h of incubation, and only approximately 20% of the added nitrate-N was recovered as N2O. For comparison, nitrite reduction was also monitored in the Nap-β mutant strain. In a treatment started with 1% O2 and 0.25 μM nitrite, the cells reduced the added nitrite rapidly with only modest levels of NO accumulation, similar to what was observed with the wild type. A second pulse of 1 mM nitrite was also rapidly reduced (Fig. 6B and D).
FIG 6.
O2 depletion and NO and N2O accumulation (A and B) and electron flow to O2 and NOx (C and D) in batch cultures of 2.4.3 Δnap-β with 1% initial O2 and 1 mM nitrate (A) or 0.25 mM nitrite (B). The e− flow rate in panel C assumes negligible accumulation of nitrite. In the culture depicted in panel B, a second pulse of 1 mM nitrite was added after 23 h and all of the available nitrite was reduced to N2O.
DISCUSSION
The data presented above indicate that Nor is the nitrogen oxide reductase most affected by the absence of NorEF (Fig. 1). This is not an unreasonable result, since Nor is a member of the HCO family and NorE is a homolog of the subunit III proteins of the HCO family. However, the biochemical function of NorEF is somewhat difficult to determine based on the observed phenotypic changes. The most significant changes in the phenotype of NorEF-deficient cells occurred in treatments with relatively high levels of nitrite. Treatments containing similar levels of nitrate did not experience runaway NO production but showed a more general slowing of NOx respiration (Table 1). Previous experiments with Agrobacterium tumefaciens, which is prone to NO accumulation, have demonstrated that NO accumulation is more severe in nitrite than in nitrate treatments when the initial O2 level is 1% (18). This suggests that nitrate reductase is rate limiting and prevents NO accumulation by limiting the size of the nitrite pool. It seems likely that Nap activity is also rate limiting in R. sphaeroides, and the nitrite pools remain small when nitrate is serving as an initial terminal oxidant. This conclusion is consistent with the observation that low levels of nitrite can be reduced by the NorEF-deficient strains without accumulation of NO (Table 2). Taken together, these results indicate that nitrogen oxide reduction is impacted under all conditions by the absence of NorEF, but significant impairment of nitrogen oxide reduction requires high levels of nitrite.
While it is unclear how the loss of NorEF impacts Nor activity, it is of interest to compare these results to those of experiments in which subunit III is removed from an aa3-type oxidase. A subunit III-deficient aa3 complex undergoes a phenomenon termed suicide inactivation, where the enzyme becomes inactivated in a turnover-dependent process (21). A similar phenomenon may occur in the NorEF-deficient strains. This would fit with the observation that Nor activity in a norEF mutant background initially was similar to that of the wild type but changed as denitrification proceeded. The particular sensitivity to nitrite may be related to the observation that nitrite levels in wild-type 2.4.3 are maintained in the low-μM levels during balanced denitrification in the presence of low-mM nitrate concentrations, suggesting Nap is rate limiting (19). Therefore, incubations with nitrite might exacerbate turnover-dependent Nor inactivation as a consequence of increased Nir activity. This would explain why NO reduction in cells exposed to high nitrite showed a significantly lower Vmax than the wild type but little change in apparent Ks.
It could be argued that since NorEF has never been found in a complex with NorCB, the former could not play a role in Nor repair or protection. However, sequence alignment of NorE with subunit III proteins reveals the conservation of a number of residues, in particular the dicyclohexylcarbodiimide (DCCD) binding glu residue found in subunit III, indicating that NorE is structurally or biochemically similar to subunit III, which has been shown to be required for oxidase stability (22, 23). Even in the aa3 oxidase family, the interaction of subunit III with subunit I is relatively weak. Mild treatments will readily remove subunit III from the purified complex without significant disruption to the enzyme's active site (21). Therefore, it is not unreasonable to suggest that current Nor purification protocols are too disruptive of what is inherently a weak interaction.
If, as proposed, NorEF is critical for mitigating turnover-dependent inactivation, it might be expected that their presence would be strongly selected for in any strain that utilizes cNor. However, this is obviously not the case, since all five of the sequenced R. sphaeroides strains contain cNor but only one, 2.4.3, has norEF. In two of these strains, 2.4.1 and 17029, the absence of norEF likely is correlated with the loss of the ability to reduce nitrate to a gaseous N-oxide, since both lack nirK (24). A lack of Nir means the main route of NO exposure will be from exogenous sources. This likely minimizes the fitness costs due to loss of norEF, since cells lacking these genes can reduce a μM bolus of exogenous NO as effectively as cells containing the wild type. Therefore, 2.4.1 and 17029 can use Nor to mitigate episodic exposure to biological or chemical sources of NO but are poorly adapted to reduce NO generated during denitrification-dependent growth.
While the loss of Nir activity can be associated with loss of norEF, this is not always the case. The other two sequenced R. sphaeroides strains, KD131 and WS8N, are distinct from 2.4.1 and 17029, since they retain nirK but lack nitrate reductase (24–26). Sequence analysis does not indicate that nirK in these strains is inactive or incapable of being expressed (J. P. Shapleigh, unpublished data). This suggests that both are able to use nitrite as a terminal electron acceptor to support anoxic growth. The absence of norEF suggests that although nitrite respiration would be problematic if concentrations were in the mM range or for long-term growth, this remains to be tested. Perhaps these strains do not routinely encounter nitrite concentrations that are sufficient to result in toxic levels of NO.
Loss of norEF in strains with cNor is not limited to R. sphaeroides. Analysis of available genome sequences has found many strains with cNor but without NorEF (J. P. Shapleigh, unpublished). Many, including Azoarcus sp. strain BH72, are denitrifiers that are only missing nitrite reductase, thereby limiting exposure to NO (27). While most of the strains lacking norEF are partial denitrifiers, there are rare examples of complete denitrifiers lacking NorEF orthologs. One example of this type of bacterium is Roseobacter sp. strain SK209-2-6. Denitrification in this strain has not been characterized, so it is unclear if nitrate or nitrite reduction is impaired. The related Roseobacter denitrificans, which has been suggested to be an aerobic denitrifier, retains norEF (28, 29).
In conclusion, the presence or absence of norEF appears to be a useful proxy for the ability of a strain to support robust, complete reduction of nitrate to nitrous oxide. This is explained by the observation that norEF is not essential for Nor activity, but its absence does affect activity under conditions where endogenous Nir activity generates prolonged exposure to NO. This protein, along with NnrS, likely are representatives of a suite of proteins that permit robust denitrification but are not always found in partial denitrifiers (30). Therefore, they may be useful markers for active denitrification in metagenomic surveys in environments where denitrification is predicted to be an important physiological process (31).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Norwegian University of Life Sciences for support allowing J.P.S. to work in Ås.
We also thank Jon Holser of the University of Mississippi Medical School for helpful discussions.
Footnotes
Published ahead of print 4 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00003-14.
REFERENCES
- 1.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. 2012. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim. Biophys. Acta 1817:629–637. 10.1016/j.bbabio.2011.09.020 [DOI] [PubMed] [Google Scholar]
- 2.Hendriks J, Gohlke U, Saraste M. 1998. From NO to OO: nitric oxide and dioxygen in bacterial respiration. J. Bioenerg. Biomembr. 30:15–24. 10.1023/A:1020547225398 [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 176:5587–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitcher RS, Watmough NJ. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655:388–399. 10.1016/j.bbabio.2003.09.017 [DOI] [PubMed] [Google Scholar]
- 5.Varanasi L, Hosler JP. 2012. Subunit III-depleted cytochrome c oxidase provides insight into the process of proton uptake by proteins. Biochim. Biophys. Acta 1817:545–551. 10.1016/j.bbabio.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumft WG. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J. Inorg. Biochem. 99:194–215. 10.1016/j.jinorgbio.2004.09.024 [DOI] [PubMed] [Google Scholar]
- 8.Kastrau DHW, Heiss B, Kroneck PMH, Zumft WG. 1994. Nitric oxide reductase from Pseudomonas stutzeri, a novel cytochrome bc complex. Eur. J. Biochem. 222:293–303. 10.1111/j.1432-1033.1994.tb18868.x [DOI] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10.Shapleigh JP. 2012. The denitrifying prokaryotes. In Rosenberg E, Stackebrandt E, Thompson F, DeLong E, Lory S. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 11.de Boer AP, van der Oost J, Reijnders WN, Westerhoff HV, Stouthamer AH, van Spanning RJ. 1996. Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur. J. Biochem. 242:592–600. 10.1111/j.1432-1033.1996.0592r.x [DOI] [PubMed] [Google Scholar]
- 12.Hartsock A, Shapleigh JP. 2010. Identification, functional studies, and genomic comparisons of new members of the NnrR regulon in Rhodobacter sphaeroides. J. Bacteriol. 192:903–911. 10.1128/JB.01026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartsock A, Shapleigh JP. 2011. Physiological roles for two periplasmic nitrate reductases in Rhodobacter sphaeroides 2.4.3 (ATCC 17025). J. Bacteriol. 193:6483–6489. 10.1128/JB.05324-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen-Bazire G, Sistrom WR, Stanier RY. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell Comp. Physiol. 49:25–68. 10.1002/jcp.1030490104 [DOI] [PubMed] [Google Scholar]
- 15.Molstad L, Dorsch P, Bakken LR. 2007. Robotized incubation system for monitoring gases (O2, NO, N2O, N2) in denitrifying cultures. J. Microbiol. Methods 71:202–211. 10.1016/j.mimet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 16.Bergaust L, Mao Y, Bakken LR, Frostegård Å. 2010. Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76:6387–6396. 10.1128/AEM.00608-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergaust L, van Spanning RR, Frostegård Å, Bakken LR. 2012. Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158:826–834. 10.1099/mic.0.054148-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergaust L, Shapleigh J, Frostegård Å, Bakken L. 2008. Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability. Environ. Microbiol. 10:3070–3081. 10.1111/j.1462-2920.2007.01557.x [DOI] [PubMed] [Google Scholar]
- 19.Jain R, Shapleigh JP. 2001. Characterization of nirV and a gene encoding a novel pseudoazurin in Rhodobacter sphaeroides 2.4.3. Microbiology 147:2505–2515 [DOI] [PubMed] [Google Scholar]
- 20.Lee DY, Ramos A, Macomber L, Shapleigh JP. 2002. The taxis response of various denitrifying bacteria to nitrate and nitrite. Appl. Environ. Microbiol. 68:2140–2147. 10.1128/AEM.68.5.2140-2147.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratton MR, Pressler MA, Hosler JP. 1999. Suicide inactivation of cytochrome c oxidase: catalytic turnover in the absence of subunit III alters the active site. Biochemistry 38:16236–16245. 10.1021/bi9914107 [DOI] [PubMed] [Google Scholar]
- 22.Haltia T, Saraste M, Wikstrom M. 1991. Subunit III of cytochrome c oxidase is not involved in proton translocation: a site-directed mutagenesis study. EMBO J. 10:2015–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varanasi L, Mills D, Murphree A, Gray J, Purser C, Baker R, Hosler J. 2006. Altering conserved lipid binding sites in cytochrome c oxidase of Rhodobacter sphaeroides perturbs the interaction between subunits I and III and promotes suicide inactivation of the enzyme. Biochemistry 45:14896–14907. 10.1021/bi061390q [DOI] [PubMed] [Google Scholar]
- 24.Choudhary M, Zanhua X, Fu YX, Kaplan S. 2007. Genome analyses of three strains of Rhodobacter sphaeroides: evidence of rapid evolution of chromosome II. J. Bacteriol. 189:1914–1921. 10.1128/JB.01498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SK, Kim SJ, Cha SH, Oh YK, Rhee HJ, Kim MS, Lee JK. 2009. Complete genome sequence of Rhodobacter sphaeroides KD131. J. Bacteriol. 191:1118–1119. 10.1128/JB.01565-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter SL, Wilkinson DA, Byles ED, Wadhams GH, Taylor S, Saunders NJ, Armitage JP. 2011. Genome sequence of Rhodobacter sphaeroides strain WS8N. J. Bacteriol. 193:4027–4028. 10.1128/JB.05257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause A, Ramakumar A, Bartels D, Battistoni F, Bekel T, Boch J, Bohm M, Friedrich F, Hurek T, Krause L, Linke B, McHardy AC, Sarkar A, Schneiker S, Syed AA, Thauer R, Vorholter FJ, Weidner S, Puhler A, Reinhold-Hurek B, Kaiser O, Goesmann A. 2006. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24:1385–1391. 10.1038/nbt1243 [DOI] [PubMed] [Google Scholar]
- 28.Doi M, Shioi Y. 1991. Enhancement of denitrifying activity in cells of Roseobacter denitrificans grown aerobically in the light. Plant Cell Physiol. 32:365-370 [Google Scholar]
- 29.Swingley WD, Gholba S, Mastrian SD, Matthies HJ, Hao J, Ramos H, Acharya CR, Conrad AL, Taylor HL, Dejesa LC, Shah MK, O'Huallachain EM, Lince MT, Blankenship RE, Beatty JT, Touchman JW. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic as opposed to photosynthetic metabolism. J. Bacteriol. 189:683–690. 10.1128/JB.01390-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern AM, Liu B, Bakken LR, Shapleigh JP, Zhu J. 2013. A novel protein protects bacterial iron-dependent metabolism from nitric oxide. J. Bacteriol. 195:4702–4708. 10.1128/JB.00836-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hewson I, Eggleston E, Doherty M, Lee DY, Owens M, Shapleigh JP, Cornwell JC, Crump BC. 2014. Metatranscriptomic analyses of plankton communities inhabiting surface and subpycnocline waters of the Chesapeake Bay during oxic-anoxic-oxic transitions. Appl. Environ. Microbiol. 80:328–838. 10.1128/AEM.02680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.