Abstract
The measurement of cytomegalovirus (CMV) IgG avidity is a powerful tool for identifying individuals with recent CMV infection. Because such patients are expected to be positive for CMV IgM, several investigators have suggested that CMV IgG-positive sera first be screened for CMV IgM and then only the IgM-reactive sera be tested for avidity. We investigated the impact of different CMV IgM assays on such a reflexing algorithm using a panel of 369 consecutive IgG-positive serum samples submitted for avidity testing. A bead-based immunofluorescent assay (BIFA) identified 105 IgM-positive serum samples, whereas an IgM-capture enzyme immunoassay (EIA) identified 48 IgM-positive serum samples; this marked difference led us to evaluate additional CMV IgM assays. An enzyme-linked immunofluorescent assay (ELFA) and a chemiluminescent immunoassay (CIA) were used to test all sera with discordant BIFA/EIA results, all sera with concordant positive results, and selected sera with concordant negative results. The findings indicated that the ELFA would identify 74 CMV IgM-positive samples and the CIA would identify 64. Of the 23 low-avidity serum samples, 2 were IgM negative by BIFA, 3 by ELFA and CIA, and 4 by EIA; of the 23 intermediate-avidity serum samples, 6 were IgM negative by BIFA, 10 by ELFA, and 15 by EIA and CIA. In both these avidity groups, BIFA IgM-negative sera were also negative by the other 3 assays. These findings demonstrate that an algorithm requiring CMV IgM reactivity as a criterion for CMV IgG avidity testing does not identify all low-avidity sera and thus misses some cases of acute CMV infection.
INTRODUCTION
Primary cytomegalovirus (CMV) infection during pregnancy can cause intrauterine infection of the fetus, leading to profound sensory and cognitive defects in the newborn (1–3). In contrast, intrauterine infection is rarely associated with CMV reactivation or reinfection during pregnancy (2, 4). Thus, laboratory tools allowing for an accurate diagnosis of primary CMV infections play an important role in managing pregnant women with suspected CMV disease (5, 6). The measurement of CMV IgG avidity has emerged as one of the most useful laboratory assays for identifying primary CMV infection; this assay also enables an estimation of the length of time that has elapsed since the infection occurred (5, 6). IgG avidity, defined as the aggregate strength of IgG binding to multiple antigenic epitopes of a given protein, gradually increases with time, reaching “high” levels by 5 to 6 months after the primary infection (1, 4, 7–9). Thus, a finding of low CMV IgG avidity in a pregnant patient, particularly during the second or third trimester, suggests that CMV infection may have occurred after conception, which carries an increased risk of fetal infection (2, 7, 10). Another sensitive laboratory tool for identifying primary CMV infection is CMV IgM detection (2, 4). However, the interpretation of a positive CMV IgM result can be problematic, since CMV IgM persists in some individuals for one or more years following primary infection; further, IgM production occurs in some patients following CMV reactivation (2, 5, 6, 11).
To enable accurate and efficient identification of primary CMV infection in pregnant women, several investigators have recommended a testing algorithm that combines the good sensitivity of CMV IgM detection with the good sensitivity and specificity of CMV IgG avidity testing (5, 8, 9, 12, 13). Per this reflexive algorithm, serum from a patient found to be positive for CMV IgG is first tested for CMV IgM, and only those sera found to be IgM reactive are tested in a CMV IgG avidity assay. However, a small number of CMV IgM-negative patients with low CMV IgG avidity have been described, raising questions about the clinical utility of this algorithm (4, 12, 14). Clearly, the success of the algorithm depends on the sensitivity and specificity of the CMV IgM assay employed (15). We thus evaluated the accuracies and efficiencies of four different CMV IgM assays for identifying sera with low or intermediate CMV IgG avidity among serum samples submitted to an esoteric reference laboratory for CMV IgG avidity testing.
MATERIALS AND METHODS
Specimens.
The study utilized 369 consecutive CMV IgG-positive serum samples submitted to the Focus Diagnostics Reference Laboratory for CMV IgG avidity testing (16); 91% of these samples were supplied by women of childbearing age (15 to 49 years old), but no clinical data were available for any of the patients who supplied the serum samples. Following release of the avidity results, the samples were deidentified before the various IgM assays were performed.
CMV IgM analysis.
Four different assays cleared by the U.S. Food and Drug Administration for measuring CMV IgM were utilized; all the assays were performed per the instructions from each manufacturer. A bead-based immunofluorescent assay (BIFA) was performed using the BioPlex system (Bio-Rad Laboratories, Hercules, CA). An IgM-capture enzyme immunoassay (EIA) was performed using a kit purchased from Diamedix Corporation (Miami, FL). An enzyme-linked immunofluorescent assay (ELFA) was performed using the Vidas system (bioMérieux, Durham, NC). A chemiluminescent immunoassay (CIA) was performed using the Liaison system (DiaSorin, Stillwater, MN). For each assay, the results were categorized as negative, equivocal, or positive per the guidelines indicated in the package insert.
CMV IgG avidity.
The serum samples were tested for CMV IgG avidity using a commercially available CMV IgG EIA kit (Wampole Laboratories, Princeton, NJ) with modifications as described in the accompanying article (16). Briefly, a given sample was diluted per the package insert and added to two microtiter wells; after incubation, one well was washed with kit-supplied wash buffer and the other well was washed with dissociating buffer (wash buffer containing 6 M urea). The assay was then finished per the instructions in the package insert, and the absorbance values were recorded. The results were expressed as an avidity index (AI), calculated using the formula AI = (absorbance for the well washed with dissociating buffer/absorbance for the well washed with kit wash buffer) × 100. AI values of ≤50% were interpreted as low avidity, 51% to 59% as intermediate avidity, and ≥60% as high avidity.
RESULTS
Agreement between BIFA and EIAs for CMV IgM.
The initial experiments focused on comparing CMV IgM detection by BIFA and EIA among the serum samples submitted for CMV IgG avidity testing. As shown in Table 1, 24 of 369 (6.5%) serum samples exhibited an equivocal result in one of these assays. Because an equivocal result represents a nonnegative result and may thus have clinical relevance, we considered an equivocal result to be positive for the purpose of this study. The number of samples positive for CMV IgM by BIFA (n = 105) was more than double the number positive by EIA (n = 48). Because of this large difference, the study was expanded to include 2 additional methodologies for CMV IgM detection, the ELFA and CIA.
TABLE 1.
CMV IgM results as determined by BIFA and EIA for 369 consecutive CMV IgG-positive serum samples submitted for CMV IgG avidity testinga
| BIFA results | EIA results |
Total | ||
|---|---|---|---|---|
| Positive | Equivocal | Negative | ||
| Positive | 35 | 11 | 46 | 92 |
| Equivocal | 0 | 0 | 13 | 13 |
| Negative | 2 | 0 | 262 | 264 |
| Total | 37 | 11 | 321 | 369 |
Results represent the number of serum samples exhibiting the indicated BIFA and EIA result set.
CMV IgM results as determined by ELFA and CIA.
All 61 serum samples with qualitatively discordant BIFA and EIA results, as well as all 46 samples with concordant positive BIFA and EIA results, were tested by ELFA and CIA. Because of cost constraints, ELFA and CIA were performed on only 26 of the 262 samples with concordant negative BIFA and EIA results; these 26 serum samples included all 8 BIFA/EIA concordant negative samples with low or intermediate IgG avidity plus 18 randomly selected concordant negative samples with high IgG avidity. As with the BIFA and EIA, equivocal ELFA or CIA results were considered positive. Table 2 presents the ELFA and CIA results in relation to the BIFA/EIA reactivity pattern for this panel of 133 serum samples. Most samples with concordant positive BIFA/EIA results were also positive by ELFA and CIA. In contrast, most samples exhibiting the discordant BIFA-positive/EIA-negative result pattern were negative by ELFA and CIA; in total, 35 serum samples positive for CMV IgM by BIFA were negative by the other three IgM assays. Of 26 concordant negative BIFA/EIA serum samples, only 1 (4%) was positive by both ELFA and CIA; this serum sample exhibited high avidity.
TABLE 2.
CMV IgM results as determined by ELFA and CIA for 133 samples selected on the basis of BIFA and EIA results
| BIFA/EIA results | No. of samples | ELFA |
CIA |
||||
|---|---|---|---|---|---|---|---|
| No. of positive | No. of negative | QNSa | No. of positive | No. of negative | QNSa | ||
| Positive/positive | 46 | 43 | 3 | 0 | 37 | 8 | 1 |
| Positive/negative | 59 | 20 | 39 | 0 | 13 | 44 | 2 |
| Negative/positive | 2 | 0 | 1 | 1 | 0 | 1 | 1 |
| Negative/negative | 26 | 1 | 25 | 0 | 1 | 25 | 0 |
QNS, quantity not sufficient for testing.
Pairwise assessment of concordance among CMV IgM assays.
The results in Table 2 were used to make pairwise assessments of concordance between the various CMV IgM assays (Table 3). For this analysis, the finding that 4% of the 26 concordant negative BIFA/EIA serum samples were positive by ELFA and CIA was extrapolated to the entire group of 262 concordant negative BIFA/EIA samples; it was thus estimated that 10 BIFA-negative/EIA-negative samples would be CMV IgM positive by ELFA or CIA. The concordance levels between the three non-BIFA assays were all >90%; in contrast, the concordance levels between the BIFA and each of the other three assays ranged from 83% to 86%. These lower concordance levels with BIFA were due to the 35 serum samples positive by BIFA but negative by the other three assays, as well as the estimated 10 samples that were IgM negative by BIFA and EIA but positive by ELFA and CIA.
TABLE 3.
Pairwise assessment of concordance between CMV IgM assays for 369 CMV IgG-positive serum samples tested for CMV IgG aviditya
| CMV IgM assays compared | No. of concordant positive results | No. of concordant negative results | Total no. (%) of concordant results |
|---|---|---|---|
| EIA vs ELFA | 44 | 291 | 335 (91) |
| EIA vs CIA | 39 | 296 | 335 (91) |
| ELFA vs CIA | 54 | 288 | 342 (93) |
| BIFA vs EIA | 46 | 262 | 308 (83) |
| BIFA vs ELFA | 63 | 253 | 316 (86) |
| BIFA vs CIA | 53 | 253 | 306 (83) |
The values for ELFA and CIA represent the calculated values based on the observation that 4% of BIFA-negative/EIA-negative samples were CMV IgM positive by ELFA or CIA, and the assumption that QNS (quantity not sufficient for testing) samples for a given assay were IgM positive by that assay.
CMV IgM detection by various assays in relation to CMV IgG avidity results.
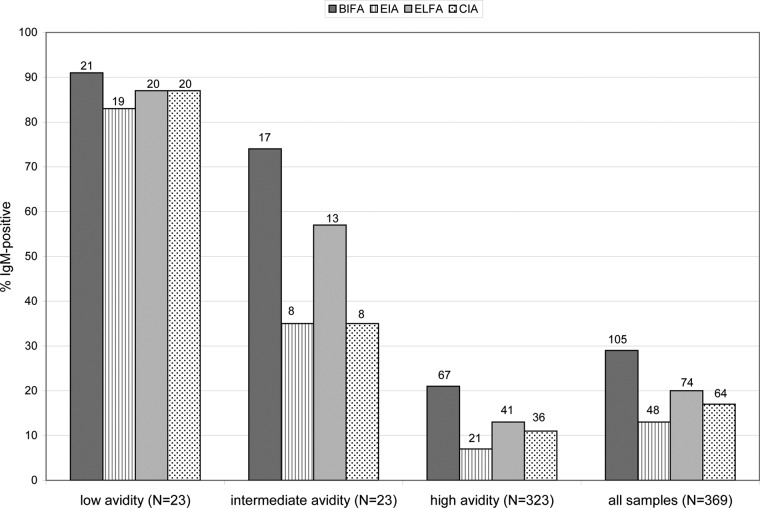
Figure 1 presents the proportions of sera that were CMV IgM positive by the 4 IgM assays in relation to the qualitative CMV IgG avidity result obtained. The sensitivities of the 4 IgM assays for detecting low-avidity samples ranged from 83% to 91% (Fig. 1). Although most serum samples with low CMV IgG avidity were CMV IgM positive in all 4 IgM assays (19 of 23 [83%]), 2 of 23 (9%) low-avidity samples were negative in all 4 CMV IgM assays. In contrast to the findings for the low-avidity serum group, the intermediate-avidity serum group exhibited a wide range of CMV IgM detection rates (35% to 74%), depending on the CMV IgM assay used. Similarly, the CMV IgM detection rates in the high-avidity serum group ranged from 7% to 21%.
FIG 1.
CMV IgM reactivity by different methodologies in relation to CMV IgG avidity results for 369 consecutive CMV IgG-positive serum samples submitted for avidity testing. The results represent the proportion of each avidity group positive for CMV IgM by the indicated methodology; the number of CMV IgM-positive samples in the group is indicated at the top of each histogram. The ELFA and CIA values for the high-avidity group and the all-samples group were calculated based on the observation that 4% of the BIFA-negative/EIA-negative samples were positive for CMV IgM by ELFA or CIA (see Table 2), and the assumption that quantity-not-sufficient (QNS) samples for a given assay were IgM positive by that assay.
DISCUSSION
We capitalized on the availability of a large panel of consecutive serum samples tested for CMV IgG avidity to assess the efficacy of an algorithm requiring CMV IgM reactivity as a criterion for IgG avidity testing. Four different CMV IgM assays, each based on a different methodology, were evaluated. We considered equivocal CMV IgM results to be positive, based on our assumption that physicians would view equivocal results as nonnegative and thus expect samples with equivocal results to reflex to CMV IgG avidity testing. An equivocal IgM result may indicate a waxing IgM response soon after infection, or alternatively, a waning IgM response several months after infection (5, 6, 11); in either of these settings, a CMV IgG avidity result may prove clinically useful in defining the postinfection time frame.
All 4 of the CMV IgM testing methodologies we evaluated successfully identified >80% of the low-avidity serum samples as IgM positive, confirming the strong link between low IgG avidity and IgM reactivity (11, 12, 14). However, none of the 4 CMV IgM assays identified all 23 serum samples exhibiting low IgG avidity; BIFA missed 2 low-avidity samples, ELFA and CIA missed 3 samples, and EIA missed 4 samples. These findings extend those of other investigators who found that a small number of low-avidity samples were IgM negative using a single IgM assay (4, 12). Collectively, our data demonstrate that if the identification of 100% of the samples exhibiting low CMV IgG avidity is the goal of a given laboratory avidity testing program, a reflexive algorithm requiring CMV IgM reactivity as a criterion for CMV IgG avidity testing should not be utilized. Rather, CMV IgG-positive sera should move directly to avidity testing.
In situations in which budgetary and staffing issues favor the employment of a reflexive algorithm, it is important that a laboratory understand the efficiency of the CMV IgM assay selected for use (15). We found that the number of samples meeting the criterion for reflex varied >2-fold depending on the CMV IgM assay employed; of 369 CMV IgG-positive serum samples, 105 samples were targeted for IgG avidity testing when tested for CMV IgM by BIFA, compared to only 48 when tested by EIA. However, the proportion of targeted sera found to exhibit low IgG avidity was 2-fold higher for the EIA-based algorithm (19/48 [40%]) than for the BIFA-based algorithm (21/105 [20%]). Thus, laboratories electing to utilize a reflexive algorithm must select a CMV IgM assay based on a balanced analysis of the number of serum samples targeted for reflex, the proportion of targeted sera exhibiting low IgG avidity, and the number of low-avidity samples missed by the algorithm due to an IgM-negative result.
While our findings demonstrate that a CMV IgM/CMV IgG avidity reflexive algorithm is effective (although not perfect) for identifying serum samples exhibiting low avidity, they clearly show that this type of algorithm is not effective for identifying sera with intermediate IgG avidity. The proportion of intermediate-avidity samples that were CMV IgM negative ranged from 26% to 65%, depending on the CMV IgM assay used. This finding is not surprising, since intermediate IgG avidity levels are expected during the 5- to 6-month postinfection time period, the same time window during which IgM levels wane. Further, the clinical significance of an intermediate CMV IgG avidity result, and thus the need to identify sera exhibiting this result, remains unclear (15, 17). Systematic prospective studies are needed to determine the value of an intermediate CMV IgG result in managing pregnant women with suspected postconception primary CMV infection.
A secondary outcome enabled by our study was an assessment of the overall concordance of the different CMV IgM assay methods employed. Although our results should be considered estimates due to the limitation that only 10% of the BIFA-negative/EIA-negative serum samples were tested by ELFA and CIA, they support previously published data from CMV IgM method comparison studies (15, 17–21). The pairwise concordance rates between EIA, ELFA, and CIA were all >90%; in contrast, the concordance rates of these 3 assays with BIFA were lower (84% to 86%), due in large part to sera that were positive by BIFA but not by the other three assays. This response pattern may reflect increased BIFA sensitivity, or alternatively, a higher BIFA false-positive rate. Discrimination between these two possibilities requires prospective studies to characterize the clinical status and changing CMV antibody profile of patients exhibiting this reactivity pattern.
ACKNOWLEDGMENT
We thank Elitza Theel for critical review of the manuscript.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Bodéus M, Feyder S, Goubau P. 1998. Avidity of IgG antibodies distinguishes primary from nonprimary cytomegalovirus infection in pregnant women. Clin. Diagn. Virol. 9:9–16. 10.1016/S0928-0197(97)10016-2 [DOI] [PubMed] [Google Scholar]
- 2.Grangeot-Keros L, Mayaux MJ, Lebon P, Freymuth F, Eugene G, Stricker R, Dussaix E. 1997. Value of cytomegalovirus (CMV) IgG avidity index for the diagnosis of primary CMV infection in pregnant women. J. Infect. Dis. 175:944–946. 10.1086/513996 [DOI] [PubMed] [Google Scholar]
- 3.Dollard SC, Grosse SD, Ross DS. 2007. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev. Med. Virol. 17:355–363. 10.1002/rmv.544 [DOI] [PubMed] [Google Scholar]
- 4.Lazzarotto T, Spezzacatena P, Pradelli P, Abate DA, Varani S, Landini MP. 1997. Avidity of immunoglobulin G directed against human cytomegalovirus during primary and secondary infections in immunocompetent and immunocompromised subjects. Clin. Diagn. Lab. Immunol. 4:469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazzarotto T, Guerra B, Lanari M, Gabrielli L, Landini MP. 2008. New advances in the diagnosis of congenital cytomegalovirus infection. J. Clin. Virol. 41:192–197. 10.1016/j.jcv.2007.10.015 [DOI] [PubMed] [Google Scholar]
- 6.Revello MG, Fabbri E, Furione M, Zavattoni M, Lilleri D, Tassis B, Quarenghi A, Cena C, Arossa A, Montanari L, Rognoni V, Spinillo A, Gerna G. 2011. Role of prenatal diagnosis and counseling in the management of 735 pregnancies complicated by primary human cytomegalovirus infection: a 20-year experience. J. Clin. Virol. 50:303–307. 10.1016/j.jcv.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 7.Lazzarotto T, Spezzacatena P, Varani S, Gabrielli L, Pradelli P, Guerra B, Landini MP. 1999. Anticytomegalovirus (anti-CMV) immunoglobulin G avidity in identification of pregnant women at risk of transmitting congenital CMV infection. Clin. Diagn. Lab. Immunol. 6:127–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro SC, Hall B, Whybin LR, Leader L, Robertson P, Maine GT, Rawlinson WD. 2005. Diagnosis of and screening for cytomegalovirus infection in pregnant women. J. Clin. Microbiol. 43:4713–4718. 10.1128/JCM.43.9.4713-4718.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enders G, Daiminger A, Bäder U, Exler S, Schimpf Y, Enders M. 2013. The value of CMV IgG avidity and immunoblot for timing the onset of primary CMV infection in pregnancy. J. Clin. Virol. 56:102–107. 10.1016/j.jcv.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 10.Bodéus M, Beulné D, Goubau P. 2001. Ability of three IgG-avidity assays to exclude recent cytomegalovirus infection. Eur. J. Clin. Microbiol. Infect. Dis. 20:248–252. 10.1007/s100960100484 [DOI] [PubMed] [Google Scholar]
- 11.Lazzarotto T, Varani S, Guerra B, Nicolosi A, Lanari M, Landini MP. 2000. Prenatal indicators of congenital cytomegalovirus infection. J. Pediatr. 137:90–95. 10.1067/mpd.2000.107110 [DOI] [PubMed] [Google Scholar]
- 12.Dollard SC, Staras SAS, Amin MM, Scott Schmid D, Cannon MJ. 2011. National prevalence estimates for cytomegalovirus IgM and IgG avidity and association between high IgM antibody titer and low IgG avidity. Clin. Vaccine Immunol. 18:1895–1899. 10.1128/CVI.05228-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macé M, Sissoeff L, Rudent A, Grangeot-Keros L. 2004. A serological testing algorithm for the diagnosis of primary CMV infection in pregnant women. Prenat. Diagn. 24:861–863. 10.1002/pd.1001 [DOI] [PubMed] [Google Scholar]
- 14.Prince HE, Leber AL. 2002. Validation of an in-house assay for cytomegalovirus immunoglobulin G (CMV IgG) avidity and relationship of avidity to CMV IgM levels. Clin. Diagn. Lab. Immunol. 9:824–827. 10.1128/CDLI.9.4.824-827.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazzarotto T, Galli C, Pulvirenti R, Rescaldani R, Vezzo R, La Gioia A, Martinelli C, La Rocca S, Agresti G, Grillner L, Nordin M, van Ranst M, Combs B, Maine GT, Landini MP. 2001. Evaluation of the Abbott AxSYM cytomegalovirus (CMV) immunoglobulin M (IgM) assay in conjunction with other CMV IgM tests and a CMV IgG avidity assay. Clin. Diagn. Lab. Immunol. 8:196–198. 10.1128/CDLI.8.1.196-198.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prince HE, Lapé-Nixon M, Novak-Weekley SM. 2014. Performance of a cytomegalovirus immunoglobulin G enzyme immunoassay kit modified to measure avidity. Clin. Vaccine Immunol. 21:808–812. 10.1128/CVI.00105-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Paschale M, Agrappi C, Manco MT, Clerici P. 2010. Positive predictive value of anti-HCMV IgM as an index of primary infection. J. Virol. Methods 168:121–125. 10.1016/j.jviromet.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 18.Binnicker MJ, Jespersen DJ, Harring JA. 2010. Multiplex detection of IgM and IgG class antibodies to Toxoplasma gondii, rubella virus, and cytomegalovirus using a novel multiplex flow immunoassay. Clin. Vaccine Immunol. 17:1734–1738. 10.1128/CVI.00332-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagrou K, Bodeus M, Van Ranst M, Goubau P. 2009. Evaluation of the new Architect cytomegalovirus immunoglobulin M (IgM), IgG, and IgG avidity assays. J. Clin. Microbiol. 47:1695–1699. 10.1128/JCM.02172-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlier P, Harika N, Bailly R, Vranken G. 2010. Laboratory evaluation of the new Access cytomegalovirus immunoglobulin M and IgG assays. J. Clin. Virol. 49:192–197. 10.1016/j.jcv.2010.07.024 [DOI] [PubMed] [Google Scholar]
- 21.Bal TA, Armstrong G, Han XY. 2012. Evaluation of the IMMULITE 2000 CMV IgM assay. Herpesviridae 3:2–6. 10.1186/2042-4280-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]