Abstract
The type III secretion system (T3SS), encoded by hrp (hypersensitive response and pathogenicity) genes in Gram-negative phytopathogenic bacteria, delivers repertoires of T3SS effectors (T3SEs) into plant cells to trigger the hypersensitive response (HR) in nonhost or resistant-host plants and promote pathogenicity in susceptible plants. The expression of hrp genes in Xanthomonas is regulated by two key regulatory proteins, HrpG and HrpX. However, the interactions between hrp gene products in directing T3SE secretion are largely unknown. Here we demonstrated that HrcT of X. oryzae pv. oryzicola functions as a T3SS component and positively regulates the expression of hrpX. Transcription of hrcT occurs via two distinct promoters; one (T1) is with the hrpB operon and the second (T3) within hrpB7 Via either promoter T1 or T3, the defect in Hrp phenotype by hrcT deletion was corrected in the presence of hrcT only from Xanthomonas species but not from other phytopathogenic bacteria. An N-terminally truncated HrcT was able to bind the hrpX promoter and activate the expression of hrpX, supporting that HrcT is a positive regulator of hrpX. A revised model showing the regulatory interactions between HrcT, HrpX, and HrpG is proposed.
INTRODUCTION
The successful infection of crop plants by Gram-negative plant-pathogenic bacteria is largely dependent on the type III secretion system (T3SS), which delivers repertoires of T3SS effectors (T3SEs) into plant cells to promote disease development (1). It has been well documented that the hrp genes encode the T3SS and control the ability of phytopathogenic bacteria to trigger the hypersensitive response (HR) in resistant-host or nonhost plants and pathogenicity in susceptible hosts (2, 3). Based on their genetic organization and transcriptional regulation, hrp gene clusters have been divided into two main groups (4, 5). The hrp genes of Erwinia amylovora, Pseudomonas syringae, and Dickeya spp. are typical representatives of group I. The expression of hrp genes in group I is modulated by the alternative sigma factor HrpL, which binds to a conserved hrp box to activate transcription (6–8). The hrp genes in group II, which includes Ralstonia solanacearum and Xanthomonas spp., are regulated by HrpG and HrpX (the latter protein is designated HrpB in Ralstonia) (9–12). HrpG is an OmpR family protein that belongs to response regulators of the two-component signal transduction system and putatively receives phosphorylation from HpaS (12, 13). HrpX is an AraC-type transcriptional activator that forms a homodimer and contains a helix-turn-helix (HTH) motif (11). The HTH motif of HrpX interacts with the plant-inducible promoter (PIP) box (TTCGC-N15-TTCGC) in hrp transcripts by binding to the TTCGC sequence (14, 15). However, evidence demonstrating that HrpX is directly regulated by HrpG is lacking from the literature.
In xanthomonads, the hrp-hrc-hpa genes are highly conserved and clustered within several sequenced genomes (16–20). Our previous studies revealed that the hrp cluster in X. oryzae pv. oryzicola, which causes bacterial leaf streak (BLS) in rice, is composed of 10 hrp, nine hrc (hrp-conserved), and eight hpa (hrp-associated) genes (3, 21). Comparative genomic analysis has revealed that at least nine hrc genes (e.g., hrcC, hrcT, hrcN, hrcJ, hrcU, hrcV, hrcQ, hrcR, and hrcS) are conserved among plant- and animal-pathogenic bacteria (22). It has been proposed that the T3SS consists of ring structures spanning the inner membrane (IM) and outer membrane (OM); these structures comprise a transport channel with an inner diameter of 2 to 3 nm (23). In Xanthomonas spp., the IM rings and export apparatus are comprised of HrcR, HrcS, HrcT, HrcU, HrcQ, and HrcV. These six proteins assemble into a structure that is connected to a predicted cytoplasmic C ring (HrcC) and an ATPase complex (HrcN) via an inner membrane protein, HrcJ (22). The core secretion apparatus is presumably associated with an extracellular Hrp pilus that serves as a transport channel for secreted T3SEs into the host cell cytosol (24–26).
hrcT is the eighth gene in the hrpB operon, which consists of hrpB1, hrpB2, hrcJ, hrpB4, hrpB5, hrcN, hrcB7, and hrcT (21, 27). Previous studies indicated that the expression of the hrpB operon is HrpX dependent because a PIP box was present in the hrpB operon promoter (11). Since the expression of HrpX is presumably controlled by HrpG, hrcT should not be transcribed in a hrpG mutant. However, our previous results showed that hrcT is expressed in the hrpG mutant of X. oryzae pv. oryzicola (2), implying a more complex regulatory paradigm for HrcT in bacterial pathogenesis.
In X. campestris pv. vesicatoria, topology analysis indicates that 41 amino acid residues at the N terminus of HrcT span the bacterial IM (1). We previously demonstrated that the hrcT mutant of X. oryzae pv. oryzicola was unable to induce HR in tobacco and failed to cause BLS in susceptible rice cultivars (2). In the present study, we conduct a more detailed analysis of hrcT expression and its contribution to pathogenicity. We also explore whether other hrp genes are regulated by HrcT and evaluate the impact of an hrcT deletion on the secretion of T3SEs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultured in LB (Luria-Bertani) medium at 37°C (28). All strains of X. oryzae pv. oryzicola and X. axonopodis pv. citri strain 306 were grown in nutrient agar (NA), nutrient broth (NB), NA without sucrose (NAN), NA with 10% sucrose (NAS), or XOM3 at 28°C (2, 29). R. solanacearum GMI1000 was grown in BG medium at 30°C (30), and P. syringae pv. tomato DC3000 was grown in King's B (KB) medium at 28°C (31). Antibiotics were used at the following final concentrations (μg/ml) when required: rifampin (Rif), 50; kanamycin (Km), 25; ampicillin (Ap), 100; spectinomycin (Sp), 50; and gentamicin (Gm), 20.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| X. oryzae pv. oryzicola | ||
| RS105 | Wild type, causal agent of bacterial leaf streak in rice, Rifr | This lab |
| RΔhrpG | hrpG deletion mutant of RS105, Rifr | 2 |
| RΔhrpX | hrpX deletion mutant of RS105, Rifr | 2 |
| RΔhrpV | hrcV deletion mutant of RS105, Rifr | 2 |
| RΔhrcT | hrcT deletion mutant of RS105, Rifr | 2 |
| RΔBP | RS105 mutant containing deletion of hrpB1 promoter and insertion of Gmr, Rifr | This study |
| C1RΔhrcT | Complemented strain of RΔhrcT with plasmid pChrcT1, Rifr Gmr | This study |
| C2RΔhrcT | Complemented strain of RΔhrcT with plasmid pChrcT3, Rifr Gmr | This study |
| C1RΔhrcTXac | Complemented strain of RΔhrcT with plasmid pC1HrcTXac, Rifr Gmr | This study |
| C1RΔhrcTRs | Complemented strain of RΔhrcT with plasmid pC1HrcTRs, Rifr Gmr | This study |
| C1RΔhrcTPst | Complemented strain of RΔhrcT with plasmid pC1HrcTPst, Rifr Gmr | This study |
| C2RΔhrcTXac | Complemented strain of RΔhrcT with plasmid pC2HrcTXac, Rifr Gmr | This study |
| C2RΔhrcTRs | Complemented strain of RΔhrcT with plasmid pC2HrcTRs, Rifr Gmr | This study |
| C2RΔhrcTPst | Complemented strain of RΔhrcT with plasmid pC2HrcTPst, Rifr Gmr | This study |
| X. axonopodis pv. citri | Strain 306 | Collected by this lab |
| R. solanacearum | Strain GMI1000 | Collected by this lab |
| P. syringae pv. tomato | Strain DC3000 | Collected by this lab |
| E. coli | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44λ− thi-1 gyrA96 relA1 | Clontech |
| BL21(DE3) | F− ompT hsdS20 gal | Novagen |
| Plasmids | ||
| pMD18-T | pUC ori, cloning vector, Apr | TaKaRa |
| pUFR034 | incW mob(p) mob+ lacZA+, PK2 replicon, Kmr | 36 |
| pBBR1MCS-5 | Broad-host-range cloning vector, Gmr | 49 |
| pKMS1 | Suicide vector derived from pK18mobGII, sacB+, Kmr | This lab |
| pT1GUS | pUFR034 expressing gusA under 350-bp promoter region of hrpB operon, Kmr | 2 |
| pT3GUS | pUFR034 expressing gusA under 500-bp promoter region of hrcT, Kmr | This study |
| pXGUS | pUFR034 expressing gusA under 300-bp promoter region of hrpX, Kmr | This study |
| pGGUS | pUFR034 expressing gusA under 490-bp promoter region of hrpG, Kmr | This lab |
| pKMSΔGm | 1,977-bp fragment containing the left and right border fragments of the hrpB operon promoter, flanks the Gm ORF in pKMS1, Kmr | This study |
| pChrcT1 | pBBR1MCS-5 expressing X. oryzae pv. oryzicola hrcT from a 350-bp promoter region derived from hrpB operon, Gmr | This study |
| pChrcT3 | pBBR1MCS-5 expressing hrcT from a 500-bp promoter region of hrcT Gmr | This study |
| pC1HrcTXac | pBBR1MCS-5 expressing hrcT from X. axonopodis pv. citri 306; promoter was derived from 350-bp promoter region derived from hrpB operon, Gmr | This study |
| pC1HrcTRs | pBBR1MCS-5 expressing hrcT from R. solanacearum GMI1000; promoter was derived from 350-bp promoter region derived from hrpB operon, Gmr | This study |
| pC1HrcTPs | pBBR1MCS-5 expressing hrcT from P. syringae pv. tomato DC3000; promoter was derived from 350-bp promoter region derived from hrpB operon, Gmr | This study |
| pC2HrcTXac | pBBR1MCS-5 expressing hrcT from X. axonopodis pv. citri 306; promoter was derived from 500-bp region of X. oryzae pv. oryzicola hrcT, Gmr | This study |
| pC2HrcTRs | pBBR1MCS-5 expressing hrcT from R. solanacearum GMI1000; promoter was derived from 500-bp region of X. oryzae pv. oryzicola hrcT, Gmr | This study |
| pC2HrcTPst | pBBR1MCS-5 expressing hrcT from P. syringae pv. tomato DC3000; promoter was derived from 500-bp region of hrcT, Gmr | This study |
| pHZWavrXa27 | AvrXa27 with Flag tag under the control of lacZ promoter, Spr, Apr | Yang Bing's lab |
| pET30a(+) | pBR322 origin, lacI, His tag/S tag; Kmr | Novagen |
| pETHrcTΔ41N | hrcT from RS105 cloned in pET30a(+) as a 711-bp fragment, His tag at C terminus, Kmr | This study |
| phrpX | The 300-bp promoter of hrpX fused to a promoterless gusA in pBI121, Kmr | This study |
| HrcT | hrcT from RS105 fused under CaMV 35S promoter in pCAMBIA1300, Kmr | This study |
| pOs8N3 | Os8N3 promoter fused to a promoterless gusA in pBI121, Kmr | 37 |
| PthXo1 | pthXo1 fused under CaMV 35S promoter in pCAMBIA1300, Kmr | 37 |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Rifr, rifampin resistance; Spr, spectinomycin resistance; Gmr, gentamicin resistance.
DNA manipulations.
DNA isolation and cloning and PCR were performed using standard procedures (32). The mobilization of plasmids into X. oryzae pv. oryzicola was performed as described previously (2). Restriction enzymes and DNA ligases were used as recommended by the manufacturer (Promega, Shanghai). Primers (see Table S1 in the supplemental material) were synthesized by Invitrogen. Ex-Taq (TaKaRa Bio Inc.) was used in PCR assays as recommended by the manufacturer.
Mutation construction in the hrpB operon promoter.
To inactivate the hrpB operon promoter, an 876-bp open reading frame (ORF) encoding gentamicin resistance (primers Gm-F/Gm-R) was inserted into the hrpB operon promoter region in the opposite direction of transcription. Two primer pairs, Gm-IF/Gm-IR and Gm-IIF/Gm-IIR (see Table S1 in the supplemental material), were used to amplify the left and right fragments flanking the hrpB operon promoter using RS105 genomic DNA as the template. PCR products were digested based on the restriction sites incorporated into primers and cloned into a vector pKMS1, resulting in the construct pKMSΔGm (Table 1), according to our previous method (33). The deletion of the hrpB operon promoter and insertion of the Gmr ORF was achieved using the sacB mutagenesis procedure described previously (33). The mutant containing a Gmr insertion in the hrpB operon promoter was verified by PCR with the primers Gm-IF/Gm-IIR (see Table S1 in the supplemental material) and named RΔBP (Table 1).
Complementation of mutants.
An 831-bp DNA fragment containing the intact hrcT gene was amplified by PCR using genomic DNA of X. oryzae pv. oryzicola RS105 as the template and primer pairs hrcT-F/hrcT-R (see Table S1 in the supplemental material). Similarly, a 350-bp fragment containing the hrpB operon promoter region (designated pT1) was amplified using primers phrcT1F/phrcT1R (see Table S1). A 500-bp fragment located upstream of the hrcT start codon (pT3) was amplified with primers phrcT3F/phrcT3R (see Table S1). After confirmation by sequence analysis, the T1 and T3 promoters and the promoterless hrcT were cloned into pBBR1MCS-5 vector at KpnI and HindIII sites to create recombinant plasmids pChrcT1 and pChrcT3, respectively (Table 1). The promoters of pT1 and pT3 were also fused with promoterless hrcT homologs from X. axonopodis pv. citri strain 306, R. solanacearum GMI1000, and P. syringae pv. tomato DC3000 to respectively create pC1HrcTXac, pC1HrcTRs, and pC1HrcTPst (with pT1) and pC2HrcTXac, pC2HrcTRs, and pC2HrcTPst (with PT3). The recombined plasmids were then transferred into the mutant RΔhrcT by electroporation, and transformants were selected on NA containing Gm. Representative transformants containing the different promoter constructs and hrcT homologs were verified by colony PCR and named C1RΔhrcT, C2RΔhrcT, C1RΔhrcTXac, C1RΔhrcTRs, C1RΔhrcTPst, C2RΔhrcTXac, C2RΔhrcTRs, andC2RΔhrcTPst (Table 1).
Determination of the hrcT promoter.
5′ rapid amplification of cDNA ends (5′-RACE) was used to determine the transcriptional start site of hrcT. X. oryzae pv. oryzicola strain RΔBP was incubated in XOM3 medium at 28°C for 16 h, and total RNA was extracted from the mutant RΔBP using an RNAiso Plus kit (TaKaRa, Dalian, China). Isolated RNA was treated with RNase-free DNase I at 37°C for 2.5 h, followed by a second purification using an RNase-free column. cDNA fragments were obtained using the 5′-Full RACE kit (TaKaRa, Dalian, China), and an anchor sequence was added to the 5′ end of the cDNA using terminal deoxynucleotide transferase. The tailed cDNA was then amplified using nested gene-specific primers hrcTO-R and hrcTI-R and RACE outer primer O-F and inner primer I-F (see Table S1 in the supplemental material). 5′-RACE products were cloned into pMD18-T and sequenced.
Pathogenicity and HR assays.
HR and pathogenicity assays were performed as described previously (3). Briefly, X. oryzae pv. oryzicola strains were grown in NB, adjusted to 3 × 108 CFU/ml (optical density at 600 nm [OD600], 0.3), and inoculated into leaves of rice seedlings (Oryza sativa cv. IR24, 2 weeks old) with needleless syringes to assess the formation of water-soaked lesions. Adult rice plants (cv. IR24, 2 months old) were inoculated by leaf needling for lesion length measurement. The T3SS mutant RΔhrcV was used as a negative control, and bacterial growth was monitored during the experiment as described previously (3). Strains (OD600 = 0.01) were also tested for the ability to elicit HR on Nicotiana benthamiana (3). All plants were maintained in a greenhouse at 25°C with a 12-h photoperiod and 75 to 80% relative humidity. Experiments were repeated three times.
qRT-PCR.
The cultivation of rice suspension cells and real-time quantitative RT-PCR (qRT-PCR) were performed as described previously (34) using the primers listed in Table S1 in the supplemental material. Total RNA was extracted from each treatment using TRIzol, and cDNA synthesis and PCR were conducted as described previously (34); gyrB was used as an internal standard. qRT-PCR was performed using the Applied Biosystems 7500 real-time PCR system and SYBR Premix Ex Taq (TaKaRa, China). The comparative threshold method was used to calculate the relative mRNA levels. All qRT-PCR experiments were performed three or more times.
GUS activity assays.
To construct transcriptional fusions to glucuronidase, the promoter regions of target genes were fused to a promoterless gusA with its ribosome binding site (35). Promoters T1 (350 bp upstream of hrpB1 translational start codon), T3 (500 bp upstream of hrcT translational start codon), and pG and pX (located 300 bp upstream of the hrpG or hrpX translational start codon) were amplified by PCR using total genomic DNA of the wild-type RS105 as the template with primer pairs phrcT1gF/phrcT1gR, phrcT3gF/phrcT3gR, phrpGgF/phrpGgR, and phrpXgF/phrpXgR (see Table S1 in the supplemental material), respectively, fused to the promoterless gusA gene, and then cloned into the EcoRI and BamHI sites of pUFR034 (36), resulting in pT1GUS, pT3GUS, pGGUS, and pXGUS (Table 1). Bacterial strains containing GUS transcriptional fusions were incubated in either NB or XOM3 at 28°C for 12 h and examined for GUS activity as described previously (2).
In vivo binding of HrcT to hrpX promoter.
To examine whether or not HrcT binds the hrpX promoter in vivo, a 300-bp promoter of hrpX (300 bp upstream of the hrpX translational start codon) was fused to a promoterless gusA in a vector, pBI121, that was transferred into tobacco leaves (N. benthamiana) mediated by Agrobacterium (37). The hrcT ORF was then fused under the CaMV 35S promoter in a vector, pCAMBIA1300, that was also used for transient expression of a tested gene in tobacco mediated by Agrobacterium (37). The primers for the above-described constructs were pXF/pXR and TF/TR (see Table S1 in the supplemental material), respectively. The PCR-amplified fragments were digested with HindIII and XbaI and cloned into HindIII/XbaI sites in pBI121 or pCAMBIA1300, respectively, generating phrpX and HrcT (Table 1). The Agrobacterium-mediated transient expression assays were performed as described previously (37, 38). For the control, the plasmids PthXo1 containing pthXo1 gene in pCAMBIA1300 (37) and pOs8N3 harboring the Os8N3 promoter (targeted by PthXo1 and fused with the promoterless gusA) in pBI121 (37) (Table 1) were used. The GUS activity was determined 2 days postinfection (dpi) by stained leaf disks (0.8 cm in diameter) with X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) as previously described (38). The experiment was repeated three times at least.
HrcTΔ41N protein production and purification.
The partial coding region of hrcT (nucleotides 124 to 831) was amplified from genomic DNA of X. oryzae pv. oryzicola RS105 by PCR using primers T-F and T-hisR (see Table S1 in the supplemental material); the latter fragment includes a hexahistidine tag code. The amplified fragment was ligated into pMD18-T and then subcloned into the EcoRI/XhoI sites of pET30a (Novagen) to generate a construct pETHrcTΔ41N (Table 1), which expresses HrcT with a C-terminal His tag. This construct was transformed into E. coli strain BL21(DE3) (Invitrogen) to generate strain BLHrcTΔ41N (Table 1). The overexpression and purification of HrcTΔ41N were performed as previously described (39).
EMSA.
DNA fragments of different sizes upstream of hrpX with respect to the translational start codon were used as probes in electrophoretic mobility shift assays (EMSA). The 3′-end of the probes was biotinylated using the biotin 3′ end DNA labeling kit (Thermo Scientific). The biotinylated DNA fragments and protein HrcTΔ41N were incubated as recommended by the LightShift chemiluminescent EMSA kit (Thermo Sci). The reaction mixture was separated by electrophoresis in a 5% polyacrylamide gel (acrylamide/bisacrylamide, 29:1 [wt/wt]) in 0.5× Tris-borate-EDTA (TBE) buffer (44.5 mM Tris base, 44.5 mM boric acid, and 1 mM EDTA, pH 8.0). Samples were electrophoresed at 100 V after prerunning the gel for 30 min, and the gel was then transferred to nylon membranes (GE Healthcare). After UV cross-linking (15 min), the biotinylated probes were detected using the chemiluminescent nucleic acid detection kit (Thermo Scientific) as described by the manual.
Type III secretion assays.
Plasmid pHZWavrXa27 containing avrXa27 with a FLAG tag code (Table 1) was introduced into X. oryzae pv. oryzicola RS105, the mutant RΔhrcT, the complemented strains C1RΔhrcT and C2RΔhrcT, and RΔhrcV, separately. Strains (OD600 = 0.3) were inoculated into rice cv. 87-15 (containing Xa27) (40), and symptoms were assessed 48 hpi. To examine secretion of AvrXa27 via the T3SS in vitro, transformants were incubated in XOM3 at 28°C for 12 h, and total cell extracts (TEs) and culture supernatants (SNs) were analyzed by immunoblotting using a Flag antibody as described previously (21).
RESULTS
hrcT is transcribed by a promoter located within the hrpB operon.
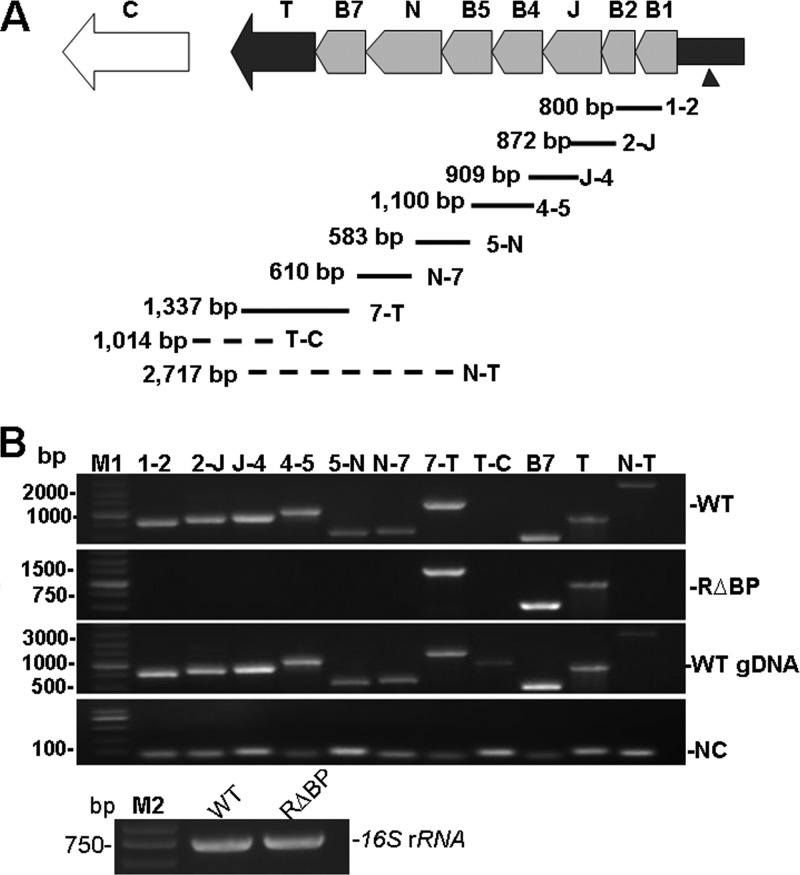
In Xanthomonas, the hrpB operon consists of eight hrp genes, hrpB1, hrpB2, hrcJ, hrpB4, hrpB5, hrcN, hrcB7, and hrcT, which are transcribed from a promoter region located upstream of hrpB1 (Fig. 1A). This promoter region contains a PIP box, and the expression of these eight genes is positively regulated by the transcriptional activator HrpX (2). Since the expression of HrpX is controlled by HrpG (12), a mutation in hrpG should result in impaired expression of these eight genes. However, the expression of hrcT in X. oryzae pv. oryzicola was still detectable in the hrpG mutant, and a putative promoter (T2) within hrpB5 was not responsible for hrcT expression (2), implying that hrcT is differentially regulated via an unknown promoter. To determine whether the hrpB operon contains an internal promoter, we generated a mutant, RΔBP (Table 1), which contains a deletion in the hrpB promoter and an insertion of the Gm gene cassette in opposition to the hrpB operon. After incubation in the hrp-inducing medium XOM3 (29) at 28°C for 12 h, mRNAs of the wild-type and mutant RΔBP strains were extracted and used for RT-PCR with primer sets designed to amplify intergenic regions within the hrpB operon (Fig. 1A; see Table S1 in the supplemental material). RT-PCR results indicated that transcription of hrpB1, hrpB2, hrcJ, hrpB4, hrpB5, and hrcN was dependent on the hrpB operon promoter (T1), since the transcription of these genes was not detected in the promoter T1 mutant (Fig. 1B). Interestingly, RT-PCR products of hrpB7 and hrcT were detected in both the wild-type and the mutant (RΔBP) strains, whereas the RT-PCR products from hrcN to hrcT were generated only in the wild type (Fig. 1B). These results indicate that the transcription of hrcT gene might be controlled by two promoters, the hrpB operon promoter (T1) and the other, unknown promoter upstream of hrcT (named T3) (Fig. 2A).
FIG 1.
Transcription of genes in the hrpB operon. (A) Graphic representation of the hrpB operon and RT-PCR products amplified using primers designed to span intergenic junctions. Arrows shaded in gray represent hrpB1 (B1), hrpB2 (B2), hrcJ (J), hrpB4 (B4), hrpB5 (B5), hrcN (N), and hrpB7 (B7). The black arrow represents hrcT (T), and the open white arrow depicts hrcC (C). The black rectangle adjacent to hrpB1 represents the hrpB operon promoter, and the black triangle indicates the insertion of the Gm cassette in opposition to the hrpB operon promoter (mutant RΔBP). The black lines indicate the sizes of RT-PCR products amplified using the intergenic primers (see Table S1 in the supplemental material). The dashed lines indicate that no RT-PCR products were generated in the mutant RΔBP. (B) RT-PCR products detected by agarose gel electrophoresis. Abbreviations: WT, cDNA from wild-type RS105; RΔBP, cDNA from the mutant in the hrpB operon promoter; WT gDNA, genomic DNA from wild-type RS105; NC, the extracted RNAs were used for PCR to ensure no residual genomic DNA in samples; M1, DL5000 DNA ladder (TaKaRa); M2, DL2000 DNA ladder (TaKaRa). 16S rRNA was used as an internal control to verify cDNA levels.
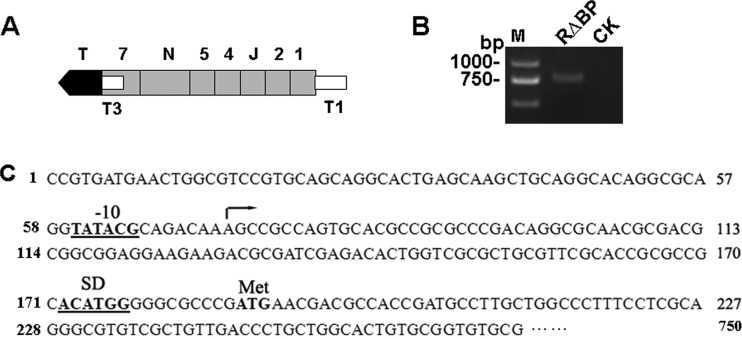
FIG 2.
Identification of the second promoter (T3) of hrcT gene by 5′-RACE-PCR. (A) Graphical representation of the hrpB operon. Gray or black regions represent eight genes in the hrpB operon as in Fig. 1A. The open rectangle T1 shows the location of the hrpB operon promoter upstream of hrpB1 (350 bp), and T3 shows the location of a promoter region (186 bp) upstream of hrcT. (B) Detection of the 750-bp 5′-RACE PCR product of hrcT in the T1 promoter mutant RΔBP by agarose gel electrophoresis. CK, RΔBP RNA used as a negative control. (C) Nucleotide sequence of T3 upstream of hrcT. Elements in the 5′ region include a −10 box (underlined), a potential transcriptional start site (arrow), and a putative ribosome binding site (Shine-Dalgarno [SD]). The boldface ATG represents the transcription start code of hrcT.
To identify the second promoter (T3) driving the expression of hrcT, 5′-RACE-PCR was explored using nested primers (see Table S1 in the supplemental material) and total RNA from the mutant RΔBP was cultured in XOM3 medium. 5′-RACE indicated that hrcT was indeed transcribed from the T3 promoter as a 750-bp PCR product (Fig. 2B). Sequence analysis of the hrcT upstream region revealed typical promoter elements, including a −10 box, a potential transcriptional start site (TSS), and a putative Shine-Dalgarno sequence (41) prior to the translation start codon (ATG) (Fig. 2C). It is noteworthy that the region upstream of hrcT lacked a PIP box.
Expression of hrcT is HrpG independent.
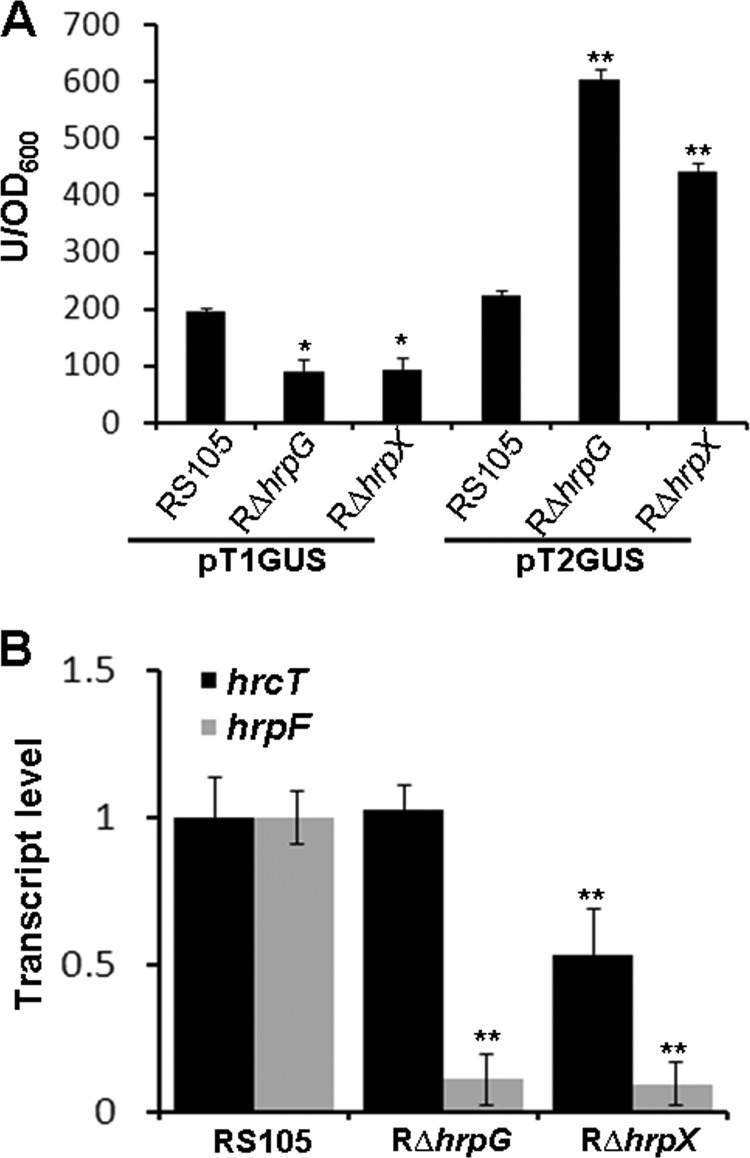
To investigate T3 activity in various backgrounds, this promoter was fused with a promoterless β-glucuronidase (gusA) gene, and the fusion (pT3GUS) (Table 1) was introduced into the wild-type RS105 and the mutants RΔhrpG and RΔhrpX. The T1 promoter fused to gusA (pT1GUS) (Table 1) was used as a control. The GUS activity of transformants containing pT1GUS or pT3GUS was measured. The results showed that GUS activity of the T1::GUS fusion was significantly lower in RΔhrpG and RΔhrpX than in the wild-type RS105 (P = 0.05, t test) (Fig. 3A), indicating that HrpG and HrpX positively regulate the expression of the hrpB operon genes via the T1 promoter. However, transcriptional activity of the T3::GUS fusion in RΔhrpG and RΔhrpX was significantly higher than that observed in the wild-type RS105 (Fig. 3A), suggesting that HrpG or HrpX may negatively regulate hrcT via the T3 promoter. To investigate this further, the posttranscript of hrcT was evaluated by qRT-PCR in the wild type, RΔhrpG, and RΔhrpX. The results showed that hrcT expression was significantly (P = 0.05, Student's t test) lower in the hrpX mutant than in both the hrpG mutant and the wild type, while the expression of hrcT in the hrpG mutant was almost the same as in the wild type (Fig. 3B). As predicted, the expression of hrpF, an hrpX-regulated gene, was significantly lower in RΔhrpG and RΔhrpX than in the wild-type RS105 (Fig. 3B). All these results together suggest that the expression of hrcT is HrpG independent.
FIG 3.
The expression of hrcT is HrpG independent. (A) β-Glucuronidase activities of transcriptional fusions designated pT1GUS and pT3GUS in the wild-type RS105, RΔhrpG, and RΔhrpX. All strains were cultured in XOM3 medium at 28°C for 12 h, and GUS activities were determined by measuring the optical density at 415 nm using 4-MUG (4-methylumbelliferyl-β-glucuronide) as a substrate. (B) Expression analysis of hrcT and hrpF by real-time quantitative RT-PCR. RNAs were isolated from cultures of X. oryzae pv. oryzicola RS105, RΔhrpG, and RΔhrpX, which were incubated in rice suspension cells at 25°C for 16 h. Data represent the means ± standard deviations of triplicate measurements. Asterisks above bars indicate significance relative to the wild type using a paired, two-tailed Student's t test. **, P = 0.01; *, P = 0.05. The experiment was repeated three times with similar results.
HrcT positively regulates the expression of hrpX but not hrpG.
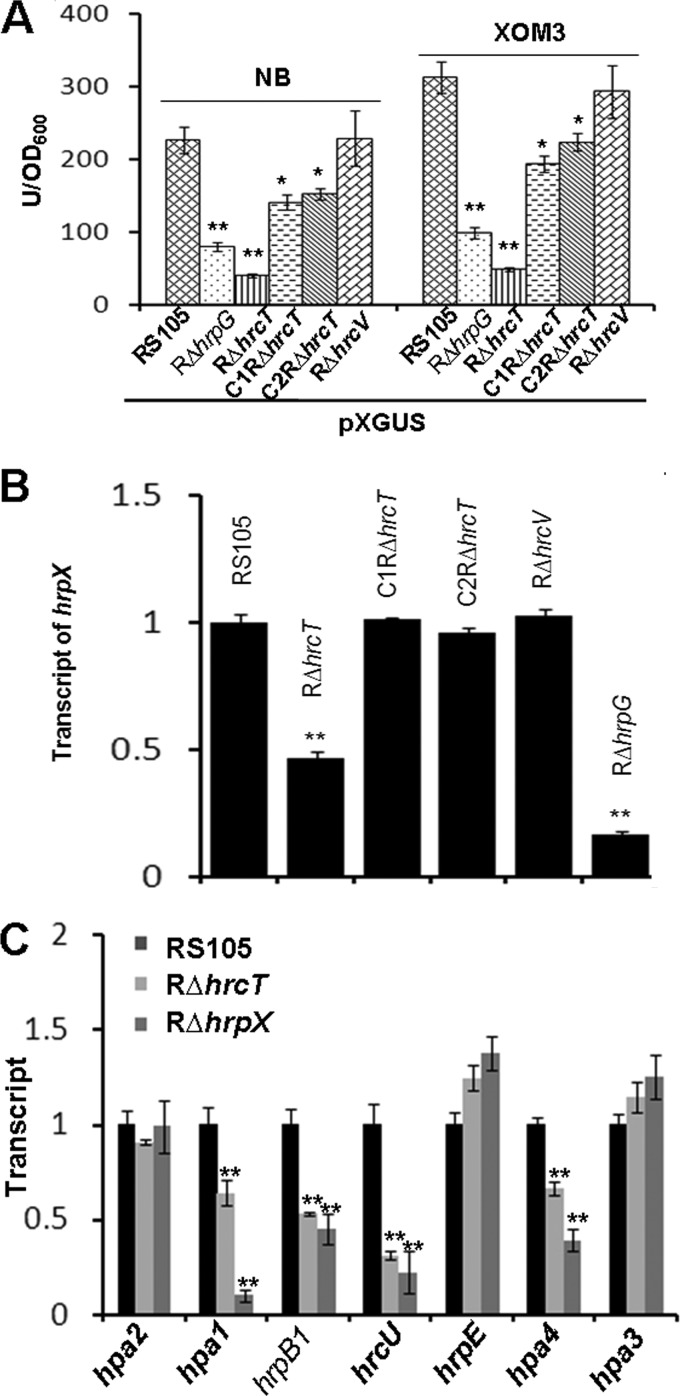
Our previous qRT-PCR data demonstrated that the deletion in hrcT dramatically affected the expression of some hrp genes in X. oryzae pv. oryzicola (unpublished data). We speculated that the possibility was that the mutation in hrcT may impact the expression of hrpG or hrpX. To test this hypothesis, the promoter activities of hrpG and hrpX were measured in the wild type and the hrcT mutant RΔhrcT using gusA as a reporter. Similar expression levels of hrpG were observed in the wild type and RΔhrcT in the hrp-inducing medium XOM3 (see Fig. S1 in the supplemental material). In addition, analysis by qRT-PCR showed no difference in the hrpG mRNA between the wild type and RΔhrcT (P ≤ 0.05, t test) (see Fig. S1 in the supplemental material). This result suggests that the expression of hrpG is not influenced by the deletion of hrcT. However, the promoter activity of hrpX was significantly lower (P = 0.01, t test) in RΔhrcT than those in the wild type grown in both NB and XOM3 (Fig. 4A). Given that HrpG is necessary for the expression of hrpX (12), the hrpG mutant RΔhrpG was included as a negative control to determine the hrpX promoter activity. The expression of hrpX in RΔhrcT could be partially restored to the wild-type levels in C1RΔhrcT and C2RΔhrcT, which contain a promoterless hrcT gene driven by promoters T1 and T3, respectively (Fig. 4A). Compared to the wild type tested by qRT-PCR, a reduction in the hrpX mRNA was detected in RΔhrcT and restored to the wild-type level in complemented strains C1RΔhrcT and C2RΔhrcT (Fig. 4B). To exclude the possibility that the lack of a functional T3SS leads to reduced expression of hrpX, we tested the promoter activity and transcript level of hrpX in another hrc deletion mutant, RΔhrcV. No difference in the promoter activity and the transcript of hrpX was observed between wild-type RS105 and RΔhrcV (Fig. 4A and B). These results suggest that the deletion of hrcT affecting the expression of hrpX is not due to the lack of the functional T3SS.
FIG 4.
HrcT functions in regulating the expression of hrpX. (A) pXGUS-mediated glucuronidase activity in X. oryzae pv. oryzicola RS105, RΔhrpG, RΔhrcT, RΔhrcV, and the complemented strains C1RΔhrcT and C2RΔhrcT. Strains were incubated in NB or XOM3 medium for 12 h at 28°C. GUS activity was determined by measuring the OD at 415 nm using 4-MUG as a substrate. (B) Expression analysis of hrpX by real-time qRT-PCR. RNAs were isolated from cultures of X. oryzae pv. oryzicola RS105, RΔhrcT, RΔhrcV, complemented strains C1RΔhrcT and C2RΔhrcT, and RΔhrpG. All strains were incubated in rice suspension cells at 25°C for 16 h, and relative mRNA levels were calculated with respect to the expression level of the corresponding transcript in the wild-type RS105. (C) Expression analysis of hrp genes by qRT-PCR. RNAs were isolated from cultures of the wild-type RS105, RΔhrcT, and RΔhrpX, which were incubated in rice suspension cells at 25°C for 16 h. The relative mRNA levels were calculated with respect to the corresponding transcript in RS105. All the experiments were repeated three times, and similar results were obtained. Data represent the means ± standard deviations of triplicate measurements. Asterisks above columns represent significance based on a paired, two-tailed Student t test relative to the wild type. **, P = 0.01; *, P = 0.05.
Since our results indicated that HrcT positively regulates the expression of hrpX, we sought to investigate whether the expression of other hrp-hrc-hpa genes is positively regulated by HrcT. The wild-type RS105 and RΔhrcT were inoculated into rice, and bacterial mRNAs were extracted and used as the templates in qRT-PCR assays 12 hpi. We chose some hrp genes as representation to confirm the conclusion by referring to the study in which the expression of some hrp genes was reported to be obviously influenced when hrpX is mutated (2). Primers for this experiment were specific for hpa2, hpa1, hrpB1, hrcU, hrpE, hpa4, and hpa3 (see Table S1 in the supplemental material). qRT-PCR results showed that the expression of hpa1, hrpB1, hrcU, and hpa4 was significantly (P ≤ 0.01, t test) reduced in RΔhrcT compared to the wild-type RS105, while the expression of hpa2, hrpE, and hpa3 was similar in RΔhrcT and RS105 (Fig. 4C). This is consistent with our previous finding that the expression of hpa1, hrpB1, hrcU, and hpa4 is positively regulated by HrpX while the expression of hpa2, hrpE, and hpa3 is not obviously influenced when hrpX is mutated (2). Thus, our current results suggest that HrcT positively regulates the expression of hrpX, which in turn controls the expression of hrp-hrc-hpa genes mentioned above.
HrcT binds the hrpX promoter and regulates the expression of hrpX.
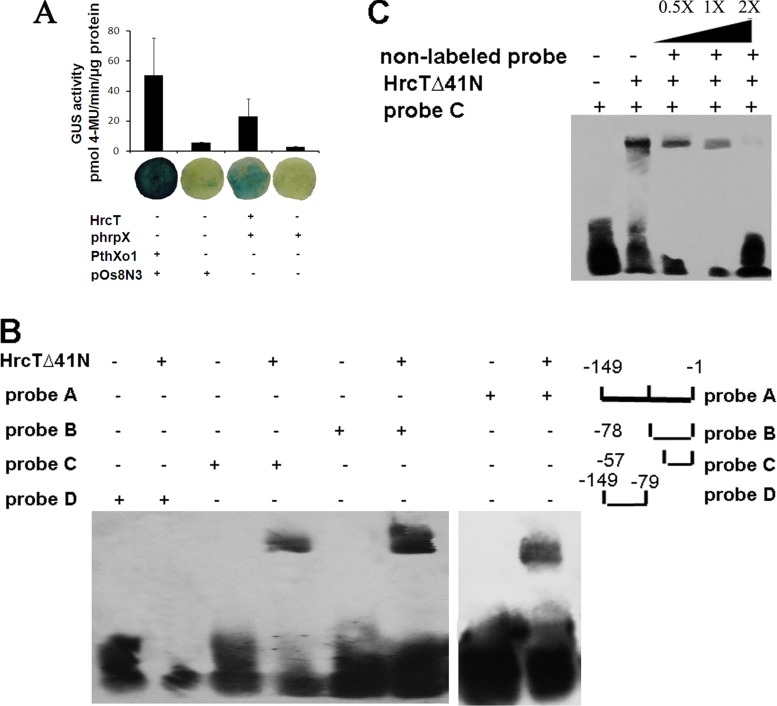
Given that HrcT regulates the expression of hrpX as demonstrated above, we speculated that HrcT may bind the hrpX promoter for regulation. The promoter of hrpX was fused to the promoterless gusA gene as a reporter. GUS activity was determined in the presence (+) or absence (−) of HrcT in N. benthamiana leaves by using an Agrobacterium-mediated transient expression system in planta (38). HrcT indeed induced stronger GUS activity when the hrpX promoter was present, like the positive control indicating that PthXo1 bound the Os8N3 promoter to activate the expression of gusA, than that when the hrpX promoter was absent (Fig. 5A). This indicates that HrcT binds the hrpX promoter in vivo. In order to find the specific region of the hrpX promoter bound by HrcT in vitro, electrophoretic mobility shift assays (EMSAs) were performed. Initially, we attempted to overproduce the entire HrcT protein in E. coli but were unsuccessful (data not shown). Topology analysis indicated that HrcT contains a 41-amino-acid (aa) region at the N terminus that spans the IM (42), which might lead to our failure in trying to overproduce the intact HrcT in E. coli. We then sought to overproduce and purify an N-terminally truncated HrcT (a 41-aa region was deleted, designated HrcTΔ41N) in E. coli. The truncated HrcT was tagged with hexahistidine at the C terminus, and the purified HrcTΔ41N-His6 was used in EMSAs. Different lengths of the target hrpX promoter were PCR amplified or synthesized, biotinylated, and used as probes. These included probes A, B, and C, which comprised 149-, 78-, and 57-bp fragments upstream of the hrpX transcription start codon (ATG), respectively, and probe D, a 70-bp fragment upstream of probe B (Fig. 5B). Each probe was incubated with purified HrcTΔ41N-His6 (1 μg) for 20 min at room temperature. After EMSA, we found that HrcTΔ41N-His6 bound to probes A, B, and C but not probe D (Fig. 5B). The observed shift was prevented by nonlabeled probe C when it competed with the labeled probe C (Fig. 5C). These results indicate that HrcT binds the hrpX promoter and binding occurs within a 57-bp region upstream of the hrpX transcript start codon.
FIG 5.
Binding of HrcT to hrpX promoter in vivo and in vitro. (A) Agrobacterium-mediated transient expression assay of hrpX activated by HrcT. GUS reporter constructs are codelivered via A. tumefaciens into N. benthamiana with (+) and without (−) the construct HrcT. PthXo1 of X. oryzae pv. oryzae and its target Os8N3 (pOs8N3) were used as a positive control. The GUS activity was determined 2 dpi by stained leaf disks (0.8 cm in diameter) with X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide). Blue indicates a positive reaction. Error bars indicate standard deviations (n = 3 samples). 4-MU, 4-methyl-umbelliferone. (B) Binding of HrcTΔ41N to the hrpX promoter by EMSA. Twenty femtomoles of biotinylated probes was used to react with 1 μg of purified HrcTΔ41N (presence indicated by +, absence by −). Different sizes of the hrpX promoter upstream of the hrpX transcriptional start codon were used as probes, displayed on the right. Probe A is 149 bp long, probe B 78 bp, probe C 57 bp, all upstream of the hrcT transcription start code, and probe D is 70 bp long, upstream of the probe B as displayed on the left. (C) Competition of biotinylated probe C with unlabeled probe C bound to HrcTΔ41N by EMSA. The unlabeled probe C with higher concentration, at 0.5×, 1×, and 2× more than 20 fmol of biotinylated probe C, was used. +, present; −, absent. The above-described experiments were repeated three times, and results from one representative experiment are shown.
The Hrp phenotype can be partially restored when hrcT is expressed under promoters T1 and T3 in the hrcT mutant.
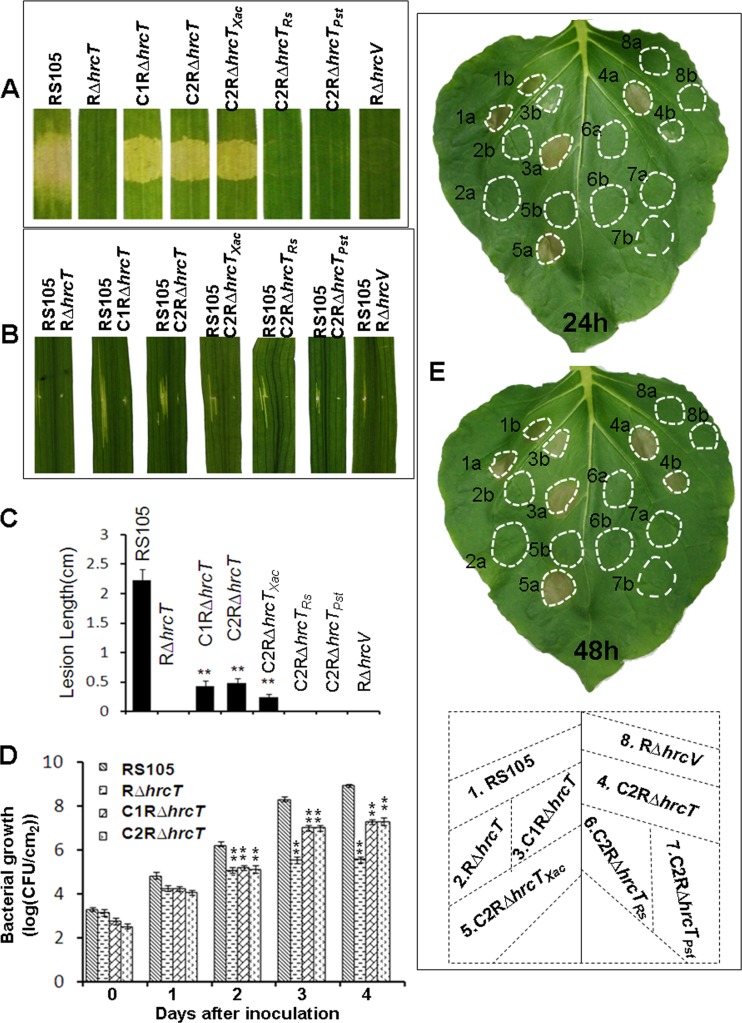
Our previous work has revealed that the hrcT mutant RΔhrcT lost the ability to trigger HR in tobacco and pathogenicity in rice (2). To investigate whether the RΔhrcT complemented strains (C1RΔhrcT and C2RΔhrcT, containing promoters T1 and T3, respectively) have the ability to trigger HR in tobacco and pathogenicity in rice, the tested strains were infiltrated into rice seedlings and inoculated into adult plants. The RΔhrcT complemented strains, C1RΔhrcT and C2RΔhrcT, induced water-soaked symptoms as the wild-type RS105 did 3 dpi (Fig. 6A), but the lesion lengths induced by C1RΔhrcT and C2RΔhrcT were significantly shorter than those induced by the wild-type RS105 (Fig. 6B). In contrast, RΔhrcT, like RΔhrcV, failed to trigger water-soaked lesions (Fig. 6A and B). Bacterial growth in rice tissue was compromised in RΔhrcT and was partially restored to the wild-type level in the complemented strains C1RΔhrcT and C2RΔhrcT (Fig. 6C). Regarding HR induction in tobacco, the hrcT-complemented strains C1RΔhrcT and C2RΔhrcT elicited strong HRs 24 hpi when the concentration of bacterial cells was adjusted to an OD600 of 0.3. However, the HR was delayed (e.g., did not appear until 48 hpi) when the OD600 was 0.01. As predicted, the T3SS mutant RΔhrcV did not elicit HR in tobacco (Fig. 6D). Collectively, these data indicate that expression of hrcT via the T1 or T3 promoter partially restores the HR in tobacco and bacterial virulence in rice.
FIG 6.
Detection of the HR in tobacco and pathogenicity in rice. (A and B) Symptoms induced by infiltration of X. oryzae pv. oryzicola strains (suspended in water, OD600 = 0.3) into leaves of rice cv. IR24 (susceptible to X. oryzae pv. oryzicola) with a needleless syringe. Photographs were taken 3 dpi. (C) BLS lesion lengths in adult rice (2 months old) inoculated by the leaf-needling method was measured 14 dpi. (D) Bacterial growth in inoculated rice leaves. Bacteria were recovered from inoculated leaves daily for a period of 4 days. (E) Assay for the HR in tobacco (N. benthamiana) inoculated with bacterial strains adjusted to the following concentrations: OD600 = 0.3 (3 × 108 CFU/ml) and OD600 = 0.01 (1 × 105 CFU/ml). Strains are identified by numbers as follows: 1, X. oryzae pv. oryzicola RS105; 2, mutant RΔhrcT; 3, hrcT-complemented strain C1RΔhrcT; 4, C2RΔhrcT; 5, C2RΔhrcTXac, RΔhrcT containing hrcT from X. axonopodis pv. citri in trans under the T3 promoter; 6, C2RΔhrcTRs, RΔhrcT containing hrcT from R. solanacearum GMI1000 in trans under the T3 promoter; 7, CRΔhrcTPst, RΔhrcT containing hrcT from P. syringae pv. tomato DC3000 in trans under the T3 promoter; 8, RΔhrcV (hrcV mutant, negative control). Tobacco leaves were inoculated with the above-listed strains by a needleless syringe, and HRs were scored 24 and 48 hpi, respectively. The experiments were repeated three times. Data represent the means ± standard deviations from three replicates. Asterisks above columns represent significance based on a paired, two-tailed Student t test relative to the wild type. **, P = 0.01; *, P = 0.05.
HrcT is functionally interchangeable between Xanthomonas spp.
HrcT, a key component of the T3SS, is conserved in both animal- and plant-pathogenic bacteria (26). Phylogenetic analysis of HrcT orthologs from various bacterial pathogens showed that HrcT proteins could be classified into four groups (see Fig. S2 in the supplemental material). Group I contained HrcT proteins from closely related Xanthomonas spp.: group II from Acidovorax citrulli, R. solanacearum, and Burkholderia sp.; group III from P. syringae pv. tomato DC3000, E. amylovora, and Dickeya dadantii 3937; and group IV from animal pathogenic Shigella boydii, Salmonella enterica, and Yersinia pseudotuberculosis (see Fig. S2 in the supplemental material). This prompted us to investigate whether HrcT proteins from other bacteria can functionally complement the X. oryzae pv. oryzicola hrcT mutant for HR and pathogenicity in plants. The promoterless hrcT genes from X. axonopodis pv. citri 306, R. solanacearum GMI1000, and P. syringae pv. tomato DC3000 were used in this experiment, and these genes were cloned as transcriptional fusions whereby the expression was driven by T1 or T3 promoter from X. oryzae pv. oryzicola hrcT (Table 1). The transcriptional fusions were introduced into RΔhrcT, and the constructs were designated C1RΔhrcTXac, C1RΔhrcTRs, C1RΔhrcTPst, C2RΔhrcTXac, C2RΔhrcTRs, and C2RΔhrcTPst (Table 1), respectively. The constructs containing hrcT from X. axonopodis pv. citri triggered HR in tobacco and BLS symptoms in rice (only the case driven by promoter T3 is shown) (Fig. 6A, B, and D), implying that hrcT is interchangeable among Xanthomonas spp. HrcT orthologs from R. solanacearum and P. syringae pv. tomato failed to functionally complement the hrcT mutation for HR or BLS symptoms in rice.
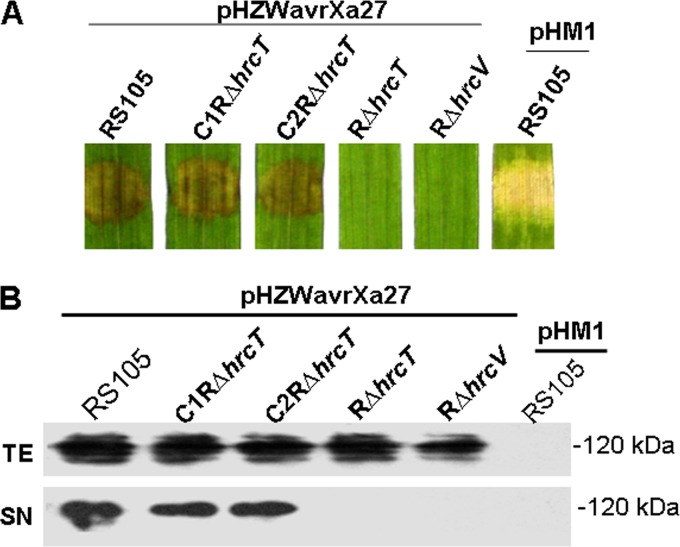
The deletion in hrcT impairs the secretion of AvrXa27.
Considering that HrcT is an inner membrane protein for the T3SS (27, 42), we speculated that an hrcT deletion might impair the secretion of T3SEs. To test this hypothesis, we used AvrXa27 to monitor secretion because AvrXa27 also induces HR in rice cv. 87-15 containing the resistance gene Xa27 (40). Plasmid pHZWavrXa27 containing avrXa27 (Table 1) was introduced into X. oryzae pv. oryzicola RS105, mutant RΔhrcT, complemented strains C1RΔhrcT and C2RΔhrcT, and RΔhrcV, respectively. Three days after infiltration (OD600 = 0.3) into rice cv. 87-15, tissues inoculated with RS105, C1RΔhrcT, and C2RΔhrcT (each containing pHZWavrXa27) exhibited dark, HR-like symptoms (Fig. 7A). In contrast, RΔhrcT and RΔhrcV containing pHZWavrXa27 did not induce visible symptoms in rice. X. oryzae pv. oryzicola RS105 harboring the empty vector pHM1 elicited typical BLS symptoms (Fig. 7A). These results suggest that HrcT functions as a key component of the T3SS and is essential for the secretion of T3SEs into plant cells.
FIG 7.
Expression of hrp genes and secretion of T3SEs in the hrcT-deleted mutant. (A) Symptoms induced in seedlings of rice cv. 87-15 (containing Xa27) inoculated with X. oryzae pv. oryzicola derivatives. Strains containing pHZWavrXa27 (suspended in water, OD600 = 0.3) were inoculated into leaves as described in Materials and Methods. (B) Western blot assays to examine the secretion of effector AvrXa27 using a monoclonal anti-FLAG antibody. Strains included X. oryzae pv. oryzicola RS105, RΔhrcT (hrcT deletion mutant), C1RΔhrcT and C2RΔhrcT (complemented strains of RΔhrcT with hrcT under the T1 or T3 promoter), and RΔhrcV, a T3SS mutant (negative control). With the exception of the empty vector control (pHM1), all strains contained pHZWavrXa27 (avrXa27 is fused to a Flag tag code).
To examine secretion of AvrXa27 via the T3SS in vitro, the strains mentioned above were incubated in XOM3 at 28°C for 12 h, and total cell extracts (TEs) and culture supernatants (SNs) were analyzed by immunoblotting using Flag antibodies. Analysis of the SN fractions revealed that AvrXa27 was secreted into the medium by X. oryzae pv. oryzicola RS105, C1RΔhrcT, and C2RΔhrcT; however, the protein was not detectable in the SNs of RΔhrcT or RΔhrcV (Fig. 7B). These results indicate that expression of hrcT via promoter T1 or T3 in trans can complement the secretion defect in the hrcT mutant.
DISCUSSION
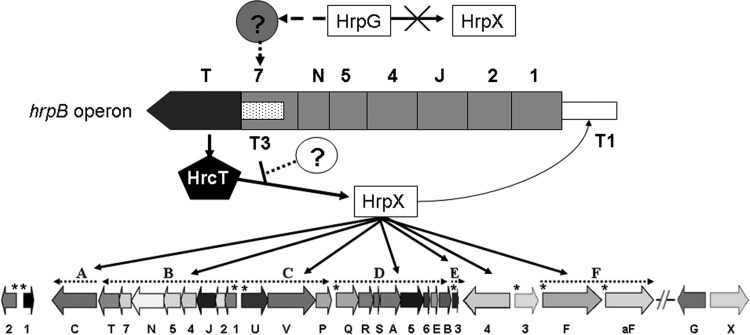
Previous studies have indicated that the regulation of hrp-hrc-hpa genes in Xanthomonas requires multiple factors. For example, HrpG regulates the expression of hrpX and the hrpA transcript, and HrpX activates the expression of the hrpB to hrpF operons (11, 12). In this study, we investigate potential regulatory roles of HrcT protein, which is a structural component of the T3SS. HrcT functioned as a positive regulator of HrpX, which adds another layer of complexity to hrp gene regulation. In our revised model (Fig. 8) (referring to our previous one, described in reference 2), an unknown regulatory factor, possibly regulated by HrpG, may switch on the expression of HrcT via promoter T3; HrcT binds to the hrpX promoter to activate hrpX transcription; HrpX then binds the PIP box promoters of multiple HrpX regulons to regulate the expression of hrp-hrc-hpa genes, including our newly identified hrp regulator gene hrpD6 (2); once the hrpB operon is activated by HrpX via promoter T1, the binding of HrcT to HrpX or the mRNA level of hrcT may possibly be degraded by another unknown factor (Fig. 8).
FIG 8.
Working model showing the proposed regulatory role of HrcT. Our results, together with our previous report (2), show that HrcT expression at the T3 promoter is HrpG independent. Thus, we propose that an unidentified regulator (question mark in a gray circle), possibly regulated by HrpG, which does not directly regulate the expression of hrpX (cross on a black arrow), promotes hrcT transcription at the T3 promoter. HrcT then positively regulates the expression of hrpX by binding the hrpX promoter. The transcriptional activator HrpX regulates multiple hrp operons (black-lined arrows) by binding to the PIP box promoter as indicated by asterisks above the hrp-hrc-hpa cluster. Another unknown protease-like protein (question mark in a white circle), possibly regulated by HrpX, may degrade the mRNA of hrcT or HrcT to repress the expression of hrpX. The dashed horizontal arrows represent hrp operons (A to F) consisting of individual hrp-hrc-hpa genes, which were assigned by choosing the last letter or number of a gene name under differentially shaded arrows, referring to references 2 and 3.
Precisely how HrcT modulates HrpX remains unclear. HrcT is a highly conserved component of the T3SS in Xanthomonas spp. (27). The N-terminal portion of HrcT presumably spans the IM of bacterial cells (42). Considering our results, we hypothesize that HrcT was possibly synthesized first in cytoplast, where it can bind the hrpX promoter to activate the expression of hrpX. Meanwhile, it is integrated into the cell membrane for the T3SS. We also show that the expression of hpa1, hrpB1, hrcU, and hpa4 is reduced in both the hrcT and hrpX mutants (Fig. 4), which is consistent with coordinated regulation via HrcT/HrpX.
Based on the fact that the expression of promoter T1 is attenuated in the hrpX mutant but that of promoter T3 is not (Fig. 3A), we propose that the expression of hrcT may be controlled by both promoters T1 and T3. The expression of hrcT under promoter T1 (containing the PIP box) was positively regulated by HrpX (2), while hrcT under promoter T3 was positively regulated by an unknown factor that may possibly be activated by HrpG (Fig. 8). Activation by an unknown regulatory protein would help explain why hrcT can be transcribed independently of the hrpB operon promoter. We speculate that HrpX binds to the PIP box in promoter T1 to produce the polycistronic hrpB operon, which would include hrcT (Fig. 1B). These speculations are consistent with the fact that the hrcT mutant can be complemented by expression via either promoter T1 or T3 as shown by partial restoration of HR and pathogenicity in planta (Fig. 6). These hypotheses remain highly speculative until the unknown regulator is identified.
Intensive studies have been undertaken to elucidate components of the Hrp regulon in Xanthomonas spp. (1, 2, 13). In addition to the regulators HrpG, HrpX, and HrpD6 (2, 11, 12), additional two-component regulatory system (TCS) proteins have been shown to modulate expression of the T3SS in Xanthomonas spp. For example, Li and coworkers (13) recently identified HpaS, the histidine protein kinase that interacts with HrpG via phosphorylation. Other regulators of the TCS include Trh and HpaR1, which were shown to positively regulate the expression of hrpG (43, 44). ColS/ColR constitute a TCS that is implicated in virulence and HR in planta; these proteins were shown to repress the expression of hrpG and the hrpC and hrpE operons but not other hrp genes (45, 46). Zur, a key regulator for zinc homeostasis, positively influenced the expression of hrp operons via hrpX but not hrpG (47). HpaR1 is a GntR family transcriptional activator that regulates the expression of all five operons in the hrp cluster via HrpG (48). It seems unlikely that the unknown regulator for HrcT is one of the preceding TCSs, particularly because the expression of hrpG is repressed by Trh, HpaR1, ColS/ColR, Zur, and HpaR, and the expression of hrcT is occasionally HrpG independent and HrpX dependent (Fig. 3B). In addition to the numerous regulatory loci that map elsewhere in the Xanthomonas genomes (16–20), our findings highlight the existence of key regulatory loci that map within the hrp-hrc-hpa genes in X. oryzae pv. oryzicola. An excellent example is the HrpD6 protein recently identified in our lab. We previously showed that the expression of hrcT is reduced in the hrpD6 mutant; furthermore, hrpD6 is positively controlled by HrpX (2). It is tempting to speculate that HrpD6 may serve as a negative regulator for the unknown regulatory factor shown in Fig. 8, and experiments to test this hypothesis are under way in our laboratory.
The expression of hrcT may be stimulated by an environmental or plant signal that is sensed by membrane-associated proteins. For example, the OM portion of HpaS presumably senses plant stimuli during the early stages of infection and transphosphorylates HrpG (13). As noted in our model, the activated form of HrpG may function via an unknown factor to activate HrcT expression via promoter T3. More detailed studies of genes that are expressed independently of HrpG and/or HrpX (e.g., HrcC, HrcT, HrpD5, HrpE, and Hpa3) will help further elucidate hrp regulatory networks in Xanthomonas.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Bing Yang (Iowa State University, USA) for providing the plasmid pHZWavrXa27 and to Chaozu He (Hainan University, China) for kindly providing rice line 87-15 for this study. We also thank Carol Bender (Oklahoma State University, USA) for her critical reading and editing of the manuscript prior to submission.
This work was supported by the State Key Basic Research and Development Project of China (2012CB114003), the Natural Science Foundation of China (31371905), and the Special Fund for Agro-scientific Research in the Public Interest of China (210303015 and 201003067-09).
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00308-14.
REFERENCES
- 1.Büttner D, Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34:107–133. 10.1111/j.1574-6976.2009.00192.x [DOI] [PubMed] [Google Scholar]
- 2.Li YR, Zou HS, Che YZ, Cui YP, Guo W, Zou LF, Chatterjee S, Biddle EM, Yang CH, Chen GY. 2011. A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola Mol. Plant Microbe Interact. 24:1086–1101. 10.1094/MPMI-09-10-0205 [DOI] [PubMed] [Google Scholar]
- 3.Zou LF, Wang XP, Xiang Y, Zhang B, Li YR, Xiao YL, Wang JS, Walmsley AR, Chen GY. 2006. Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72:6212–6224. 10.1128/AEM.00511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano JR, Collmer A. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683–1698. 10.1105/tpc.8.10.1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan L, Deng X, Zhou J, Tang X. 2006. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS Mol. Plant Microbe Interact. 19:976–987. 10.1094/MPMI-19-0976 [DOI] [PubMed] [Google Scholar]
- 6.Wei ZM, Beer SV. 1995. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J. Bacteriol. 177:6201–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson SW. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y, Hutcheson SW. 1994. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 176:3089–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito B, Aldon D, Barberis P, Boucher C, Genin S. 2002. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant Microbe Interact. 15:109–119. 10.1094/MPMI.2002.15.2.109 [DOI] [PubMed] [Google Scholar]
- 10.Genin S, Gough CL, Zischek C, Boucher CA. 1992. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6:3065–3076. 10.1111/j.1365-2958.1992.tb01764.x [DOI] [PubMed] [Google Scholar]
- 11.Wengelnik K, Bonas U. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wengelnik K, Van den Ackerveken G, Bonas U. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant Microbe Interact. 9:704–712. 10.1094/MPMI-9-0704 [DOI] [PubMed] [Google Scholar]
- 13.Li RF, Lu GT, Li L, Su HZ, Feng GF, Chen Y, He YQ, Jiang BL, Tang DJ, Tang JL. 2013. Identification of a putative cognate sensor kinase for the two-component response regulator HrpG, a key regulator controlling the expression of the hrp genes in Xanthomonas campestris pv. campestris. Environ. Microbiol. 10.1111/1462-2920.12207 [DOI] [PubMed] [Google Scholar]
- 14.Furutani A, Nakayama T, Ochiai H, Kaku H, Kubo Y, Tsuge S. 2006. Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a −10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae FEMS Microbiol. Lett. 259:133–141. 10.1111/j.1574-6968.2006.00265.x [DOI] [PubMed] [Google Scholar]
- 15.Koebnik R, Kruger A, Thieme F, Urban A, Bonas U. 2006. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J. Bacteriol. 188:7652–7660. 10.1128/JB.00795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, Patil PB, Van Sluys MA, Ryan RP, Meyer DF, Han SW, Aparna G, Rajaram M, Delcher AL, Phillippy AM, Puiu D, Schatz MC, Shumway M, Sommer DD, Trapnell C, Benahmed F, Dimitrov G, Madupu R, Radune D, Sullivan S, Jha G, Ishihara H, Lee SW, Pandey A, Sharma V, Sriariyanun M, Szurek B, Vera-Cruz CM, Dorman KS, Ronald PC, Verdier V, Dow JM, Sonti RV, Tsuge S, Brendel VP, Rabinowicz PD, Leach JE, White FF, Salzberg SL. 2011. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193:5450–5464. 10.1128/JB.05262-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jalan N, Aritua V, Kumar D, Yu F, Jones JB, Graham JH, Setubal JC, Wang N. 2011. Comparative genomic analysis of Xanthomonas axonopodis pv. citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity. J. Bacteriol. 193:6342–6357. 10.1128/JB.05777-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BM, Park YJ, Park DS, Kang HW, Kim JG, Song ES, Park IC, Yoon UH, Hahn JH, Koo BS, Lee GB, Kim H, Park HS, Yoon KO, Kim JH, Jung C, Koh NH, Seo JS, Go SJ. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577–586. 10.1093/nar/gki206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo YS, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys MA, Leach JE, White FF, Bogdanove AJ. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. 10.1186/1471-2164-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Buttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, McHardy AC, Meyer F, Mittenhuber G, Nies DH, Niesbach-Klosgen U, Patschkowski T, Ruckert C, Rupp O, Schneiker S, Schuster SC, Vorholter FJ, Weber E, Puhler A, Bonas U, Bartels D, Kaiser O. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254–7266. 10.1128/JB.187.21.7254-7266.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YR, Che YZ, Zou HS, Cui YP, Guo W, Zou LF, Biddle EM, Yang CH, Chen GY. 2011. HpaII required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Appl. Environ. Microbiol. 77:3809–3818. 10.1128/AEM.02849-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76:262–310. 10.1128/MMBR.05017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desvaux M, Hébraud M, Talon R, Henderson IR. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139–145. 10.1016/j.tim.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 24.Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795. 10.1128/MMBR.68.4.771-795.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He SY, Jin Q. 2003. The Hrp pilus: learning from flagella. Curr. Opin. Microbiol. 6:15–19. 10.1016/S1369-5274(02)00007-3 [DOI] [PubMed] [Google Scholar]
- 26.He SY, Nomura K, Whittam TS. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181–206. 10.1016/j.bbamcr.2004.03.011 [DOI] [PubMed] [Google Scholar]
- 27.Fenselau S, Bonas U. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc. Spa, and Fli secretion systems. Mol. Plant Microbe Interact. 8:845–854. 10.1094/MPMI-8-0845 [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Xiao YL, Li YR, Liu ZY, Xiang Y, Chen GY. 2007. Establishment of the hrp-inducing systems for the expression of the hrp genes of Xanthomonas oryzae pv. oryzicola. Wei Sheng Wu Xue Bao 47:396–401 (In Chinese) [PubMed] [Google Scholar]
- 30.Cheng J, Ma J, Lin J, Fan ZC, Cronan JE, Wang H. 2012. Only one of the five Ralstonia solanacearum long-chain 3-ketoacyl-acyl carrier protein synthase homologues functions in fatty acid synthesis. Appl. Environ. Microbiol. 78:1563–1573. 10.1128/AEM.07335-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King EO, Ward NK, Raney DE. 1954. Two simple media for the demonstration of pyrocyanin and fluorescein. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 32.Sambrook J, Gething MJ. 1989. Protein structure. Chaperones, paperones. Nature 342:224–225 [DOI] [PubMed] [Google Scholar]
- 33.Jiang J, Zou H, Li Y, Chen G. 2009. Expression of the hrcC, hrpE and hpa3 genes is not regulated by the hrpG and hrpX genes in a rice pathogen Xanthomonas oryzae pv. oryzicola. Wei Sheng Wu Xue Bao 49:1018–1025 [PubMed] [Google Scholar]
- 34.Guo W, Zou LF, Li YR, Cui YP, Ji ZY, Cai LL, Zou HS, Hutchins WC, Yang CH, Chen GY. 2012. Fructose-bisphophate aldolase exhibits functional roles between carbon metabolism and the hrp system in rice pathogen Xanthomonas oryzae pv. oryzicola. PLoS One 7:e31855. 10.1371/journal.pone.0031855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, Ueno K, Mochizuki A, Tanimoto H, Tsugawa H, Otsuki Y, Ohashi Y. 1996. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37:49–59. 10.1093/oxfordjournals.pcp.a028913 [DOI] [PubMed] [Google Scholar]
- 36.DeFeyter R, Kado CI, Gabriel DW. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65–72. 10.1016/0378-1119(90)90060-5 [DOI] [PubMed] [Google Scholar]
- 37.Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B. 2010. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22:3864–3876. 10.1105/tpc.110.078964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Zou L, Ye G, Xiong L, Ji Z, Zakria M, Hong N, Wang G, Chen G. 7 January 2014. A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Mol. Plant 10.1093/mp/sst176 [DOI] [PubMed] [Google Scholar]
- 39.Zou HS, Song X, Zou LF, Yuan L, Li YR, Guo W, Che YZ, Zhao WX, Duan YP, Chen GY. 2012. EcpA, an extracellular protease, is a specific virulence factor required by Xanthomonas oryzae pv. oryzicola but not by X. oryzae pv. oryzae in rice. Microbiology 158:2372–2383. 10.1099/mic.0.059964-0 [DOI] [PubMed] [Google Scholar]
- 40.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. 2005. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435:1122–1125. 10.1038/nature03630 [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42.Berger C, Robin GP, Bonas U, Koebnik R. 2010. Membrane topology of conserved components of the type III secretion system from the plant pathogen Xanthomonas campestris pv. vesicatoria. Microbiology 156:1963–1974. 10.1099/mic.0.039248-0 [DOI] [PubMed] [Google Scholar]
- 43.Tsuge S, Nakayama T, Terashima S, Ochiai H, Furutani A, Oku T, Tsuno K, Kubo Y, Kaku H. 2006. Gene involved in transcriptional activation of the hrp regulatory gene hrpG in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 188:4158–4162. 10.1128/JB.00006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei K, Tang DJ, He YQ, Feng JX, Jiang BL, Lu GT, Chen B, Tang JL. 2007. hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris. J. Bacteriol. 189:2055–2062. 10.1128/JB.01331-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Q, Wang N. 2011. The ColR/ColS two-component system plays multiple roles in the pathogenicity of the citrus canker pathogen Xanthomonas citri subsp. citri J. Bacteriol. 193:1590–1599. 10.1128/JB.01415-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang SS, He YQ, Xu LM, Chen BW, Jiang BL, Liao J, Cao JR, Liu D, Huang YQ, Liang XX, Tang DJ, Lu GT, Tang JL. 2008. A putative colR(XC1049)-colS(XC1050) two-component signal transduction system in Xanthomonas campestris positively regulates hrpC and hrpE operons and is involved in virulence, the hypersensitive response and tolerance to various stresses. Res. Microbiol. 159:569–578. 10.1016/j.resmic.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 47.Huang DL, Tang DJ, Liao Q, Li XQ, He YQ, Feng JX, Jiang BL, Lu GT, Tang JL. 2009. The Zur of Xanthomonas campestris is involved in hypersensitive response and positively regulates the expression of the hrp cluster via hrpX but not hrpG Mol. Plant Microbe Interact. 22:321–329. 10.1094/MPMI-22-3-0321 [DOI] [PubMed] [Google Scholar]
- 48.An SQ, Lu GT, Su HZ, Li RF, He YQ, Jiang BL, Tang DJ, Tang JL. 2011. Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol. Plant Microbe Interact. 24:1027–1039. 10.1094/MPMI-08-10-0180 [DOI] [PubMed] [Google Scholar]
- 49.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.