Abstract
The environment is a reservoir of nontuberculous mycobacteria and is considered a source of infection for animals and humans. Mycobacteria can persist in different types of environments for a relatively long time. We have studied their possible internalization into plant tissue through intact, as well as damaged, root systems of different types of plants grown in vitro and under field conditions. The substrate into which plants were seeded was previously contaminated with different strains of Mycobacterium avium (108 to 1010 cells/g of soil) and feces from animals with paratuberculosis. We detected M. avium subsp. avium, hominissuis, and paratuberculosis in the stems and leaves of the plants by both culture and real-time quantitative PCR. The presence of mycobacteria in the plant tissues was confirmed by microscopy. The concentration of mycobacteria found inside plant tissue was several orders of magnitude lower (up to 104 cells/g of tissue) than the initial concentration of mycobacteria present in the culture medium or substrate. These findings led us to the hypothesis that plants may play a role in the spread and transmission of mycobacteria to other organisms in the environment.
INTRODUCTION
Nontuberculous mycobacteria cause a wide range of diseases in animals and immunocompromised individuals. Mycobacterial infection is acquired mainly through the respiratory and gastrointestinal tracts. Mycobacteria are ubiquitously distributed, and some are present in high numbers in natural and man-made environments; thus, they pose a constant risk to susceptible species of animals and immunocompromised humans. The diversity of mycobacteria in the environment was studied with a combination of molecular biology methods (1). This enabled qualitative and quantitative analysis and detection of sequences of pathogenic mycobacteria in all types of tested soil. Nontuberculous mycobacteria have been described as causal agents in different types of diseases, most often pulmonary, skin, and soft tissue infections (2). Although members of the Mycobacterium avium complex are usually associated with pulmonary disease, colonization and infection of the gastrointestinal tracts of AIDS patients have also been described (3). However, the route of transmission is usually unclear. Water has been proposed as a main reservoir (4), while infection through aerosol from soil has also been described (5).
Little information regarding the possible contamination of plants or food of vegetable origin with mycobacteria is available. Likewise, only a small number of studies have been concerned with food as a source of infection in humans. A study of food as the source of exposure of HIV-positive patients to mycobacteria detected mycobacteria in 7 out of 121 samples examined (6). A later study compared the genotypes of M. avium isolates from patients and foods by using PCR-restriction fragment length polymorphism and demonstrated a link between them (7). Nontuberculous mycobacteria were isolated from salads, leeks, lettuce, mushrooms, and other vegetables, as well as apple juice. Twenty-nine isolates were obtained from 46 samples, with the predominantly isolated species being M. avium (8).
Studies investigating the contamination of vegetables with mycobacteria have not proven whether mycobacteria can be present inside plant tissue. In a few studies, mycobacteria were identified in or on the surfaces of different plants (9, 10, 11). Zwielehner et al. (11) studied the microbial communities present in the phylosphere of lettuce leaves. After denaturing gradient gel electrophoresis and sequencing analyses, sequences of members of the genus Mycobacterium were found on leaves, as well as in soil samples. The sequence obtained from conventionally grown lettuce was most similar to M. alvei. Also, M. avium subsp. paratuberculosis was detected in grass samples by quantitative PCR (qPCR) (12).
Plants in aquatic environments are also known to harbor mycobacteria. M. avium was detected in a reed bed sample from a constructed wetland, and plants were also selected as possible reservoirs of M. ulcerans in the environment (9, 13).
The penetration of plant tissues by bacteria, namely, Salmonella and Escherichia coli, has been studied previously (14, 15, 16, 17). It was shown that motile bacteria can enter the plant through roots or even hydathodes on the leaves of tomato plants (18).
The persistence of bacteria inside plant tissue most probably depends on the conditions inside the plant. The survival of Salmonella in basil was limited to a few days (17). To the best of our knowledge, there is no record of internalization of mycobacteria through the intact root system of plants and their distribution inside the plant itself.
The aims of this study were to investigate whether mycobacteria present in culture medium or in feces from infected cattle can penetrate the intact or damaged tissue of two different plant species through the root system. To this end, we first analyzed the presence of mycobacteria in in vitro-grown plants under sterile conditions. Subsequently, we performed a field experiment in which plants were grown in a phytotron. We used beans and tomatoes, as both produce edible parts that are not in contact with contaminated soil. Moreover, both plants can be routinely cultured under laboratory conditions.
MATERIALS AND METHODS
In vitro experiment.
M. avium subsp. hominissuis (field isolate obtained from infected swine) was grown under laboratory conditions on Middlebrook broth (M7H9) with enrichment (oleic acid-albumin-dextrose-catalase [OADC]; Becton Dickinson) with constant shaking for 2 weeks. M. avium subsp. hominissuis was chosen for its rapid availability and fast growth in vitro. The concentration of IS1245 was quantified by qPCR (19). To 20 ml of Murashige and Skoog agar (Duchefa, Haarlem, The Netherlands) at a temperature of 50°C, the suspension was added at 109 cells/ml. Cultivation was performed in 250-ml cylindrical glass vessels. Surface sterilization of pinto bean (Phaseolus vulgaris) seeds was performed by submersion in 3% sodium hypochlorite for 3 min, followed by three subsequent washes in sterile water (for 5 min each). Sterilized seeds were planted on agar and grown under laboratory conditions for 2 weeks. Samples for microscopy, mycobacterial culture, and qPCR were collected from leaves and stems with sterile forceps and scissors.
Field experiment.
For the field experiment, we used M. avium subsp. paratuberculosis (reference strain CAPM 6381) and M. avium subsp. avium (reference strain CAPM 5889). These subspecies were chosen because of their possible presence in soil after fertilization with feces from infected animals. Both strains were grown on Middlebrook broth (M7H9) with enrichment (OADC; Becton Dickinson) and mycobactin J (Allied Monitor) with constant shaking for 1 month.
Tomato (Solanum lycopersicum) seeds were planted in pots containing 1 liter of a commercially available substrate of potting soil. The substrate was tested by qPCR, and it was negative for the presence of M. avium subsp. avium and paratuberculosis prior to inoculation. A total of 24 plants were tested; 9 of them were grown in soil contaminated with M. avium subsp. avium, 12 were grown in soil contaminated with M. avium subsp. paratuberculosis, and 3 served as negative controls. Three groups of plants were grown in soil contaminated with M. avium subsp. avium, and four groups were grown in soil contaminated with M. avium subsp. paratuberculosis. Each group contained three plants. In the first group, the substrate was contaminated with an M. avium subsp. avium (108 cells/liter of potting soil) or M. avium subsp. paratuberculosis (1010 cells/liter of potting soil) suspension immediately before seeding. Ten milliliters of the suspension was added to the soil, and the soil was mixed with a glass spatula. In the second and third groups, the suspensions were added 2 weeks after seeding, in the proximity of the main roots, with the difference that the roots of the third group were mechanically damaged with a syringe needle. The fourth group consisted of plants seeded in the substrate mixed with feces from a cow clinically ill with paratuberculosis. The concentration of cells in the substrate after the addition of feces was 106 cells/liter of potting soil (quantified by qPCR).
The plants were grown in a phytotron with a daytime temperature of 22°C, a humidity of 29%, and a CO2 concentration of 630 ppm. There were three collection points. Leaf, stem, and fruit samples (where available) were collected 2, 4, and 8 weeks after seeding (for group 1) and after contamination (for groups 2 and 3).
Additionally, we performed distribution analysis of two selected plants, one grown on M. avium subsp. avium-contaminated soil and the other grown on M. avium subsp. paratuberculosis-contaminated soil. After 8 weeks, we analyzed stems and leaves taken from the plants at 5-cm height intervals, as well as root, fruit, and pollen samples (pollen samples were collected from several plants during the flowering phase).
Sample examination. (i) Light and fluorescence microscopy.
Microscopy of histological sections (15 to 20 μm thick, made with a cryostat) of stem, root, and leaf tissues was performed after staining by the Ziehl-Neelsen method. Slides were analyzed at a magnification of ×1,000 with an Olympus BX41 microscope. Slides were prepared for fluorescence microscopy with a primary polyclonal rabbit anti-Mycobacterium antibody and a Cy3-labeled anti-rabbit secondary antibody (ExBio, Czech Republic). Briefly, the protocol included fixation in acetone, followed by incubation with a blocking solution (Dako). The primary antibody was diluted to 10 μg/ml, and incubation was performed overnight (12 to 16 h) at 4°C. After three washes in phosphate-buffered saline (PBS), the secondary antibody was applied for 1 h at room temperature. After the final wash in PBS, slides were mounted with mounting medium and analyzed at a magnification of ×1,000 with an Olympus BX41 microscope (Olympus, Japan). For the samples stained with fluorescent antibody, the 510- to 550-nm filter was used.
(ii) Electron microscopy.
Ultrathin sections of plant tissue were fixed in 3% glutaraldehyde in cacodylate buffer; postfixed in 1% OsO4 solution in cacodylate buffer; dehydrated in 50, 70, 90, and 100% acetone; and embedded in an Epon 812 (Serva, Germany)-Durcupan (ACM Fluka, Switzerland) mixture. The sections were stained with 2% uranyl acetate and 2% lead citrate and observed at 80 kV in a Philips EM 208 transmission electron microscope (Philips, The Netherlands).
Cultivation of mycobacteria.
Samples from stems and leaves were washed in 3% sodium hypochlorite for 3 min and sterile water (three times for 5 min each time) prior to culture to avoid surface contamination. Samples were cut from tomatoes aseptically, and the inside part of the fruit was cultured. Cultivation of 1 g of each sample (homogenized in 2 ml of PBS) without any decontamination and an additional 1 g with decontamination was performed according to Fischer et al. (20). Briefly, the homogenized sample was treated with 1 M HCl for 20 min and subsequently neutralized with 2 M NaOH. The material was inoculated onto four different culture media (Table 1), Herrold egg yolk medium, Leslie medium, and Middlebrook M7H11 with a PANTA (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin) antibiotic mixture or without antibiotics. Middlebrook M7H11 was obtained from BD Diagnostics (Denmark). The rest of the media were prepared in our laboratory as described previously (21). When culturing the samples from the tomato plants, it was necessary to perform a decontamination step because of the high rate of contamination.
TABLE 1.
qPCR and culture examinations of stem and leaf samples from in vitro-grown bean plantsa
| Plant part and no. or parameter | No. of cells/gb | No. of CFU/g after cultivation: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Without decontamination |
With decontamination |
||||||||
| MB + PANTAc | MBd | HEYMe | Leslief | MB + PANTA | MB | HEYM | Leslie | ||
| Stem | |||||||||
| 1 | 6.31 × 106 | >1,000 | >1,000 | 100 | 0 | 150 | 0 | 0 | 0 |
| 2 | 1.54 × 107 | 100 | 0 | 0 | 200 | 20 | 0 | 0 | 0 |
| 3 | 7.74 × 106 | 1,000 | >200 | >1,000 | 200 | 10 | 10 | 0 | 0 |
| Mean (SD) | 9.81 × 106 (4.88 × 106) | 3,700 (5,474) | 3,400 (5,716) | 366 (550.7) | 133.33 (115.5) | 60 (78.1) | 3.33 (5.7) | 0 (0) | 0 (0) |
| Leaf | |||||||||
| 1 | 6.70 × 106 | 200 | >200 | 1,000 | 0 | 0 | 100 | 100 | 10 |
| 2 | 1.74 × 106 | >200 | >200 | 500 | 100 | 0 | 0 | 0 | 0 |
| 3 | 3.49 × 106 | >100 | >200 | >1,000 | 0 | 0 | 100 | 10 | 100 |
| Mean (SD) | 3.97 × 106 (2.51 × 106) | 166 (57.73) | 200 (0) | 3,833 (5,346) | 33.33 (57.7) | 0 (0) | 66.67 (57.7) | 36.67 (55) | 36.67 (55) |
| Negative control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The input concentration of M. avium subsp. hominissuis culture added to the agar was 109 cells/ml.
Determined by qPCR assay for IS1245.
MB + PANTA, Middlebrook M7H11 plus PANTA.
MB, Middlebrook M7H11 without antibiotics.
HEYM, Herrold egg yolk medium.
Leslie, Leslie medium.
DNA isolation and qPCR.
Several commercially available kits for DNA isolation from plant material were tested for efficiency. Because of the expected low number of bacterial cells in the plant tissue, we attempted to use as much starting material as possible. The best results for 0.25 g of tissue were achieved with the commercially available PowerFood Microbial DNA isolation kit (MoBio, USA) with certain modifications of the original protocol. Initial homogenization of the samples was done in a MagnaLyser (Roche, Germany) at 6,400 rpm for 2 min after the addition of four 3.2-mm chrome steel beads (Biospec, USA), as well as the beads provided in the kit. An increased volume of lysis buffer (700 μl) was used, and an additional step of heating at 65°C for 10 min with shaking at 1,400 rpm was included. The remaining steps were performed according to the manufacturer's recommendations. DNA was eluted in 100 μl of preheated Tris-EDTA buffer (Amresco, USA) and used subsequently in qPCR assays.
Triplex qPCR for simultaneous detection of IS1245 and IS901 was performed for every sample in duplicate for the plants contaminated with M. avium subsp. avium and hominissuis, according to Slana et al. (19). For the tomato plants contaminated with M. avium subsp. paratuberculosis, qPCR for detection of IS900 was performed as described earlier (22). The qPCR results were transformed to numbers of cells per gram by calculating the mean copy number of insertion sequences per cell (25 copies of IS1245 in M. avium subsp. hominissuis, 15 copies of IS901 in M. avium subsp. avium, and 15 copies of IS900 in M. avium subsp. paratuberculosis).
RESULTS
In vitro experiment.
The qPCR analyses showed that mycobacteria were present in all of the leaf and stem samples from the plants grown on artificially contaminated medium. The quantity of mycobacteria was 3 orders of magnitude smaller than the quantity present in the substrate medium. The negative control gave no signal.
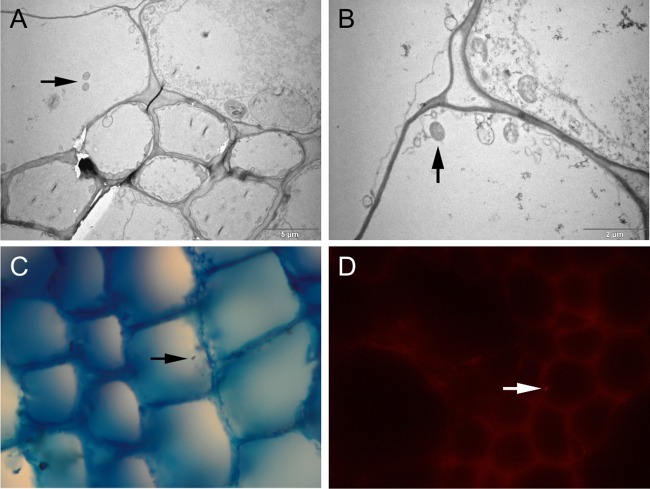
The culture results are presented in Table 1. The decontamination method clearly resulted in much lower yields in culture than did sample processing without the decontamination step. The comparison of four different culture media also shows that the best recovery was obtained with Middlebrook M7H11 agar. The addition of PANTA antibiotics had no adverse effect on mycobacterial growth. Using electron microscopy, we observed structures similar to bacterial cells in the plant tissues (Fig. 1). Using Ziehl-Neelsen microscopy of plant tissue sections 15 to 20 nm thick, we were able to observe acid-fast rods. We made similar observations by fluorescence microscopy (Fig. 1). Mycobacteria were observed by microscopy only in the bean plants grown in vitro, where the number of cells per gram exceeded 106.
FIG 1.
Microscopy of bean plant stems sections containing M. avium subsp. avium. (A, B) Transmission electron microscopy. Arrows indicate structures similar to bacterial cells. (C) Ziehl-Neelsen staining of bean plant stem tissue. A mycobacterium is stained red against a blue background. (D) Specific-antibody-labeled M. avium subsp. avium inside bean plant stem tissue. Fluorescent rods were observed inside plant transport cells.
Field experiment.
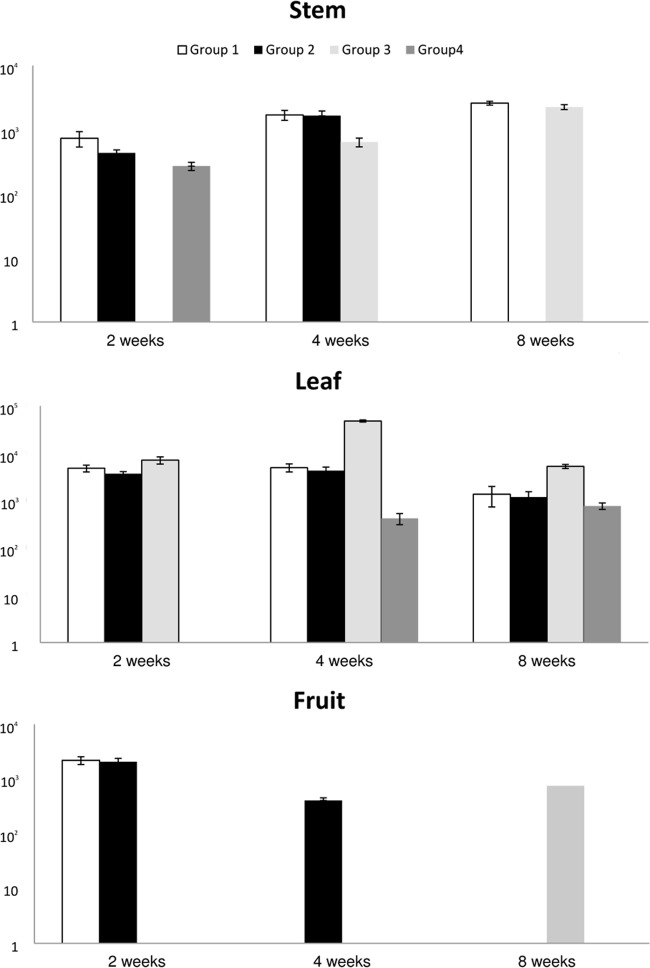
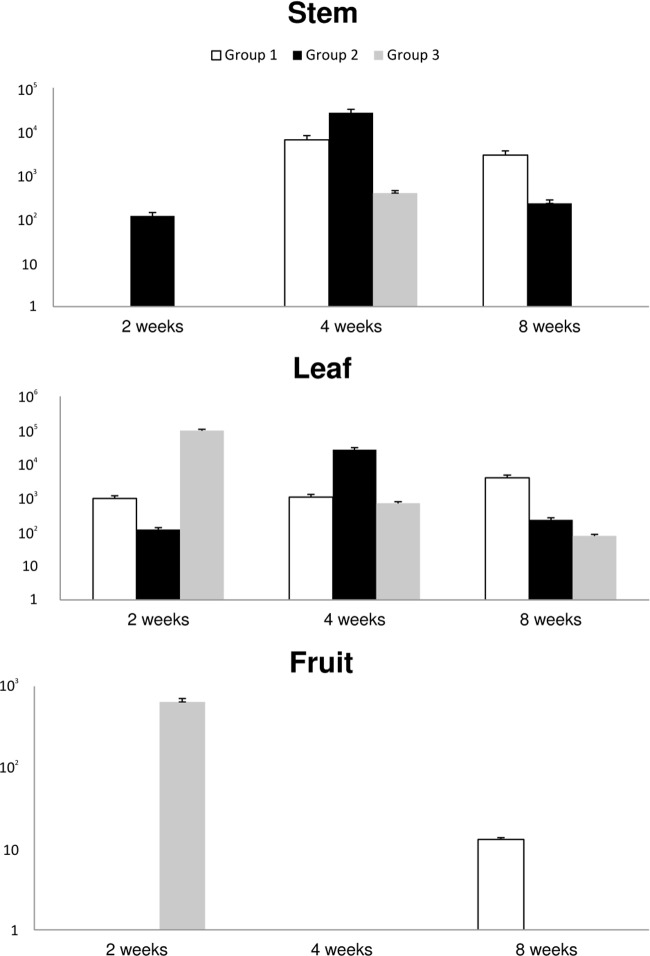
In the field experiment with the tomato plants, the results from qPCR are shown in Fig. 2 and 3. M. avium subsp. paratuberculosis DNA was present in the entire stem and leaf samples from the plants in group 1 at the three sampling times. The fruit samples from this group were positive only at the first sampling point 2 weeks postinoculation. The samples from group 2 gave similar results, although leaf stem samples were negative 8 weeks after inoculation, but DNA was detected in fruit samples at 4 weeks postcontamination. Stem samples from group 3 (with damaged roots) were positive 4 and 8 weeks after contamination, but leaf samples were positive at all three time points. Fruit samples were positive 8 weeks after contamination with mycobacteria. The quantities were similar in all of the samples and ranged from 102 to 104 cells/g of tissue. Leaf and stem tissue samples from group 4 were positive, but fruit samples were not. The quantity of IS900 reached up to 103/g of tissue (Fig. 2). The samples from tomato plants grown in soil contaminated with M. avium subsp. avium gave similar results. Stem samples from group 1 were positive at 4 and 8 weeks after seeding, but leaf samples were positive at all sampling points. All of the stem and leaf samples from group 2 were positive, but none of the fruit samples were. Leaf samples from group 3 were positive at all of the time points tested, and fruit samples were positive at 2 weeks after contamination. The quantity of M. avium subsp. avium-specific DNA ranged from 101 to 105 IS901 copies/g of tissue (Fig. 3).
FIG 2.
Detection of M. avium subsp. paratuberculosis DNA inside tomato plants.
FIG 3.
Detection of M. avium subsp. avium DNA inside tomato plants.
We also obtained three isolates from cultivation. Two of the isolates (M. avium subsp. avium) from stems of plants in groups 2 and 3 were obtained at the 2-week sampling time, and one isolate (M. avium subsp. paratuberculosis) from group 2 was obtained 1 month after seeding. The samples from the control group were negative. The qPCR results of the distribution of mycobacteria inside the plant are shown in Fig. 4. Mycobacteria were concentrated mostly in the root samples, and their quantity decreased through the fruit, although we also detected the target sequence in pollen samples.
FIG 4.
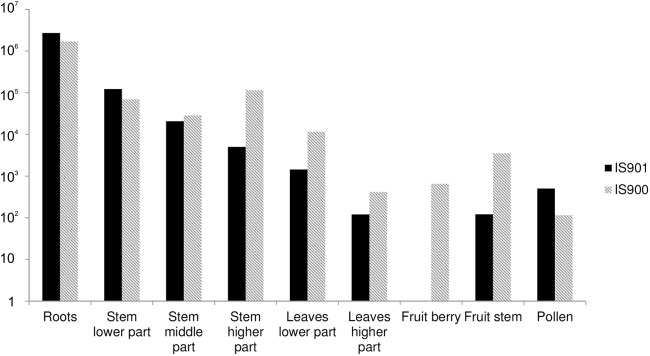
Distribution of M. avium subsp. avium and paratuberculosis DNA inside tomato plants.
DISCUSSION
The food safety of vegetables has been of increasing concern since recent outbreaks of Salmonella and E. coli were traced back to vegetables and sprouts. Much research has focused on these two pathogens and their potential for penetration versus surface contamination of vegetables (23, 24). Solomon et al. (25) described the migration of E. coli O157:H7 from contaminated soil into the tissue of lettuce. There have also been studies on the internalization of Salmonella into tomato plants through roots or even hydathodes (18). Although mycobacteria were detected in vegetables previously, the present study is the first to confirm their internalization inside plant tissue. The large numbers of M. avium cells used in our experiment may have biased the results; however, a recent study showed that the concentration of mycobacteria in soil, as well as their diversity, is high (1).
Plants or vegetables have been suspected as possible sources of food-borne mycobacterial diseases (6, 8). Typing of M. avium isolates from food and patients showed the same DNA patterns (8). Our results demonstrate that mycobacteria can be taken up by the root systems of plants, even plants with intact roots. The persistence of mycobacteria in soil, manure, and different parts of the environment has been demonstrated previously (12, 26). M. avium subsp. paratuberculosis remained viable in manure after 55 weeks (26). Manure from domestic animals is used as fertilizer in fields where crops or vegetables are grown. Therefore, it is plausible that because of its presence inside plants, there might be a risk of infection of grazing animals or of humans. Although in the present study we have proven the presence of mycobacteria inside plant tissue, we have not performed any experiments regarding its pathogenicity. Future research should test the pathogenicity of mycobacteria after their internalization in plants. The next step would be to feed animals such plants to see whether this route of transmission is plausible.
In our study, mycobacteria were present inside plant tissue for at least 2 months after the contamination of potting soil. This may be due to the properties of mycobacteria, as well as the environment inside the plant tissue. A study of the survival of Salmonella in basil showed a decline after only 3 days (17), although other studies have detected Salmonella in tomato fruit samples 49 days after inoculation (27).
Regarding the distribution of mycobacteria inside the plants, M. avium subsp. avium and paratuberculosis DNA quantities were highest in the roots and gradually decreased along the height of the plant. The presence of mycobacterial DNA in the fruit and pollen samples is noteworthy regarding food safety and further spread of mycobacteria. However, we did not obtain an isolate from these samples by culture.
In conclusion, we have demonstrated the internalization of mycobacteria into different types of plants; furthermore, their distribution within the plants was found to be even. However, the concentration of mycobacteria found inside plant tissue was several orders of magnitude smaller than the initial concentration of mycobacteria present in the culture medium or substrate. Mycobacteria are probably passively taken inside the roots rather than actively penetrate the root epidermis. This passive intake could be facilitated by the relatively small size of mycobacterial cells. Although mycobacteria inside plant tissue pose a possible risk of transmission, we suspect that the subsequent handling of vegetables and secondary surface contamination with mycobacteria might play a bigger role in the transmission of the infectious agent.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Agriculture (QH81065 and MZe0002716202). The results of the project LO1218 were obtained with financial support from the Ministry of Education, Youth, and Sports (MEYS) of the Czech Republic under the NPU I program.
Footnotes
Published ahead of print 18 April 2014
REFERENCES
- 1.Pontiroli A, Khera TT, Oakley BB, Mason S, Dowd SE, Travis ER, Erenso G, Aseffa A, Courtenay O, Wellington EM. 2013. Prospecting environmental mycobacteria: combined molecular approaches reveal unprecedented diversity. PLoS One 8:e68648. 10.1371/journal.pone.0068648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkinham JO. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun HY, Chen MY, Wu MS, Hsieh SM, Fang CT, Hung CC, Chang SC. 2005. Endoscopic appearance of GI mycobacteriosis caused by the Mycobacterium avium complex in a patient with AIDS: case report and review. Gastrointest. Endosc. 61:775–779. 10.1016/S0016-5107(04)02786-5 [DOI] [PubMed] [Google Scholar]
- 4.Whiley H, Keegan A, Giglio S, Bentham R. 2012. Mycobacterium avium complex—the role of potable water in disease transmission. J. Appl. Microbiol. 113:223–232. 10.1111/j.1365-2672.2012.05298.x [DOI] [PubMed] [Google Scholar]
- 5.Kaevska M, Slana I, Kralik P, Reischl U, Orosova J, Holcikova A, Pavlik I. 2011. “Mycobacterium avium subsp. hominissuis” in neck lymph nodes of children and their environment examined by culture and triplex quantitative real-time PCR. J. Clin. Microbiol. 49:167–172. 10.1128/JCM.00802-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argueta C, Yoder S, Holtzman AE, Aronson TW, Glover N, Berlin OGW, Stelma GN, Froman S, Tomasek P. 2000. Isolation and identification of nontuberculous mycobacteria from foods as possible exposure sources. J. Food Prot. 63:930–933 [DOI] [PubMed] [Google Scholar]
- 7.Yajko DM, Chin DP, Gonzalez PC, Nassos PS, Hopewell PC, Reingold AL, Horsburgh CR, Yakrus MA, Ostroff SM. 1995. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:176–182 [PubMed] [Google Scholar]
- 8.Yoder S, Argueta C, Holtzman A, Aronson T, Berlin OGW, Tomasek P, Glover N, Froman S, Stelma G. 1999. PCR comparison of Mycobacterium avium isolates obtained from patients and foods. Appl. Environ. Microbiol. 65:2650–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durnez L, Stragier P, Roebben K, Ablordey A, Leirs H, Portaels F. 2009. A comparison of DNA extraction procedures for the detection of Mycobacterium ulcerans, the causative agent of Buruli ulcer, in clinical and environmental specimens. J. Microbiol. Methods 76:152–158. 10.1016/j.mimet.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 10.Drewe JA, Mwangi D, Donoghue HD, Cromie RL. 2009. PCR analysis of the presence and location of Mycobacterium avium in a constructed reed bed, with implications for avian tuberculosis control. FEMS Microbiol. Ecol. 67:320–328. 10.1111/j.1574-6941.2008.00618.x [DOI] [PubMed] [Google Scholar]
- 11.Zwielehner J, Handschur M, Michaelsen A, Irez S, Demel M, Denner EBM, Hasiberger AG. 2008. DGGE and real-time PCR analysis of lactic acid bacteria in bacterial communities of the phyllosphere of lettuce. Mol. Nutr. Food Res. 52:614–623. 10.1002/mnfr.200700158 [DOI] [PubMed] [Google Scholar]
- 12.Pribylova R, Slana I, Kaevska M, Lamka J, Babak V, Jandak J, Pavlik I. 2011. Soil and plant contamination with Mycobacterium avium subsp. paratuberculosis after exposure to naturally contaminated mouflon feces. Curr. Microbiol. 62:1405–1410. 10.1007/s00284-011-9875-7 [DOI] [PubMed] [Google Scholar]
- 13.McIntosh M, Williamson H, Benbow ME, Kimbirauskas R, Quaye C, Boakye D, Small P, Merritt R. 2014. Associations between Mycobacterium ulcerans and aquatic plant communities of West Africa: implications for Buruli ulcer disease. EcoHealth 10.1007/s10393-013-0898-3 [DOI] [PubMed] [Google Scholar]
- 14.Guo XA, van Iersel MW, Chen JR, Brackett RE, Beuchat LR. 2002. Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl. Environ. Microbiol. 68:3639–3643. 10.1128/AEM.68.7.3639-3643.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heaton JC, Jones K. 2008. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J. Appl. Microbiol. 104:613–626. 10.1111/j.1365-2672.2007.03587.x [DOI] [PubMed] [Google Scholar]
- 16.Mitra R, Cuesta-Alonso E, Wayadande A, Talley J, Gilliland S, Fletcher J. 2009. Effect of route of introduction and host cultivar on the colonization, internalization, and movement of the human pathogen Escherichia coli O157:H7 in Spinach. J. Food Prot. 72:1521–1530 [DOI] [PubMed] [Google Scholar]
- 17.Gorbatsevich E, Sela S, Pinto R, Bernstein N. 2013. Root internalization, transport and in-planta survival of Salmonella enterica serovar Newport in sweet basil. Environ. Microbiol. Rep. 5:151–159. 10.1111/1758-2229.12008 [DOI] [PubMed] [Google Scholar]
- 18.Gu G, Cevallos-Cevallos JM, van Bruggen AH. 2013. Ingress of Salmonella enterica Typhimurium into tomato leaves through hydathodes. PLoS One 8:e53470. 10.1371/journal.pone.0053470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slana I, Kaevska M, Kralik P, Horvathova A, Pavlik I. 2010. Distribution of Mycobacterium avium subsp. avium and M. a. hominissuis in artificially infected pigs studied by culture and IS901 and IS1245 quantitative real time PCR. Vet. Microbiol. 144:437–443. 10.1016/j.vetmic.2010.02.024 [DOI] [PubMed] [Google Scholar]
- 20.Fischer O, Matlova L, Dvorska L, Svastova P, Bartl J, Melicharek I, Weston RT, Pavlik I. 2001. Diptera as vectors of mycobacterial infections in cattle and pigs. Med. Vet. Entomol. 15:208–211. 10.1046/j.1365-2915.2001.00292.x [DOI] [PubMed] [Google Scholar]
- 21.Balazova T, Makovcova J, Sedo O, Slany M, Faldyna M, Zdrahal Z. 2014. The influence of culture conditions on the identification of Mycobacterium species by MALDI-TOF MS profiling. FEMS Microbiol. Lett. 10.1111/1574-6968.12408 [DOI] [PubMed] [Google Scholar]
- 22.Slana I, Kralik P, Kralova A, Pavlik I. 2008. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128:250–257. 10.1016/j.ijfoodmicro.2008.08.013 [DOI] [PubMed] [Google Scholar]
- 23.Deering AJ, Mauer LJ, Pruitt RE. 2012. Internalization of E. coli O157:H7 and Salmonella spp. in plants: a review. Food Res. Int. 45:567–575. 10.1016/j.foodres.2011.06.058 [DOI] [Google Scholar]
- 24.Standing TA, du Plessis E, Duvenage S, Korsten L. 2013. Internalisation potential of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar Typhimurium and Staphylococcus aureus in lettuce seedlings and mature plants. J. Water Health 11:210–223. 10.2166/wh.2013.164 [DOI] [PubMed] [Google Scholar]
- 25.Solomon EB, Yaron S, Matthews KR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397–400. 10.1128/AEM.68.1.397-400.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittington RJ, Marshall DJ, Nicholls PJ, Marsh AB, Reddacliff LA. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989–3004. 10.1128/AEM.70.5.2989-3004.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X, Chen J, Brackett RE, Beuchat LR. 2001. Survival of salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 10:4760–4764. 10.1128/AEM.67.10.4760-4764.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]