Abstract
The sanitary quality of recreational waters that may be impacted by sewage is assessed by enumerating fecal indicator bacteria (FIB) (Escherichia coli and enterococci); these organisms are found in the gastrointestinal tracts of humans and many other animals, and hence their presence provides no information about the pollution source. Microbial source tracking (MST) methods can discriminate between different pollution sources, providing critical information to water quality managers, but relatively little is known about factors influencing the decay of FIB and MST genetic markers following release into aquatic environments. An in situ mesocosm was deployed at a temperate recreational beach in the Mississippi River to evaluate the effects of ambient sunlight and biotic interactions (predation, competition, and viral lysis) on the decay of culture-based FIB, as well as molecularly based FIB (Entero1a and GenBac3) and human-associated MST genetic markers (HF183 and HumM2) measured by quantitative real-time PCR (qPCR). In general, culturable FIB decayed the fastest, while molecularly based FIB and human-associated genetic markers decayed more slowly. There was a strong correlation between the decay of molecularly based FIB and that of human-associated genetic markers (r2, 0.96 to 0.98; P < 0.0001) but not between culturable FIB and any qPCR measurement. Overall, exposure to ambient sunlight may be an important factor in the early-stage decay dynamics but generally was not after continued exposure (i.e., after 120 h), when biotic interactions tended to be the only/major influential determinant of persistence.
INTRODUCTION
Contamination of environmental waters by sewage poses a serious threat to human health; hence, identifying sources of fecal pollution is vital for water quality management, remediation efforts, and estimation of their impacts, aided by quantitative microbial risk assessment (QMRA) (1–3). Combined sewer overflows (CSOs), sanitary sewer overflows (SSOs), sewage spills, and faulty septic systems are all common sources through which high concentrations of untreated or partially treated sewage can enter environmental waters.
Recreational water quality is typically assessed using culture-based enumeration of fecal indicator bacteria (FIB) (e.g., Escherichia coli and enterococci) worldwide and, more recently, quantitative real-time PCR (qPCR) in the United States (4), neither of which provides information about the pollution source, since these organisms are commensal inhabitants of the gastrointestinal tracts of mammals, birds, and various insects (5–14). Microbial source tracking (MST) has emerged in response to the increasing need to identify and manage sources of fecal pollution, and it aims to remedy the situation through identification of contributing fecal sources by targeting genetic markers thought to be closely associated with particular animal hosts (15). A large subset of human-associated MST methods target genetic markers harbored in Bacteroides spp. (recently reviewed in reference 15) via endpoint PCR and qPCR. These methods have been successfully employed to identify likely human fecal pollution in many different water types (16–22).
However, relatively little is known about the fate of various MST genetic markers in the environment. Most decay studies to date either have been conducted in the laboratory under simulated environmental conditions or have used closed-microcosm systems (e.g., glass beakers or aquarium tanks) ex situ to mimic ambient conditions (23–27). Nonetheless, the decay of various Bacteroides host-associated genetic markers has been reported to increase with higher temperatures representative of summer conditions (23, 26, 28, 29), and it appears to be more rapid in freshwater compared to marine waters (25, 26, 30). The effect of ambient sunlight, however, remains ambiguous. Some researchers report no significant difference in the decay of Bacteroides host-associated genetic markers with or without sunlight exposure (24, 25, 31), while others note that decay under light conditions is significantly faster (32). It has also been suggested that exposure to sunlight is more detrimental to live cells (as assessed by propidium monoazide treatment) than it is to DNA, implying that the disagreement in the reported literature may be due to the physiological state rather than to a direct effect of sunlight (33). Considerably less is known about the impact of biotic interactions (e.g., predation, competition, and viral lysis); existing laboratory-scale and mesocosm studies suggest that these interactions can play an important role (23, 24, 26, 28), but this assertion has not been confirmed in field studies.
Successful management of recreational waterways requires a thorough understanding of FIB, MST genetic marker, and pathogen decay dynamics in aquatic habitats, as well as the effects that different environmental factors have on their decline. Here we present the effects of ambient sunlight and biotic interactions on decay of sewage-born FIB, measured by both culture and qPCR techniques, as well as selected human-associated MST genetic markers. In order to simulate accidental release of high concentrations of untreated sewage and capture the complexities of the changing environmental conditions, we utilized an in situ mesocosm device consisting of multiple diffusion bags filled with mixtures of primary treated sewage and river water (34).
MATERIALS AND METHODS
Site description.
Experiments were conducted on the Upper Mississippi River at Buffalo Shores Beach (GPS coordinates 41°27′11.23″N and 90°44′33.33″W) during October 2011. The average water temperature and turbidity range during the study were 14.1 ± 0.7°C and 14.5 to 19.8 nephelometric turbidity units (NTU), respectively. The monthly average solar insolation incident on a horizontal surface (3.08 kW h m−2 day−1) and monthly average daylight cloud (61.9%) data for the Buffalo Shores Beach were obtained from NASA Langley Research Center (http://eosweb.larc.nasa.gov/).
Submersible aquatic mesocosm.
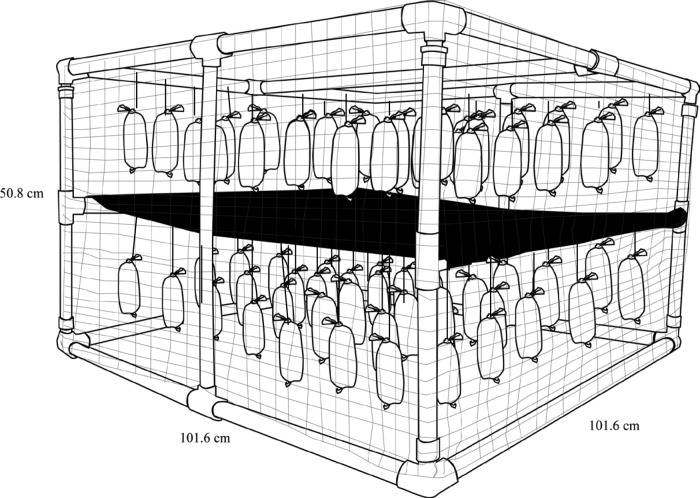
A submersible mesocosm device (Fig. 1) was constructed as previously described (34). Briefly, the device consisted of a polyvinyl chloride (PVC) pipe (diameter, 3/4 to 1 in.) frame wrapped in plastic mesh wire to prevent floating debris from damaging the encased 200-ml dialysis tubing diffusion bags. The mesocosm was designed to evaluate the effect of sunlight irradiation, so the lower half of the device was covered using a heavy-duty black plastic sheet (dark treatments) and the top half of the device left uncovered (light treatments) (Fig. 1). The mesocosm was deployed so that the light treatment dialysis bags were submerged approximately 5 to 10 cm below the water surface. The potential sunlight attenuation by the dialysis bag was evaluated by measuring the strength of UV light (solar power meter by Ambient Weather, Chandler, AZ, and UVX radiometer by UVP, LLC, Upland, CA) with and without the dialysis bag cover. The difference in UV readings was minor (i.e., <10%).
FIG 1.
Schematic diagram of the submersible aquatic mesocosm.
Sample mixtures (200 ml) were contained within dialysis bags consisting of 75-mm-flat-width, 13- to 14-kDa-pore-size regenerated cellulose dialysis tubing (Spectrum Laboratories, Rancho Dominguez, CA); bags were affixed to the device using fishing line and fishing snap swivels. Three replicate dialysis bags for each treatment (treatments 1 to 4, described below) were collected approximately every other day for a period of 7 days. Upon harvest, dialysis bags were placed into marked ziplock bags, filled partially with ambient water at the site (to avoid desiccation), and transported on ice within an hour to Iowa State Hygienic Laboratory located in Coralville, IA. Dialysis bags were vigorously shaken to mix the contents and opened using sterile scissors.
Study design.
In order to determine the effects of environmental variables on the decay of culture-based and molecularly based FIB, as well as human-associated MST genetic markers, experimental treatments were as follows: 1, exposure to ambient sunlight and indigenous river microbiota; 2, exposure to indigenous river microbiota only; 3, exposure to ambient sunlight only; and 4, exposure to neither ambient sunlight nor indigenous river microbiota. For all of the treatments, dialysis bags were filled with 100 ml of Mississippi River water and 100 ml of primary sewage effluent from a local wastewater treatment plant (City of Davenport Water Pollution Control Plant; GPS coordinates 41°29′31.83″N and 90°37′41.71″W). A 1:1 ratio was selected to mimic sewage spills such as CSO events, where estimated loads from sanitary sewage can be greater than 50% and pollutants such as fecal indicator organisms are reported to be up to 10 times higher than in treated sewage discharges (35–40), and is consistent with earlier decay studies (23–25, 34, 41, 42). Both river water and primary sewage effluent were collected in the morning on the same day that the mesocosm was deployed. For treatments 3 and 4, indigenous microbiota were removed by filtering the river water through 0.45-μm and then 0.22-μm nitrocellulose membranes, followed by processing through an electropositive 6-inch NanoCeram cartridge filter (2 to 3 μm; Argonide, Stanford, FL) for virus removal (43). Residual culturable enterococci, E. coli, and aerobic/facultatively anaerobic heterotrophs in filtered river water were tested on mEI (44), modified mTEC (45), and tryptic soy agar (TSA), respectively, with negligible detection (i.e., <10 CFU per 1 ml of river water.
Culture-based FIB enumeration.
Membrane filtration on mEI and modified mTEC agar was used to enumerate culturable enterococci and E. coli according to standard protocols (44, 45). In the earlier stages of the experiment (e.g., 0 h and 72 h), decimal dilution series were used, while 10-ml dialysis bag aliquots were processed for the later time points (e.g., 120 h and 168 h). All data were log10 transformed and expressed as log10 CFU 100 ml−1.
qPCR assays.
A 50-ml aliquot from each dialysis bag was filtered through a 0.45-μm nitrocellulose membrane, and the filter was stored at −80°C until further processing (<6 months). In order to minimize the holding time, samples were divided into two sets (or batches), each containing an even number of filters. DNA was extracted from filters using the PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA). The manufacturer's recommended protocol was used except for the following: (i) an additional 10-min incubation of bead beating tube containing filter and C1 reagent at 65°C followed by (ii) utilization of a FastPrep homogenizer (MP Biomedicals, Santa Ana, CA) for 1 min at 60 ms−1 instead of vortexing. Extracted DNA was stored at 4°C for no longer than 24 h prior to qPCR testing. The total mass of extracted DNA was quantified using the Quant-iT Pico Green double-stranded DNA (dsDNA) assay kit (Life Technologies, Grand Island, NY) on a SpectraMax Paradigm Multi-Mode microplate detection platform (Molecular Devices, LLC, Sunnyvale, CA) according to the manufacturer's instructions.
GenBac3, HF183, HumM2, and Sketa22 qPCR assays were performed in simplex format, while Entero1a was performed in multiplex format (the targets were the enterococcal 23S rRNA gene and an internal amplification control [IAC]), as described previously (46). All qPCR assays were performed within 24 h after DNA extraction. Table 1 lists the TaqMan probes (Life Technologies, Grand Island, NY) and primers used. Environmental MasterMix (Life Technologies, Grand Island, NY) was used for qPCRs. Simplex reaction mixtures contained 0.2 mg ml−1 bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 1 μM each forward and reverse primers, 80 nM 6-carboxyfluorescein (FAM)-labeled TaqMan probe (Life Technologies, Grand Island, NY), and 2 μl of template DNA, and the volume was brought up to 25 μl using ultrapure H2O (Sigma-Aldrich, St. Louis, MO). Multiplex reaction mixtures were prepared using the same reaction conditions as described above except with the addition of 80 nM VIC-labeled UCP1 TaqMan probe, and 2 μl of IAC template (50 copies). For calibration curve reactions, 2 μl of linearized plasmid constructs ranging from 10 to 106 copies per reaction were used as templates. All qPCRs were performed in triplicate using a 7900 HT Fast real-time sequence detector (Life Technologies, Grand Island, NY). The thermal cycling profile for all assays was as follows: 2 min at 95°C followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. The threshold was adjusted manually to either 0.03 (GenBac3, Entero1a, HF183, and Sketa22) or 0.08 (HumM2) and quantification cycle (Cq) values were exported to Microsoft Excel for further analyses.
TABLE 1.
Summary of master calibration curve characteristics and TaqMan probes and primers used
| Assay | Target(s) | Equation | E | r2 | LLOQb | Reference(s) |
|---|---|---|---|---|---|---|
| Entero1aa | Enterococcal 23S rRNA gene and internal amplification control | y = 38.4 − 3.38x | 0.94 | 0.99 | 0.84 | 67, 68 |
| GenBac3 | Bacteroidales 16S rRNA gene | y = 38.9 − 3.50x | 0.94 | 0.99 | 0.89 | 29 |
| HF183 | Human-associated Bacteroidales 16S rRNA gene | y = 38.4 − 3.30x | 0.94 | 0.98 | 0.98 | 69 |
| HumM2 | Bacteroidales-like putative σ factor | y = 39.6 −3.30x | 0.94 | 0.98 | 0.84 | 70 |
Multiplex format.
The lower limit of quantification (LLOQ), expressed as a log10 copy number, represents the upper bound of the 95% confidence interval for the average of 10 copy dilutions from six independent standard curves (n = 18).
Quality assurance/quality controls (QA/QC).
A new working stock of the salmon DNA sample processing control (final concentration, 0.2 μg ml−1) was prepared for each DNA extraction batch and added to each bead beating tube containing nitrocellulose filter and to extraction blanks (five replicates per DNA extraction batch). The concentration of salmon DNA was estimated using the Sketa22 qPCR assay (4, 46–48). Acceptance thresholds were calculated for each batch as extraction blank mean Cq + 3, as previously used (46, 49).
Potential amplification interference was specifically estimated by spiking 50 copies of the IAC into Entero1a test reaction mixtures as previously described (47). The interference threshold was established by repeated readings of the IAC (n = 20) in buffer only. Evidence of amplification interference was defined as Cq values exceeding the control IAC mean Cq ± 1.5 (49–52). In order to estimate contamination by extraneous DNA, a minimum of five no-template controls (NTCs) were included with each instrument run.
Data analyses.
Master calibration curves (generated from six independent standard curves), lower limit of quantification (LLOQ), and concentrations estimates of qPCR genetic markers were calculated using a Bayesian Markov chain Monte Carlo (MCMC) approach on the publicly available software WinBUGS, version 1.4.1 (53). The upper bound of the 95% confidence interval for the highest dilution (i.e., 10 copies) of the plasmids used to generate master calibration curves served as the LLOQ. Amplification efficiencies (E) were calculated using the formula E = 10(−1/slope) − 1, and concentration estimates are reported as log10 copy number 100 ml−1.
Changes in target bacteria estimates were calculated as log10 (C/C0), where C0 was the estimate at the beginning of the experiment and C was the estimate remaining after 72 or 120 h. The 72-h time point was selected to represent early stages of the decay, and 120 h was selected as the final time point because estimates for some of the targets at the 168-h time point were below the respective LLOQ.
Two-way analysis of variance (ANOVA) (StatMate version 2.0 for Windows; GraphPad, San Diego, CA) was used to characterize effects of exposure to indigenous river microbiota or ambient sunlight on culture-based FIB, qPCR-based FIB, and human-associated MST measurements. Data for the analyses were organized in a 2-by-2 block design with “indigenous microbiota” and “sunlight” as fixed factors, each with two levels (i.e., presence and absence). The number of data points used in each two-way ANOVA ranged from 12 (culture based) to 36 (qPCR). The Pearson correlation coefficient (StatMate version 2.00 for Windows; GraphPad, San Diego, CA) was used to determine whether statistically significant relationships between decay patterns existed for the different indicator types.
RESULTS
QA/QC and performance metrics.
Master calibration curve equations and associated performance metrics are listed in Table 1. The total DNA mass per reaction ranged from 5.5 ng to 45 ng, depending on the sample. Addition of salmon DNA and subsequent Sketa22 qPCR analysis was used as a sample processing control, and all samples were within acceptance values for the first and second batches of DNA extractions, being 24 ± 3.0 Cq and 28 ± 3.0 Cq, respectively (46, 49). DNA extraction batch-specific thresholds were determined for the IAC to identify potential amplification interferences, and these values were 31.1 + 1.5 Cq and 33.9 + 1.5 Cq for the first and second batches, respectively (49, 50, 52). No amplification interference was detected. A total of 108 qPCR amplifications (sum of NTCs and extraction blanks) were used to assess potential contamination by extraneous DNA, and none contained Cq values within the quantification range of the qPCR assays (Table 1). Variability between triplicate qPCR measurements was calculated as percent coefficient of variation (CV), and it ranged from 0.12 to 4.06 at 0 h, from 0.09 to 3.82 at 72 h, and from 0.54 to 19.7 at 120 h for all assays. For the last time point (168 h), the percent CV for the GenBac3 and Entero1a assays ranged from 0.34 to 5.18, as these two targets remained within our range of quantification, but it was considerably higher for HF183 and HumM2 (1.57 to 71.0), since a number of samples were below the LLOQ at this time point.
Culturable enterococci and E. coli.
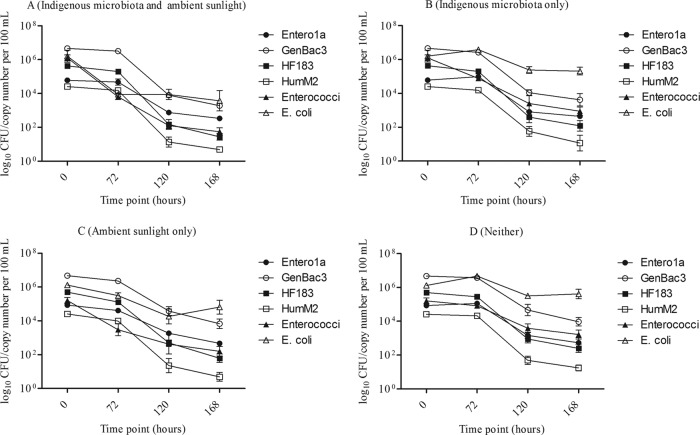
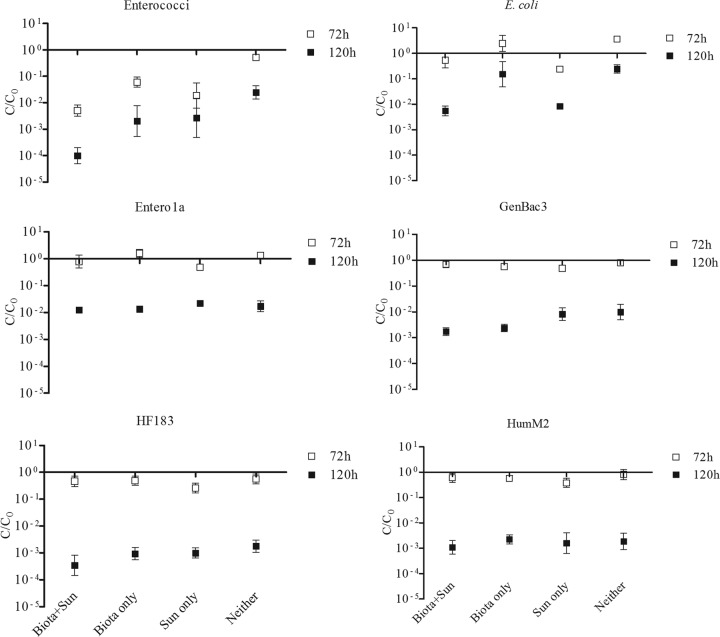
Standard membrane filtration methods were used to enumerate enterococci and E. coli for each time point and treatment. Both groups exhibited a largely biphasic decay pattern, with slight decreases in concentrations observed within 72 h followed by greater reductions between 72 and 120 h and apparent leveling off during the last 48 h (Fig. 2). Overall, the highest reductions of culturable FIB occurred when dialysis bags were exposed to both indigenous microbiota and ambient sunlight, while selective exclusion of either factor generally resulted in prolonged survival (Fig. 2 and 3; Tables 2 and 3). For enterococci, both factors were important in early decay dynamics (i.e., after 72 h), although sunlight appeared to be a more important contributor (66% of total variation) than microbiota (24%) (Tables 2 and 3). At later time intervals (i.e., after 120 h), the importance of sunlight decreased (36% of total variation), while the contribution from microbiota became more important (43%) (Fig. 3; Tables 2 and 3).
FIG 2.
Changes in concentration estimates over time for culture-based and molecularly based FIB, as well as genetic human-associated MST markers, when exposed to different treatment variables. Data are averages from replicate dialysis bags (n = 3); error bars represent standard deviations.
FIG 3.
Effects of treatment variables on decay rates of culture- and molecularly based FIB, as well as human-associated MST genetic markers, after 72 h and 120 h. Data are averages from replicate dialysis bags (n = 3); error bars represent standard deviations.
TABLE 2.
Effect of environmental variables on decline of culture-based FIB
| Assay | Time point (h) | Biotic interactions |
Sunlight |
Interaction |
|||
|---|---|---|---|---|---|---|---|
| % Contribution | P value | % Contribution | P value | % Contribution | P value | ||
| Enterococci | 72 | 24 | 0.002 | 66 | <0.0001 | 1.4 | 0.30 |
| 120 | 44 | 0.003 | 36 | 0.005 | 0.8 | 0.60 | |
| E. coli | 72 | 0.8 | 0.51 | 80 | <0.0001 | 6.4 | 0.08 |
| 120 | 1.6 | 0.27 | 89 | <0.0001 | 0.01 | 0.93 | |
TABLE 3.
Post hoc tests of effect of biotic interactions in the presence or absence of sunlight for culture-based FIB
| Assay | Time point (h) | Sunlight | P value |
|---|---|---|---|
| Enterococci | 72 | Present | >0.05 |
| Absent | <0.01 | ||
| 120 | Present | <0.05 | |
| Absent | >0.05 | ||
| E. coli | 72 | Present | >0.05 |
| Absent | >0.05 | ||
| 120 | Present | >0.05 | |
| Absent | >0.05 |
In contrast to the case for culturable enterococci, exposure to ambient sunlight was the only significant factor impacting culturable E. coli and contributing more than 80% to total variability, irrespective of the time point (Fig. 3; Tables 2 and 3). Interaction of variables was not statistically significant for either enterococci or E. coli (Tables 2 and 3).
FIB (Entero1a and GenBac3) measured by qPCR.
FIB measured by qPCR followed biphasic decay patterns similar to those described for culturable enterococci and E. coli (Fig. 2), although the decrease in concentrations was less pronounced. Furthermore, both Entero1a and GenBac3 appeared to decrease less than the human-associated genetic MST markers (Fig. 2). Exposure to ambient sunlight was a significant factor in the early decay of Entero1a signal, contributing 56% to the variation, but its importance diminished as time progressed (Fig. 3; Tables 4 and 5). The opposite was the case in the presence of indigenous aquatic microbiota; while only a marginally important contributor to early stages of decay (8.0% contribution to variability), it became the only significant contributor after 120 h, accounting for 34% of the variability (Fig. 3; Tables 4 and 5). There was no statistically significant interaction between variables in this data set (Tables 4 and 5).
TABLE 4.
Effect of environmental variables on decline of molecularly based FIB
| Assay | Time point (h) | Biotic interactions |
Sunlight |
Interaction |
|||
|---|---|---|---|---|---|---|---|
| % Contribution | P value | % Contribution | P value | % Contribution | P value | ||
| Entero1a | 72 | 8.0 | 0.01 | 56 | <0.0001 | 1.9 | 0.20 |
| 120 | 34 | 0.0002 | 1.5 | 0.38 | 5.0 | 0.11 | |
| GenBac3 | 72 | 0.04 | 0.89 | 5.7 | 0.09 | 35 | 0.0001 |
| 120 | 70 | <0.0001 | 2.0 | 0.14 | 0.1 | 0.70 | |
TABLE 5.
Post hoc tests of effect of biotic interactions in the presence or absence of sunlight for molecularly based FIB
| Assay | Time point (h) | Sunlight | P value |
|---|---|---|---|
| Entero1a | 72 | Present | <0.05 |
| Absent | >0.05 | ||
| 120 | Present | <0.001 | |
| Absent | >0.05 | ||
| GenBac3 | 72 | Present | <0.01 |
| Absent | <0.05 | ||
| 120 | Present | <0.0001 | |
| Absent | <0.0001 |
Neither variable was a statistically significant contributor to the early decay dynamics of GenBac3; however, after 120 h, the importance of the presence of indigenous aquatic microbiota became more apparent, as it was the only statistically significant contributor to decay for that time point, accounting for 70% of the variation (Fig. 3; Tables 4 and 5). Interaction of variables was statistically significant in the early stages of decay, but not after 120 h, suggesting that the effect of the presence of indigenous aquatic microbiota on GenBac3 was not dependent on sunlight in the later stages of the decay.
Human-associated MST genetic markers (HF183 and HumM2).
Overall, the decay patterns of human-associated MST markers were similar to those observed for qPCR-based FIB (Fig. 2 and 3). This was especially evident for HF183, where early decay dynamics were significantly impacted by sunlight only (17% contribution to variation), with the importance of indigenous river microbiota presence becoming more evident after 120 h (28% contribution to variability) (Fig. 3; Tables 6 and 7). Interaction of variables was significant only for decay patterns observed after 72 h, suggesting that the impact of sunlight within that time frame was dependent on the presence of indigenous aquatic microbiota.
TABLE 6.
Effect of environmental variables on decline of human-associated MST genetic markers
| Assay | Time point (h) | Biotic interactions |
Sunlight |
Interaction |
|||
|---|---|---|---|---|---|---|---|
| % Contribution | P value | % Contribution | P value | % Contribution | P value | ||
| HF183 | 72 | 4.5 | 0.15 | 17 | 0.008 | 14 | 0.02 |
| 120 | 28 | 0.0002 | 23 | 0.0006 | 1.5 | 0.33 | |
| HumM2 | 72 | 0.3 | 0.71 | 15 | 0.01 | 19 | 0.006 |
| 120 | 0.6 | 0.71 | 9.5 | 0.13 | 3.7 | 0.33 | |
TABLE 7.
Post hoc tests of effect of biotic interactions in the presence or absence of sunlight for human-associated MST genetic markers
| Assay | Time point (h) | Sunlight | P value |
|---|---|---|---|
| HF183 | 72 | Present | <0.05 |
| Absent | >0.05 | ||
| 120 | Present | <0.01 | |
| Absent | >0.05 | ||
| HumM2 | 72 | Present | <0.05 |
| Absent | >0.05 | ||
| 120 | Present | >0.05 | |
| Absent | >0.05 |
Similar to the early decay pattern of HF183, sunlight was the only statistically significant factor impacting decline of the HumM2 marker after 72 h of exposure, and statistically significant interaction of variables suggests that the observed effect of sunlight was dependent on the presence of indigenous aquatic microbiota (Fig. 3; Tables 6 and 7). However, unlike for HF183, neither environmental variable was an important contributor to decay after 120 h of exposure (Fig. 3; Tables 6 and 7). It is important to note that the HumM2 marker was near the LLOQ for some of the treatments at this time point, which could introduce higher variability.
Correlations in decay of different indicator types.
There was a moderately strong, positive correlation between decay of the two culturable FIB examined, irrespective of the treatment type, but there was no statistically significant correlation between culturable FIB and qPCR-based FIB/human-associated genetic marker decay patterns (Table 8). The decay of qPCR-based FIB (Entero1a and GenBac3) and human-associated genetic markers (HF183 and HumM2) was strongly correlated among each other and between the two categories (Table 8).
TABLE 8.
Correlations in decay patterns of culture-based/qPCR-based FIB and human-associated genetic MST markers
| Indicator | Correlation (P value) with: |
|||||
|---|---|---|---|---|---|---|
| Enterococci | E. coli | Entero1a | GenBac3 | HF183 | HumM2 | |
| Enterococci | 0.76 (0.005) | 0.50 (0.05) | 0.55 (0.04) | 0.53 (0.04) | 0.47 (0.06) | |
| E. coli | 0.64 (0.02) | 0.62 (0.02) | 0.60 (0.01) | 0.65 (0.02) | ||
| Entero1a | 0.96 (<0.0001) | 0.98 (<0.0001) | 0.98 (<0.0001) | |||
| GenBac3 | 0.98 (<0.0001) | 0.96 (<0.0001) | ||||
| HF183 | 0.99 (<0.0001) | |||||
DISCUSSION
Earlier epidemiological studies established a reasonable correlation between levels of culturable FIB and incidence of gastrointestinal illness in recreational bathers (54, 55), but more recent data support improved health risk prediction from qPCR targets (Entero1a more so than GenBac3) in municipal wastewater-impacted freshwater (56) and marine water (57) settings. Our in situ decay study was designed to investigate the effects of ambient sunlight and biotic interactions on culture-based enterococci and E. coli, FIB measured by qPCR, and select human-associated MST genetic markers.
Both exposure to ambient sunlight and biotic interactions (viral lysis, predation, and/or competition) are factors known to adversely affect survival of culturable FIB (enterococci and E. coli) in the environment (58–60). However, recent studies have shown that the magnitude of that effect is likely to vary among FIB in different habitats and even in different locations within the same habitat (i.e., water column and sediments) (58, 60). Our results indicate that persistence of culturable, sewage-borne enterococci in the Mississippi River water was affected by both ambient sunlight and biotic interactions. Sunlight was a more important determinant of survival in the early stages, while biotic interactions became more influential in the later stages. This finding is similar to the results of Wanjugi and Harwood (60), who showed that the survival of culturable enterococci was negatively affected by indigenous river microbiota; however, they used a single strain of Enterococcus faecalis, while we utilized enterococci present in primary sewage effluent. The effect of sunlight on culturable, sewage-borne enterococci also corroborates a previous report indicating a significantly greater reduction when exposed to sunlight compared to dark conditions (42).
Unlike enterococci, culturable sewage-borne E. coli did not appear to be affected by biotic interactions. Instead, exposure to sunlight was the only significant factor in decay in both early and later stages of the experiment. The effect of sunlight exposure on E. coli is not surprising (42, 61, 62), but the lack of an effect from biotic interactions was unexpected. Recent studies (34, 58, 60) have shown that the indigenous aquatic microbiota is an important determinant of E. coli survival in both marine and freshwater habitats. While some of these studies utilized laboratory-grown strains and were conducted in outdoor mesocosms (58, 60), which may overestimate the effects of experimental variables, others were conducted using the same in situ mesocosm and primary sewage effluent as the inoculum (34). The most notable differences between these studies were temperate versus subtropical rivers (Mississippi River, IA, versus Hillsborough River, FL) used as receiving waters and different (local) primary sewage effluents, suggesting that associated variables such as indigenous microbial populations (from sewage and ambient water) may play an important role. In the current study, we noted a much stronger effect of sunlight exposure, which was not apparent in our earlier subtropical river work (34), possibly due at least in part to the higher turbidity of the latter (17.0 versus 66.6 NTU). Previous studies have shown that bacterivorous protozoa prefer metabolically active prey over damaged or dead cells (63–65), which suggests that the lack of a biotic interaction effect in the current study may be partly due to the cellular damage caused by sunlight exposure.
As previously reported (25, 27), we also observed extended persistence of the Entero1a qPCR signal compared to that of culturable enterococci and slower decay of qPCR-based FIB (Entero1a and GenBac3) than of human-associated MST genetic markers (24, 25) but relatively similar overall decay profiles among all qPCR targets (25, 30), at least partly due to comparable responses to treatments tested in this study. The detrimental effects of sunlight and biotic treatments on the culturable fecal indicator bacteria compared to DNA molecules targeted by qPCR suggest that the underlying mechanisms of decay were different. However, similarities in decay trends across all qPCR assays suggest that the mechanism for deterioration of the primer/probe region may be the same. Additional research is needed to characterize how robust these trends are under other conditions and how these differences help or hinder water quality applications such as recreation water quality monitoring, fecal source allocation, and subsequent QMRA modeling.
The existing literature on the effect of ambient sunlight on decay of genetic MST markers in surface freshwaters (groundwater and lake, creek, and river water) generally supports the point that sunlight is not an important factor in decay (25, 27, 30, 66). These studies employed modeling of the decay rates and usually did not examine decay patterns at early time points. Our work supports the idea to describe in situ decay profiles during the period when the effect of experimental variables appeared to be the strongest. Thus, while our results suggesting that ambient sunlight does not appear to be an important contributor after 120 h of wastewater release agree with previous findings, the significant effect of sunlight on early-stage decay dynamics appears to be less well understood.
Only a limited number of studies have examined the effects of biotic interactions (predation, viral lysis, and competition) on the persistence of (q)PCR targets, such as Bacteroides and/or enterococci in freshwater environments (23, 24, 26, 28). The earlier work supports the view that survival of a laboratory strain of Bacteroides fragilis (23) and feces-derived Bacteroides distasonis (28) was negatively impacted by the combined effects of predation and elevated temperatures. Extended persistence of sewage- and/or fecally derived indicators (measured by qPCR) and human-associated MST genetic markers is documented for autoclaved (24) and filter-sterilized (26) river water. Our study expands on these findings, as it was not conducted either under laboratory conditions (24) or in a fully closed outdoor mesocosm (23, 26); rather, we attempted greater environmental interaction by using in situ submersed diffusion bags and primary sewage effluent as an inoculum.
Although these experiments advance our understanding of fecal indicator decay in a freshwater system, it is important to note that this study focused on pollution events where high concentrations of untreated (or partially treated) sewage are typical and may not be representative of pollution events from treated sewage. Furthermore, it is also likely that biotic effects will be influenced by the proportion of pollution source to ambient river water. Additional research is warranted to determine if the trends identified in this study are consistent under these different scenarios.
In summary, our results indicate that biotic interactions and exposure to ambient sunlight are both important factors in the decay of sewage-borne culturable and qPCR-based FIB, as well as human-associated MST genetic markers. In general, sunlight-induced decay was often a key factor in the early stages of decomposition (<72 h after wastewater release), while biotic interactions played a larger role during the later stages. Future studies are needed to explore the range of influence that different water types and associated indigenous microbiota can have on the decay of fecal pollution markers, combined with characterization of microbial community members to identify key taxa involved in these important biotic interactions.
ACKNOWLEDGMENTS
The U.S. Environmental Protection Agency through its Office of Research and Development funded and managed the research described here.
The work described here has been subjected to the U.S. Environmental Protection Agency's administrative review and approved for publication. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 18 April 2014
REFERENCES
- 1.Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291. 10.1021/es903523q [DOI] [PubMed] [Google Scholar]
- 2.Schoen ME, Soller JA, Ashbolt NJ. 2011. Evaluating the importance of faecal sources in human-impacted waters. Water Res. 45:2670–2680. 10.1016/j.watres.2011.02.025 [DOI] [PubMed] [Google Scholar]
- 3.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44:4674–4691. 10.1016/j.watres.2010.06.049 [DOI] [PubMed] [Google Scholar]
- 4.U.S. Environmental Protection Agency. 2010. Method A: enterococci in water by TaqMan quantitative polymerase chain reaction (qPCR) assay. EPA-821-R-10-004. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 5.Barata A, Malfeito-Ferreira M, Loureiro V. 2012. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153:243–259. 10.1016/j.ijfoodmicro.2011.11.025 [DOI] [PubMed] [Google Scholar]
- 6.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. 2012. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 76:685–706. 10.1128/MMBR.00023-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doud CW, Zurek L. 2012. Enterococcus faecalis OG1RF:pMV158 survives and proliferates in the house fly digestive tract. J. Med. Entomol. 49:150–155. 10.1603/ME11167 [DOI] [PubMed] [Google Scholar]
- 8.Harwood VJ, Butler J, Parrish D, Wagner V. 1999. Isolation of fecal coliform bacteria from the diamondback terrapin (Malaclemys terrapin centrata). Appl. Environ. Microbiol. 65:865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Nan X, Wei C, He H. 2012. The gut bacteria associated with Camponotus japonicus Mayr with culture-dependent and DGGE methods. Curr. Microbiol. 65:610–616. 10.1007/s00284-012-0197-1 [DOI] [PubMed] [Google Scholar]
- 10.Pourcher AM, Devriese LA, Hernandez JF, Delattre JM. 1991. Enumeration by a miniaturized method of Escherichia coli, Streptococcus bovis and enterococci as indicators of the origin of faecal pollution of waters. J. Appl. Bacteriol. 70:525–530. 10.1111/j.1365-2672.1991.tb02752.x [DOI] [PubMed] [Google Scholar]
- 11.Ramarao N, Nielsen-Leroux C, Lereclus D. 2012. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis Exp. 70:e4392. 10.3791/4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedlacek I, Holochova P, Maslanova I, Kosina M, Sproer C, Bryndova H, Vandamme P, Rudolf I, Hubalek Z, Svec P. 2013. Enterococcus ureilyticus sp. nov. and Enterococcus rotai sp. nov., two urease-producing enterococci from the environment. Int. J. Syst. Evol. Microbiol. 63:502–510. 10.1099/ijs.0.041152-0 [DOI] [PubMed] [Google Scholar]
- 13.Souza V, Rocha M, Valera A, Eguiarte LE. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitman RL, Byers SE, Shively DA, Ferguson DM, Byappanahalli M. 2005. Occurrence and growth characteristics of Escherichia coli and enterococci within the accumulated fluid of the northern pitcher plant (Sarracenia purpurea L.). Can. J. Microbiol. 51:1027–1037. 10.1139/w05-091 [DOI] [PubMed] [Google Scholar]
- 15.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 38:1–40. 10.1111/1574-6976.12031 [DOI] [PubMed] [Google Scholar]
- 16.Ahmed W, Goonetilleke A, Powell D, Chauhan K, Gardner T. 2009. Comparison of molecular markers to detect fresh sewage in environmental waters. Water Res. 43:4908–4917. 10.1016/j.watres.2009.09.047 [DOI] [PubMed] [Google Scholar]
- 17.Chase E, Hunting J, Staley C, Harwood VJ. 2012. Microbial source tracking to identify human and ruminant sources of faecal pollution in an ephemeral Florida river. J. Appl. Microbiol. 113:1396–1406. 10.1111/jam.12007 [DOI] [PubMed] [Google Scholar]
- 18.Gordon KV, Brownell M, Wang SY, Lepo JE, Mott J, Nathaniel R, Kilgen M, Hellein KN, Kennedy E, Harwood VJ. 2013. Relationship of human-associated microbial source tracking markers with enterococci in Gulf of Mexico waters. Water Res. 47:996–1004. 10.1016/j.watres.2012.10.032 [DOI] [PubMed] [Google Scholar]
- 19.Korajkic A, Badgley BD, Brownell MJ, Harwood VJ. 2009. Application of microbial source tracking methods in a Gulf of Mexico field setting. J. Appl. Microbiol. 107:1518–1527. 10.1111/j.1365-2672.2009.04351.x [DOI] [PubMed] [Google Scholar]
- 20.Korajkic A, Brownell MJ, Harwood VJ. 2011. Investigation of human sewage pollution and pathogen analysis at Florida Gulf coast beaches. J. Appl. Microbiol. 110:174–183. 10.1111/j.1365-2672.2010.04869.x [DOI] [PubMed] [Google Scholar]
- 21.Staley C, Reckhow KH, Lukasik J, Harwood VJ. 2012. Assessment of sources of human pathogens and fecal contamination in a Florida freshwater lake. Water Res. 46:5799–5812. 10.1016/j.watres.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 22.Staley ZR, Chase E, Mitraki C, Crisman TL, Harwood VJ. 2013. Microbial water quality in freshwater lakes with different land use. J. Appl. Microbiol. 115:1240–1250. 10.1111/jam.12312 [DOI] [PubMed] [Google Scholar]
- 23.Balleste E, Bonjoch X, Belanche LA, Blanch AR. 2010. Molecular indicators used in the development of predictive models for microbial source tracking. Appl. Environ. Microbiol. 76:1789–1795. 10.1128/AEM.02350-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl. Environ. Microbiol. 76:3255–3262. 10.1128/AEM.02636-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green HC, Shanks OC, Sivaganesan M, Haugland RA, Field KG. 2011. Differential decay of human fecal Bacteroides in marine and freshwater. Environ. Microbiol. 13:3235–3249. 10.1111/j.1462-2920.2011.02549.x [DOI] [PubMed] [Google Scholar]
- 26.Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890–901. 10.1007/s00253-006-0714-x [DOI] [PubMed] [Google Scholar]
- 27.Walters SP, Field KG. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ. Microbiol. 11:1410–1421. 10.1111/j.1462-2920.2009.01868.x [DOI] [PubMed] [Google Scholar]
- 28.Kreader CA. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl. Environ. Microbiol. 64:4103–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249–259. 10.1111/j.1462-2920.2004.00702.x [DOI] [PubMed] [Google Scholar]
- 30.Bae S, Wuertz S. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 43:4850–4859. 10.1016/j.watres.2009.06.053 [DOI] [PubMed] [Google Scholar]
- 31.Savichtcheva O, Okayama N, Okabe S. 2007. Relationships between Bacteroides 16S rRNA genetic markers and presence of bacterial enteric pathogens and conventional fecal indicators. Water Res. 41:3615–3628. 10.1016/j.watres.2007.03.028 [DOI] [PubMed] [Google Scholar]
- 32.Walters SP, Yamahara KM, Boehm AB. 2009. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters. Water Res. 43:4929–4939. 10.1016/j.watres.2009.05.047 [DOI] [PubMed] [Google Scholar]
- 33.Bae S, Wuertz S. 2012. Survival of host-associated bacteroidales cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl. Environ. Microbiol. 78:922–932. 10.1128/AEM.05157-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korajkic A, McMinn BR, Shanks OC, Harwood VJ, Fout GS, Ashbolt NJ. 2013. Differential decay of enterococci and Escherichia coli originating from two fecal pollution sources. Appl. Environ. Microbiol. 79:2488–2492. 10.1128/AEM.03781-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasperi J, Rocher V, Gilbert S, Azimi S, Chebbo G. 2010. Occurrence and removal of priority pollutants by lamella clarification and biofiltration. Water Res. 44:3065–3076. 10.1016/j.watres.2010.02.035 [DOI] [PubMed] [Google Scholar]
- 36.Musolff A, Leschik S, Reinstorf F, Strauch G, Schirmer M. 2010. Micropollutant loads in the urban water cycle. Environ. Sci. Technol. 44:4877–4883. 10.1021/es903823a [DOI] [PubMed] [Google Scholar]
- 37.Phillips PJ, Chalmers AT, Gray JL, Kolpin DW, Foreman WT, Wall GR. 2012. Combined sewer overflows: an environmental source of hormones and wastewater micropollutants. Environ. Sci. Technol. 46:5336–5343. 10.1021/es3001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rechenburg A, Koch C, Classen T, Kistemann T. 2006. Impact of sewage treatment plants and combined sewer overflow basins on the microbiological quality of surface water. Water Sci. Technol. 54:95–99. 10.2166/wst.2006.454 [DOI] [PubMed] [Google Scholar]
- 39.Soonthornnonda P, Christensen ER. 2008. Source apportionment of pollutants and flows of combined sewer wastewater. Water Res. 42:1989–1998. 10.1016/j.watres.2007.11.034 [DOI] [PubMed] [Google Scholar]
- 40.Weyrauch P, Matzinger A, Pawlowsky-Reusing E, Plume S, von Seggern D, Heinzmann B, Schroeder K, Rouault P. 2010. Contribution of combined sewer overflows to trace contaminant loads in urban streams. Water Res. 44:4451–4462. 10.1016/j.watres.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 41.Bordalo AA, Onrassami R, Dechsakulwatana C. 2002. Survival of faecal indicator bacteria in tropical estuarine waters (Bangpakong River, Thailand). J. Appl. Microbiol. 93:864–871. 10.1046/j.1365-2672.2002.01760.x [DOI] [PubMed] [Google Scholar]
- 42.Noble RT, Lee IM, Schiff KC. 2004. Inactivation of indicator micro-organisms from various sources of faecal contamination in seawater and freshwater. J. Appl. Microbiol. 96:464–472. 10.1111/j.1365-2672.2004.02155.x [DOI] [PubMed] [Google Scholar]
- 43.Ikner LA, Gerba CP, Bright KR. 2012. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 4:41–67. 10.1007/s12560-012-9080-2 [DOI] [PubMed] [Google Scholar]
- 44.U.S. Environmental Protection Agency. 2002. Method 1600: enterococci in water by membrane filtration using membrane-enterococcus indoxyl-B-d-glucoside agar (mEI). EPA 821-R-02-022. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 45.U.S. Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA 821-R-02-023. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 46.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst. Appl. Microbiol. 33:348–357. 10.1016/j.syapm.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 47.Haugland RA, Siefring S, Lavender J, Varma M. 2012. Influences of sample interference and interference controls on quantification of enterococci fecal indicator bacteria in surface water samples by the qPCR method. Water Res. 46:5989–6001. 10.1016/j.watres.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 48.U.S. Environmental Protection Agency. 2010. Method B: Bacteroidales in water by TaqMan quantitative polymerase chain reaction (qPCR) assay EPA-822-R-10-003. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 49.Shanks OC, Sivaganesan M, Peed L, Kelty CA, Blackwood AD, Greene MR, Noble RT, Bushon RN, Stelzer EA, Kinzelman J, Anan'eva T, Sinigalliano C, Wanless D, Griffith J, Cao Y, Weisberg S, Harwood VJ, Staley C, Oshima KH, Varma M, Haugland RA. 2012. Interlaboratory comparison of real-time PCR protocols for quantification of general fecal indicator bacteria. Environ. Sci. Technol. 46:945–953. 10.1021/es2031455 [DOI] [PubMed] [Google Scholar]
- 50.Kelty CA, Varma M, Sivaganesan M, Haugland RA, Shanks OC. 2012. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl. Environ. Microbiol. 78:4225–4232. 10.1128/AEM.07819-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shanks OC, White K, Kelty CA, Hayes S, Sivaganesan M, Jenkins M, Varma M, Haugland RA. 2010. Performance assessment PCR-based assays targeting Bacteroidales genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 76:1359–1366. 10.1128/AEM.02033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, Varma M, Haugland RA. 2010. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ. Sci. Technol. 44:6281–6288. 10.1021/es100311n [DOI] [PubMed] [Google Scholar]
- 53.Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC. 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9:120. 10.1186/1471-2105-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabelli VJ, Dufour AP, Levin MA, McCabe LJ, Haberman PW. 1979. Relationship of microbial indicators to health effects at marine bathing beaches. Am. J. Public Health 69:690–696. 10.2105/AJPH.69.7.690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabelli VJ, Dufour AP, McCabe LJ, Levin MA. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606–616 [DOI] [PubMed] [Google Scholar]
- 56.Wade TJ, Calderon RL, Sams E, Beach M, Brenner KP, Williams AH, Dufour AP. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Persp. 114:24–28. 10.1289/ehp.8273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, Wymer L, Rankin CC, Love D, Li Q, Noble R, Dufour AP. 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 9:66. 10.1186/1476-069X-9-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korajkic A, Wanjugi P, Harwood VJ. 2013. Indigenous microbiota and habitat influence Escherichia coli survival more than sunlight in simulated aquatic habitats. Appl. Environ. Microbiol. 79:5329–5337. 10.1128/AEM.01362-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menon P, Billen G, Servais P. 2003. Mortality rates of autochthonous and fecal bacteria in natural aquatic ecosystems. Water Res. 37:4151–4158. 10.1016/S0043-1354(03)00349-X [DOI] [PubMed] [Google Scholar]
- 60.Wanjugi P, Harwood VJ. 2013. The influence of predation and competition on the survival of commensal and pathogenic fecal bacteria in aquatic habitats. Environ. Microbiol. 15:517–526. 10.1111/j.1462-2920.2012.02877.x [DOI] [PubMed] [Google Scholar]
- 61.Davies-Colley RJ, Bell RG, Donnison AM. 1994. Sunlight inactivation of enterococci and fecal coliforms in sewage effluent diluted in seawater. Appl. Environ. Microbiol. 60:2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinton LW, Hall CH, Lynch PA, Davies-Colley RJ. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 68:1122–1131. 10.1128/AEM.68.3.1122-1131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boenigk J, Matz AC, Jurgens K, Arndt H. 2001. Confusing selective feeding with differential digestion in bacterivorous nanoflagellates. J. Eukaryot. Microbiol. 48:425–432. 10.1111/j.1550-7408.2001.tb00175.x [DOI] [PubMed] [Google Scholar]
- 64.Landry MR, Lehner-Fournier JM, Sundstrom JA, Fagerness VL, Selph KE. 1991. Discrimination between living and heat-killed prey by a marine zooflagellate Paraphysomonas vestita (Stokes). J. Exp. Mar. Biol. Ecol. 146:139–152. 10.1016/0022-0981(91)90021-N [DOI] [Google Scholar]
- 65.Massana R, Unrein F, Rodriguez-Martinez R, Forn I, Lefort T, Pinhassi J, Not F. 2009. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 3:588–596. 10.1038/ismej.2008.130 [DOI] [PubMed] [Google Scholar]
- 66.Sokolova E, Astrom J, Pettersson TJ, Bergstedt O, Hermansson M. 2012. Decay of Bacteroidales genetic markers in relation to traditional fecal indicators for water quality modeling of drinking water sources. Environ. Sci. Technol. 46:892–900. 10.1021/es2024498 [DOI] [PubMed] [Google Scholar]
- 67.Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562. 10.1016/S0723-2020(00)80030-2 [DOI] [PubMed] [Google Scholar]
- 68.Siefring S, Varma M, Atikovic E, Wymer L, Haugland RA. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health 6:225–237. 10.2166/wh.2008.022 [DOI] [PubMed] [Google Scholar]
- 69.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574. 10.1128/AEM.66.10.4571-4574.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 75:5507–5513. 10.1128/AEM.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]