Abstract
Carnocyclin A (CCLA) is an antimicrobial peptide produced by Carnobacterium maltaromaticum ATCC PTA-5313, which can be used to control the growth of Listeria monocytogenes in ready-to-eat meat products. The aim of this research was to elucidate the cellular responses of L. monocytogenes 08-5923 exposed to a sublethal dose of CCLA. Microarray, quantitative reverse transcription-PCR, tandem mass spectrometry, and electron microscopy were used to investigate the alteration in gene expression, protein production, and morphological changes in cells of Listeria following treatment with CCLA. The genes involved in metabolism (baiE, trn, and pykA), cell wall synthesis (murZ and dacB2), and cell division (clpE and divIVA) were upregulated following a 15-min exposure to CCLA as a result of stress responses. Genes involved in cell division, cell wall synthesis, flagellar synthesis, and metabolism were downregulated after 4 h as a result of adaptation. Analysis of total soluble proteins confirmed the downregulation of pykA and gnd after 4 h of exposure to CCLA. The absence of flagella was observed in L. monocytogenes following 30 h of exposure to CCLA. A sublethal dose of CCLA induced adaptation in L. monocytogenes 08-5923 by inhibition of expression of genes and proteins critical for synthesis of cell wall structures and maintaining metabolic functions. Both the mannose- and cellobiose-specific phosphotransferase systems could be targets for CCLA.
INTRODUCTION
Listeria monocytogenes has been responsible for numerous food-borne illness outbreaks as a result of consumption of contaminated ready-to-eat (RTE) meat products (1). A listeriosis outbreak from a single manufacturer in Canada in 2008 resulted in 22 deaths and 57 confirmed positive cases (http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/listeria-monocytogenes-eng.php).
To control the growth of L. monocytogenes, bacteriocin-containing antimicrobials such as Micocin (Griffith Laboratories, Canada) can be used in RTE meat products. One active compound in Micocin, carnocyclin A (CCLA), is a 5.9-kDa cyclic, class IIc bacteriocin (2) that has strong antilisterial activity. CCLA forms ion channels in the membrane and results in dissipation of membrane potential, which leads to cell death (3). Neither the stress response of Listeria to CCLA nor the development of resistance in Listeria have been elucidated. Previous research focused on class IIa bacteriocins, which cause cell surface alterations in Listeria (4) and, in particular, result in cell lysis by targeting the mannose-specific phosphotransferase (PTS) system (5, 6). The disruption of the mannose-specific PTS system plays a role in bacteriocin resistance (7, 8). Alternative sigma factors, such as σB (sigB) (9–11) and σ54 (rpoN) (12), are also involved in resistance to class IIa bacteriocins. These alternative sigma factors are also involved in other environmental stresses, such as cold shock, acid, and osmotolerance in Listeria (13, 14).
It has been postulated that CCLA can trigger alteration in the expression levels of genes that are also regulated by σB/σ54. The differential expression of genes and proteins and the morphology of L. monocytogenes 08-5923, one of the two strains that were involved in the 2008 listeriosis outbreak in Canada (GenBank accession number NC_013768) (15), in the presence and absence of CCLA was analyzed.
MATERIALS AND METHODS
Bacterial strains and cultures.
L. monocytogenes 08-5923 was grown in tryptic soy broth (TSB; Becton, Dickinson, Ontario, Canada) at 21°C for 24 h prior to use. Carnobacterium maltaromaticum ATCC PTA-5313 (formerly Carnobacterium maltaromaticum UAL307) was grown in all-purpose Tween (APT; BD-Canada) broth at 21°C.
CCLA isolation and purification.
CCLA was isolated from an overnight culture of C. maltaromaticum ATCC PTA-5313 grown at 21°C and purified according to previous methods (2).
RNA isolation.
Twenty-five ml of log-phase L. monocytogenes 08-5923 (optical density at 600 nm [OD600] of 0.2) was grown in TSB or in TSB with 7 μg/ml CCLA (1/10 the MIC on L. monocytogenes 08-5923) for 4 h at 21°C (final OD600, 0.5) to obtain cells that were at the mid-log phase of growth. RNAprotect bacterial reagent (Qiagen Inc., Ontario, Canada) was added to the cell culture according to the manufacturer's instructions. Total RNA was isolated using the RNeasy minikit (Qiagen Inc.) and treated with DNase I (New England BioLabs Ltd., Ontario, Canada) on column according to the manufacturers' protocol. RNA quantity was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Ontario, Canada), and the samples were stored at −80°C. RNA quality and quantity were assessed on an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Ontario, Canada) using an Agilent Nano 6000 assay, and an RNA with an integrity number (RIN) of >7.0 was used for subsequent microarray experiments. For quantitative reverse transcriptase real-time PCR (qRT-PCR), total RNA was isolated as described above from cells in lag phase (15 min), mid-log phase (4 h), and early stationary phase (30 h).
Gene expression microarrays and data analysis.
The L. monocytogenes EGD-e 385 K gene expression microarray from Roche NimbleGen, including all of the 2,857 annotated open reading frames (ORFs) of the genome, was used. The cDNA was synthesized from RNA extracted from three independent biological repetitions (SOP#M007 and SOP#M008; J. Craig Venter Institute [http://pfgrc.jcvi.org/index.php/microarray/protocols]) and labeled with Cy3 monoreactive dye (GE Healthcare Life Sciences, Ontario, Canada). Hybridization of the labeled cDNA probes using the NimbleGen hybridization station and washing were carried out according to the NimbleGen protocol. Microarray raw data were preprocessed and normalized by NimbleScan (version 2.6; Roche NimbleGen).
GeneSifter software (trial version; Geospiza, Inc.) (16) was used for the analysis of normalized results. The statistical analyses were the averages from three independent samples (CCLA-treated L. monocytogenes 08-5923 versus untreated) with the cutoff being a 2-fold change in expression level and P values of <0.05 to determine differentially expressed (DE) genes. Genes that showed 2-fold or greater changes in expression levels were selected for subsequent qRT-PCR.
Total cytosoluble protein identification.
L. monocytogenes 08-5923 was grown in TSB or in TSB with CCLA at 21°C for 4 h. Cells were pelleted by centrifugation at 10,600 × g for 1 min and washed once with 1× SigmaFAST protease inhibitor cocktail tablet (Sigma-Aldrich Ltd., Ontario, Canada) reconstituted in double-distilled water (ddH2O). Cell pellets were resuspended in the fresh cocktail, mixed 1:1 (vol/vol) with 0.1-mm Zirconia-silica beads (BioSpec Inc., OK), and lysed by bead beating for 45 s a total of 3 times. The cell envelope was removed by centrifugation at 10,600 × g for 2 min. The concentration of the total soluble protein was determined by Bio-Rad protein assay by following the manufacturer's instructions (Bio-Rad Laboratories Ltd., Ontario, Canada).
The total soluble proteins were separated by SDS-PAGE with 16% polyacrylamide (17). Five μg of proteins from the untreated and treated Listeria samples were loaded onto each lane and subjected to electrophoresis at 130 V for 80 min. The gel was fixed (45 min in a solution containing 50% methanol and 10% acetic acid), and protein bands were visualized by EZBlue gel staining (Sigma-Aldrich Ltd.). The density of protein bands was measured by AlphaEase FluorChem densitometry (Alpha Innotech; Fisher Scientific) to identify differentially expressed proteins.
The differentially expressed proteins in treated and untreated L. monocytogenes 08-5923 samples were analyzed by the Mass Spectrometry Facility, Department of Chemistry, University of Alberta. Protein bands that were differentially expressed were excised from the SDS-PAGE gel, digested in gel by trypsin (Sigma-Aldrich Ltd.), and analyzed by ultraperformance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer (UPLC-ESI-Q-TOF-MS) (Q-TOF Premier; Waters, MA). Proteins were identified from mass spectrometry data using the Mascot search engine (Matrix Science).
qRT-PCR and data analysis.
qRT-PCR experiments were performed to validate the differential expression of genes at different times of exposure to CCLA. The experimental design included three biological replicates and two technical replicates within each biological replicate. Genes that were ≥2-fold up- or downregulated (P < 0.05) from the microarray experiments, as well as genes encoding the differentially expressed proteins, were quantified using SYBR green (Invitrogen, Ontario, Canada). Primers (Tables 1 and 2) were designed (using Primer3 [http://frodo.wi.mit.edu/primer3/]) and tested for specificity with genomic DNA prior to the analysis. Total RNA was isolated and reverse transcribed into cDNA using SuperScript III reverse transcriptase (Promega, WI) according to the manufacturer's protocol. Quantifications of the transcripts were carried out (7500 Fast real-time PCR system; Applied Biosystems, CA) using a QuantiFast SYBR green PCR kit (Qiagen Inc.). Relative quantification values were obtained using a comparative threshold cycle method (ΔΔCT) (18). The housekeeping gene rpoB was selected as the reference. The CT slope method was used to validate the internal control (rpoB). rpoB was amplified from 10-fold serial dilutions of genomic DNA prepared from L. monocytogenes. The CT slope was linear, and the qRT-PCR efficiency of rpoB was 100.9%. A one-tailed unpaired t test was performed for determination of the significance (P < 0.05) in differential expression of genes.
TABLE 1.
Primers used in qRT-PCR for confirmation of the genes found differentially expressed in microarray experiments
| Gene | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| ftsE | TATGCGATGGAAGTGGTTGA | CTCACCACCGGAAAGTTCAT |
| ftsX | AATGGTTGGATGACCTTTGC | CGTTGCAAGCTTGTTCATGT |
| rpoB | GAAATCTGGGTTCGTCGTGT | GCTACGTTTGGACGTTGGTT |
| fliF | TCTACATGAACACGCCCAAA | AAAATGTTGCCGCTTTTGTC |
| clpE | AGCAAACTTTGGGTCGAATG | GTTCACGGTTTGCTTGGTTT |
| baiE | ACAAGCAAAGGCAGACATCC | CAATAAAGCCATGCCCAGTT |
| deoR | CAGGCCTTGAGCAAAATGAT | ATAATACGCGCGATTGGAAG |
| hisC | TGCGAAACGTTTTGAACAAG | CAGCACCAGAACGCGTAATA |
| flgD | AAATGGCGCAACTTTCCTTA | CGCCGTTTAGTGAAACACCT |
| yitT | TGGATTTTCCAGCGAATACC | ATCACTACCGCCAGTTGTCC |
| tgl | GACAAGCCGAGTTTCAAAGC | CGGCATCTGTGACAGTTTGT |
| lmo0867 | CAATCGGCTATCTCGCTTTT | TGCGATAATTGCCATCGTTA |
| ywzB | ACTTTTTGGGCACTTCAAGC | CCTAACACAATCGAAATAATCACAA |
| lmo1796 | CACCCGGAAGACGTTGTTAT | ACGAATCGCTTTCGCAATAC |
| ftsW | GGGATCGCTAGTCTGATTGC | TACCAAGCATCATCGACAGC |
| lmo2311 | GCTCAATGCGTTTATGACGA | CGCTGGCAAATAATCTCGTT |
| trn | CCGAAAAGAGTCGCGATAAT | TGCTGCATAGGTGGATGCTA |
| hrcA | GGGATATTCCCGATGGTCTT | TCTTGTGGGCCCTAGGAGTA |
| abc | AATGAGAGCGCCAATTATGG | TGCGCGATACCTTCTACAAA |
| fmt | AGAAGCCGACCTGCTTGTAA | GTCAAGCAGCGCGTAGTGTA |
| lmo2317 | CCCTTTTCAACCGTGATGTT | GCTCACGAACCCTTTCCATA |
| mfs | GGGGAAAAAGCTGGAATTGT | TTGCAATCGTTGGCATGTAT |
| malP | CAATTGGATGGAGCGAAAAT | CGCCATTACGGTGAATTTCT |
| mptC | ATTCCAGCTGCAGCACTTTT | CAACGGCAACTACCATTCCT |
| mbl | GCAATTCGTGAAGTTGCTGA | ACCGGAAGGCTCAAAAATCT |
| gidA | GGCGTGTGTAAAGGCGTTAT | GGTTATTTGGACCGCTTGAA |
| rodA | CTTCCGCTTGGTCTTGTAGC | CAATGAGCGCAATCGAACTA |
| divIVA | TCCGTGAGCGTTTACGTATG | CGCAAGTTCTGTCGCATCTA |
| murZ | GCGATTTATTTACGGGCAGA | TTGCACGAACAGCTGCTAAC |
| fliM | GTGTGACATCACCGAACGAC | TCTTCCACCGAGAAAAATGG |
| motB | TTCATGGAAGCGATCATTCA | CGCACCATGATTTCTACACG |
| mptAB | ACGTTTCGGCAATACCAAAG | ACAGAGTGAGCCATGGAACC |
| murE | TTATGGATCTCCGACCCAAG | ACGATACATCGTGCCAATCA |
| pdp | AGTGAAAATCGCACCGAAAG | GCAATAATTCCGAGGCATGT |
| agrB | CTCCGGCAGACACAGAAAGT | TGCGAATGGTATTAGCAACG |
| bglK | CCGAAAAATGGCTTGGTAAA | CCGCGAACTAATTCACCATT |
| cheA | CGACGTTCCGAATTGAGATT | CGTGAACGTGTTGAATGTCC |
| celC | TTGCAGCTATTCGTGACACC | CATACCAGATCCGCCGTAGT |
| dacB2 | ACCAAAAAGCGAGAGAAGCA | GGCATTTTCTTGCAACCATT |
| celA | ATTTCGAGGAAGCAGAAGCA | TTCCACTTTTTCACCGGAAG |
| celB | TGGATATCGTGATGCTTGGA | CGGCTTTCCCGTTTAACATA |
TABLE 2.
Primers used in qRT-PCR for confirmation of the downregulation of proteins
| Gene name | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| alaS | GGTAGCTGCTGGAAACGAAG | TTTCGCTCTCGATTGTTCCT |
| adhE | CAAGCTGGCTTCAAAGTTCC | ACGATCGAATGCTTCTGCTT |
| pflA | CCCACTTAACCCAGAAGCAG | TACCACCGTTGATAGCACCA |
| pnpA | TATTCACTCGTGGCCAAACA | GCCAGTTTCCCCAACACTAA |
| pykA | AAGTGCTGCAGTTGTTGTGG | TTGCATCTTTAGCACCAACG |
| gnd | GGAAGAGAATGCGGACAAAA | ATCTGTAGCATCGCCAGCTT |
| fri | GGCGAACAAATGGATGAAGT | GCTTCTTCTACGCTCGCATT |
| rpsK | TACTCGTAAACGCCGTGTGA | TAGGGAACCTGCACTTGACC |
| rpsH | AGTAAGCCAGGTTTGCGTGT | TTTAGCACGGGCTTCTTTGT |
| rplU | TGAAACAGGTGGGAAACAAA | TAGCGGAATCTCCACCTACG |
| rplL | AATTTGGCGTAACTGCTGCT | ACCAGTGATTTCACGAACCA |
| hup | GCAGCGAAAGCAGTAGAAGC | GTACGAGGGTTACGGCCTTT |
| rpsT | TTGATGAAGCAGCTGCAAAC | CGAGCAGCATTGTTTTTGTG |
| csp | GAAAAAGGCTTCGGTTTCATC | ATTCAACGCTTTGACCTTCG |
Cell morphology examination.
Culture of L. monocytogenes 08-5923 was fixed with 10% (vol/vol) formaldehyde and pelleted by centrifugation at 11,000 × g for 15 s. The cell pellet was resuspended in 0.1 M phosphate buffer containing 2.5% glutaraldehyde and 2% paraformaldehyde, stained with 2% sodium phosphotungstate (PTA), and examined using a transmission electron microscope (TEM; Philips Morgagni 268; FEI, OR) operating at 80 kV.
Microarray data accession number.
The complete microarray data set generated in this study is deposited for public access in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE56558.
RESULTS
Functional classification of DE genes.
cDNA microarray experiments were performed to compare the expression levels of genes in untreated and treated (CCLA for 4 h) L. monocytogenes. Global differential gene expression changes in response to CCLA were determined using a high-density NimbleGen microarray for L. monocytogenes of 2,857 genes. Forty genes were found to be differentially expressed with a minimum threshold of a 2-fold difference (P < 0.05) (Table 3). These genes were grouped into several functional categories, including cell division, cell wall synthesis, motility, transport, and translation (Table 3).
TABLE 3.
Identification of differentially expressed genes in CCLA-treated L. monocytogenes 08-5923a
| Gene name and function | Gene annotation | Product | Fold change |
|---|---|---|---|
| Cell division | |||
| clpE | lmo0997 | ATP-dependent protease | +3.69 |
| ftsW | lmo1071 | Cell division protein FtsW | −2.14 |
| divIVA | lmo2020 | Cell division initiation protein | −2.01 |
| rodA | lmo2428 | Similar to cell division protein RodA | −2.64 |
| ftsX | lmo2506 | Cell division transport system permease protein | −2.08 |
| ftsE | lmo2507 | Cell division transport system ATP-binding protein | −2.32 |
| mbl | lmo2525 | Rod shape-determining protein MreB | −2.03 |
| Cell wall synthesis | |||
| pdp | lmo0835 | Peptidoglycan binding protein | −2.21 |
| murE | lmo2038 | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | −2.02 |
| murZ | lmo2552 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | −2.41 |
| dacB2 | lmo1855 | d-Alanyl-d-alanine carboxypeptidase | −2.41 |
| Membrane functions | |||
| abc | lmo0767 | ABC transporter, permease protein | +2.13 |
| mfs | lmo1250 | Major facilitator superfamily | +2.05 |
| ywzB | lmo2527 | Similar to B. subtilis YwzB protein | +2.37 |
| mptAB | lmo0096 | PTS system, mannose-specific IIA and IIB component | +2.14 |
| mptC | lmo0097 | PTS system, mannose-specific IIC component | −2.29 |
| celA | lmo2765 | PTS system, cellobiose-specific IIA component | +2.35 |
| celB | lmo2762 | PTS system, cellobiose-specific IIB component | +2.10 |
| celC | lmo2763 | PTS system, cellobiose-specific IIC component | +2.81 |
| Gene regulation | |||
| deoR | lmo2107 | Similar to deoR family transcriptional regulator | +2.28 |
| hrcA | lmo1475 | Heat-inducible transcription repressor | +2.43 |
| agrB | lmo0048 | Similar to Staphylococcus two-component sensor histidine kinase AgrB | +3.18 |
| Nucleotide and protein synthesis | |||
| fmt | lmo1823 | Methionyl-tRNA formyltransferase | −2.07 |
| hisC | lmo1925 | Histidinol-phosphate aminotransferase | −2.26 |
| gidA | lmo2810 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme GidA | −2.61 |
| Metabolism | |||
| tgl | lmo0717 | Transglycosylase | −2.01 |
| baiE | lmo0754 | Bile acid 7-alpha dehydratase | −2.74 |
| trn | lmo1903 | Thioredoxin | −2.21 |
| yitT | lmo1909 | YitT family protein | −2.05 |
| malP | lmo2121 | Maltose phosphorylase | +2.01 |
| bglK | lmo2764 | Beta-glucoside kinase | +2.85 |
| Motility | |||
| flgD | lmo0696 | Flagellar basal body rod modification protein | −2.70 |
| fliF | lmo0713 | Flagellar MS-ring protein | −2.20 |
| motB | lmo0686 | Chemotaxis protein | −2.23 |
| fliM | lmo0699 | Flagellar motor switch protein | −2.26 |
| cheA | lmo0692 | Two-component sensor histidine kinase | −2.82 |
| Unknown | |||
| lmo0867 | lmo0867 | Hypothetical protein | −2.39 |
| lmo1796 | lmo1796 | Hypothetical protein | −2.33 |
| lmo2311 | lmo2311 | Hypothetical protein | +2.25 |
| lmo2317 | lmo2317 | Hypothetical protein | +2.07 |
Identification of differentially expressed genes in CCLA-treated (4 h) L. monocytogenes 08-5923 is represented as fold change (n = 3; P < 0.05). +, upregulation; −, downregulation.
Differential expression of total soluble proteins.
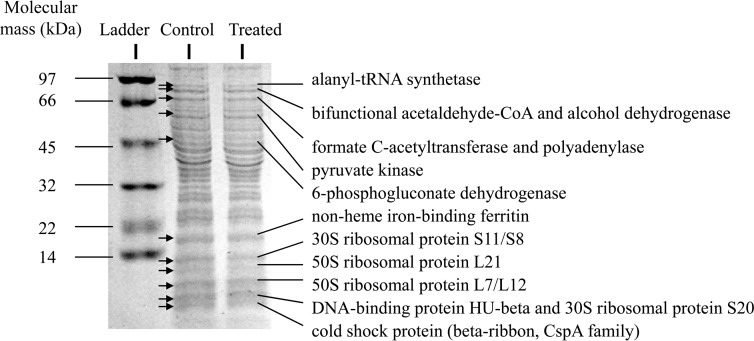
To gain further insight, the protein expression patterns of whole-cell lysate from L. monocytogenes with and without exposure to CCLA (4 h) were compared using SDS-PAGE. Out of 11 protein bands that were visibly different in density (Fig. 1) between the lysate from the CCLA-treated and untreated cells, there were 14 proteins identified by UPLC-ESI-Q-TOF-MS. The proteins were involved in glycolysis (PykA), pyruvate metabolism (AdhE and PflA), stress response (Csp protein), DNA replication (Hup), translation (Rps and Rpl proteins), and purine/pyrimidine biosynthesis (PnpA) (Table 4). The downregulation of PykA in treated L. monocytogenes confirmed the observed downregulation of the gene encoding this protein as shown in Fig. 2G.
FIG 1.
Differential production of proteins in L. monocytogenes 08-5923; treated (CCLA for 4 h) and control samples are shown on an SDS-PAGE gel. Proteins expressed differentially are indicated by arrows. CoA, coenzyme A.
TABLE 4.
Identification of differentially expressed proteins by MS
| Protein name | Product | Accession no. | Calculated pI | Nominal molecular mass (Da) | Differential expression |
|---|---|---|---|---|---|
| AlaS | Alanyl-tRNA synthetase | YP_003416804 | 5.13 | 92,938 | Down |
| AdhE | Bifunctional acetaldehyde-coenzyme A/alcohol dehydrogenase | YP_003416936 | 6.48 | 95,135 | Down |
| PflA | Formate C-acetyltransferase | CAK20840 | 5.41 | 83,950 | Down |
| PnpA | Polyadenylase | YP_003416626 | 5.23 | 79,780 | Down |
| PykA | Pyruvate kinase | YP_003416871 | 5.39 | 62,673 | Down |
| Gnd | 6-Phosphogluconate dehydrogenase | YP_003416672 | 5.11 | 52,497 | Down |
| Fri | Nonheme iron-binding ferritin | YP_003416184 | 4.86 | 18,036 | Down |
| RpsK | 30S ribosomal protein S11 | YP_003415461 | 11.40 | 13,834 | Down |
| RpsH | 30S ribosomal protein S8 | YP_003415450 | 9.48 | 14,635 | Down |
| RplU | 50S ribosomal protein L21 | YP_003416842 | 9.57 | 11,207 | Down |
| RplL | 50S ribosomal protein L7/L12 | YP_003415499 | 4.54 | 12,462 | Down |
| Hup | DNA-binding protein HU-beta | YP_003417289 | 9.65 | 9,876 | Down |
| RpsT | 30S ribosomal protein S20 | YP_003416777 | 10.70 | 9,163 | Down |
| Csp | Cold shock protein, CspA family | ADB71297 | 4.45 | 7,261 | Down |
FIG 2.
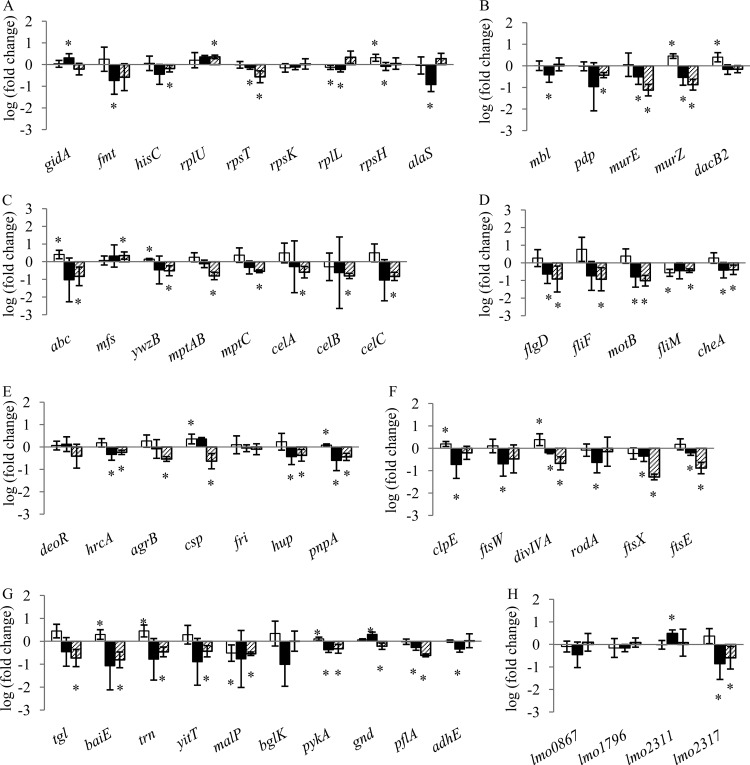
qRT-PCR measurement of DE genes in L. monocytogenes 08-5923 exposed to CCLA for 15 min (□), 4 h (■), and 30 h (▨). Untreated samples were used as a reference. Genes were grouped according to their biological function. (A) Nucleotide/protein synthesis; (B) cell wall synthesis; (C) membrane functions; (D) motility; (E) transcription regulation; (F) cell division; (G) metabolism; (H) unknown functions. Note the different y axes for panels C and D. Data are expressed as log values (fold change). Means ± standard deviations (n = 3) are given. An asterisk indicates genes significantly up- or downregulated at P < 0.05.
Confirmation of DE genes in L. monocytogenes.
To validate results from microarray experiments and to investigate the gene expression changes over time, a total of 54 genes (40 DE genes from microarray experiments and 14 DE genes from SDS-PAGE) (Fig. 2) were selected for qRT-PCR, and gene expression levels were quantified following treatment with CCLA after 15 min (lag phase), 4 h (log phase), and 30 h (early stationary phase). For a number of the genes, expression was significantly (P < 0.05) upregulated after 15 min of exposure to CCLA and downregulated after 4 h and 30 h.
The upregulation of genes encoding proteins involved in metabolism and membrane functions was observed upon 15 min of exposure to a sublethal dose of CCLA. Genes encoding proteins involved in metabolism, including bile acid 7-alpha dehydratase (BaiE), thioredoxin (Trn), and pyruvate kinase (PykA), were significantly upregulated (P < 0.05). The genes involved in cell wall synthesis (murZ and dacB2) were upregulated (P < 0.05). Genes involved in cell division, including clpE and divIVA, were upregulated, while malP was downregulated in cells exposed to CCLA for 15 min. After 4 h of exposure to CCLA, genes involved in cell division (clpE, ftsW, divIVA, rodA, ftsX, and ftsE), cell wall synthesis (mbl, murE, and murZ), flagellar synthesis (flgD, motB, and cheA), and metabolism (pykA and gnd) were significantly downregulated. Upregulation of lmo2311, which is regulated by σ54 (19), was observed. After the cells were exposed to CCLA for 30 h, more genes were downregulated compared to those after 4 h of exposure. These genes were involved in cell division, cell wall synthesis, membrane function, transcription regulation, metabolism, and motility (Fig. 2). Both the mannose-specific PTS genes (mpt genes) and the cellobiose-specific PTS genes, such as celA, celB, and celC, were downregulated (P < 0.05).
Cell morphology.
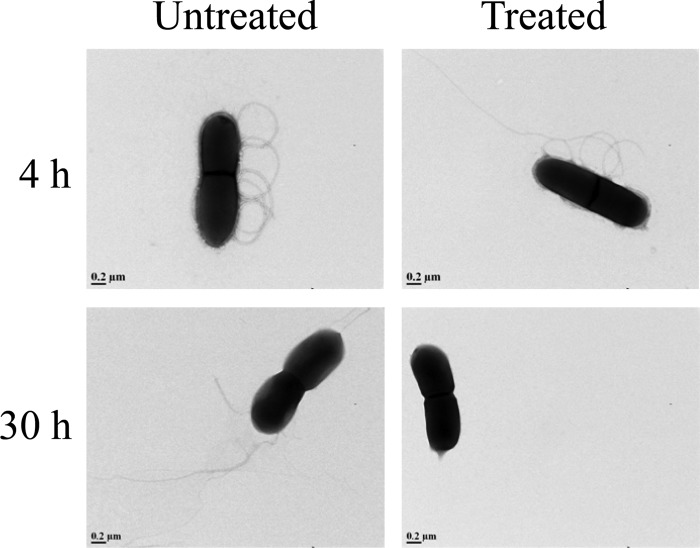
The morphologies of untreated and treated (CCLA for 4 h or 30 h) L. monocytogenes 08-5923 samples were compared using TEM to examine whether exposure to CCLA could result in visible damage to the cell structure. No morphological changes were observed after 4 h of treatment of L. monocytogenes 08-5923 with CCLA (Fig. 3). However, after 30 h the cells had no flagella attached to the cell surface (Fig. 3). This corresponds to the downregulation of motility genes observed previously.
FIG 3.
Transmission electron micrographs of cells of L. monocytogenes 08-5923 in the presence (treated, 4 h and 30 h) and absence (untreated) of CCLA (28,000× magnification).
DISCUSSION
Commercial preservatives that are used to control the growth of L. monocytogenes in RTE meats include Micocin, which contains carnocyclin A along with two class IIa bacteriocins, carnocyclin BM1 and piscicolin 126. Although the stress responses of L. monocytogenes to class IIa bacteriocins have been characterized, the response to class IIc cyclic bacteriocins has not been studied to date. Our study described the cellular alteration and adaptation of L. monocytogenes when exposed to a sublethal dose of a class IIc bacteriocin, which provides insight into the general stress response and resistance mechanisms to bacteriocins.
Treatment of L. monocytogenes 08-5923 with CCLA resulted in differential expression of genes that are known to be involved in cell division, cell wall synthesis, membrane function, transcriptional regulation, nucleotide and protein synthesis, metabolism, and cell motility. The PTS function, cell wall synthesis, motility, and cell division genes were affected the most. The downregulation of these genes was not because of inhibition of cell growth, as at each time point the OD600 values of treated and untreated cells were comparable. No filamentation of cells was observed. This indicates that the downregulation of these genes was due to the adaptation to CCLA.
The upregulation of σ54-regulated mptAB and cel detected by cDNA microarray after 4 h of exposure to CCLA indicates an immediate adaptive response of Listeria cells, as the mannose-specific (7) and cellobiose-specific PTS systems are receptors for bacteriocins (20), and the upregulation of mannose-specific PTS systems has been observed in bacteriocin-sensitive cells (21, 22). Gong et al. (3) demonstrated that CCLA forms ion-selective channels in lipid bilayers, which indicates that CCLA interaction with membranes is not receptor mediated. However, our microarray results suggested that both the mannose-specific and cellobiose-specific PTS systems are receptors for CCLA. The downregulation of the mannose-specific and cellobiose-specific PTS systems observed by qRT-PCR analysis after 30 h of exposure to CCLA demonstrates that Listeria limits the expression of genes to minimize the production of the PTS targets to CCLA. This downregulation of the PTS targets could play a role in adaptation to CCLA, as these targets have been suggested to play a role in resistance to class II bacteriocins (6, 23, 24).
The expression of genes involved in cell wall synthesis (mur) followed the same pattern as that of the PTS systems after 30 h of exposure to CCLA. Although the function of Mur proteins in the antimicrobial stress response of Listeria has not been described in the literature, Mur is the target of antibiotics in Staphylococcus aureus (25).
Absence of flagella in Listeria cells exposed to CCLA for 30 h could be a result of inhibition of flagellar synthesis, because the basal body of the flagella may not develop properly as genes encoding various components of flagella, including the MS ring (fliF), motor switch (fliM), motor rotation (motB), and rod synthesis (flgD), were downregulated. In addition, cheA, a chemotaxis gene responsible for virulence (26), was downregulated. Flagellar motility is also important for biofilm formation of L. monocytogenes (27). Others have found that stress from antimicrobials can inhibit the synthesis of flagella in E. coli (28), but the impact of bacteriocins on flagellar synthesis in L. monocytogenes has not been described in the literature. Lack of flagella can inhibit adhesion of Listeria (29) and may affect its ability to form biofilms.
The cell division gene ftsE was upregulated when L. monocytogenes was exposed to CCLA for 15 min. High pressure for 15 min also upregulates ftsE in Listeria (30). After 4 h of exposure to CCLA, ftsE was downregulated, along with ftsX and ftsW. Gene ftsE in L. monocytogenes is regulated by σB (31), which is a general stress response regulator. Various stresses, such as liquid smoke (32), cold (33), antibiotic (31, 34), and heat (35), also downregulate fts genes in Listeria. The up- or downregulation of fts genes may depend on the type of stress and time of exposure.
In summary, a sublethal dose of carnocyclin A targeted the PTS systems of Listeria and cells began to repair injury by upregulation of genes involved in cell wall synthesis. After 4 h, cells gradually became desensitized to CCLA and started to decrease activities such as cell division, cell wall synthesis, and motility. After 30 h, cells adapted to CCLA through a reduction of several metabolic processes and motility functions. Adaptation to bacteriocins could affect the sensitivity to antimicrobials that are used to control the growth of L. monocytogenes.
ACKNOWLEDGMENTS
This project was supported by funding from the Alberta Livestock and Meat Agency and Alberta Innovates BioSolutions.
We thank Jing Zheng of the Mass Spectrometry Facility, Department of Chemistry, University of Alberta, for mass spectrometry analysis and Arlene Oatway of the Advanced Microscopy Facility, Department of Biological Sciences, for technical support in TEM analysis.
Footnotes
Published ahead of print 18 April 2014
REFERENCES
- 1.Hof H. 2003. History and epidemiology of listeriosis. FEMS Immunol. Med. 35:199–202. 10.1016/S0928-8244(02)00471-6 [DOI] [PubMed] [Google Scholar]
- 2.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756–4763. 10.1128/AEM.00817-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong X, Martin-Visscher LA, Nahirney D, Vederas JC, Duszyk M. 2009. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim. Biophys. Acta 1788:1797–1803. 10.1016/j.bbamem.2009.05.008 [DOI] [PubMed] [Google Scholar]
- 4.Vadyvaloo V, Arous S, Gravesen A, Héchard Y, Chauhan-Haubrock R, Hastings JW, Rautenbach M. 2004. Cell-surface alterations in class IIa bacteriocin-resistant Listeria monocytogenes strains. Microbiology 150:3025–3033. 10.1099/mic.0.27059-0 [DOI] [PubMed] [Google Scholar]
- 5.Tessema GT, Møretrø T, Kohler A, Axelsson L, Naterstad K. 2009. Complex phenotypic and genotypic responses of Listeria monocytogenes strains exposed to the class IIa bacteriocin sakacin P. Appl. Environ. Microbiol. 75:6973–6980. 10.1128/AEM.00608-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl. Environ. Microbiol. 77:3335–3342. 10.1128/AEM.02602-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diep DB, Skaugen MSZ, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389. 10.1073/pnas.0608775104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jansch L, Hechard Y, Hastings JW, Knochel S. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361–2369 http://mic.sgmjournals.org/content/148/8/2361 [DOI] [PubMed] [Google Scholar]
- 9.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2010. Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains? Appl. Environ. Microbiol. 76:4216–4232. 10.1128/AEM.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sue D, Fink D, Wiedmann M, Boor KJ. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843–3855. 10.1099/mic.0.27257-0 [DOI] [PubMed] [Google Scholar]
- 11.Wemekamp-Kamphuis HH, Wouters JA, de Leeuw PPLA, Hain T, Chakraborty T, Abee T. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457–3466. 10.1128/AEM.70.6.3457-3466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robichon D, Gouin E, Débarbouillé M, Cossart P, Cenatiempo Y, Héchard Y. 1997. The rpoN (σ54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591–7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira A, O'Byrne CP, Boor KJ. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454–4457. 10.1128/AEM.67.10.4454-4457.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. 2006. The sigma factor RpoN (σ54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol. Lett. 263:54–60. 10.1111/j.1574-6968.2006.00405.x [DOI] [PubMed] [Google Scholar]
- 15.Gilmour MW, Graham M, van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. 10.1186/1471-2164-11-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reimers M. 2010. Making informed choices about microarray data analysis. PLoS Comput. Biol. 6:e1000786. 10.1371/journal.pcbi.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analyt. Biochem. 166:368–379. 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 19.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hébraud M, Héchard Y. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581–1590. 10.1099/mic.0.26860-0 [DOI] [PubMed] [Google Scholar]
- 20.Campelo AB, Gaspar P, Roces C, Rodríguez A, Kok J, Kuipers OP, Neves AR, Martínez B. 2011. The Lcn972-bacteriocin plasmid pBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 77:7576–7585. 10.1128/AEM.06107-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjos M, Nes IF, Diep DB. 2009. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 155:2949–2961. 10.1099/mic.0.030015-0 [DOI] [PubMed] [Google Scholar]
- 22.Ramnath M, Arous S, Gravesen A, Hastings JW, Héchard Y. 2004. Expression of mptC of Listeria monocytogenes induces sensitivity to class IIa bacteriocins in Lactococcus lactis. Microbiology 150:2663–2668. 10.1099/mic.0.27002-0 [DOI] [PubMed] [Google Scholar]
- 23.Williams T, Joseph B, Beier D, Goebel W, Kuhn M. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252:287–298. 10.1016/j.femsle.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 24.Giotis ES, Muthaiyan A, Blair IS, Wilkinson BJ, McDowell DA. 2008. Genomic and proteomic analysis of the alkali-tolerance response (AlTR) in Listeria monocytogenes 10403S. BMC Microbiol. 8:102. 10.1186/1471-2180-8-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill AJ, Lindsay JA, Gould K, Hinds J, Chopra I. 2009. Transcriptional signature following inhibition of early-stage cell wall biosynthesis in Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1701–1704. 10.1128/AAC.01309-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dons L, Eriksson E, Jin Y, Rottenberg ME, Kristensson K, Larsen CN, Bresciani J, Olsen JE. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72:3237–3244. 10.1128/IAI.72.6.3237-3244.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemon KP, Higgins DE, Kolter R. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189:4418–4424. 10.1128/JB.01967-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burt SA, van der Zee R, Koets AP, de Graaff AM, van Knapen F, Gaastra W, Haagsman HP, Veldhuizen EJ. 2007. Carvacrol induces heat shock protein 60 and inhibits synthesis of flagellin in Escherichia coli O157:H7. Appl. Environ. Microbiol. 73:4484–4490. 10.1128/AEM.00340-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tresse O, Lebret V, Benezech T, Faille C. 2006. Comparative evaluation of adhesion, surface properties, and surface protein composition of Listeria monocytogenes strains after cultivation at constant pH of 5 and 7. J. Appl. Microbiol. 101:53–62. 10.1111/j.1365-2672.2006.02968.x [DOI] [PubMed] [Google Scholar]
- 30.Bowman JP, Bittencourt CR, Ross T. 2008. Differential gene expression of Listeria monocytogenes during high hydrostatic pressure processing. Microbiology 154:462–475. 10.1099/mic.0.2007/010314-0 [DOI] [PubMed] [Google Scholar]
- 31.Shin JH, Kim J, Kim SM, Kim S, Lee JC, Ahn JM, Cho JY. 2010. σB-dependent protein induction in Listeria monocytogenes during vancomycin stress. FEMS Microbiol. Lett. 308:94–100. 10.1111/j.1574-6968.2010.01998.x [DOI] [PubMed] [Google Scholar]
- 32.Guilbaud M, Chafsey I, Pilet MF, Leroi F, Prévost H, Hébraud M, Dousset X. 2008. Response of Listeria monocytogenes to liquid smoke. J. Appl. Microbiol. 104:1744–1753. 10.1111/j.1365-2672.2008.03731.x [DOI] [PubMed] [Google Scholar]
- 33.Mattila M, Somervuo P, Rattei T, Korkeala H, Stephan R, Tasar T. 2012. Phenotypic and transcriptomic analyses of sigma L-dependent characteristics in Listeria monocytogenes EGD-e. Food Microbiol. 32:152–164. 10.1016/j.fm.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 34.Nielsen PK, Andersen AZ, Mols M, van der Veen S, Abee T, Kallipolitis BH. 2012. Genome-wide transcriptional profiling of the cell envelope stress response and the role of LisRK and CesRK in Listeria monocytogenes. Microbiology 158:963–974. 10.1099/mic.0.055467-0 [DOI] [PubMed] [Google Scholar]
- 35.van der Veen S, Hain T, Wouters JA, Hossain H, de Vos WM, Abee T, Chakraborty T, Wells-Bennik MH. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593–3607. 10.1099/mic.0.2007/006361-0 [DOI] [PubMed] [Google Scholar]