Abstract
Salmonella accounts for approximately 50% of produce-associated outbreaks in the United States, several of which have been traced back to contamination in the produce production environment. To quantify Salmonella diversity and aid in identification of Salmonella contamination sources, we characterized Salmonella isolates from two geographically diverse produce-growing regions in the United States. Initially, we characterized the Salmonella serotype and subtype diversity associated with 1,677 samples collected from 33 produce farms in New York State (NYS). Among these 1,677 samples, 74 were Salmonella positive, yielding 80 unique isolates (from 147 total isolates), which represented 14 serovars and 23 different pulsed-field gel electrophoresis (PFGE) types. To explore regional Salmonella diversity associated with production environments, we collected a smaller set of samples (n = 65) from South Florida (SFL) production environments and compared the Salmonella diversity associated with these samples with the diversity found among NYS production environments. Among these 65 samples, 23 were Salmonella positive, yielding 32 unique isolates (from 81 total isolates), which represented 11 serovars and 17 different PFGE types. The most common serovars isolated in NYS were Salmonella enterica serovars Newport, Cerro, and Thompson, while common serovars isolated in SFL were Salmonella serovars Saphra and Newport and S. enterica subsp. diarizonae serovar 50:r:z. High PFGE type diversity (Simpson's diversity index, 0.90 ± 0.02) was observed among Salmonella isolates across both regions; only three PFGE types were shared between the two regions. The probability of three or fewer shared PFGE types was <0.000001; therefore, Salmonella isolates were considerably different between the two sampled regions. These findings suggest the potential for PFGE-based source tracking of Salmonella in production environments.
INTRODUCTION
The genus Salmonella is divided into two species, Salmonella enterica and Salmonella bongori, and represents approximately 2,600 known serovars. S. enterica has six subspecies, enterica (I) salamae (II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV), and indica (V), and accounts for over 99% of Salmonella strains isolated worldwide (1). S. enterica subsp. enterica is the leading cause of bacterial food-borne illnesses, hospitalizations, and deaths in the United States (2). Approximately 95% of S. enterica infections in the United States are a result of consumption of contaminated foods (3, 4). S. enterica is estimated to be responsible for half of the produce-associated illnesses and a majority of produce-associated outbreaks in the United States (4–7). As a result, there is a need to identify likely sources of Salmonella contamination throughout the farm-to-fork continuum.
Subtyping is an important tool to detect food-borne outbreaks and to identify outbreak sources (8), as well as a powerful tool to investigate the diversity of food-borne pathogens in various hosts and environments. Even the most basic of subtyping methods (e.g., serotyping) can yield information on likely reservoirs for specific food-borne pathogens (9–11). For instance, S. enterica serovars Dublin and Choleraesuis are routinely associated with cattle and swine hosts, respectively (9, 12). In other studies (1, 13), S. enterica subsp. salamae, arizonae, diarizonae, houtenae, and indica were predominantly associated with cold-blooded animal hosts. S. enterica subsp. diarizonae was isolated most frequently from reptiles and amphibians obtained from the central coast of California (USA) (14). Another study (15) found that approximately 81% of pet snake fecal samples collected in Germany were positive for S. enterica subsp. diarizonae serovars. Other serovars are associated with a broad host range and a number of geographically diverse regions. Salmonella serovars Typhimurium and Enteritidis are two of the most common serovars reported among human Salmonella isolates worldwide (16). A study conducted in Great Britain on wild-bird populations was able to identify two host-adapted Salmonella serovar Typhimurium strains by use of pulsed-field gel electrophoresis (PFGE) and phage typing (PT) (17), demonstrating that host-associated subtypes can be identified even in broad-host-range serovars.
While subtyping can yield information on likely potential hosts of Salmonella, studies (18–22) have also demonstrated the application of subtyping to track specific food-borne pathogen subtypes in the environment. One study (22) investigating the source of fecal pollution on a Japanese beach showed a strong association between Enterococcus faecium isolated in samples from the beach and one of the suspected contamination sources (a river that drains into the beach) by PFGE typing. Cooley et al. (18) used PFGE typing to track Escherichia coli O157:H7 in a California produce-growing region and found that the same PFGE type of E. coli O157:H7 was isolated from feral swine, cattle, surface water, sediment, and soil from one of the spinach farms that had been implicated in the 2006 spinach-borne E. coli O157:H7 outbreak. In another study, Patchanee et al. (20) reported that distinct Salmonella PFGE types were recovered from water samples collected from sites near swine production or forestry, residential/industrial sites, and agriculture cropland, further supporting that subtyping methods, such as PFGE typing, can be used to track Salmonella in the environment and to identify specific contamination sources.
Studies (20, 23–25) have characterized the distribution and diversity of Salmonella isolates from a number of different environments; however, there is minimal information on Salmonella in the produce production environment, and no one to our knowledge has compared Salmonella isolates obtained from two geographically diverse produce-growing regions using the same sample collection, detection, and isolation schemes. The purpose of this study was to characterize Salmonella isolates obtained from environmental samples collected in produce production environments in New York State (NYS) and South Florida (SFL). We used both qualitative and quantitative methods to examine the distribution and diversity of Salmonella isolates from each region, as well as to compare the distribution of subtypes between these two regions. Additionally, Salmonella isolate subtype data were used to suggest potential sources of Salmonella contamination.
MATERIALS AND METHODS
Description of isolates used in this study.
A total of 228 Salmonella isolates (147 isolates from NYS and 81 isolates from SFL) were assembled for this study using five Salmonella data sets, consisting of two published, two in preparation, and one reported here (Table 1; see File S1 in the supplemental material for further detailed descriptions). Four data sets (I, II, IV, and V) representing NYS production environments had been collected to explore the association between Salmonella prevalence in produce fields and geographical and/or management factors (74 of 1,677 samples were Salmonella positive), whereas one data set (III) obtained from SFL was specifically collected for this study (23 of 65 samples were Salmonella positive) (Table 1; see also file S1). All sample collection and preparation (i.e., preparation of samples for Salmonella enrichment) were performed using the same methodology as previously described (26, 27). Detection and isolation of Salmonella were performed using a modified version of the Food and Drug Administration Bacteriological Analytic Manual (FDA BAM) method (28). Briefly, samples were diluted 1:10 with tryptic soy broth (TSB; Becton, Dickinson, Franklin Lakes, NJ) and incubated for 24 h at 35 ± 2°C. Enrichment aliquots of 1.0 and 0.1 ml were transferred to tetrathionate (TT; Oxoid; Cambridge, United Kingdom) and Rappaport-Vassiliadis (RV; Oxoid) broths, respectively, and incubated for 24 h in a shaking water bath at 42 ± 2°C. TT and RV cultures were plated onto xylose-lysine-deoxycholate agar (XLD; Neogen, Lansing, MI) and Salmonella chromogenic agar (CHROMagar; CHROMagar Company, Paris, France) and incubated for 24 and 48 h at 35 and 37 ± 2°C, respectively. Presumptive Salmonella colonies (up to four colonies per isolation scheme, i.e., TT-XLD, RV-XLD, TT-CHROMagar, and RV-CHROMagar) were substreaked to brain heart infusion (BHI; Becton, Dickinson) agar and incubated for 24 h at 37 ± 2°C. One isolated colony was selected from each BHI agar plate and tested by a PCR assay for invA, which is specific to Salmonella (29). All PCR-confirmed Salmonella isolates were preserved at −80°C in 15% glycerol.
TABLE 1.
Summary of study data sets and Salmonella isolatesa
| Data set no. | Regionb | Isolation year(s) | Sample group or type | No. of samples | Frequency of positive samples (%) | No. of isolates subtypedc | No. of representative isolatesd | No. of serovars | No. of PFGE types |
|---|---|---|---|---|---|---|---|---|---|
| I | NYS | 2009–2011 | Total group | 588 | 27 (4.6)e | 57 | 27 | 7 | 11 |
| Soil | 178 | 4 (2.8) | |||||||
| Drag swab | 175 | 3 (1.7) | |||||||
| Water | 174 | 16 (9.2) | |||||||
| Fecal | 61 | 4 (6.6) | |||||||
| II | NYS | 2012 | Total group | 600 | 26 (4.3) | 35 | 29 | 9 | 11 |
| Soil | 263 | 13 (4.9) | |||||||
| Drag swab | 263 | 5 (1.9) | |||||||
| Water | 74 | 8 (10.8) | |||||||
| Fecal | NCf | ||||||||
| III | SFL | 2010 | Total group | 65 | 23 (35.4) | 81 | 32 | 11 | 17 |
| Soil | 8 | 5 (62.5) | |||||||
| Drag swab | 8 | 3 (37.5) | |||||||
| Water | 40 | 15(37.5) | |||||||
| Fecal | 9 | 0 (0) | |||||||
| IV | NYS | 2011 | Total group | 429 | 17 (4.0) | 44 | 19 | 7 | 9 |
| Soil | 90 | 2 (2.2) | |||||||
| Drag swab | 219 | 4 (1.8) | |||||||
| Water | 120 | 11 (9.2) | |||||||
| Fecal | NC | ||||||||
| V | NYS | 2010 | Total group | 60 | 5 (8.3) | 11 | 5 | 4 | 4 |
| Soil | 20 | 2 (10) | |||||||
| Drag swab | 20 | 0 (0) | |||||||
| Water | 13 | 1 (7.7) | |||||||
| Fecal | 7 | 2 (28.6) | |||||||
| All | Total group | 1,742 | 98 (5.8) | 228 | 112 | 20 | 37 | ||
| Soil | 559 | 26 (5.1) | |||||||
| Drag swab | 685 | 15 (2.2) | |||||||
| Water | 421 | 51 (12.1) | |||||||
| Fecal | 77 | 6 (7.8) |
Data set I Salmonella isolates, serovars, and PFGE types have been previously described (26); data set II Salmonella isolates and serovars have been previously described (27); data set III is reported here; data set IV and V Salmonella isolates and serovars are unpublished. See File S1 in the supplemental material for further data set descriptions.
NYS, New York State; SFL, South Florida.
PFGE (using the restriction enzyme XbaI) was performed on one isolate per Salmonella positive sample for each isolation scheme (up to four isolates could be selected for the schemes RV-XLD, TT-XLD, RV-CHROMagar, and TT-CHROMagar).
Only isolates that were representative(s) of the Salmonella isolated in that sample were kept for further analyses (i.e., isolates from the same sample with identical serovars and PFGE types were excluded).
Isolates from one Salmonella-positive sample were unavailable for subtyping.
NC, not collected (a certain sample type was not collected for that data set).
Isolates selected for molecular characterization.
One isolate per isolation scheme (i.e., TT-XLD, RV-XLD, TT-CHROMagar, and RV-CHROMagar) was selected for serotyping and PFGE typing (147 isolates from NYS and 81 isolates from SFL, for a total of 228 isolates). This approach was used to capture all potential strains of Salmonella that may be present in a sample as several studies (14, 17, 23, 27, 30, 31) have shown that multiple Salmonella strains may be isolated from the same sample.
Traditional serotyping.
Serotyping was performed on all 228 isolates by the Wadsworth Center, New York State Department of Health (Albany, NY), using the White-Kauffman-Le Minor scheme (1).
Pulsed-field gel electrophoresis.
PFGE typing was performed on all 228 isolates using the standard U.S. Centers for Disease Control and Prevention (CDC) PulseNet protocol (32). Briefly, Salmonella cells were embedded in 1% SeaKem Gold agarose (Lonza, Rockland, ME), lysed, washed, and digested with 50 U/plug of XbaI (Roche Applied Science, Indianapolis, IN) at 37°C. Separation of the restricted DNA fragments was performed by a Chef Mapper XA (Bio-Rad, Hercules, California) for 18 to 20 h in 1% agarose gels. Voltage was set to 6 V/cm with an initial switch time of 2.16 s and a final switch time of 63.8 s. Salmonella serovar Braenderup (strain H9812) was used as the reference standard to allow normalization and comparison of gel images (33). Gel images were captured by a Bio-Rad Gel Doc using Multi-Analyst software, version 1.1 (Bio-Rad). PFGE images were analyzed by BioNumerics software, version 5.1 (Applied Maths, Sint-Martens-Latem, Belgium). Similarity clustering analyses were performed using the unweighted pair group method with arithmetic mean algorithm (UPGMA) based on Dice coefficients with a maximum space tolerance of 1.5%. PFGE types were named using Cornell University Food Safety Laboratory (CUFSL) nomenclature (34): laboratory location of New York Cornell University (NYCU), genus Salmonella (JAA), enzyme XbaI (X01), and pattern code (four-digit number). Four-digit pattern codes were assigned by comparison of patterns to an internal reference database (CUFSL, Ithaca, NY) comprised of approximately 6,000 Salmonella isolates. Lastly, PFGE was used to predict the serovar of a given isolate by comparison to the CUFSL database using a similarity cluster analysis (as described above) (35).
Molecular serotyping.
Molecular serotyping was performed on 12 isolates where serovars reported by traditional serotyping did not match serovars predicted by PFGE. It has been reported in one study (36) that approximately 10% of S. enterica subsp. enterica isolates (n = 754 tested) were incorrectly serotyped by traditional serotyping, and thus molecular serotyping may aid in identification (37). Serovar was confirmed by PCR detection of the Salmonella O serogroup genes by a multiplex PCR assay that simultaneously targets genes for five Salmonella O antigens (B, C1, C2–C3, D1, and E1), as described by Ranieri et al. (35) and Herrera-Léon et al. (38); in addition, PCR amplification and sequencing of the genes encoding the H1 and H2 antigens were performed. Primer sets, DNA amplification, and PCR conditions for fliC (encodes H1 antigen) and fljB (encodes H2 antigen) have been previously described by Imre et al. (39) and Mortimer et al. (40). DNA sequence data were compared to sequences in an internal database (CUFSL) of H1 and H2 sequences, as previously described (35). Molecular serotyping results were able to resolve serovar conflicts for the 12 isolates with different serovars identified by traditional serotyping and PFGE. Discrepancies between traditional serotyping and molecular-based methods can be due to a variety of reasons, including auto-agglutination or cross-reactivity of antisera, or can occur in mucoid strains (where the O antigen may be masked) (35, 41, 42). In all cases, the serovar predicted by PFGE matched the serovar identified by molecular serotyping results.
Final Salmonella isolate data set.
Upon completion of serotyping and PFGE typing, only isolates that were “unique” (i.e., defined as isolates from a given sample with different serotype-PFGE pattern combinations) were kept for further analyses; isolates from the same sample with identical serovars and PFGE types were excluded from further analysis (Table 1 gives isolate details, such as the number of isolates subtyped and the number of representative isolates yielded). This approach yielded 112 unique Salmonella isolates (80 from NYS and 32 from SFL).
Statistical analysis.
All analyses were performed in the statistical computing environment R, version 3.0.2 (43). Diversity of PFGE types between the NYS and SFL regions was assessed using two assessment tools: a diversity index and probability simulations.
Simpson's index of diversity (D) was calculated for PFGE types found among all unique isolates and among unique isolates for each region (44), with 95% confidence intervals (45). D values closest to 1 indicate a high diversity, while D values closest to 0 indicate a low diversity. To test if isolates from NYS and SFL were drawn from two distinct populations, we performed simulations to quantify the likelihood of three or fewer shared subtypes occurring between the regions, given that all the Salmonella isolates were drawn from one population. Briefly, we randomly permuted the isolates across the two regions and computed the number of shared PFGE types. The simulation was performed 1,000,000 times, and the probability of simulations (i.e., permutations) with three or fewer shared subtypes was calculated.
Data access.
Isolate information and subtyping data from this study are archived and available through the Cornell University Food Microbe Tracker database (http://www.foodmicrobetracker.com).
RESULTS AND DISCUSSION
This study is one of the first to characterize Salmonella distribution and diversity associated with the produce production environment in NYS. In addition, we assembled a smaller set of Salmonella isolates from SFL that were characterized to examine the regional distribution of subtypes between NYS and SFL. In the NYS data set, 74 of 1,677 previously collected environmental samples were Salmonella positive (Table 1). In the smaller SFL data set, 23 of 65 previously collected environmental samples were Salmonella positive (Table 1). The difference in Salmonella prevalence rates between the NYS and SFL data sets (approximately 4.5 and 35%, respectively) may be due to several factors, such as differences in environmental pressures, agricultural practices, wildlife, and/or weather. While neither our study nor previous studies (14, 24, 46, 47) were designed to identify the exact reasons for these differences in Salmonella prevalences on a regional scale, these findings suggest the need for future studies to investigate what factors may influence differences in pathogen prevalences between regions. Upon completion of serotyping and PFGE typing, the combined NYS and SFL data sets consisted of 112 unique Salmonella isolates (Table 2), specifically, 80 isolates from NYS and 32 isolates from SFL. Overall, multiple Salmonella serovars were isolated from a number of individual NYS and SFL samples, further supporting the importance of using multiple enrichment and plating schemes for detection and isolation of Salmonella. Additionally, across both NYS and SFL data sets, we observed a number of different serovars that previously have been associated with certain hosts, suggesting in some cases that Salmonella contamination in production environments could be linked to a probable source through use of subtyping. Lastly, we showed a distinct difference between Salmonella subtypes isolated from the sampling areas in two produce-growing regions (NYS and SFL), suggesting that regional characteristics, such as specific landscapes, climates, and/or wildlife populations influence the Salmonella subtype diversity found in different produce production environments.
TABLE 2.
Serovars and PFGE types found among Salmonella isolates from study samples collected in New York State and South Florida U.S. produce production environments
| Salmonella organisma | PFGE typeb | No. of isolates by region and data set: |
|||||
|---|---|---|---|---|---|---|---|
| NYS |
SFL III | ||||||
| I | II | IV | V | Total | |||
| S. enterica subsp. enterica serovars | |||||||
| Agona | NYCU.JAAX01.1131 | 3 | 3 | ||||
| Baildon | NYCU.JAAX01.0345 | 2 | |||||
| Braenderup | NYCU.JAAX01.1196 | 1 | |||||
| Cerro | NYCU.JAAX01.0213 | 10 | 5 | 1 | 16 | ||
| Enteritidis | NYCU.JAAX01.1225 | 4 | 4 | ||||
| Gaminara | NYCU.JAAX01.1202 | 1 | |||||
| Give | NYCU.JAAX01.1215 | 1 | 1 | ||||
| NYCU.JAAX01.1216 | 1 | 2 | 3 | ||||
| NYCU.JAAX01.1217 | 1 | 1 | |||||
| NYCU.JAAX01.1218 | 1 | 1 | |||||
| Infantis | NYCU.JAA.X01.1203 | 2 | 2 | ||||
| Litchfield | NYCU.JAAX01.1197 | 1 | |||||
| NYCU.JAAX01.1198 | 1 | ||||||
| Newport | NYCU.JAAX01.0121 | 6 | 6 | ||||
| NYCU.JAAX01.0126 | 1 | 1 | |||||
| NYCU.JAAX01.0296 | 2 | 2 | 2 | 1 | 7 | ||
| NYCU.JAAX01.1212 | 1 | 4 | 5 | ||||
| NYCU.JAAX01.1213 | 1 | ||||||
| NYCU.JAAX01.1221 | 3 | 3 | 2 | ||||
| NYCU.JAAX01.1222 | 3 | ||||||
| NYCU.JAAX01.1223 | 1 | ||||||
| Rubislaw | NYCU.JAAX01.1201 | 3 | |||||
| NYCU.JAAX01.1220 | 1 | 1 | |||||
| Saphra | NYCU.JAAX01.1194 | 7 | |||||
| Senftenberg | NYCU.JAAX01.1005 | 1 | 1 | ||||
| Tennessee | NYCU.JAAX01.1214 | 1 | 1 | ||||
| Thompson | NYCU.JAAX01.0157 | 4 | 5 | 1 | 2 | 12 | 1 |
| NYCU.JAAX01.1199 | 1 | 1 | 1 | ||||
| NYCU.JAAX01.1200 | 1 | ||||||
| Typhimurium | NYCU.JAAX01.0072 | 1 | |||||
| NYCU.JAAX01.1207 | 1 | 1 | |||||
| NYCU.JAAX01.1208 | 1 | 1 | |||||
| 4,5,12:i:− | NYCU.JAAX01.1209 | 2 | |||||
| 6,8:i:− | NYCU.JAAX01.0096 | 1 | 1 | ||||
| S. enterica subsp. diarizonae serovar | |||||||
| 50:r:z | NYCU.JAAX01.1210 | 3 | 3 | ||||
| NYCU.JAAX01.1211 | 3 | ||||||
| S. enterica subsp. houtenae serovar | |||||||
| 40:z4,z32:− | NYCU.JAAX01.1219 | 2 | 2 | 1 | 5 | ||
Serotyping was performed by agglutination at the Wadsworth Center, New York State Department of Health.
PFGE was performed in accordance with the standard CDC PulseNet protocol for Salmonella using the restriction enzyme XbaI (32). PFGE types were named with Cornell University Food Safety Laboratory (CUFSL) nomenclature and were assigned by comparison of PFGE patterns to an internal reference database (CUFSL, Ithaca, NY) comprised of approximately 6,000 Salmonella isolates. PFGE types in boldface were found in both NYS and FL.
Multiple serovars were isolated from Salmonella-positive samples.
Serotyping and PFGE typing were performed on one isolate per isolation scheme from the 97 Salmonella-positive samples (74 and 23 Salmonella-positive samples from NYS and SFL, respectively); while our data set represented 98 Salmonella-positive samples (Table 1), isolates from one Salmonella-positive sample from NYS data set I were unavailable for subtyping (preservation failed). Among the 97 Salmonella-positive samples evaluated, 83 samples yielded one serovar (92 and 65% of Salmonella-positive samples from NYS and SFL, respectively, yielded only one serovar), and 14 samples yielded two or more serovars. The 14 Salmonella-positive samples that yielded two or more serovars represented 6 samples from NYS and 8 samples from SFL (Table 3); for each of the 6 samples from NYS, two different serovars were isolated. Among the SFL samples, two serovars were isolated from seven samples, and three serovars (Salmonella serovar Newport, Salmonella serovar Saphra, and S. enterica subsp. diarizonae serovar 50:r:z) were isolated from one sample (Table 3). While our study found multiple serovars in some samples by subtyping only one isolate per isolation scheme, it may be warranted in future studies to test multiple colonies per isolation scheme to further investigate potential polyclonal contamination.
TABLE 3.
Two or more serovars that were isolated from Salmonella-positive samples by the four different detection and isolation schemes
| Sample region and no.a | Sample type |
S. enterica serovar or subsp. by isolation schemeb |
|||
|---|---|---|---|---|---|
| RV |
TT |
||||
| CHROMagar | XLD | CHROMagar | XLD | ||
| NYS | |||||
| 1 | Soil | Newport | Thompson | ||
| 2 | Soil | Senftenberg | Newport | ||
| 3 | Drag swab | Tennessee | Agona | ||
| 4 | Water | Typhimurium | Give | ||
| 5 | Water | Enteritidis | diarizonae | ||
| 6 | Water | Newport | diarizonae | ||
| SFL | |||||
| 7 | Soil | Braenderup | Litchfield | ||
| 8 | Soil | Newport | Saphra | ||
| 9 | Drag swab | Thompson | Litchfield | ||
| 10 | Drag swab | Baildon | Thompson | ||
| 11 | Water | Newport | Saphra | ||
| 12 | Water | diarizonae | Thompson | ||
| 13 | Water | Rubislaw | Gaminara | ||
| 14 | Water | Saphra | diarizonae | Newport | |
NYS, New York State; SFL, South Florida.
Enrichment media are Rappaport-Vassiliadis (RV) and tetrathionate (TT) broths; plating media are Salmonella chromogenic agar (CHROMagar) and xylose-lysine-deoxycholate agar (XLD).
Several studies (14, 17, 23, 47) have isolated multiple Salmonella serovars from one sample. For instance, Jokinen et al. (23) isolated more than one serovar from all Salmonella-positive water samples (n = 29) collected from a Canadian watershed. In our study, 50% of the Salmonella-positive samples with multiple serovars represented water samples (3/6 and 4/8 positive samples from NYS and SFL, respectively). Our water sample volume tested was 250 ml, while in the Jokinen et al. study the water sample volume tested was 500 ml. The number of serovars isolated from each sample may be influenced by the sample volume tested; however, further studies are required to correlate the volume of water tested and the likely number of serovars. Isolation of multiple serovars from water samples may also be more likely because Salmonella is more uniformly dispersed in water than in other sample types in our study (e.g., soil samples; 25 g).
Multiple Salmonella enrichment and plating schemes were used here to facilitate detection of Salmonella with atypical phenotypic characteristics as previous studies (30, 31, 48–51) have shown that detection of certain serovars of Salmonella may be influenced by enrichment and plating media. In our study, the 14 Salmonella-positive samples where multiple serovars of Salmonella were isolated yielded 14 different serovars (Table 3). Overall, no apparent association was observed between isolation of a Salmonella serovar and a specific enrichment and plating scheme (Table 3). For example, S. enterica subsp. diarizonae serovar 50:r:z was isolated from three of the four enrichment and plating schemes (RV-CHROMagar, RV-XLD, and TT-XLD). Similarly, Salmonella serovar Saphra was also isolated from three of the four schemes (TT-CHROMagar, RV-CHROMagar, and TT-XLD). Other serovars that were isolated more than once were also isolated from two or more schemes. Select strains of four of the serovars isolated here (Salmonella serovars Thompson, Enteritidis, and Typhimurium and S. enterica subsp. diarizonae) have been reported to be weak H2S producers. No evident association was observed between serovars that have been reported as weak H2S producers and a specific scheme (Table 3). In this study, we employed one plating medium that does (XLD) and one that does not (CHROMagar) use the production of H2S as an indicator of Salmonella (49, 52). Select strains of six of the serovars isolated here (Salmonella serovars Tennessee, Newport, Agona, Senftenberg, and Typhimurium and S. enterica subsp. diarizonae) have been reported to ferment lactose (53–55). No apparent association was observed here between the serovars that have been reported to ferment lactose and a specific enrichment and plating scheme (Table 3). Even though our sample size was small (14 cases of isolation of multiple Salmonella serovars from one sample), our results further support the idea that the use of different enrichment and plating schemes is needed to detect and isolate different Salmonella subtypes that may be present in a sample.
Salmonella serovars isolated from produce-growing regions were diverse.
We identified 20 different serovars among the 112 subtyped unique isolates from NYS and SFL (Table 2). Specifically, 14 and 11 serovars were identified among the 80 NYS and 32 SFL isolates, respectively; 5 serovars were found among isolates from both regions. S. enterica subspecies identified across both NYS and SFL data sets were enterica (18 serovars), diarizonae (1 serovar), and houtenae (1 serovar) (Table 2). While S. enterica subsp. enterica serovars are often associated with warm-blooded animal hosts and are the most commonly associated with human Salmonella infections linked to foods (56–58), a few documented cases have linked cold-blooded animal hosts (e.g., reptiles and amphibians) to S. enterica subsp. enterica infections (9, 59, 60). Serovars of S. enterica subsp. diarizonae and S. enterica subsp. houtenae are primarily associated with cold-blooded animal hosts (1, 13, 14) and are most commonly associated with human Salmonella infections linked to reptiles (15, 61). In the NYS data set, isolates identified as S. enterica subsp. enterica represented Salmonella serovars Newport (22 isolates), Cerro (16 isolates), Thompson (13 isolates), Give (6 isolates), Enteritidis (4 isolates), Agona (3 isolates), Typhimurium (2 isolates), and Infantis (2 isolates) as well as 6,8:i:−, Rubislaw, Senftenberg, and Tennessee (1 isolate each); in addition S. enterica subsp. diarizonae serovar 50:r:z (3 isolates) and S. enterica subsp. houtenae serovar 40:z4,z32:− (5 isolates) were identified (Table 2). In the SFL data set, isolates identified as S. enterica subsp. enterica represented Salmonella serovars Newport (7 isolates), Saphra (7 isolates), Rubislaw (3 isolates), Thompson (3 isolates), 4,5,12:i:− (2 isolates), Litchfield (2 isolates), and Baildon (2 isolates), as well as Salmonella serovars Braenderup, Gaminara, and Typhimurium (1 isolate each); in addition S. enterica subsp. diarizonae serovar 50:r:z (3 isolates) was identified (Table 2). Salmonella serovars Newport, Cerro and Thompson were isolated most frequently in NYS and represented approximately 28, 20, and 16% of the NYS isolates (Table 2). Salmonella serovars Newport and Saphra were isolated most frequently in SFL, and each represented 22% of the SFL isolates (Table 2). All other serovars represented <10% of the NYS and SFL isolates.
Among the Salmonella-positive samples obtained from NYS and SFL, Salmonella serovar Newport was isolated the most (Fig. 1, green line) (22/80 isolates NYS and 7/32 isolates SFL). Salmonella serovar Newport has been isolated from a broad range of hosts (e.g., dairy cattle, snakes, and hedgehogs) and from a number of diverse environmental sources (e.g., soil and water) (20, 62). Additionally, Salmonella serovar Newport is one of the most frequently reported serovars among human Salmonella isolates in North America, Europe, and Latin America (16, 63) and is associated with a wide range of foods and animals (e.g., beef and poultry) (58, 64). Produce-borne outbreaks of Salmonella serovar Newport have been routinely traced back to the produce production environment (65, 66) and have been associated with numerous commodities, including lettuce, mangoes, melons, alfalfa sprouts, and tomatoes (4, 67). For example, in 2005, tomatoes from Virginia were implicated as the food vehicle for an outbreak of Salmonella serovar Newport, with an estimated 72 illness cases across 16 states. The source of this outbreak was traced back to a pond that served as an irrigation source for the tomato fields (66). The identification of Salmonella serovar Newport in both regions highlights the widespread nature of this serovar; furthermore, the number of Salmonella serovar Newport outbreaks associated with different produce commodities emphasizes the importance of this serovar in produce production environments.
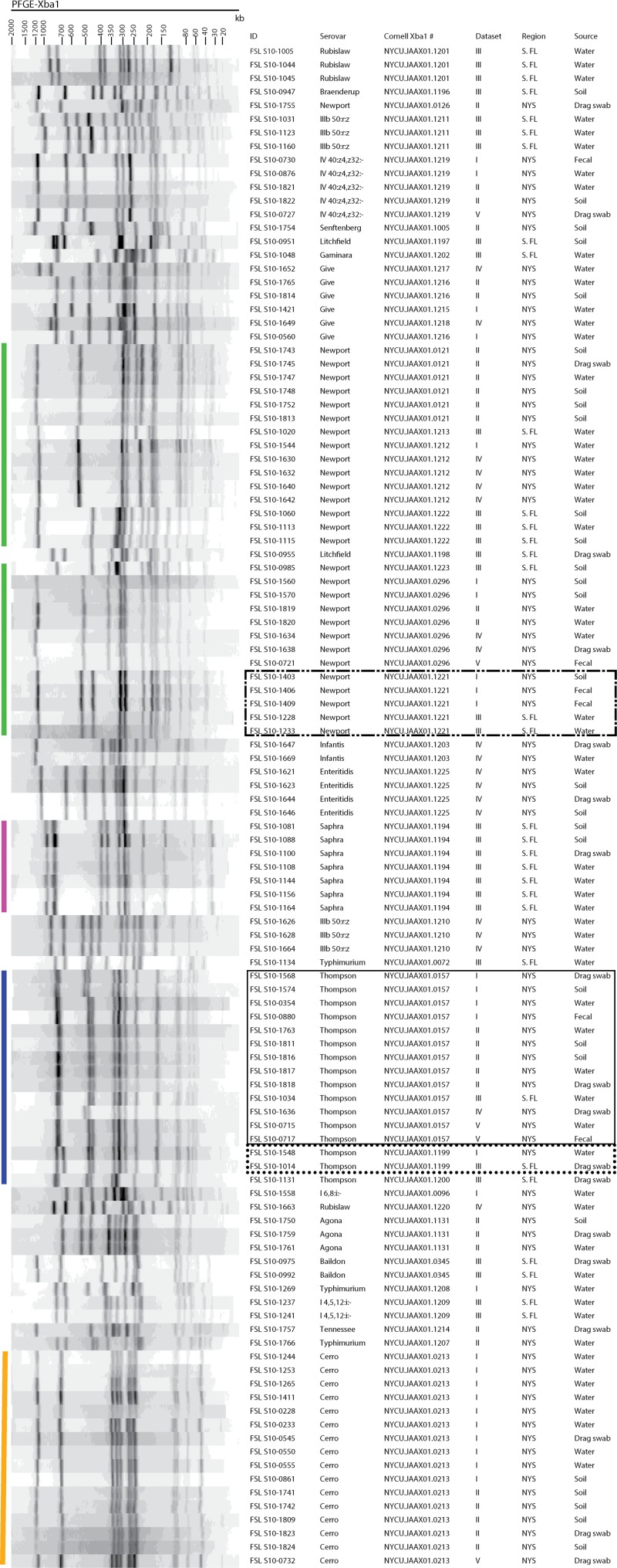
FIG 1.
XbaI PFGE patterns for the representative 112 Salmonella isolates from environmental samples obtained from New York State and South Florida produce production environments. Band sizes (kb) are displayed at the top of the PFGE pattern images. The PFGE pattern order displayed is the result of BioNumerics similarity analyses using the unweighted pair group method with arithmetic mean algorithm (UPGMA) and the Dice correlation coefficient with a maximum space tolerance of 1.5%. ID is the isolate designation, serovar is the serovar confirmed by the traditional or molecular method, Cornell XbaI # is the PFGE type assigned by comparison of PFGE patterns to the Cornell University Food Safety Laboratory (CUFSL; Ithaca, NY) database of 6,000 Salmonella isolates, data set indicates the study origin for the isolate, region is New York State (NYS) or South Florida (SFL), and source is the type of sample for the isolate. Salmonella serovars discussed in the text are labeled by color: green, Newport; purple, Saphra, blue, Thompson; orange, Cerro. Boxes represent isolates from NYS and SFL that share PFGE types: dashed box, NYCU.JAAX01.1221 (Salmonella serovar Newport); solid box, NYCU.JAAX01.0157 (Salmonella serovar Thompson); dotted box, NYCU.JAAX01.1199 (Salmonella serovar Thompson).
Salmonella serovar Cerro was isolated exclusively from NYS (Fig. 1, orange line) (16/80 isolates). Previous studies (68–70) have indicated that Salmonella serovar Cerro is prevalent in Pennsylvania (USA) and NYS, especially in dairy production environments. This suggests that Salmonella serovar Cerro may be a likely contaminant in produce production environments near dairy cattle operations. Yet Salmonella serovar Cerro has rarely been associated with human Salmonella infections and has been isolated from asymptomatic, healthy children in India (71). Of the few documented Salmonella serovar Cerro outbreaks, the most notable occurred in New Mexico (USA) (29 illnesses and 7 hospitalizations) and was linked to consumption of contaminated carne seca (71).
Salmonella serovar Thompson was isolated from both NYS and SFL (Fig. 1, blue line) (13/80 isolates from NYS and 3/32 isolates from SFL). One field study (72) conducted in NYS found 10% (13/129) of surface water samples tested positive for Salmonella serovar Thompson. Of the 13 isolates identified as Salmonella serovar Thompson from NYS in our study here, 5 were isolated from surface water samples. This serovar has been associated with two produce-borne outbreaks, a 1999 outbreak with 41 reported cases linked to the consumption of contaminated cilantro in California (USA) (73) and a 2004 outbreak with 21 cases linked to rucola lettuce in Norway (74). Salmonella serovar Thompson is common among human salmonellosis cases, as evident by its rank of 13 in the CDC's top 20 Salmonella isolates from human sources (56).
Salmonella serovar Saphra was isolated exclusively from SFL and represented 22% of the SFL Salmonella isolates (Fig. 1, purple line) (7/32 isolates). This serovar is not commonly isolated from humans or animals, and minimal information has been reported on its ecology in the environment. Two studies, conducted in Argentina (75) and Brazil (76), isolated Salmonella serovar Saphra from surface water and animal drinking water samples. There has been only one documented outbreak of Salmonella serovar Saphra in the United States, with 24 illnesses and 5 hospitalizations, due to consumption of contaminated cantaloupe (77), purchased by a single distributor who obtained the cantaloupe from a specific region in Mexico.
We identified a number of Salmonella serovars that have previously been linked to produce-borne outbreaks; however, we also identified serovars that are rarely linked to human cases of salmonellosis (e.g., Salmonella serovar Cerro). This finding suggests a need for future studies to investigate the differences in virulence between different Salmonella serovars (and in some cases virulence between different strains of the same serovar). In addition, we observed several serovars that are associated with specific hosts, such as S. enterica subsp. diarizonae serovar 50:r:z, which is commonly associated with reptile and amphibian hosts. Additional studies are needed to further our understanding of various Salmonella serovars and their association with different hosts in produce production environments, especially geographical locations in the United States and abroad.
Salmonella PFGE types show significant differences between regions.
To explore the regional Salmonella diversity associated with the produce production environment, we also performed PFGE on the Salmonella isolates from NYS (n = 80) and SFL (n = 32). We identified 37 unique XbaI-PFGE types among these 112 Salmonella isolates (Table 2). Isolates from NYS and SFL represented 23 and 17 different PFGE types, respectively; only 3 PFGE types were found among isolates from both regions (Table 2 and Fig. 1). A high level of PFGE type diversity was observed among all isolates (D = 0.90 ± 0.02) as well as within each region (D = 0.92 ± 0.03 and D = 0.93 ± 0.05 for NYS and SFL, respectively). The largest number of PFGE types was observed among Salmonella serovar Newport isolates (eight unique PFGE types among 29 Salmonella serovar Newport isolates). Of the eight Salmonella serovar Newport PFGE types, four and three types each were observed exclusively in the NYS and SFL regions, respectively, and one PFGE type was shared between the two regions (Table 2 and Fig. 1, dashed box). Previously, a specific Salmonella serovar Newport subtype had been repeatedly isolated over a 10-year span from the eastern shore of Virginia (USA) (66). This Salmonella serovar Newport subtype has been associated with at least two known outbreaks, linked to tomatoes harvested in Virginia (66), in addition to being isolated from several waterfowl and non-waterfowl fecal samples collected from the eastern shore of Virginia (78). These findings suggest that specific Salmonella serovar Newport subtypes may be associated with certain regions. Additionally, in our study, different PFGE types of S. enterica subsp. diarizonae serovar 50:r:z were identified from the NYS and SFL regions sampled (Fig. 1). These data may indicate carriage of different Salmonella strains by reptile or amphibian populations in these regions; further studies could explore whether different reptile or amphibians in these regions carry distinct Salmonella subtypes. One study conducted in Mississippi (USA) observed a strong association between patients infected with Salmonella serovar Javiana and contact with amphibian species endemic to the southeastern United States (79). Gorski et al. (14) also found several Salmonella strains, all with the same PFGE type, repeatedly isolated from cold-blooded vertebrates and surface water in the same region in California. Overall, a number of studies, including our data reported here, suggest that some Salmonella subtypes may be more prevalent in certain areas, possibly due to persistence in animal and human host populations or in abiotic environments or due to their adaptation to specific hosts or environments found in a given region.
Only three PFGE types were shared between the two regions, Salmonella serovar Thompson patterns NYCU.JAAX01.0157 (Fig. 1, solid box) and NYCU.JAAX01.1199 (Fig. 1, dotted box) and Salmonella serovar Newport pattern NYCU.JAAX01.1221 (Fig. 1, dashed box). A-1,000,000 iteration simulation calculated the probability of three or fewer shared PFGE types being observed by chance between the two regions as <0.000001, indicating a distinct difference in Salmonella PFGE types between isolates recovered in the NYS and SFL regions sampled. Salmonella isolates from NYS were obtained over a 3-year period (2010 to 2012) and represented a number of produce production environments across the state and fields planted with a number of crop types (e.g., cabbage, peppers, summer squash, and melons) (see File S1 the supplemental material). Salmonella isolates from SFL were obtained at one time point (December 2010) and represented a small subset of produce production environments and a single crop type (leafy greens) (see File S1). As our study did not specifically examine the association between specific produce production environments, including crop types, farm-specific management practices, or sampling time and Salmonella subtype diversity, future studies are needed to determine how these and other factors may influence regional Salmonella ecology and serotype diversity. Therefore, one cannot easily extrapolate from our findings that show subtype differences between the two sampled regions, NYS and SFL, with distinct overall environmental conditions to other sampling areas or regions. Other studies have also reported meteorological events (23, 26, 46) and/or field management practices (27, 80, 81) to influence pathogen prevalence in produce production environments; whereas the effects of these factors on subtype diversity still remain to be explored, it is feasible that subtypes isolated may differ by season (e.g., as season may have effects on movement or presence of wildlife hosts). While many factors can clearly affect Salmonella subtype diversity, it is still important to note that a number of previous studies (23–25, 46, 47) in regions within the United States and Canada have reported Salmonella serovar diversity distinct from that reported here, supporting the idea that Salmonella subtype diversity may differ by region and sampled area. For example, one study (25) found that Salmonella isolated from water samples in the Upper Suwannee River Basin in North Florida (30 isolates, representing 8 serovars) represented several different serovars from those reported here for isolates from SFL. Other studies found that S. enterica subsp. arizonae represented 40% of the Salmonella isolates from a watershed in Georgia (USA) (47) and that Salmonella serovar Rubislaw was isolated most frequently from water samples obtained from a Canadian watershed (23). Furthermore, a study investigating the distribution and diversity of Salmonella in a produce-growing region in California primarily isolated Salmonella serovar Give (46). Our findings, in concert with previously discussed studies, demonstrate the potential for PFGE-based source tracking of Salmonella as Salmonella subtypes can be highly associated with certain hosts (e.g., Salmonella serovar Cerro and cattle) and environments (e.g., Salmonella serovar Newport and the eastern shore of Virginia). These data also suggest that food safety in the produce production environment may require efforts to tailor preventive controls that account for unique food safety hazards and contamination routes associated with different areas and regions.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by (i) Agriculture and Food Research Initiative Competitive grant 2012-67011-19875 (awarded to L.K.S. and M.W.) from the U.S. Department of Agriculture (USDA) National Institute of Food and Agriculture and (ii) National Integrated Food Safety Initiative grant 2008-51110-04333 (awarded to R.W.W.) from the USDA.
We thank Lorraine D. Rodriguez-Rivera for her time in discussion of Salmonella and PFGE. Additionally, we are grateful for the technical assistance of Emily Wright and Anna Sophia Harrand and the editing assistance of Andrea Moreno Switt.
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00348-14.
REFERENCES
- 1.Grimont PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frenzen PD, Riggs TL, Buzby JC, Breuer T, Roberts T, Voetsch D, Reddy S. 1999. Salmonella cost estimate updated using FoodNet data. Food Rev. 22:10–15 [Google Scholar]
- 4.Hanning IB, Nutt JD, Ricke SC. 2009. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog. Dis. 6:635–648. 10.1089/fpd.2008.0232 [DOI] [PubMed] [Google Scholar]
- 5.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342–2353 [DOI] [PubMed] [Google Scholar]
- 6.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 19:407–415. 10.3201/eid1903.111866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould LH, Nisler AL, Herman KM, Cole DJ, Williams IT, Mahon BE, Griffin PM, Hall AJ. 2011. Surveillance for foodborne disease outbreaks—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 60:1197–1202 [PubMed] [Google Scholar]
- 8.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389. 10.3201/eid0703.017303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, Casadesus J, Platt DJ, Olsen JE. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229–255. 10.1017/S0950268899004379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pires SM, Hald T. 2010. Assessing the differences in public health impact of Salmonella subtypes using a Bayesian microbial subtyping approach for source attribution. Foodborne Pathog. Dis. 7:143–151. 10.1089/fpd.2009.0369 [DOI] [PubMed] [Google Scholar]
- 11.Pires SM, Evers EG, van Pelt W, Ayers T, Scallan E, Angulo FJ, Havelaar A, Hald TMed-Vet-Net Workpackage 28 Working Group 2009. Attributing the human disease burden of foodborne infections to specific sources. Foodborne Pathog. Dis. 6:417–424. 10.1089/fpd.2008.0208 [DOI] [PubMed] [Google Scholar]
- 12.Smith HW, Jones JET. 1967. Observations on experimental oral infection with Salmonella dublin in calves and Salmonella choleraesuis in pigs. J. Pathol. Bacteriol. 93:141–156. 10.1002/path.1700930114 [DOI] [PubMed] [Google Scholar]
- 13.de Sa IVA, Solari CA. 2001. Salmonella in Brazilian and imported pet reptiles. Braz. J. Microbiol. 32:293–297. 10.1590/S1517-83822001000400007 [DOI] [Google Scholar]
- 14.Gorski L, Jay-Russell MT, Liang AS, Walker S, Bengson Y, Govoni J, Mandrell RE. 2013. Diversity of pulsed-field gel electrophoresis pulsotypes, serovars, and antibiotic resistance among Salmonella isolates from wild amphibians and reptiles in the California central coast. Foodborne Pathog. Dis. 10:540–548. 10.1089/fpd.2012.1372 [DOI] [PubMed] [Google Scholar]
- 15.Schroter M, Roggentin P, Hofmann J, Speicher A, Laufs R, Mack D. 2004. Pet snakes as a reservoir for Salmonella enterica subsp. diarizonae (serogroup IIIb): a prospective study. Appl. Environ. Microbiol. 70:613–615. 10.1128/AEM.70.1.613-615.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galanis E, Wong D, Patrick ME, Binsztein N, Cieslik A, Chalermchaikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization Global Salm-Surv. 2006. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 12:381–388. 10.3201/eid1203.050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson B, Hughes LA, Peters T, de Pinna E, John SK, Macgregor SK, Cunningham AA. 2011. Pulsed-field gel electrophoresis supports the presence of host-adapted Salmonella enterica subsp. enterica serovar Typhimurium strains in the British garden bird population. Appl. Environ. Microbiol. 77:8139–8144. 10.1128/AEM.00131-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooley M, Carychao D, Crawford-Miksza L, Jay MT, Myers C, Rose C, Keys C, Farrar J, Mandrell RE. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One 2:e1159. 10.1371/journal.pone.0001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uesugi AR, Danyluk MD, Mandrell RE, Harris LJ. 2007. Isolation of Salmonella Enteritidis phage type 30 from a single almond orchard over a 5-year period. J. Food Prot. 70:1784–1789 [DOI] [PubMed] [Google Scholar]
- 20.Patchanee P, Molla B, White N, Line DE, Gebreyes WA. 2010. Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathog. Dis. 7:1113–1120. 10.1089/fpd.2010.0572 [DOI] [PubMed] [Google Scholar]
- 21.Ram JL, Ritchie RP, Fang JW, Gonzales FS, Selegean JP. 2004. Sequence-based source tracking of Escherichia coli based on genetic diversity of beta-glucuronidase. J. Environ. Qual. 33:1024–1032. 10.2134/jeq2004.1024 [DOI] [PubMed] [Google Scholar]
- 22.Furukawa T, Yoshida T, Suzuki Y. 2011. Application of PFGE to source tracking of faecal pollution in coastal recreation area: a case study in Aoshima Beach, Japan. J. Appl. Microbiol. 110:688–696. 10.1111/j.1365-2672.2010.04918.x [DOI] [PubMed] [Google Scholar]
- 23.Jokinen C, Edge TA, Ho S, Koning W, Laing C, Mauro W, Medeiros D, Miller J, Robertson W, Taboada E, Thomas JE, Topp E, Ziebell K, Gannon VP. 2011. Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res. 45:1247–1257. 10.1016/j.watres.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 24.Walters SP, Gonzalez-Escalona N, Son I, Melka DC, Sassoubre LM, Boehm AB. 2013. Salmonella enterica diversity in central Californian coastal waterways. Appl. Environ. Microbiol. 79:4199–4209. 10.1128/AEM.00930-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajabi M, Jones M, Hubbard M, Rodrick G, Wright AC. 2011. Distribution and genetic diversity of Salmonella enterica in the Upper Suwannee River. Int. J. Microbiol. 2011:461321. 10.1155/2011/461321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Grohn YT, Worobo RW, Wiedmann M, Bergholz PW. 2013. Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl. Environ. Microbiol. 79:588–600. 10.1128/AEM.02491-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strawn LK, Grohn YT, Warchocki S, Worobo RW, Bihn EA, Wiedmann M. 2013. Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79:7618–7627. 10.1128/AEM.02831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews WH, Hammack T. 2001. Bacteriological analytical manual: detection and enumeration of Salmonella, p 105–128 U.S. Food and Drug Administration, Silver Spring, MD [Google Scholar]
- 29.Kim JS, Lee GG, Park JS, Jung YH, Kwak HS, Kim SB, Nam YS, Kwon ST. 2007. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J. Food Prot. 70:1656–1662 [DOI] [PubMed] [Google Scholar]
- 30.Michael GB, Simoneti R, da Costa M, Cardoso M. 2003. Comparison of different selective enrichment steps to isolate Salmonella sp. from feces of finishing swine. Braz. J. Microbiol. 34:138–142. 10.1590/S1517-83822003000200009 [DOI] [Google Scholar]
- 31.Schonenbrucher V, Mallinson ET, Bulte M. 2008. A comparison of standard cultural methods for the detection of foodborne Salmonella species including three new chromogenic plating media. Int. J. Food Microbiol. 123:61–66. 10.1016/j.ijfoodmicro.2007.11.064 [DOI] [PubMed] [Google Scholar]
- 32.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 33.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Rivera LD, Wright EM, Siler JD, Elton M, Cummings KJ, Warnick LD, Wiedmann M. 2014. Subtype analysis of Salmonella isolated from subclinically infected dairy cattle and dairy farm environments reveals the presence of both human- and bovine-associated subtypes. Vet. Microbiol. 170:307–316. 10.1016/j.vetmic.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranieri ML, Shi CL, Switt AIM, den Bakker HC, Wiedmanna M. 2013. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J. Clin. Microbiol. 51:1786–1797. 10.1128/JCM.03201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wattiau P, Van Hessche M, Schlicker C, Vander Veken H, Imberechts H. 2008. Comparison of classical serotyping and PremiTest assay for routine identification of common Salmonella enterica serovars. J. Clin. Microbiol. 46:4037–4040. 10.1128/JCM.01405-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi C, Singh P, Ranieri ML, Wiedmann M, Moreno Switt AI. 14 November 2013. Molecular methods for serovar determination of Salmonella. Crit. Rev. Microbiol. 10.3109/1040841X.2013.837862 [DOI] [PubMed] [Google Scholar]
- 38.Herrera-Leon S, Ramiro R, Arroyo M, Diez R, Usera MA, Echeita MA. 2007. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res. Microbiol. 158:122–127. 10.1016/j.resmic.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 39.Imre A, Olasz F, Nagy B. 2005. Development of a PCR system for the characterisation of Salmonella flagellin genes. Acta Vet. Hung. 53:163–172. 10.1556/AVet.53.2005.2.2 [DOI] [PubMed] [Google Scholar]
- 40.Mortimer CKB, Peters TM, Gharbia SE, Logan JMJ, Arnold C. 2004. Towards the development of a DNA-sequence based approach to serotyping of Salmonella enterica. BMC Microbiol. 4:31. 10.1186/1471-2180-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin K, Lingohr EJ, Yoshida C, Anjum M, Bodrossy L, Clark CG, Kropinski AM, Karmali MA. 2011. Rapid genoserotyping tool for classification of Salmonella serovars. J. Clin. Microbiol. 49:2954–2965. 10.1128/JCM.02347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida C, Franklin K, Konczy P, McQuiston JR, Fields PI, Nash JH, Taboada EN, Rahn K. 2007. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J. Microbiol. Methods 70:261–271. 10.1016/j.mimet.2007.04.018 [DOI] [PubMed] [Google Scholar]
- 43.R Development Core Team. 2004. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 44.Simpson EH. 1949. Measurement of diversity. Nature 163:688. 10.1038/163688a0 [DOI] [Google Scholar]
- 45.Grundmann H, Hori S, Tanner G. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190–4192. 10.1128/JCM.39.11.4190-4192.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorski L, Parker CT, Liang A, Cooley MB, Jay-Russell MT, Gordus AG, Atwill ER, Mandrell RE. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734–2748. 10.1128/AEM.02321-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haley BJ, Cole DJ, Lipp EK. 2009. Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl. Environ. Microbiol. 75:1248–1255. 10.1128/AEM.01648-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrente M, Madio A, Friedrich KG, Greco G, Desario C, Tagliabue S, D'Incau M, Campolo M, Buonavoglia C. 2004. Isolation of Salmonella strains from reptile feces and comparison of different culture media. J. Appl. Microbiol. 96:709–715. 10.1111/j.1365-2672.2004.02186.x [DOI] [PubMed] [Google Scholar]
- 49.Rall VLM, Rall R, Aragon LC, da Silva MG. 2005. Evaluation of three enrichment broths and five plating media for Salmonella detection in poultry. Braz. J. Microbiol. 36:147–150. 10.1590/S1517-83822005000200009 [DOI] [Google Scholar]
- 50.Vassiliadis P. 1983. The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of Salmonellas—an overview. J. Appl. Bacteriol. 54:69–74. 10.1111/j.1365-2672.1983.tb01302.x [DOI] [PubMed] [Google Scholar]
- 51.Papavassiliou J, Samaraki-Lyberopoulou V, Piperakis G. 1969. Production of tetrathionate reductase by Salmonella. Can. J. Microbiol. 15:238–240. 10.1139/m69-041 [DOI] [PubMed] [Google Scholar]
- 52.Mallinson ET, Miller RG, de Rezende CE, Ferris KE, DeGraft-Hanson J, Joseph SW. 2000. Improved plating media for the detection of Salmonella species with typical and atypical hydrogen sulfide production. J. Vet. Diagn. Invest. 12:83–87. 10.1177/104063870001200119 [DOI] [PubMed] [Google Scholar]
- 53.McDonough PL, Shin SJ, Lein DH. 2000. Diagnostic and public health dilemma of lactose-fermenting Salmonella enterica serotype Typhimurium in cattle in the northeastern United States. J. Clin. Microbiol. 38:1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puhr N, O'Hara C, Miller M, Bopp C, Wells J. 1994. Evaluation of methods for isolation and identification of lactose-fermenting Salmonella, abstr 557. Abstr. 94th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 55.Tyc Z, Szych J, Kaluzewski S. 1989. Selection of methods of the detection of lactose-fermenting Salmonella strains and evaluation of their usefulness in diagnostic studies. Med. Dosw. Mikrobiol. 41:106–114 (In Polish) [PubMed] [Google Scholar]
- 56.Centers for Disease Control and Prevention. 2008. Salmonella surveillance: annual summary, 2006. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 57.Callaway TR, Edrington TS, Anderson RC, Byrd JA, Nisbet DJ. 2008. Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J. Anim. Sci. 86:E163–E172. 10.2527/jas.2007-0457 [DOI] [PubMed] [Google Scholar]
- 58.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 19:1239–1244. 10.3201/eid1908.121511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Centers for Disease Control and Prevention. 2003. Reptile-associated salmonellosis—selected states, 1998–2002. MMWR Morb. Mortal. Wkly. Rep. 52:1206–1209 [PubMed] [Google Scholar]
- 60.Centers for Disease Control and Prevention. 2010. Multistate outbreak of human Salmonella Typhimurium infections associated with aquatic frogs—United States, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:1433–1436 [PubMed] [Google Scholar]
- 61.Ackman DM, Drabkin P, Birkhead G, Cieslak P. 1995. Reptile-associated salmonellosis in New York State. Pediatr. Infect. Dis. 14:955–959. 10.1097/00006454-199511000-00006 [DOI] [PubMed] [Google Scholar]
- 62.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet. Res. 42:34. 10.1186/1297-9716-42-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendriksen RS, Vieira AR, Karlsmose S, Wong D, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 8:887–900. 10.1089/fpd.2010.0787 [DOI] [PubMed] [Google Scholar]
- 64.Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, Didelot X, Paglietti B, Rabsch W, Brisse S, Weill FX, Roumagnac P, Achtman M. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476. 10.1128/JB.00969-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Food and Drug Administration. 2013. Environmental assessment: factors potentially contributing to the contamination of fresh whole cantaloupe implicated in a multi-state outbreak of salmonellosis FDA. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/Food/RecallsOutbreaksEmergencies/Outbreaks/ucm341476.htm [Google Scholar]
- 66.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 136:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris LJ, Farber JN, Beuchat LR, Parish ME, Suslow TV, Garrett EH, Busta FF. 2003. Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh cut produce. Compr. Rev. Food Sci. Food Saf. 2:78–141. 10.1111/j.1541-4337.2003.tb00031.x [DOI] [Google Scholar]
- 68.Van Kessel JA, Karns JS, Wolfgang DR, Hovingh E. 2013. Regional distribution of two dairy-associated Salmonella enterica serotypes. Foodborne Pathog. Dis. 10:448–452. 10.1089/fpd.2012.1380 [DOI] [PubMed] [Google Scholar]
- 69.Cummings KJ, Warnick LD, Elton M, Rodriguez-Rivera LD, Siler JD, Wright EM, Groehn YT, Wiedmann M. 2010. Salmonella enterica serotype Cerro among dairy cattle in New York: an emerging pathogen? Foodborne Pathog. Dis. 7:659–665. 10.1089/fpd.2009.0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoelzer K, Cummings KJ, Wright EM, Rodriguez-Rivera LD, Roof SE, Switt AIM, Dumas N, Root T, Schoonmaker-Bopp DJ, Grohn YT, Slier JD, Warnick LD, Hancock DD, Davis MA, Wiedmann M. 2011. Salmonella Cerro isolated over the past twenty years from various sources in the US represent a single predominant pulsed-field gel electrophoresis type. Vet. Microbiol. 150:389–393. 10.1016/j.vetmic.2011.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mammina C, Cannova L, Carfi Pavia S, Nastasi A. 2000. Endemic presence of Salmonella enterica serotype Cerro in southern Italy. Euro Surveill. 5:84–86 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=28 [DOI] [PubMed] [Google Scholar]
- 72.Dondero NC, Thomas CT, Khare M, Timoney JF, Fukui GM. 1977. Salmonella in surface waters of central New York State. Appl. Environ. Microbiol. 33:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell JV, Mohle-Boetani J, Reporter R, Abbott S, Farrar J, Brandl M, Mandrell R, Werner SB. 2001. An outbreak of Salmonella serotype Thompson associated with fresh cilantro. J. Infect. Dis. 183:984–987. 10.1086/319254 [DOI] [PubMed] [Google Scholar]
- 74.Nygard K, Lassen J, Vold L, Andersson Y, Fisher I, Lofdahl S, Threlfall J, Luzzi I, Peters T, Hampton M, Torpdahl M, Kapperud G, Aavitsland P. 2008. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathog. Dis. 5:165–173. 10.1089/fpd.2007.0053 [DOI] [PubMed] [Google Scholar]
- 75.Benassi FO, Martinez Vazquez F, Eiguer T, Bendersky S, Martos MA. 1985. Isolation of new serovars of salmonella from streams. Rev. Argent. Microbiol. 17:149–156 [PubMed] [Google Scholar]
- 76.de Souza LC, Laria ST, Paim GV. 1992. Salmonellas and fecal coliforms in animal drinking water. Rev. Saude Publica 26:321–327 (In Portuguese) [DOI] [PubMed] [Google Scholar]
- 77.Mohle-Boetani JC, Reporter R, Werner SB, Abbott S, Farrar J, Waterman SH, Vugia DJ. 1999. An outbreak of Salmonella serogroup Saphra due to cantaloupes from Mexico. J. Infect. Dis. 180:1361–1364. 10.1086/314995 [DOI] [PubMed] [Google Scholar]
- 78.Gruszynski K, Pao S, Kim C, Toney D, Wright K, Ross PG, Colon A, Levine S. 2014. Evaluating wildlife as a potential source of Salmonella serotype Newport (JJPX01.0061) contamination of tomatoes on the eastern shore of Virginia. Zoonoses Public Health 61:202–207. 10.1111/zph.12061 [DOI] [PubMed] [Google Scholar]
- 79.Srikantiah P, Lay JC, Hand S, Crump JA, Campbell J, Van Duyne MS, Bishop R, Middendor R, Currier M, Mead PS, Molbak K. 2004. Salmonella enterica serotype Javiana infections associated with amphibian contact, Mississippi, 2001. Epidemiol. Infect. 132:273–281. 10.1017/S0950268803001638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukhejee A, Speh D, Diez-Gonzalez F. 2007. Association of farm management practices with risk of Escherichia coli contamination in pre-harvest produce grown in Minnesota and Wisconsin. Int. J. Food Microbiol. 120:296–302. 10.1016/j.ijfoodmicro.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 81.Park S, Navratil S, Gregory A, Bauer A, Srinath I, Jun M, Szonyi B, Nightingale K, Anciso J, Ivanek R. 2013. Generic Escherichia coli contamination of spinach at the preharvest stage: effects of farm management and environmental factors. Appl. Environ. Microbiol. 79:4347–4358. 10.1128/AEM.00474-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.