Abstract
SUMMARY
Viruses were defined as one of the two principal types of organisms in the biosphere, namely, as capsid-encoding organisms in contrast to ribosome-encoding organisms, i.e., all cellular life forms. Structurally similar, apparently homologous capsids are present in a huge variety of icosahedral viruses that infect bacteria, archaea, and eukaryotes. These findings prompted the concept of the capsid as the virus “self” that defines the identity of deep, ancient viral lineages. However, several other widespread viral “hallmark genes” encode key components of the viral replication apparatus (such as polymerases and helicases) and combine with different capsid proteins, given the inherently modular character of viral evolution. Furthermore, diverse, widespread, capsidless selfish genetic elements, such as plasmids and various types of transposons, share hallmark genes with viruses. Viruses appear to have evolved from capsidless selfish elements, and vice versa, on multiple occasions during evolution. At the earliest, precellular stage of life's evolution, capsidless genetic parasites most likely emerged first and subsequently gave rise to different classes of viruses. In this review, we develop the concept of a greater virus world which forms an evolutionary network that is held together by shared conserved genes and includes both bona fide capsid-encoding viruses and different classes of capsidless replicons. Theoretical studies indicate that selfish replicons (genetic parasites) inevitably emerge in any sufficiently complex evolving ensemble of replicators. Therefore, the key signature of the greater virus world is not the presence of a capsid but rather genetic, informational parasitism itself, i.e., various degrees of reliance on the information processing systems of the host.
INTRODUCTION
Viruses were originally defined as “filterable disease agents,” i.e., infectious agents that are small enough to pass through bacterial filters. The recent discoveries of giant viruses that infect protists and bacteria and that do not pass through porcelain filters traditionally used for collection of bacteria have put this size-centered definition to rest (1–5). One of the latest attempts to define viruses on the basis of more fundamental criteria was undertaken by Raoult and Forterre (6). Under their proposal, viruses are capsid-encoding organisms as opposed to ribosome-encoding cellular organisms. This “capsidocentric” perspective on the virus world is buttressed by observations on the extremely wide spread of certain capsid protein (CP) structures that are shared by an enormous variety of viruses, from the smallest to the largest ones, that infect bacteria, archaea, and all divisions of eukaryotes. The foremost among such conserved capsid protein structures is the so-called jelly roll capsid (JRC) protein fold, which is represented, in a variety of modifications, in extremely diverse icosahedral (spherical) viruses that infect hosts from all major groups of cellular life forms (7–9). In particular, the presence of the double-beta-barrel JRC (JRC2b) in a broad variety of double-stranded DNA (dsDNA) viruses infecting bacteria, archaea, and eukaryotes has been touted as an argument for the existence of an “ancient virus lineage,” of which this type of capsid protein is the principal signature (9). Under this approach, viruses that possess a single-beta-barrel JRC (JRC1b)—primarily RNA viruses and single-stranded DNA (ssDNA) viruses—could be considered another major viral lineage. A third lineage is represented by dsDNA viruses with icosahedral capsids formed by the so-called HK97-like capsid protein (after bacteriophage HK97, in which this structure was first determined), with a fold that is unrelated to the jelly roll fold. This assemblage of viruses is much less expansive than those defined by either JRC2b or JRC1b, but nevertheless, it unites dsDNA viruses from all three domains of cellular life (10, 11).
In more general terms, the morphospace of viral capsids and capsid proteins appears to be severely constrained by requirements of symmetry and stability allowing for only 20 or so distinct, commonly adopted structural designs (12, 13). Certainly, the existence of such constraints does not imply that the structural space of the virus world has already been explored fully. For instance, the diversity of virion structures among the viruses infecting hyperthermophilic Crenarchaeota organisms is astounding, and the discovery of additional novel forms can readily be anticipated (14, 15). Moreover, the study of the pleomorphic viruses of Haloarchaea (16, 17) and the recent remarkable discovery of the humongous pandoraviruses (18) show that some viruses possess virions but not typical proteinaceous capsids. Nevertheless, although numerous “exotic” and relatively rare structures undoubtedly remain to be discovered, it appears most likely that the truly common capsid shapes and capsid protein folds are not too numerous and are largely known.
The capsid-based definition of a virus does capture a quintessential distinction between the two major empires of life forms, i.e., viruses and cellular life forms (19), and the JRC (and, to a lesser extent, other capsid protein folds) indeed is extremely common among viruses, but the capsid-centered paradigm of the virus world appears to be substantially incomplete. The essence of this fundamental incompleteness is that numerous groups of typical viruses share a common evolutionary history with genetic elements that lack a capsid protein gene and are never encapsidated or, in some cases, encapsidated in the virions of “host” viruses. Capsidless relatives have been identified for viruses that employ different replication-expression strategies and infect diverse hosts, and their evolution does not seem to be a one-way street, as some appear to have evolved from typical viruses that lost the genes for virion proteins, whereas others are likely ancestors of the respective viruses. In this article, we review the evolutionary relationships between typical viruses with different replication-expression strategies and capsidless genetic elements, and we propose a paradigm of virus world evolution that does not focus on any particular gene but rather is rooted in the concept of genetic, informational parasitism.
PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES
The gene repertoires of viruses are enormously diversified, with the great majority of the genes represented only in narrow groups of viruses (20, 21). However, several “viral hallmark genes” that encode proteins responsible for key functions in virion formation and genome replication are shared by numerous, diverse viruses with different strategies of genome replication and expression (22) (Fig. 1). In viruses with small genomes, such as most of the RNA viruses and ssDNA viruses, the hallmark genes account for all or most of the genetic capacity, whereas in larger viruses, with dsDNA genomes, these genes occupy only a small portion of the genome. Extensive exchange and reassortment of gene modules are the key features in the evolution of the virus world, so the hallmark genes often occur in different combinations in different groups of viruses.
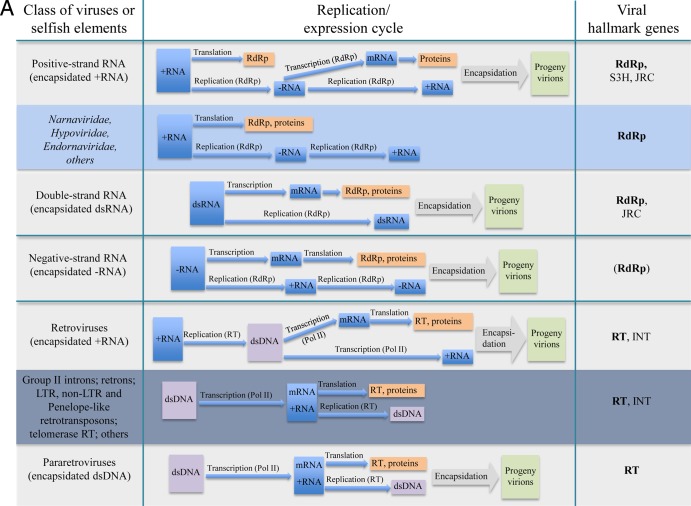
FIG 1.
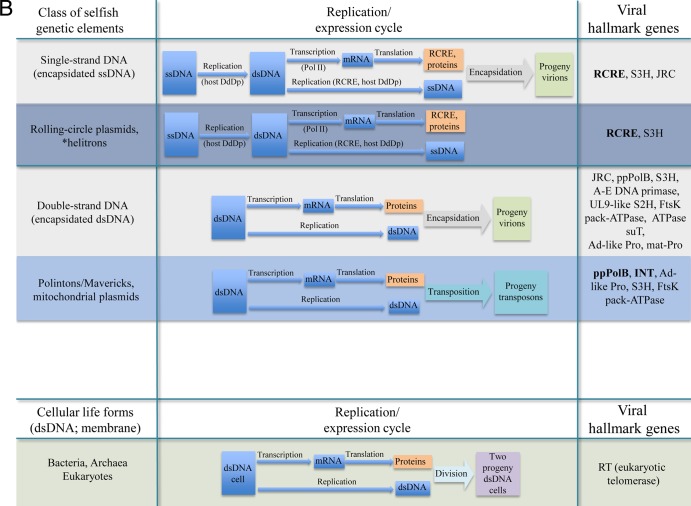
Replication-expression classes of viruses and homologous, capsidless selfish elements. (A) RNA and reverse-transcribing elements. (B) DNA elements. The three shades of the blue background denote approximate relative prevalences of capsidless selfish elements in the respective Baltimore class (i.e., low for ssRNA genomes, moderate for dsDNA genomes, and high for retroelements and ssDNA genomes; so far, there are no capsidless elements with negative-strand RNA or dsRNA genomes). The abbreviations for the virus hallmark genes are as follows: RdRp, RNA-dependent RNA polymerase; S3H, superfamily 3 helicase; JRC, jelly roll capsid protein; RT, reverse transcriptase; INT, retro-type integrase; RCRE, rolling circle replication endonuclease; A-E DNA primase, archaeo-eukaryotic DNA primase; UL9-like S2H, UL9-like superfamily 2 helicase; FtsK pack-ATPase, FtsK-family packaging ATPase; ATPase suT, ATPase subunit of terminase; ppPolB, protein-primed DNA polymerase B; Ad-like Pro, adeno-like protease; and mat-Pro, maturation protease. The hallmark genes that are present in all known members of the given class are rendered in bold. For negative-strand RNA viruses, the RdRp is indicated in parentheses to emphasize the tentative relationship between the RNA polymerases of these viruses and the RdRp/RT. Helitrons are marked by an asterisk because of their distinct replication cycle: unlike other RCRE-encoding ssDNA selfish elements, helitrons are transposed as dsDNA. DdDp, DNA-dependent DNA polymerase.
The JRC (defined as one hallmark protein, with single- and double-beta-barrel proteins lumped together) appears to be the most common viral protein (Fig. 1). However, two hallmark genes that encode key proteins involved in genome replication, namely, RNA-dependent RNA polymerase (RdRp)/reverse transcriptase (RT) (these two enzyme families are homologous, with the relationship readily detectable through highly significant sequence similarity, and some of the RdRps might even have evolved from RTs [23–25], so they are naturally combined) and superfamily 3 helicase (S3H), are not too far behind with regard to their spread through the virus world (Fig. 1).
Furthermore, given that the JRC proteins are extremely divergent at the sequence level, with the similarity between distant forms (particularly between the single-beta-barrel and double-beta-barrel classes) detectable only through structural comparison (8, 9, 14), a fairer comparison might involve a coarser classification of the other hallmark genes. Under this approach, RdRp/RT would be unified with the B family DNA polymerases and archaeo-eukaryotic DNA primases because all these proteins contain the so-called palm domain (26, 27). Similarly, S3H would be combined with numerous families of ATPases containing the P-loop domain, including at least two additional viral hallmark genes, encoding superfamily 2 helicases and packaging ATPase (28, 29). Such broader classes of hallmark proteins appear to be even more common among viruses than the JRC. Other genes that we define as hallmarks, e.g., the maturation proteases or the endonucleases involved in rolling circle DNA replication, are less widespread but nevertheless are highly virus specific and shared by groups of viruses that substantially differ with respect to genome size, structure, and host range (Fig. 1).
The replication-associated viral hallmark genes are often found in viruses that lack the JRC but rather possess other, unrelated capsid structures, such as the filamentous capsids of numerous positive-strand RNA viruses and some ssDNA viruses, or nucleocapsids of retroviruses. Even more importantly, most of the genes for proteins involved in replication are shared between viruses and capsidless genetic elements, such as plasmids or transposons (Fig. 1). This key observation indicates that some defining features of the virus world reach beyond the range of “capsid-encoding organisms” (6) and prompted us to examine the relationships between bona fide viruses and capsidless elements in greater detail. We discuss these relationships in the subsequent sections, after presenting the classification scheme of viral genome replication and expression strategies.
CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS
Viruses are not monophyletic in the traditional sense, i.e., all extant viruses did not evolve from a single ancestral virus (22, 30, 31). Instead, the virus world is a complex, modular network of genomes that are linked through shared genes, some of which (the hallmark genes) are present in highly diverse viruses and thus connect different modules of the network (20, 32). The high-level classification of viruses that was first delineated in the seminal work of Baltimore (33, 34) is traditionally based on the strategy of genome replication and expression (Fig. 1) (henceforth termed Baltimore classification). In this system, the replication-expression scheme that is common to all cellular life forms represents only a single, even if the largest, class of viruses (Fig. 1B), whereas the other classes embody replication-expression schemes that are not found in cellular organisms. Evolutionary relationships between bona fide viruses and capsidless elements are prominent in the four most common replication-expression classes, namely, positive-strand RNA viruses, retroelements (Fig. 1A), single-stranded DNA viruses, and double-stranded DNA viruses (Fig. 1B). In the following sections, we attempt to reconstruct the evolutionary scenarios that link viruses and capsidless elements within each of these classes.
EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS
Capsidless Derivatives of Positive-Strand RNA Viruses
Positive-strand RNA viruses represent the majority of the viruses of eukaryotes, with three major superfamilies and numerous families infecting animals, plants, and diverse unicellular forms, whereas in prokaryotes only a single family with a rather narrow host range is known (35, 36). A considerable variety of capsidless forms of positive-strand RNA viruses have been identified (37). In more precise terms, these agents are virus-like elements, but because they are traditionally classified as viruses, here we use these terms interchangeably. Only one gene is shared by all positive-strand and double-stranded RNA (dsRNA) viruses (with the exception of some satellite viruses), namely, the RNA-dependent RNA polymerase (RdRp) gene (36), and this is also the only gene that is represented in all capsidless RNA genomes. Beyond the RdRp, the capsidless elements show a broad range of gene repertoires (Fig. 2), and the analysis of these genes suggests different evolutionary scenarios.
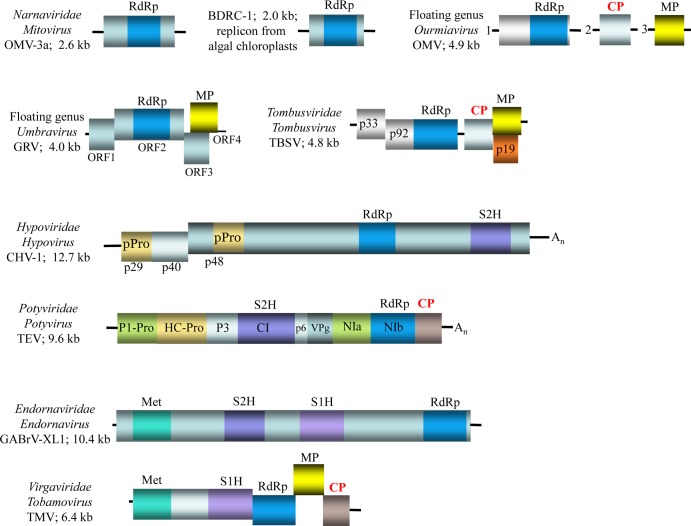
FIG 2.
Genome architectures of capsidless positive-strand RNA selfish elements and related viruses. Genome architectures for a subset of capsidless selfish elements and related positive-strand RNA viruses (for which CP is shown in bold red for clarity) are drawn roughly to scale. Black lines correspond to noncoding regions; rectangles denote open reading frames (ORFs), with identified protein domains color coded. RdRp, RNA-dependent RNA polymerase; Met, methyltransferase/capping enzyme; S2H and S1H, superfamily 2 and 1 helicases, respectively; CP, capsid protein; MP, movement protein; p-Pro, papain-like protease; P1-Pro, protein 1 trypsin-like protease; HC-Pro, helper component–papain-like protease; P3, protein 3; CI, cylindrical inclusion protein; VPg, virus protein, genome-linked; NIa, nuclear inclusion a, trypsin-like protease; NIb, nuclear inclusion b protein. Virus names are as follows: OMV-3a, Ophiostoma mitovirus 3a; BDRC-1, Bryopsis cinicola dsRNA replicon from chloroplasts; OMV, Ourmia melon virus; GRV, Groundnut rosette virus; TBSV, Tomato bushy dwarf virus; CHV-1, Cryphonectria parasitica hypovirus 1; TEV, Tobacco etch virus; GABrV-XL1, Gremmeniella abietina type B RNA virus XL1; TMV, Tobacco mosaic virus.
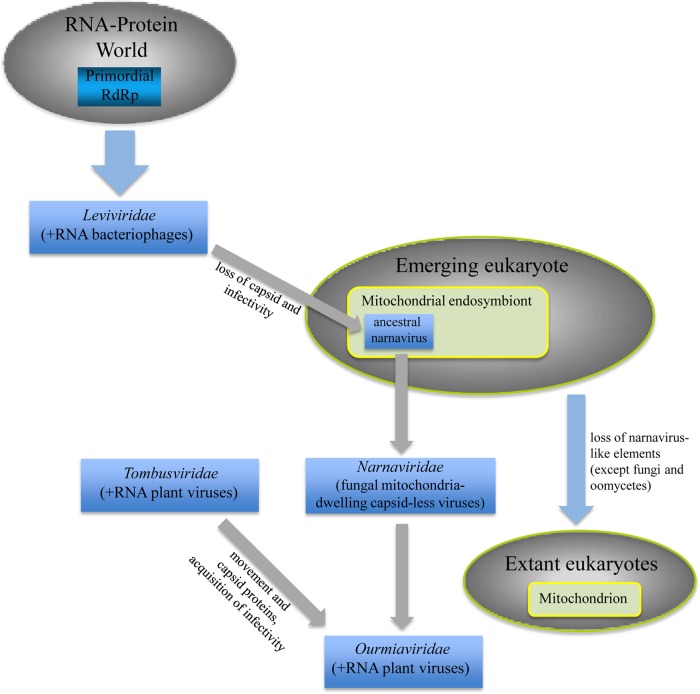
The fungal, capsidless elements of the family Narnaviridae (naked RNA viruses) possess ∼2.5-kb genomes that encode a single functional protein responsible for the synthesis of the minus and plus strands of the viral genome, i.e., RdRp (Fig. 2) (38–42). Phylogenetic analysis of these “minimal” RNA viruses clearly indicates that the closest, although still rather distant, relatives of the narnavirus RdRps are the RdRps of RNA bacteriophages of the family Leviviridae (Fig. 3) (43–45). The phylogenetic tree of the RdRps is based on an alignment of highly diverged protein sequences, and accordingly, the “star topology” of this tree has to be interpreted with caution (see further discussion below). However, many of the affinities within the major branches are reliable, and in this section, we discuss only these relationships. The intriguing evolutionary connection with the leviviruses implies that narnaviruses evolved from an ancestral bacterial virus (Fig. 4). This hypothesis is compatible with the unusual lifestyle of the narnaviruses of the genus Mitovirus, whose entire reproduction cycle occurs within fungal mitochondria (38).
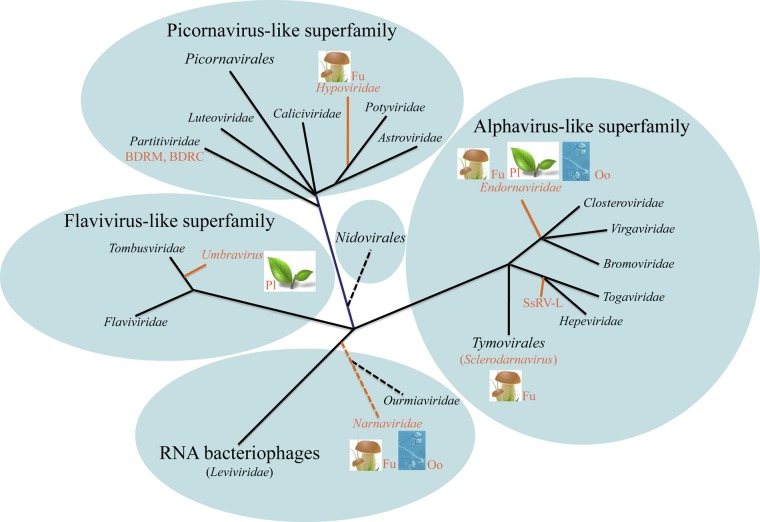
FIG 3.
Schematic phylogeny of the RdRps of positive-strand RNA viruses and their capsidless derivatives. The tree topology is based on those described previously (23, 36, 37, 201). The orange lines denote capsidless RNA replicons. Abbreviations: Fu, fungi; Pl, plants; Oo, oomycetes; BDRM, Bryopsis cinicola dsRNA replicon from mitochondria; BDRC, Bryopsis cinicola dsRNA replicon from chloroplasts; SsRV-L, Sclerotinia sclerotiorum RNA virus L.
FIG 4.
Evolutionary scenario for narnaviruses and ourmiaviruses.
The simplest evolutionary scenario for the origin of mitoviruses would posit that the protomitochondrial endosymbiont of eukaryotes brought with it an RNA bacteriophage that became resigned to replicating within the mitochondria in the evolving eukaryotic cell. Reductive evolution of this intramitochondrial parasite that paralleled the genomic reduction of the mitochondrion itself resulted in a loss of capsid and infectivity, i.e., the ability to infect new cells via extracellular routes. The extant mitoviruses conform with this noninfectious lifestyle: they spread only via intracellular pathways that include cell division, asexual and sexual spores (conidia and ascospores, respectively), and horizontal transmission via hyphal fusion (anastomosis) (46, 47). Given that all extant eukaryotes appear to possess mitochondria or their degraded derivatives and that mitochondria are thought to be monophyletic (48, 49), the RNA bacteriophage apparently was lost in most of the eukaryotes and retained only in several lineages of fungi, plants, and, possibly, protists.
Unlike mitoviruses that infect a wide variety of the plant-pathogenic and symbiotic (mycorrhizal) fungi, two currently recognized members of the Narnavirus genus are “RdRp-only” viruses that replicate in the cytoplasm of baker's yeast, Saccharomyces cerevisiae (41). Given the scarcity of Narnavirus species, the most likely scenario for the origin of this genus is escape of an ancestral mitovirus from fungal mitochondria and adaptation to cytoplasmic replication.
Deeper sampling of diverse eukaryotes and metagenomes is likely to yield additional narnaviruses and allow more detailed evolutionary insight. Indeed, recent research in this area produced two surprising findings. One of these is the discovery of a Narnavirus-like agent that replicates in an oomycete (43), a protist with a fungus-like lifestyle that is, however, evolutionarily distinct from fungi. Oomycetes belong to the eukaryotic supergroup Chromalveolata, whereas fungi, along with animals, Amoebozoa, and some other protists, comprise the Unicont supergroup (51, 52). The origin of the oomycete narnavirus could be attributed either to an ancestor shared with fungal narnaviruses or to more recent horizontal virus transfer between a plant-pathogenic fungus and an oomycete. The scenario of the origin of Narnaviridae from a bacterial virus carried over to the proto-eukaryotic host by the mitochondrial endosymbiont implies that these agents are ancestral in eukaryotes and thus favors an ancient common origin of fungal and oomycete narnaviruses (Fig. 4).
The second unexpected finding comes from phylogenomic analysis of Narnavirus, a distinct family of plant viruses. The tripartite ourmiavirus genomes appear to have evolved via combination of the narnavirus-like RdRp gene with two additional RNA segments that carry capsid and movement protein genes most similar to those of plant RNA viruses of the family Tombusviridae (45) (Fig. 2 to 4). The history of this group of viruses is remarkable in that a capsidless, degenerate virus-like agent apparently regained the active virus lifestyle by forming a composite genome with segments from very distantly related viruses.
In contrast to the small narnavirus genomes, fungal viruses of the family Hypoviridae (Hypo, from hypovirulence) possess relatively large genomes of up to 13 kb (47, 53). These genomes encode polyproteins that harbor several protein domains involved in polyprotein processing (papain-like proteases), RNA replication (RdRp and RNA helicase), and several aspects of virus-host interaction (Fig. 2). Phylogenomic analysis reliably links hypoviruses to Potyviridae, a family of filamentous plant viruses that belong to the picornavirus-like superfamily of positive-strand RNA viruses (54) (Fig. 3). The hypovirus-potyvirus connection is supported by phylogenies of RdRp, RNA helicase, and papain-like protease. Moreover, hypoviruses and potyviruses are unique among the members of the picornavirus-like superfamily in possessing a superfamily 2 helicase instead of a superfamily 3 helicase as is typical of this vast virus group (23). Both potyviral and hypoviral proteases are involved in suppression of the host RNA interference defense (55, 56), in agreement with the common ancestry of these virus families.
Currently, the family Hypoviridae includes only four recognized viruses, isolated from the same chestnut blight fungus (26), whereas Potyviridae is the largest family of plant viruses, infecting a broad variety of flowering plants (50). Given the likely common origin but dramatically different ecologies of hypoviruses and potyviruses, it seems most plausible that the ancestor of the hypoviruses was a potyvirus that crossed the species barrier to a plant-pathogenic fungus and adopted the capsidless persistent infection style common among fungal viruses.
The family Endornaviridae (Endorna, from endo, within, and RNA) includes capsidless, persistent, nontransmissible viruses infecting plants, fungi, and oomycetes (57). The polyprotein-encoding RNA genomes of endornaviruses range from 14 to over 17 kb (Fig. 2) (58). Phylogenetic analysis of the RdRps confidently demonstrates the evolutionary affinity of endornaviruses with the alphavirus-like superfamily of positive-strand RNA viruses (Fig. 3) (59, 60). Interestingly, some endornaviruses also encode a superfamily 1 helicase typical of the alphavirus-like superfamily, whereas others encode a superfamily 2 helicase related to the helicases present in flaviviruses, potyviruses, and hypoviruses (36, 58). One of the fungal endornaviruses even harbors a tandem of superfamily 1 and 2 helicases, an unprecedented case among RNA viruses (Fig. 2) (61). Other domains identifiable in the endornavirus polyproteins show patchy distributions (58). The RNA-capping methyltransferase domain conserved in the alphavirus-like superfamily was detected in only some endornaviruses. Intriguingly, many endornaviruses also encode one or two glycosyltransferase domains, whose function(s) in virus reproduction remains unknown (58, 62).
The phylogeny of endornavirus RdRps is largely incongruent with their host ranges, showing clustering of some plant endornaviruses with viruses infecting fungi rather than other plants (58). This phylogenetic pattern implies horizontal virus transfer between plants and fungi as an important route of endornavirus evolution. It seems likely that given the phylogenetic affinity between the RdRps, methyltransferases, and RNA helicases of the endornaviruses and alphavirus-like viruses, the ancestral endornavirus originated from an alphavirus-like virus via capsid loss. The mosaic distribution of the RNA helicase domains among endornaviruses suggests a lineage-specific loss and nonorthologous displacement of the helicase gene via recombination with coinfecting viral genomes.
Several diverse fungal viruses that belong to the alphavirus-like superfamily appear to have evolved as a result of horizontal transfer of viruses from plants to plant-pathogenic fungi (46, 63). One of these is the capsidless Sclerodarnavirus, which shows phylogenetic affinity to Alphaflexiviridae, a family of filamentous plant viruses (64). The Botrexvirus genus within the same family includes a single fungal virus that possesses a filamentous capsid but does not encode a movement protein typical for plant viruses (65). The single known representative of the family Gammaflexiviridae, Botrytis virus F, possesses a similar genome organization related to plant flexiviruses (66). These few known filamentous fungal viruses most likely are evolutionary intermediates between full-fledged plant viruses and the “nude” fungal viruses that have fully resigned to nontransmissible persistent infections. An additional recent example of such viruses is the capsidless fungal virus SsRV-L, which encodes methyltransferase, RNA helicase, and RdRp domains that are most closely related to the respective domains of the rubiviruses (genus Rubivirus, family Togaviridae) and the family Hepeviridae within the alphavirus-like superfamily (Fig. 3) (60).
Another group of capsidless elements consists of dsRNAs isolated from the green alga Bryopsis. One of these elements, BDRM, is ∼4.5 kb long and replicates exclusively in the algal mitochondria (67). Another element is even smaller (∼2 kb) and localizes to Bryopsis chloroplasts (Fig. 2) (68). Both these RNA elements encode RdRps that are closely related to those of Partitiviridae, a family of dsRNA viruses that persistently infect plants, fungi, and some protists and are incapable of extracellular transmission (Fig. 3) (69). Because of the evolutionary affinity of their RdRps, partitiviruses have been included in the expanded picornavirus-like superfamily, although the virions of most partitiviruses contain multiple segments of dsRNA (23). Conceivably, the capsidless elements from Bryopsis evolved from partitiviruses via capsid loss and switching to exclusive reproduction within organelles.
The floating genus Umbravirus (Umbra, shadow in Latin) consists of plant RNA viruses whose lifestyle is distinct from that of the bona fide capsidless viruses discussed above (70). Although umbravirus genomes do not encode capsid proteins (Fig. 2), they borrow a capsid from helper viruses of the family Luteoviridae. This “cuckoo” strategy provides for plant-to-plant transmission by the insect vectors of luteoviruses, i.e., aphids. The evolutionary provenance of the umbravirus RdRp confidently links this genus to the family Tombusviridae, within the flavivirus-like superfamily of positive-strand RNA viruses (Fig. 3). Therefore, the parsimonious scenario for the origin of umbraviruses derives their ancestor from a tombusvirus that lost the capsid protein gene and switched from the conventional viral lifestyle to borrowing a helper virus capsid. Since umbraviruses retain the ability to spread between hosts, unlike true naked viruses, they could be considered an intermediate between bona fide viruses and capsidless persistent elements.
The very existence of capsidless viruses challenges the Baltimore classification (Fig. 1). Among the RNA viruses with no DNA phase, three classes are distinguished under that scheme: positive-strand, negative-strand, and double-stranded RNA viruses. However, all RNA viruses produce each of these three types of RNA during their intracellular replication, so the classification is based on the RNA form that is incorporated into the virions, a criterion that becomes moot in the case of capsidless elements. Not surprisingly, there is certain confusion about assigning these elements to the Baltimore classes. The Narnaviridae are currently classified as positive-strand RNA viruses, whereas the Endornaviridae are classified as dsRNA viruses (71). The Hypoviridae were initially considered dsRNA viruses but are currently classified as positive-strand RNA viruses. In large part, this confusion stems from experimental isolation of viral dsRNA from infected cells. However, because dsRNA can also be isolated from cells infected with many capsid-possessing RNA viruses and because the genes of the Narnaviridae, Hypoviridae, and Endornaviridae show clear phylogenetic affinities for counterparts among positive-strand RNA viruses, it appears that all three families should be classified as positive-strand RNA viruses. Furthermore, the positive-strand RNA logically should be considered the primary genomic form of capsidless viruses, because this is the only type of viral RNA that can initiate the reproduction cycle by translation into viral proteins, in particular the RdRp. Indeed, so far, capsidless derivatives have not been identified for negative-strand RNA viruses and dsRNA viruses other than the Partitiviridae. It appears likely that the intrinsically virion-confined lifestyle of these two classes of RNA viruses that incorporate RdRp into their virions would preclude the transition to a capsidless reproduction cycle.
Phylogenomics of naked RNA elements provides further insight into the routes of virus evolution. The previously developed general concept of the origin of eukaryotic viruses is that their entire diversity evolved via mixing and matching genes derived from bacteriophages, viruses of archaea, endosymbiotic bacteria, and the emerging eukaryotes (22, 72). The common origin of the RdRps of fungal narnaviruses, plant ourmiaviruses, and positive-strand RNA bacteriophages of the Leviviridae family is a clear case in point for this scenario (45), especially given that the majority of currently known narnaviruses reproduce in mitochondria. This confinement to an endosymbiotic organelle appears to be a legacy of the distant bacterial past of these viruses that is preserved in their lifestyle. The RNA phages and the Narnaviridae and Narnavirus appear to represent an ancient lineage of positive-strand RNA viruses that crossed the boundary between the bacterial and eukaryotic domains of cellular life (Fig. 4). While the Narnaviridae failed to become bona fide infectious viruses in eukaryotes, the Narnavirus have reached this status via the acquisition of capsid protein and movement protein genes from a plant virus. The origin of ourmiaviruses via reassortment of disparate genetic elements is a striking illustration of genomic mixing and matching that appears to be the major trend in the evolution of viruses (45).
In contrast to the apparently ancient origin of the Narnaviridae from RNA bacteriophages, the likely scenario for the origin of the Hypoviridae and Endornaviridae involves more recent evolution from plant-infecting potyvirus-like and alphavirus-like ancestors, respectively. The ancestral viruses probably were transferred from plants to plant-pathogenic fungi as a result of the intimate host-parasite association. This transfer was followed by capsid loss and genome rearrangement favoring the intracellular lifestyle of fungal viruses. A similar evolutionary scenario involving horizontal transfer of viruses can be proposed for several other, flexivirus-like fungal viruses, both those that have lost the capsid and those that still possess it.
Another way in which capsidless RNA viruses illuminate the intricate evolutionary relationships between viruses and cells is the bidirectional virus-cell gene flow. Given that glycosyltransferases are common in prokaryotes and eukaryotes but, among RNA viruses, are encoded only by some representatives of the Hypoviridae and Endornaviridae, horizontal gene transfer from cells to viruses appears to be a certainty in this case. The opposite direction of gene transfer, from virus to host, is equally obvious for Mitovirus-like RdRp genes that are present in mitochondrial genomes of some plants (44, 73). These RdRp genes could have been acquired by plant mitochondrial genomes from either extinct or as yet unidentified plant mitoviruses or from fungal mitoviruses.
In summary, the phylogenetic affinity of the RdRps of each known group of capsidless positive-strand RNA viruses with distinct groups of bona fide viruses of this class leaves no doubt that the capsidless elements evolved from viruses on multiple, independent occasions. We are currently aware of a single case of apparent evolution in the opposite direction, namely, the origin of ourmiaviruses from narnaviruses as a result of acquisition of the genes for the capsid protein and the movement protein from tombusviruses.
Retroelements and Retroviruses: Viruses as Derived Forms
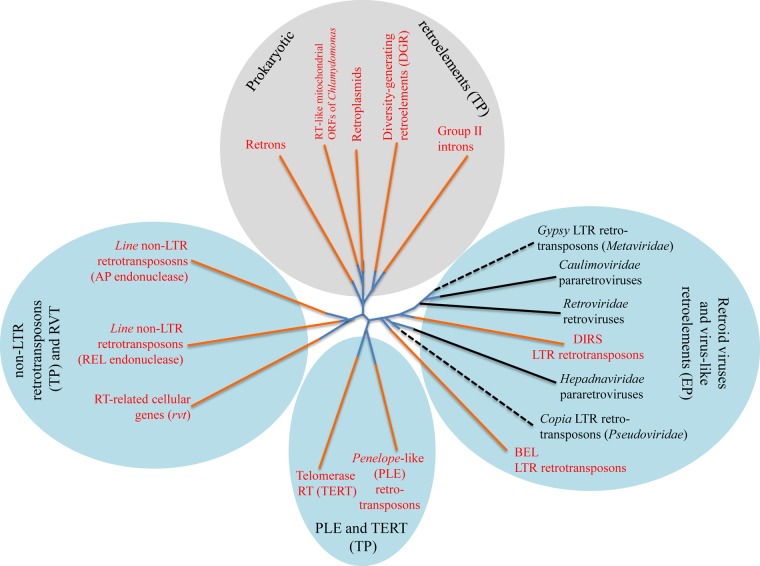
The vast class of retroelements is united by a single conserved gene, the RT gene, which also defines the key feature of their reproduction cycle, reverse transcription (74). In many retroelements, the RT gene is the only gene which makes it difficult to distinguish between such minimal retroelements and stand-alone RT genes on the basis of genome comparison (25). Phylogenetic analysis divides the RTs into four major branches that can roughly be described as follows: (i) retroelements from prokaryotes, (ii) LINE (long interspersed nuclear element)-like elements, (iii) Penelope-like elements (PLE), and (iv) reverse-transcribing viruses and related retrotransposons that contain long terminal repeats (LTRs) (75) (Fig. 5). Historically, all retroelements, with the exception of reverse-transcribing viruses and their relatives, are often called non-LTR retrotransposons. However, this negative definition lumps together highly diverse elements. A more rational classification was proposed recently whereby the retroelements are divided into extrachromosomally primed (EP) ones (the LTR retrotransposons) and target-primed (TP) ones (most, but not all, of the non-LTR retrotransposons), based on the differences in replication and integration mechanisms (see below) (76).
FIG 5.
Schematic phylogeny of the RTs of retroelements and the derivative retroviruses. Four major groups of prokaryotic retroelements (gray oval), as well as eukaryotic retroelements and related viruses (blue ovals), are shown. Orange branches represent capsidless retroelements, whereas black branches represent retroviruses, pararetroviruses, and virus-like noninfectious retrotransposons (Metaviridae and Pseudoviridae; dashed black lines). The two large categories of the retroelements are the extrachromosomally primed ones (EP or LTR) and target-primed ones (TP or non-LTR). (Adapted from reference 75 with permission.)
As with almost all phylogenetic trees of highly diverse, ancient protein families, the deepest branchings in the RT tree are not particularly reliable, and the four main branches effectively form a star topology (Fig. 5). It is difficult to ascertain the relative contributions to this characteristic pattern of the loss of information over the course of evolution, resulting in the failure of the evolution models underlying phylogenetic methods, and actual explosive, Big Bang-like evolution (compressed cladogenesis) associated with such major radiations (23, 77–80). Regardless, it is impossible to rely fully on the RT tree to reconstruct the evolution of retroelements. For example, it is impossible to rule out that all eukaryotic retroelements evolved from a single prokaryotic group.
The archaeal and bacterial retroelements (Fig. 6A) that comprise one of the four major subtrees in the RT tree (Fig. 5) include three (relatively) well-characterized classes, namely, group II introns, retrons, and diversity-generating retroelements (DGRs), and another tentatively characterized group, the abortive-infection-associated RTs (81, 82). The RTs of these three biologically distinct groups form well-supported branches in the prokaryotic part of the RT tree (Fig. 5). The fourth group in this subtree consists of the RTs of the so-called retroplasmids that replicate in fungal mitochondria and that, given the endosymbiotic origin of the mitochondria, are likely to be of bacterial origin (Fig. 5) (83, 84), analogously to the narnaviruses (see above). In addition, analysis of archaeal and bacterial genomes revealed many RTs of unclear provenance that are likely to constitute or be derived from uncharacterized retroelements (85). Notably, the sequence variability of the prokaryotic RTs is extremely high, with only the essential motifs of the RT domain conserved throughout, by far exceeding the variance among the eukaryotic retroelements (85). This greater sequence diversity of the RTs in prokaryotes than those in eukaryotes, despite their relatively low abundance, seems to be compatible with the origin of all eukaryotic retroelements from a distinct prokaryotic group (see below).
FIG 6.
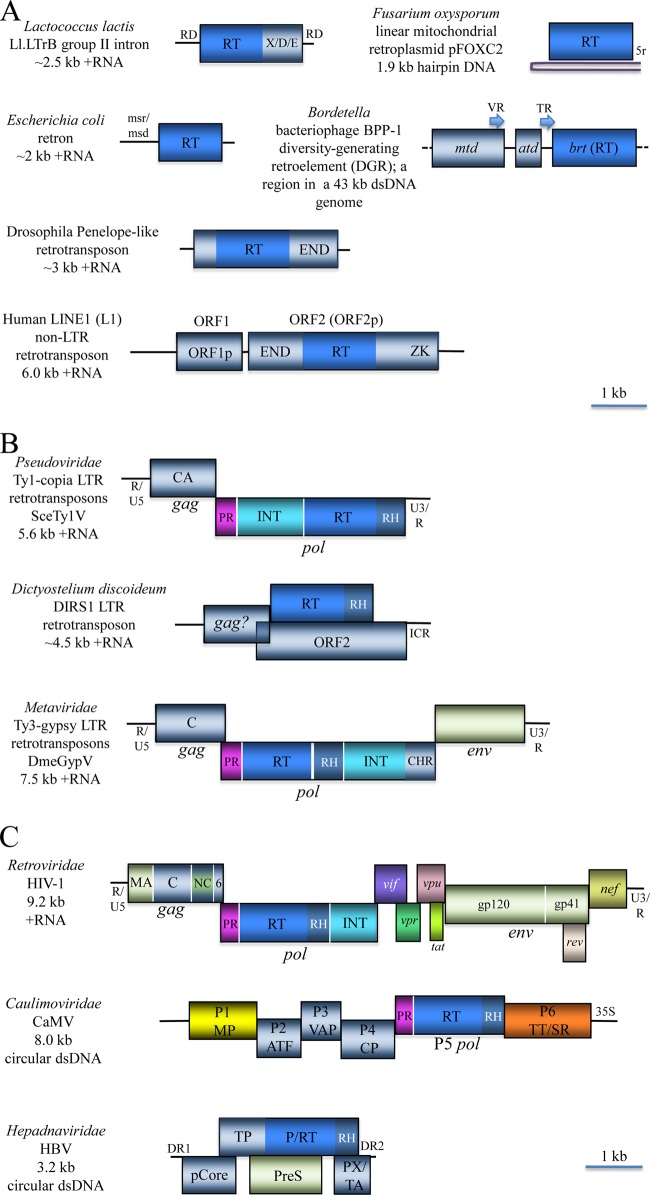
Representative genome architectures of retroelements and the derivative retroviruses. (A) Prokaryotic and eukaryotic capsidless retroelements. Group II introns are scattered in genomes of diverse bacteria and some archaea, as well as mitochondrial and chloroplast genomes of many eukaryotes. Retrons are typical of bacteria, whereas Penelope-like and non-LTR retrotransposons are widespread in diverse eukaryotes. The diversity-generating retroelements (DGR) are present in a narrow range of tailed DNA bacteriophages and in some bacteria. Linear mitochondrial retroplasmids are present in some fungi. RD, RNA domains involved in the splicing of intron RNA; X/D/E, maturase, DNA binding, and endonuclease domains, respectively, of the intron-encoded protein; msr/msd, regions encoding RNA and DNA components, respectively, of the satellite msDNA; 5r, a telomere-like iteration of a 5-nucleotide sequence; VR, variable repeat; TR, template repeat; mtd, major tropism determinant; atd, accessory tropism determinant; brt, bacteriophage reverse transcriptase; LINE, long interspersed nucleotide elements; ORF1p and ORF2p, ORF1 and 2 proteins; END, endonuclease; ZK, zinc knuckle. (B) The LTR (long terminal repeat) retrotransposons are ubiquitous in eukaryotes. Because many of them form primarily noninfectious, virion-like particles encoded by the gag (group-specific antigen) and env (envelope) ORFs, two classes of these retrotransposons are recognized as viral families Metaviridae and Pseudoviridae. The pol (polymerase) ORF encodes a complete or partial complement of the aspartate protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (INT) domains and, in Metaviridae, a chromodomain (CHR). The sites of Pol processing by PR are shown as vertical white lines. ICR, internal complementarity region. Viral name acronyms: DmeGypV, Drosophila melanogaster gypsy virus; SceTy1V, Saccharomyces cerevisiae Ty1 virus. (C) Reverse-transcribing (retroid) viruses. The genomes are shown as RNA or primarily double-stranded DNA that is circular but rendered linear for the sake of comparison. In HIV-1, both gag and pol are processed (vertical white lines) by PR, whereas env is processed by the host proteases. MA, matrix protein; C, capsid protein; NC, nucleocapsid; 6, 6-kDa protein; vif, vpr, vpu, tat, rev, and nef, regulatory proteins encoded by spliced mRNAs (only the main parts of the coding regions are shown); gp120 and gp41, the 120- (surface) and 41-kDa (transmembrane) glycoproteins; ATF, aphid transmission factor; VAP, virion-associated protein; CP, capsid protein; TT/SR, translation trans-activator/suppressor of RNA interference; 35S, 35S RNA polymerase Pol II promoter; pCore, capsid (core) protein; TP, terminal protein; P, polymerase; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; DR1 and DR2, direct repeat sequences; HIV-1, Human immunodeficiency virus 1; CaMV, Cauliflower mosaic virus; HBV, Hepatitis B virus.
In stark contrast to the prokaryotic retroelements, which are sparsely represented among bacteria and archaea and do not reach high copy numbers, except in some organellar genomes of plants and fungi, diverse eukaryotic genomes are replete with integrated retroelements of different varieties. By conservative estimates, retroelement-derived sequences account for over 60% of mammalian genomes (86, 87) and up to 90% of some plant genomes (e.g., maize) (88, 89). Although usually not reaching such an extravagant excess, retroelements are also abundant in genomes of diverse unicellular eukaryotes (90). Similar to the prokaryotic retroelements, eukaryotic retrotransposons and reverse-transcribing viruses share only a single gene, the RT gene. However, compared to their counterparts in prokaryotes, the RT sequences of eukaryotic retroelements are highly conserved, in sharp contrast with the enormous diversity in genome organizations and reproduction strategies (Fig. 6A).
PLE are the simplest eukaryotic retroelements, typically encoding a single large protein that in the originally discovered group of PLE is a fusion of the RT with a GIY-YIG endonuclease (Fig. 6A) (91). So far, this complete form of PLE has been identified only in animals. A shorter version of PLE that lacks the endonuclease is integrated in subtelomeric regions of chromosomes in a broad variety of eukaryotes (92). In the phylogenetic tree for RT, the PLE confidently cluster with the RT subunit of the telomerase (TERT), a pan-eukaryotic enzyme that is essential for replication of the ends of the linear chromosomes of eukaryotes (75). This relationship implies that the PLE-TERT branch of retroelements predates the last common ancestor of the extant eukaryotes, although complete, endonuclease-encoding PLE, so far detected only in animals, might have evolved later.
The LINE-like elements comprise a group of simple retroelements that typically consist of two genes, one of which encodes an RT-endonuclease fusion protein and the other of which encodes a protein containing the RNA-binding domain that is required for transposition (93, 94). The RTs of the LINEs form two distinct branches in the phylogenetic tree (Fig. 5) that also differ by the nature of the endonuclease encoded in the element. The “classic” LINEs, including all mammalian forms, encode an apurinic/apyrimidinic (AP) endonuclease that also possesses RNase H (RH) activity (95, 96) (Fig. 6A). A subset of LINEs from diverse eukaryotes, however, encode a bona fide RNase H enzyme. Although some phylogenetic analyses suggest that RNase H is a late acquisition in the history of TP retroelements (97), it cannot be ruled out that this is the ancestral architecture among LINEs. Overall, the LINEs could be the most abundant family of retroelements on earth, as they reach extremely high copy numbers in vertebrate genomes.
In the phylogenetic tree for RT (Fig. 5), the LINEs cluster (albeit not with full confidence) with a recently discovered distinct group of elements, denoted the RVT group, that contain no identifiable domains other than the RT and are not currently known to behave as mobile elements but are present in a single copy in the genomes of diverse eukaryotes, suggestive of some still unknown function(s) in eukaryotic cells (75). Members of the RVT group have also been identified in several bacterial genomes, but the evolutionary scenario here is unclear: given the much wider spread of the RVT genes in eukaryotes than in prokaryotes, horizontal gene transfer from eukaryotes to bacteria (a rare event, in general) has been suggested (75).
Among the RT elements, bona fide viruses, with genomes encased in virus particles and typical, productive infection cycles, are a minority. Importantly, capsidless retroelements are found in all major divisions of cellular organisms and, by inference, should be considered ancestral to this entire class of genetic elements. In contrast, reverse-transcribing viruses appear to be derived forms that are represented only in animals, fungi, plants, and algae and apparently evolved at an early stage in the evolution of eukaryotes.
Numerous retroviruses have been isolated from all major groups of vertebrates but not, so far, from any nonvertebrate hosts (98, 99). The reproduction strategy of the retroviruses (family Retroviridae) partly resembles that of RNA viruses, combining aspects analogous to both positive-strand RNA viruses and negative-strand RNA viruses (Fig. 1). The positive RNA strand (typically, two copies of the genome) is packed into virions that, unlike the case for the positive-strand RNA viruses but similar to that for negative-strand RNA viruses, incorporate the product of the virus pol gene, i.e. the RT fused to RNase H and integrase (INT) domains (Fig. 6B and C). The RT catalyzes the synthesis of the negative DNA strand and then, after hydrolysis of the parental RNA catalyzed by the viral RNase H, the positive DNA strand, yielding a dsDNA provirus that integrates into the host genome. The progeny virus RNA is then transcribed by the host RNA polymerase, which employs the viral long terminal repeat as the promoter. The progeny RNA is packaged within new virions, along with a host tRNA that is used as the primer for provirus DNA synthesis. Thus, in terms of the replication-expression strategy, retroviruses can be viewed as RNA viruses that have adopted a DNA intermediate and, accordingly, a second mode of replication within the host genome. In addition to the typical infectious retroviruses, vertebrate genomes carry numerous “endogenous” retroviruses that are largely transmitted vertically and are often inactivated by mutations but, until that happens, maintain the potential to become activated and yield infectious virus (100, 101).
The other two families of reverse-transcribing viruses, the Hepadnaviridae, infecting animals, and the Caulimoviridae, infecting plants, have ventured farther into the DNA world: these viruses package the DNA form of the genome (or a DNA-RNA hybrid, in the case of hepadnaviruses) into the virions but retain the reverse transcription stage in the reproduction cycle (Fig. 1 and 6C) (102–104). In contrast to the case with retroviruses, integration into the host genome is not an essential stage in the reproduction cycle of these viruses, although integration is common among caulimoviruses, many of which can persist as endogenous viruses (105, 106).
The remaining two families of reverse-transcribing viruses, Metaviridae and Pseudoviridae, include RT-encoding elements that are traditionally not even considered viruses but rather retrotransposons, because they are not known to infect new cells. Nevertheless, these elements, such as Gypsy/Ty3-like elements (Metaviridae) or Copia/Ty1-like elements (Pseudoviridae), which are widely represented in invertebrates, fungi, and some protists, encode virion proteins (Fig. 6B) and form particles, thus meeting the definition of a virus (107–109).
Among the retroelements, the reverse-transcribing viruses possess the most complex genomes (Fig. 6C). All retroviruses share three major genes that are traditionally denoted pol, gag, and env, and many also encompass additional, variable genes. As emphasized above, the RT gene is the only gene that is conserved in all retroelements. In retroviruses, the RT is a domain of the Pol polyprotein (Fig. 6B and C). In the entire viral branch of retroelements, the conserved module consists of the RT together with another domain, the RNase H domain, that is essential for the removal of the RNA strand during the synthesis of the DNA form of the viral genome. Two other domains, the aspartic protease (PR) and integrase (INT) domains, are found in only a subset of Pol polyproteins. However, superposition of the domain architectures of the Pol polyproteins (Fig. 6B and C) over the phylogenetic tree for the RTs (Fig. 5) implies that the common ancestor of the reverse-transcribing viruses encoded the complex form of Pol, most likely one with the PR-RT-RH-INT arrangement that is shared between retroviruses and metaviruses. The phylogenies of the RT, RH, and INT domains of reverse-transcribing viruses appear to be concordant and clearly cluster metaviruses with retroviruses, to the exclusion of pseudoviruses, in agreement with the RT phylogeny in Fig. 5 and in support of the inference of a complex ancestral form (110). Under this scenario of evolution, caulimoviruses have lost the integrase domain, whereas hepadnaviruses have lost both the integrase and the protease but acquired the terminal protein domain that is involved in the initiation of DNA synthesis. A more complete phylogenetic analysis of RH that also involved TP (non-LTR) retroelements of the LINE branch, as well as bacterial and eukaryotic RNH I, implies that the TP retroelements in eukaryotes are older than the EP elements (111).
The INT domain of the EP retroelements (reverse-transcribing viruses) is a member of the DDE (named after the distinct catalytic triad) family of transposases that mediate the transposition of numerous DNA transposons in prokaryotes and eukaryotes (112–114). Therefore, the founder of the EP (LTR) retrotransposon branch most likely evolved through recombination between a TP (non-LTR) retrotransposon and a DNA transposon (115, 116). The aspartic protease of the EP retroelements is homologous to the pan-eukaryotic protein DDI1, an essential, ubiquitin-dependent regulator of the cell cycle, and DDI1, in turn, appears to have been derived from a distinct group of bacterial aspartyl proteases (117, 118). Thus, strikingly, the ancestral Pol polyprotein of the EP retroelements seems to have been assembled from four distinct components, only one of which, the RT, is derived from a preexisting retroelement.
Apart from the case of the Pol polyprotein, the relationships between genes in different groups of reverse-transcribing viruses are convoluted (Fig. 6B and C). The capsid protein domain of the Gag polyprotein is conserved between retroviruses and the Ty3/Gypsy metaviruses. The conserved region of the capsid protein consists of a distinct C2HC Zn-knuckle module that, at least in retroviruses, is involved in RNA and DNA binding. In addition, the capsid proteins contain a conserved α-helical domain, known as SCAN, that mediates protein dimerization (119, 120). Phylogenetic analysis of the conserved portion of Gag suggests that three classes of retroviruses evolved from three distinct lineages of metaviruses, as captured in the “three kings” hypothesis (121). However, it is unclear whether the Gag-like protein of Copia/Ty1 (pseudoviruses) is homologous to those of retroviruses and metaviruses, nor is it clear where the ultimate origin of this protein outside the retroelements is located; homologs of the Ty3/Gypsy Gag proteins have been identified in eukaryotes but appear to have evolved by “domestication” of the respective viral genes (122). A common origin has been claimed for the env genes of retroviruses and Ty3/Gypsy (123), but this relationship is based on extremely weak sequence similarity and is difficult to ascertain.
Caulimoviruses and, especially, hepadnaviruses are highly derived forms that apparently have lost and/or displaced several genes of the ancestral reverse-transcribing virus, with the exception of the RT and RH genes, and the PR gene in the case of caulimoviruses (Fig. 6C). In addition, the capsid proteins of caulimoviruses share the distinct C2HC Zn-knuckle module with the CPs of retroviruses and metaviruses (124). Thus, at least part of the ancestral capsid protein of reverse-transcribing viruses survives in caulimoviruses; whether or not the remaining portions of the capsid proteins are homologous remains unclear: both divergence beyond straightforward recognition and displacement by an unrelated domain(s) cannot be ruled out. In contrast, the core protein of hepadnaviruses shows no similarity to capsid proteins of retroviruses or caulimoviruses and appears to be a displacement of uncertain provenance.
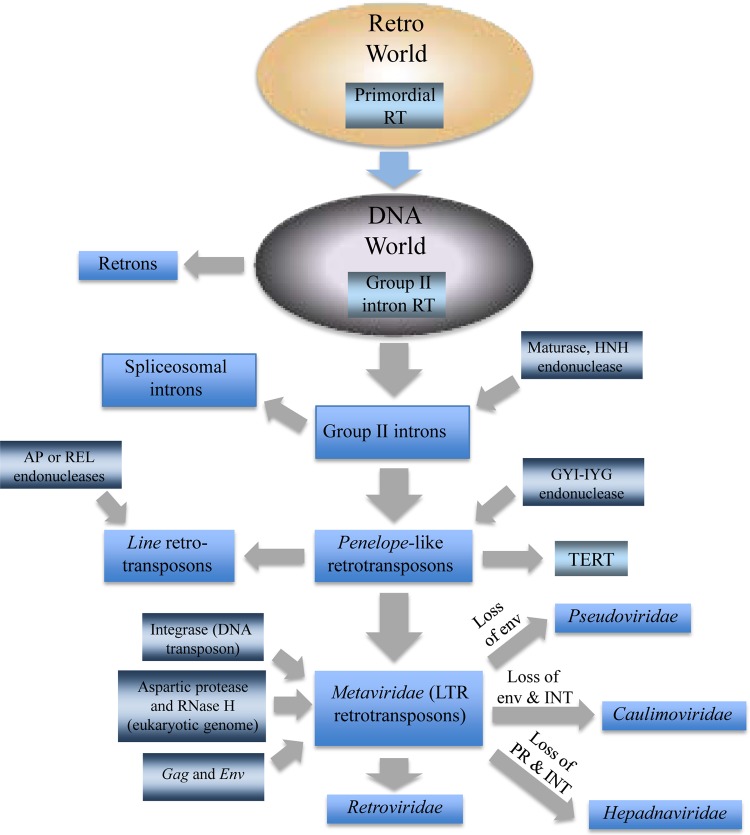
Most likely, retroelements have been an integral part of biological systems since the stage of the primordial replicators, when they actually gave rise to the first DNA genomes (125). However, in prokaryotes, these elements maintain a relatively low profile and never attain complex genomic architectures (Fig. 6A). In eukaryotes, the fortunes of retroelements completely changed: they proliferated dramatically and have become a key factor of genome evolution. It appears likely that group II introns, by far the most common retroelements in prokaryotes and the only class of prokaryotic retroelements with demonstrated horizontal mobility, are the ancestors of all eukaryotic retroelements (Fig. 7). Conceivably, at an early stage of eukaryotic evolution, recombination between group II introns and genes (including transposons) encoding unrelated nucleases led to the emergence of four major classes of eukaryotic retroelements, namely, PLE, LINE-AP, LINE-REL, and reverse-transcribing viruses (Fig. 7). The evolution of the retroelement-derived viruses involved additional recombination events that resulted in the acquisition of several proteins and domains, most importantly, the capsid protein, whose origin remains uncertain. The wide spread of each of the major groups of retroelements in eukaryotes implies that the principal events in the evolution of retroelements occurred at an early stage of eukaryotic evolution, before the radiation of the eukaryotic supergroups. The evolutionary connection between PLE and the RT subunit of TERT that is conserved in all eukaryotes provides additional evidence in favor of this scenario. Given the star topology of the RT tree (Fig. 5), the exact sequence of these events is difficult or outright impossible to infer. It appears likely that similar to the evolution of the supergroups of eukaryotes themselves and of the major groups of eukaryotic viruses (22, 23, 52, 126), different classes of eukaryotic retroelements evolved rapidly in a Big Bang-like event(s), perhaps from different group II introns (although, taking into account the inherent problems of deep phylogenies, it would be incorrect to claim that the star topology of the tree “supports” the Big Bang scenario).
FIG 7.
Evolutionary scenario for the evolution of retroelements and the origin of retroviruses.
Among all the Baltimore classes of viruses and virus-like agents, the retroelements are the “most nonviral” ones, whereby the bulk of the diversity and the ancestral state are represented by nonencapsidating, autonomous genetic elements, with bona fide viruses representing only one derived branch, even if a diversified and successful one. Conceivably, the dominance of capsidless elements in this class of selfish replicons stems from the early evolution of the coupling of reverse transcription with integration into host genomes. Such coupling provides the selfish elements of this class the opportunity to propagate by occupying new sites on the host chromosomes and then reproducing with the host. This facile reproduction strategy, which is maintained even by some of the retroviruses, apparently weakens the evolutionary pressure for the acquisition of genes for capsid proteins and the ensuing transition to the viral lifestyle.
Rolling Circle Replicons: Multiple Transitions from Viruses to Plasmids and Back?
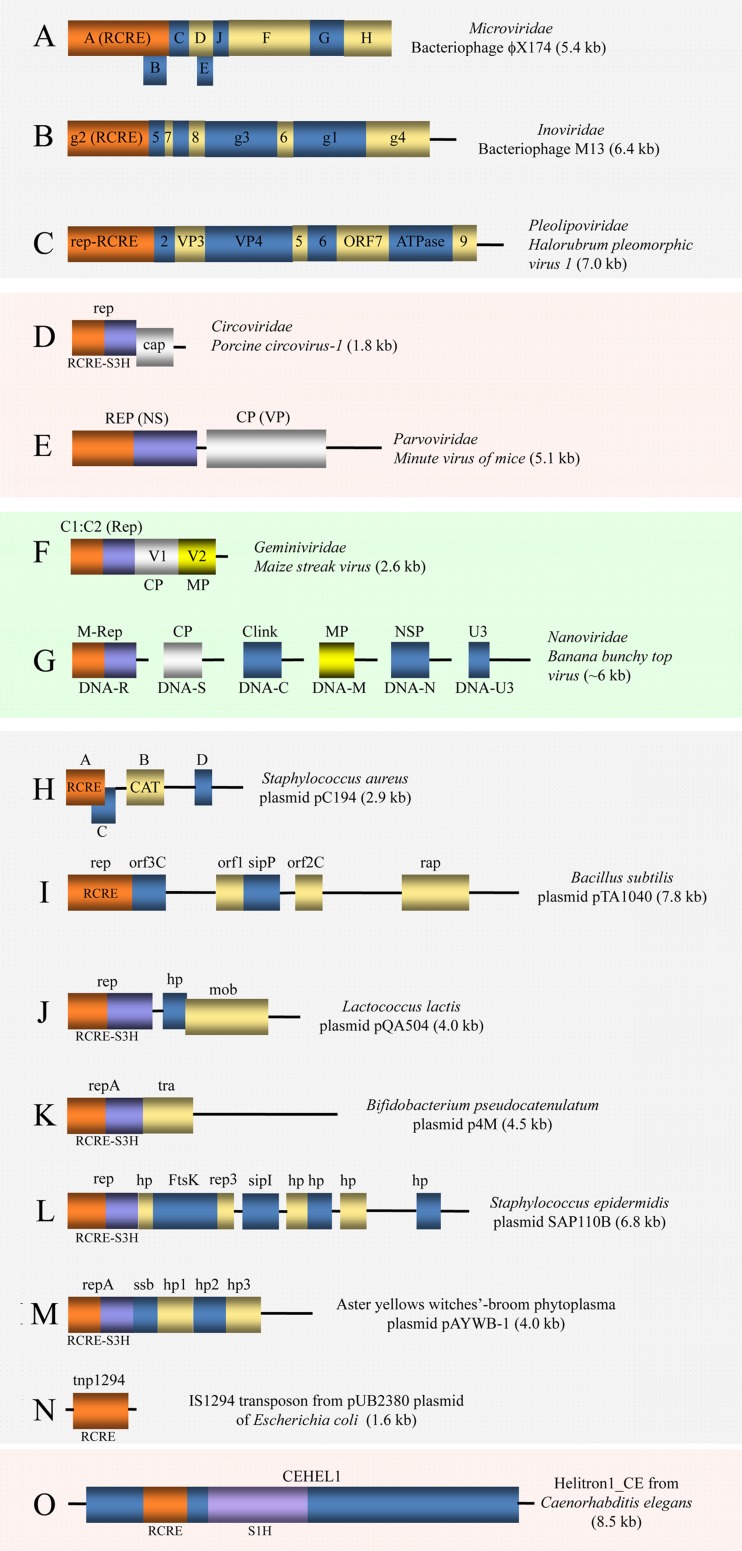
The rolling circle replication (RCR) mechanism (127) unifies several groups of ssDNA viruses and plasmids (Fig. 8). The viruses that replicate via RCR include the families Microviridae (isometric ssDNA phages), Inoviridae (filamentous ssDNA phages), Pleolipoviridae (archaeal ssDNA viruses), Parvoviridae, Circoviridae, Nanoviridae, Geminiviridae (Fig. 8A to G), and several unclassified viruses. The viruses of the latter 4 families infect diverse eukaryotes, including plants, animals, fungi, and protists. The families Polyomaviridae and Papillomaviridae (formerly a single family, the Papovaviridae) include small dsDNA viruses of animals that are related to RCR replicons, notwithstanding their having a different replication mechanism.
FIG 8.
Genome architectures of single-stranded DNA viruses and homologous plasmids. All genomes are shown as linear diagrams, although most of them are circular ssDNAs, except for those of Parvoviridae, which are linear ssDNAs with terminal repeats. The colors of RCRE and S3H domains reflect homology; other colors were chosen arbitrarily. The background color code is light gray for the viruses and plasmids of prokaryotes, light pink for the viruses of animals, and light green for the viruses of plants. (A) The functions of the encoded proteins are as follows: A, replication initiation; B, internal scaffolding protein; C, ssDNA synthesis; D, external scaffolding protein; E, cell lysis; F, major capsid protein (JRC); G, major spike protein; H, minor spike protein; J, DNA binding. (B) The functions of the encoded proteins are as follows: g2, replication initiation; g5, genome replication; g7, g9, g8, g3, and g6, capsid proteins; g1 and g4, virion morphogenesis. (C) VP3 and VP4, viral proteins present in the lipoprotein coat of this capsidless virus of a haloarchaeon. (D) rep, replication protein, a fusion of the RCRE and S3H domains; cap, capsid protein, JRC fold. (E) REP (NS), replication-associated, nonstructural protein; CP (VP), capsid or virion protein, JRC fold. (F) C1:C2 (Rep), complementary strand-encoded replication proteins 1 and 2 (RCRE-S3H); V1 (CP) and V2 (MP), virion strand-encoded capsid and movement proteins, respectively. (G) M-Rep, master replication initiator protein (RCRE-S3H); CP, capsid protein; Clink, cell cycle regulator protein; MP, movement protein; NSP, nonstructural protein; U3, DNA U3-encoded protein. (H) CAT, chloramphenicol acetyltransferase (antibiotic resistance). (I) sipP, signal peptidase; rap, response regulator aspartyl phosphatase. (J) hp, hypothetical protein; mob, mobilization relaxase. (K) tra, protein involved in plasmid mobilization. (L) FtsK, ATPase involved in plasmid segregation, homolog of viral packaging ATPases; rep3, uncharacterized protein involved in plasmid replication; sipI, signal peptidase. (M) repA, replication protein A; ssb, ssDNA-binding protein; hp1 to -3, hypothetical proteins 1 to 3. (N) tnp1294, a transposase that possesses an RCRE domain. (O) CEHEL1, large protein encoded by Caenorhabditis elegans Helitron1 and possessing RCRE and superfamily 1 helicase (S1H) domains.
With the exception of the narrowly distributed and poorly characterized families Bidnaviridae and Anelloviridae, all the ssDNA viruses of eukaryotes encode a signature protein domain, the endonuclease involved in the initiation of RCR (RCRE), that in all eukaryotic RCR replicons is fused to a viral hallmark domain, the S3H domain (128–130). In contrast, both bacterial and archaeal ssDNA viruses encode only the RCRE, not the S3H domain. The RCRE-S3H fusion (known as the vRep protein) is the only large, replication-related protein encoded in the genomes of the eukaryotic ssDNA viruses, which additionally encode a single capsid protein and, in some cases, several small accessory proteins.
In addition to the eukaryotic ssDNA viruses, the RCRE-S3H fusion protein is encoded in numerous bacterial plasmids (Fig. 8J to M), many of which appear to be integrated into bacterial genomes (30). Furthermore, some of these plasmids show a phylogenetic affinity with homologs from a particular group of viruses (30). The presence of this signature Rep protein architecture in both plasmids and ssDNA viruses infecting eukaryotes but not prokaryotes immediately suggests that the two classes of RCR replicons share an evolutionary relationship.
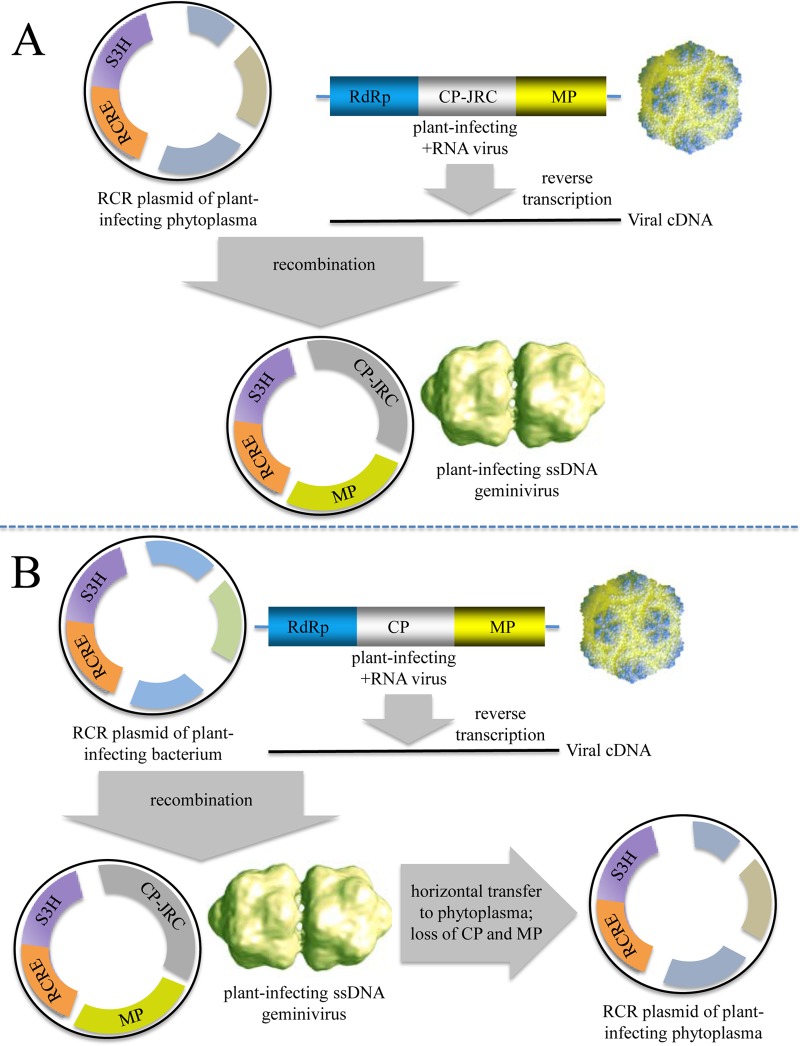
The most conspicuous case is that of the Geminiviridae, which appear to have evolved by recombination between a plasmid and a plant positive-strand RNA virus (Fig. 9). This possibility was first proposed on the basis of limited sequence similarity of the vRep protein to the Rep proteins of the pLS1 family of plasmids from Gram-positive bacteria (131). Subsequently, plasmids encoding Rep proteins with a much greater similarity to geminivirus homologs were reported for Phytoplasma organisms, the mollicute parasites of plants (132). Phylogenetic analysis of the vRep proteins confirmed that geminiviruses formed a clade with a family of phytoplasmal plasmid Reps. In contrast, the capsid proteins of geminiviruses that adopt the JRC fold are highly similar to the capsid proteins of satellite tobacco necrosis virus and other positive-strand RNA viruses of plants. These findings implied a plasmid-to-virus scenario for the evolution of geminiviruses. This scenario was contested in a more recent analysis because the phytoplasmal plasmids encoding Rep proteins related to those of geminiviruses appear to comprise an isolated group with limited similarity to Reps from other bacterial plasmids, suggesting that these particular plasmids actually evolved from geminiviruses (133). The fact remains, however, that geminivirus genomes consist of two parts of different provenances: the vRep gene, with closest homologs in plasmids, and the JRC gene, with closest homologs among plant RNA viruses. Even if the particular group of phytoplasmal plasmids that was singled out by Krupovic et al. (132) as the likely ancestors of the geminiviruses actually derived from geminiviruses, the ultimate origin of the geminivirus ancestor via recombination between a plasmid and a DNA copy of an RNA virus genome appears most likely (Fig. 9).
FIG 9.
Two alternative scenarios for the evolution of geminiviruses and related plasmids.
The above scenario for the evolution of geminiviruses strikingly parallels the results of recent metagenomic studies that have been highly productive in the identification of many novel ssDNA viruse genomes and have led to a substantial expansion of the family Microviridae (134), as well as to the discovery of numerous viruses that are related to circoviruses and nanoviruses and probably infect unicellular eukaryotes (135, 136). In addition, novel genomic architectures of ssDNA viruses have been discovered. Some of these unexpected genome organizations seem to result from recombinational events whereby a circovirus-like vRep protein combines with a geminivirus-like capsid protein, or even with a capsid protein related to those of positive-strand RNA viruses (in particular, tombusviruses). The latter novel entity, denoted an RNA-DNA hybrid virus (RDHV), probably evolved via a route parallel to that of the evolution of geminiviruses (137). A recent exhaustive analysis of metagenomic sequences led to the discovery of multiple hybrid genomes of putative novel viruses that appear to have evolved via recombination between different groups of positive RNA viruses that provide the CP gene and RCR replicons that are the source of the Rep gene (138).
Comparative analysis of the Rep protein sequences of circoviruses and nanoviruses reveals a complex network of relationships between viruses, plasmids, and transposons (30, 135, 139). A plausible hypothesis has been proposed that recombination events between ssDNA genetic elements and RNA viruses are pervasive in the evolution of ssDNA replicons and underlie the evolutionary success of this class of selfish elements (140).
The genomes of several pleomorphic viruses of Haloarchaea that contain either ssDNA or dsDNA in their virions and encode a Rep protein with an RCRE domain (Fig. 8C) show a close relationship with the genome of the plasmid pHK2 from Haloferax lucentense (16, 17). Actually, the genome of this plasmid encodes homologs of viral proteins present in the lipoprotein membrane that encloses naked viral DNA. Thus, pHK2 appears to be a provirus that exists in an episomal state and probably serves as an intermediate between viruses and integrated proviruses that are abundant in Halobacteria (17, 141). It has been noticed that the pleomorphic viruses resemble lipid vesicles that are secreted by some archaea and, by capturing plasmid DNA, apparently contribute to horizontal gene transfer (30, 142). Thus, these unusual virus forms could be intermediates on the evolutionary path from plasmids to typical viruses.
In addition to plasmids and ssDNA viruses, RCR replicons include prokaryotic and eukaryotic transposons. The bacterial transposons of the IS91-like family encode a single protein that is homologous to RCRE (Fig. 8N) and, in particular, to the Rep proteins of plasmids from Gram-positive bacteria and is involved in transposition via RCR (143, 144). The eukaryotic RCR transposons are known as helitrons because they encode a Rep protein that is a fusion of the RCRE domain with a helicase domain; this helicase domain, however, belongs to superfamily 1 and is unrelated to the S3H domain of viral and plasmid Rep proteins (Fig. 8O) (145–147). The helitrons are present in plants, animals, and diverse protists and reach extremely high copy numbers in some genomes.
Homologs of the Rep proteins, or even entire ssDNA virus genomes, apparently acquired as a result of integration of RCR replicons, have also been detected in the genomes of large DNA viruses (such as canarypoxvirus and Phaeocystis globosa virus), several unicellular eukaryotes, plants, and animals (140, 148). It cannot be ruled out that some of these integrated RCR replicons will evolve to become bona fide transposable elements; indeed, this possibility appears most likely, because some of these “endogenous ssDNA viruses” are associated with transposases (148).
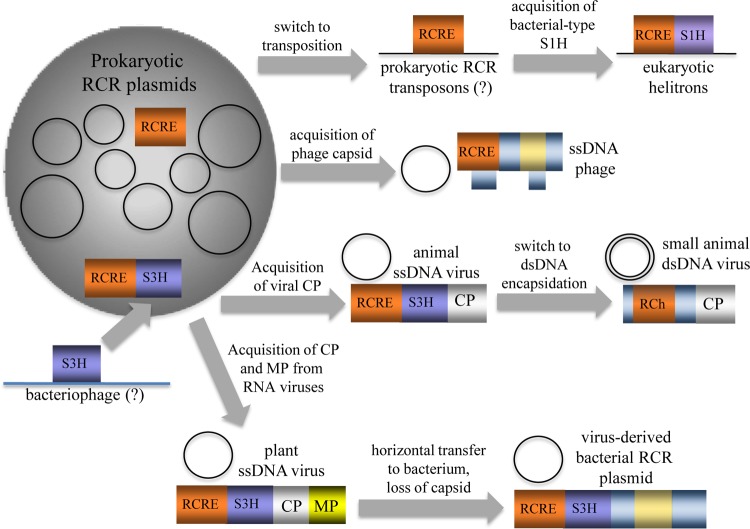
Taken together, the numerous lines of evidence on the evolutionary connections between different classes of RCR replicons suggest polyphyletic origins for ssDNA viruses that encode a Rep-RCRE protein. This scenario envisages a pool of RCR plasmids that independently gave rise to different groups of ssDNA viruses as a result of recombination or genome segment reassortment with various preexisting viruses, including those with RNA genomes (30) (Fig. 10). The reverse transition, from viruses to plasmids, also probably occurred on multiple occasions, as might be the case for geminiviruses and phytoplasmal plasmids. The evolutionary scenario(s) for the RCR transposons is less clear, but a similar orgination from different plasmids appears most plausible.
FIG 10.
General scheme for the origin of ssDNA viruses from capsidless RCR replicons. RCh, RCRE homolog in small dsDNA polyomaviruses.
Notably, hallmark proteins of RCR have also been identified outside the realm of typical RCR replicons. In particular, highly diverged RCREs are encoded in the genomes of the archaeal dsDNA viruses of the family Rudiviridae (149). These viruses possess dsDNA genomes of approximately 35 kb with a covalently closed hairpin at each end, and the RCRE protein has been shown to initiate replication by introducing a nick near the hairpin apex and then to reseal the DNA molecule during concatemer resolution (149). The presence of an RCRE that mediates an RCR-like replication mechanism in these relatively large dsDNA viruses suggests that some of the large viral genomes, particularly those of archaeal viruses, might have evolved from small RCR elements via gene accretion. Similarly, RCREs are encoded in the genomes of some dsDNA bacteriophages, such as the corticovirus PM2 (150), as well as certain members of the order Caudovirales, such as P2-like bacteriophages (130, 151), which, at least under some conditions, replicate via RCR.
dsDNA Viruses: from Viruses to Self-Replicating Transposons and Back
dsDNA viruses, primarily bacteriophages, comprise the majority of virus particles in the biosphere, and conceivably also the bulk of the genomic diversity of viruses (152, 153). Furthermore, this is the only one of the Baltimore classes that includes viruses with large genomes that reach 2 Mb and thus encroach on the characteristic range of the genome sizes of bacteria and archaea (3, 18, 154). The dsDNA viruses show the most spectacular evidence of the capsid structure conservation, in particular in the “ancient lineage” of JRC2b, that encompasses diverse viruses infecting each of the three domains of cellular life (9, 13, 14). The evolutionary relationships and transitions between bona fide viruses and capsidless genetic elements appear to be less conspicuous among dsDNA viruses than they are among retroelements or RCR elements. Nevertheless, substantial evidence of such relationships exists.
The most compelling case in point is the relationship between the large, self-replicating transposons, known as polintons (mavericks), and dsDNA viruses, primarily the virophages. Polintons are scattered across genomes of diverse eukaryotes and reach extremely high abundances in some protists, such as Trichomonas vaginalis. The transposons of this class have long been considered virus-like because of their large size (>20 kb) and the presence of several genes that are common in viruses but not other transposable elements (Fig. 1) (145, 155–157).
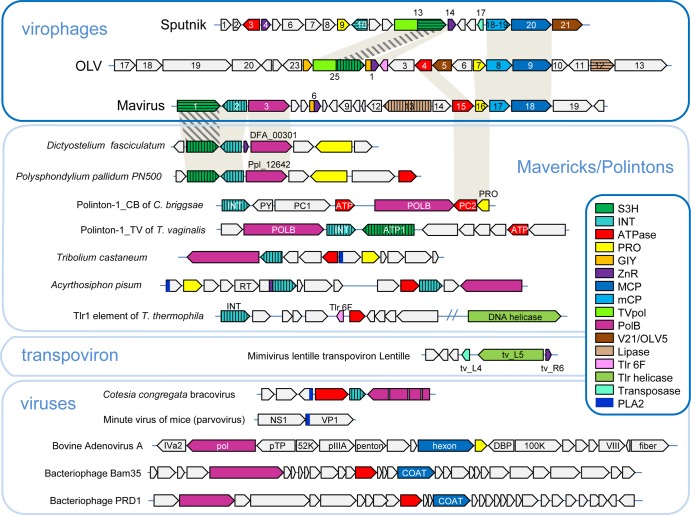
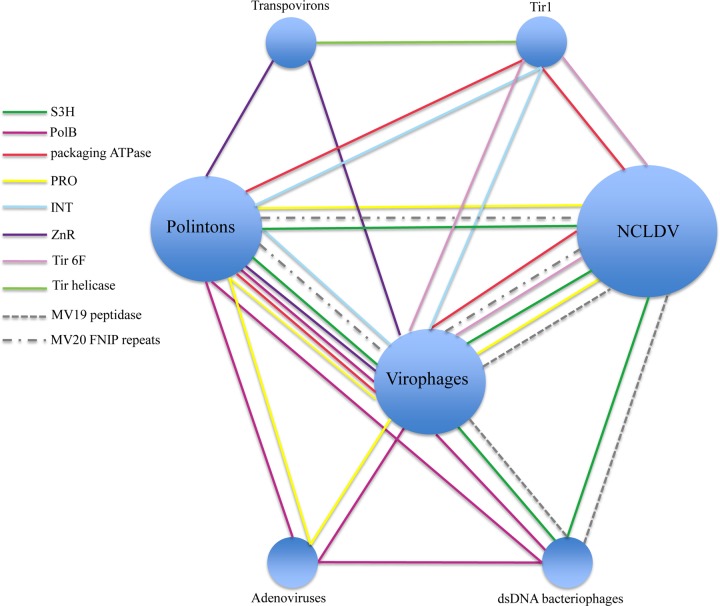
Unexpectedly, multiple connections have been detected between polintons and a recently discovered class of dsDNA viruses, the virophages, which are relatively small viruses with circular dsDNA genomes of 20 to 26 kb that depend on the giant dsDNA viruses of the family Mimiviridae for replication (158–164). The mimiviruses themselves belong to the vast assemblage of nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes that comprise the proposed viral order Megavirales (126, 154, 165, 166). Genome analysis of mavirus, a virophage that parasitizes Cafeteria roenbergensis virus (CroV) infecting a marine flagellate (160), resulted in the unexpected discovery that this virophage shared 5 homologous genes (DNA polymerase PolB, retrovirus-type integrase, packaging ATPase, distinct thiol protease, and S3H genes) with the polintons (Fig. 11). Mavirus shows the closest affinity with the polintons among the currently known viruses, by far, and accordingly, it has been proposed that the polintons evolved from the virophages (160). The other virophages lack PolB and integrase and thus share only three genes with mavirus and the polintons. However, these virophages possess additional genes in common with mavirus, in particular a tandem of genes encoding the major and minor capsid proteins (Fig. 11). Phylogenetic analysis of each of the conserved genes placed mavirus within the polinton group, suggesting that a specific group of polintons actually gave rise to mavirus, possibly via recombination with an ancestral virophage (32).
FIG 11.
Comparison of the genome architectures of virophages and polinton-like transposable elements. Homologous genes are color coded. Different hatching patterns are used to mark nonorthologous primase-helicase, integrase, and lipase genes. Homologous regions are shaded. Reference sequences were extracted from GenBank, using the following accession numbers: Dictyostelium fasciculatum, GI:328871053; Polysphondylium pallidum, GI:281202948; Tribolium castaneum, GI:58197573; Acyrthosiphon pisum, GI:156713484; Mimivirus lentille transpoviron Lentille, GI:374110342; Cotesia congregata bracovirus, GI:326937614; minute virus of mice, GI:9626993; bovine adenovirus A, GI:52801677; bacteriophage Bam35, GI:38640293; and bacteriophage PRD1, GI:159192286. Some sequences were extracted from Repbase (Polinton-1_CB and Polinton-1_TV [156, 202]). PLA2, phospholipase A2 domain of the parvovirus capsid protein. Other color key abbreviations are the same as those used throughout the text. (Adapted from reference 32, published under a Creative Commons license.)
The subsequent exhaustive comparative genomic analysis of the virophages, polintons, and related genetic elements, including, among others, linear plasmids known as transpovirons (167), whose replication depends on giant viruses, revealed a complex network of evolutionary relationships (Fig. 11 and 12) (32). In this network, different classes of agents, either bona fide viruses or capsidless elements, are connected through shared homologous genes which, in many cases, comprise distinct families of virus hallmark genes, such as S3H, maturation thiol protease, or JRC2b (Fig. 12). Clearly, among dsDNA viruses and related capsidless elements, the evolutionary process is much more complex than that among small viruses and virus-like elements, being confounded by numerous gene exchanges and acquisition of genes from diverse sources. Nevertheless, the overall evolutionary trend appears to be the same as that outlined above for the RCR replicons, namely, multiple transitions from viruses to capsidless genetic elements and vice versa.
FIG 12.
Network of evolutionary connections between mavericks, virophages, and other viruses and capsidless elements. Bacteriophage groups that are involved in the network connections are as follows: Tectiviridae (PolB), Caudovirales (tailed bacteriophages) (S3H and GIY-YIG), and cyanophages (MV19 peptidase). Groups of NCLDV that are involved in the network connections are as follows: irido-, mimi-, pox-, and marseilleviruses (mavirus S3H helicase); marseillevirus (OLV S3H helicase and MV19 peptidase); Phaeocystis globosa virus and invertebrate iridescent virus 6 (GIY-YIG); Phycodnaviridae (Tir 6F); Poxviridae and Asfarviridae (ATPase), and Mimiviridae (MV20 FNIP repeats). (Adapted from reference 32, published under a Creative Commons license.)
Genetic elements that are hybrids between viruses (members of the family Fuselloviridae, such as SSV2) and plasmids have been discovered in hyperthermophilic archaea (168). Moreover, it has been shown that the virus-related plasmids can be packed into virions and effectively behave as satellite viruses (168, 169). These and related findings indicate that, at least in prokaryotes, viruses and capsidless elements (plasmids) form integrated “ecosystems” in which different types of elements are linked both genetically and functionally (31).
Another case of evolutionary connections between dsDNA viruses and capsidless elements involves linear and circular dsDNA plasmids that replicate inside mitochondria of plants and fungi (84, 170). Most of these plasmids are small dsDNA molecules of 3 to 12 kb that typically encode a DNA polymerase of the PolB family and a single-subunit, phage-type RNA polymerase, along with carrying several uncharacterized genes. Both the DNA polymerase and RNA polymerase genes of these plasmids are most closely related to homologs from bacteriophages that contain proteins attached to the genomic DNA termini, such as PRD1 and phi29 (171). Moreover, the linear plasmids encode terminal proteins whose origin remains unclear. In an obvious parallel with the origin of narnaviruses from RNA phages (see above), the mitochondrial DNA plasmids appear to have evolved from dsDNA bacteriophages of the bacterial endosymbionts that gave rise to the mitochondria.
A distinct group of capsidless genetic elements includes linear dsDNA plasmids that replicate in the cytoplasm of Ascomycete fungi (172). These 12- to 13-kb plasmids encompass several genes, including those for DNA polymerase, two RNA polymerase subunits, capping enzyme, and a helicase, that are most closely related to the respective homologs from NCLDV (173, 174). The evolutionary scenario in this case is unclear, especially because NCLDV are not known to infect fungi and because the plasmids possess a terminal protein suggestive of a phage contribution to their origin. Nevertheless, the common theme in the evolution of all these capsidless elements appears to be the substantial genomic reduction of dsDNA viruses en route to capsidless derivatives.
Although dsDNA bacteriophages are prototypical viruses and generally are unrelated to capsidless elements, the transition between encapsidating (lytic) and nonencapsidating (lysogenic) lifestyles is the key feature of numerous phages. In most cases, lysogenic phages integrate into the host bacterial chromosome as prophages that can be transmitted through many bacterial generations and often become defective and get “stuck” in the chromosome. However, for several phages, such as P4 or N15, nonintegrating, stably inherited plasmid forms of prophages have been described (175, 176), which is suggestive of the evolutionary transition between viruses and nonviral selfish elements. Similar observations have been reported for viruses of Haloarchaea (177) and for eukaryotic viruses such as herpesviruses (178).
UBIQUITY OF SELFISH GENETIC ELEMENTS AND ORIGIN OF VIRUSES FROM CAPSIDLESS GENETIC PARASITES
Host-parasite arms races are a major formative factor in all evolution of life (72, 179). Genetic parasites, i.e., viruses and virus-like selfish elements, seem to be truly ubiquitous: some such elements apparently are associated with all cellular life forms. Mathematical models of the evolution of replicator systems aimed at the reconstruction of the first stages in the history of life invariably reveal partitioning into hosts and parasites (180–183). This fundamental separation emerges as soon as the evolving systems reach a minimum complexity whereby dedicated replication devices, such as polymerases, evolve and thus can be hijacked by “cheaters,” the first parasites (184). Such primitive parasites would have emerged even in the hypothetical primordial RNA world, inasmuch as there existed ribozyme polymerases capable of replicating other RNA molecules in trans.
The simplest genomic parasites might have been small RNA molecules that encoded no proteins and consisted primarily of cis signals for replication (polymerase recognition). Such molecules are the end products of in vitro evolution experiments, starting with the classic early experiments of Spiegelman and coworkers (185–188). Among the parasites of modern organisms, viroids that cause many diseases of plants and satellites of plant RNA viruses show a striking resemblance to the putative primary parasites. Viroids are highly structured circular RNA molecules of approximately 400 nucleotides that are replicated by the host DNA-dependent RNA polymerase II; satellite RNAs that are structurally similar to viroids are replicated by the viral RdRps (189, 190). Given that viroids so far have been identified only in plants, it appears unlikely that they are direct descendants of the primordial parasites. Nevertheless, viroids seem to recapitulate the principal features of the selfish elements from the ancient RNA world. Hepatitis delta virus (HDV) appears to be a derivative of a viroid that encodes a protein required for replication and virion formation and is encapsidated into particles that consist of the capsid protein of the helper hepatitis B virus (191, 192). Most likely, HDV evolved from a viroid-like ancestor by acquiring a protein-encoding gene from a still unknown source and adapting to use the capsid protein of the helper virus. This special case of evolution of a virus from the simplest known variety of capsidless selfish elements might be relatively recent but, again, appears to mimic the likely primordial stages of virus evolution.
The precellular stages of the evolution of life are murky, to say the least. Nevertheless, although the details are extremely difficult to decipher, a “virus-like” stage at which the primordial life forms consisted of ensembles of small replicons, initially RNA molecules, appears to be a logical inevitability (22, 72, 193). As pointed out above, these populations of primordial replicons are predicted to have segregated into hosts and parasites. In this scenario (Fig. 13), the first selfish elements did not encode a capsid protein (and probably no proteins at all). The first protein-encoding selfish elements most likely encoded their own replicase, rather than a capsid protein, and exploited the translation systems encoded in other genomes. Moreover, capsidless progenitors of today's retroelements could have played the key role in the origin of DNA genomes, leading eventually to the origin of protocells. Capsids, most likely spherical ones formed by the JRC, undoubtedly emerged at an early stage of evolution, perhaps concomitant with the origin of the first cells, marking the origin of bona fide viruses (Fig. 13). Nevertheless, there is little doubt that capsidless selfish elements came first and gave rise to viruses.
FIG 13.
Conceptual scheme of the coevolution of selfish elements/genetic parasites with their hosts, spanning the entire history of life.
Evolution of viruses from capsidless selfish elements may be considered the central trend of virus evolution. Although two of the largest Baltimore classes, positive-strand RNA and dsDNA viruses, are clearly dominated by bona fide viruses, it appears most likely that they originally evolved via the same scenario as the other two major classes, i.e., retroelements and ssDNA replicons. Moreover, as with the ssDNA viruses, multiple origins of viruses resulting from acquisition of capsid protein genes, in some cases nonhomologous ones, by different capsidless elements are apparent. In the case of positive-strand RNA viruses, the primary candidates for distinct origins would involve leviviruses (RNA bacteriophages) and the rest of the positive-strand RNA viruses that infect eukaryotes. Indeed, the capsid proteins of leviviruses are unrelated to those of any of the eukaryotic viruses (194), and the RdRps are only distantly related (23). Actually, the relationships between the three superfamilies of eukaryotic positive-strand RNA viruses, the picornavirus-like, alphavirus-like, and flavivirus-like viruses, are distant as well, so independent origins cannot be ruled out (23). In the case of dsDNA viruses, several highly diverged groups of bacteriophages, particularly viruses of hyperthermophilic archaea (see above), could also be candidates for independent origination from plasmids.
CONCLUSIONS
The pervasive evolutionary connections between viruses and capsidless selfish genetic elements that are traceable for all major classes of viruses reveal the limitations of the capsidocentric perspective on the evolution of viruses. Evolutionary transitions between viruses and capsidless elements appear to have occurred on multiple occasions in the history of life. The relationships between viruses and capsidless elements differ between different classes of selfish elements. The evolution of two classes, the retroelements and the ssDNA replicons, apparently started from capsidless forms and involved a single and multiple origins of viruses, respectively. Similar evolutionary scenarios also cannot be ruled out for the origins of different major groups of positive-strand RNA and dsDNA elements, even though these classes are presently dominated by viruses. Moreover, it appears almost certain that at the earliest, precellular stages of life's evolution, capsidless genetic parasites evolved first and then gave rise to viruses.
In a seminal article, Raoult and Forterre (6), although duly recognizing the existence of evolutionary relationships between some viruses and “orphan replicons,” such as plasmids and transposons (also see references 195 and 196), classified life forms into ribosome-encoding organisms (cellular) and capsid-encoding organisms (viruses). This classification rightly emphasizes the distinct and fundamental status of viruses among life forms but only partially reflects the central, perennial division of life forms into informationally self-sufficient organisms that encode the basic functional systems required for their reproduction (cellular life forms) and genetic (informational) parasites. It may be useful to emphasize that the distinction between the capsidocentric perspective and the “greater virus world” concept is not primarily semantic but rather has to do with the early differentiation of two fundamental, complementary evolutionary strategies.
The two biological worlds strongly interacted over the entire history of life, but each retained its autonomy, which is manifest, in particular, in the ubiquity of the monophyletic translation system in the cellular world and the existence of a network of hallmark genes uniting the virus world (22, 197). Numerous cases of reductive evolution of cellular life forms led to parasites that possess drastically shrunken genomes, which are often smaller than genomes of giant viruses, and commensurate, dramatically diminished repertoires of cellular functions (198, 199). However, this cellular reduction ultimately may yield organelles, some of which, such as hydrogenosomes and mitosomes, have even lost their genome altogether (200), but to the best of our current knowledge, it never produces virus-like entities. It appears that the fundamental divide between cells and genetic parasites was never crossed during the entire history of life, compatible with the hypothesis that genetic parasites coevolved with the first replicating entities. Among these parasites, capsid-encoding organisms, or viruses, represent only one, even if extremely successful, evolutionary strategy. In contrast, taken as a whole, the greater virus world of informational parasites that exploit a variety of evolutionary strategies represents an equal partner to informationally self-sufficient cellular life forms during the entire history of life.
ACKNOWLEDGMENTS
We thank Mart Krupovic for a critical reading of the manuscript and for helpful discussions.
E.V.K.'s research is supported by intramural funds of the U.S. Department of Health and Human Services (to the National Library of Medicine).
Biographies

Eugene Koonin is the leader of the Evolutionary Genomics Group at the National Center for Biotechnology Information. He received his Ph.D. in Molecular Biology in 1983 from the Department of Biology, Moscow State University, joined the NCBI in 1991, and became a Senior Investigator in 1996. His group is pursuing several research directions in evolutionary genomics of prokaryotes, eukaryotes, and viruses. Dr. Koonin is the author of Sequence-Evolution-Function: Computational Approaches in Comparative Genomics (2003; with Michael Galperin) and The Logic of Chance: the Nature and Origin of Biological Evolution (2011). He is the founder and editor in chief (with Laura Landweber and David Lipman) of Biology Direct, an open-access, open peer-review journal. Dr. Koonin is a Fellow of the American Academy of Arts and Sciences, American Academy of Microbiology, and American College of Medical Informatics, a Foreign Associate of the European Molecular Biology Organization, and Doctor Honoris Causa of Universite Aix-Marseille.

Valerian V. Dolja, Ph.D., D.Sc., graduated from Moscow State University (Russia) in 1974 and received his Ph.D. and D.Sc. degrees from the same university. He moved to the United States in 1991 as a Visiting Scientist at Texas A&M University (College Station, TX). In 1994, he joined the faculty of the Department of Botany and Plant Pathology at Oregon State University (Corvallis, OR) and was promoted to Full Professor in 2001. His laboratory studies functional genomics of plant RNA viruses, virus gene expression and RNA interference vectors, and mechanisms of membrane transport in plant cells. In addition, his long-term interest is evolution and origins of viruses, a research direction on which he has collaborated with Eugene Koonin (NIH) for over two decades. Dr. Dolja is a Fellow of American Academy of Microbiology and a member of the editorial boards of Journal of Virology, Virology, Biology Direct, and Frontiers in Plant Science.
ADDENDUM IN PROOF
After this paper was accepted for publication, an in-depth analysis of the protein sequences encoded in the genomes of large transposons of the polinton/maverick family has shown that most of these elements encode distinct variants of the major, double beta-barrel capsid protein and the minor, single beta-barrel capsid protein (M. Krupovic, D. H. Bamford, and E. V. Koonin, Biol. Direct, in press). Thus, the majority of the polintons appear to combine salient features of genuine viruses and transposable elements; accordingly, it has been proposed that these elements should be renamed polintoviruses. However, a distinct family of polintons appears to consist of capsidless transposons. The life style of the polintoviruses appears to be analogous to that of metaviruses and pseudoviruses discussed in this article. These findings emphasize the continuity of viruses and capsidless elements and parallel evolutionary processes in different parts of the greater virus world.
REFERENCES
- 1.Claverie JM, Abergel C. 2010. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 26:431–437. 10.1016/j.tig.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 2.Claverie JM, Abergel C, Ogata H. 2009. Mimivirus. Curr. Top. Microbiol. Immunol. 328:89–121 [DOI] [PubMed] [Google Scholar]
- 3.Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. 2006. Mimivirus and the emerging concept of “giant” virus. Virus Res. 117:133–144. 10.1016/j.virusres.2006.01.008 [DOI] [PubMed] [Google Scholar]
- 4.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. 10.1126/science.1081867 [DOI] [PubMed] [Google Scholar]
- 5.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. 10.1126/science.1101485 [DOI] [PubMed] [Google Scholar]
- 6.Raoult D, Forterre P. 2008. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 6:315–319. 10.1038/nrmicro1858 [DOI] [PubMed] [Google Scholar]
- 7.Bamford DH. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231–236. 10.1016/S0923-2508(03)00065-2 [DOI] [PubMed] [Google Scholar]
- 8.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655–663. 10.1016/j.sbi.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 9.Krupovic M, Bamford DH. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941–948. 10.1038/nrmicro2033 [DOI] [PubMed] [Google Scholar]
- 10.Hendrix RW. 2005. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv. Virus Res. 64:1–14. 10.1016/S0065-3527(05)64001-8 [DOI] [PubMed] [Google Scholar]
- 11.Pietila MK, Laurinmaki P, Russell DA, Ko CC, Jacobs-Sera D, Hendrix RW, Bamford DH, Butcher SJ. 2013. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc. Natl. Acad. Sci. U. S. A. 110:10604–10609. 10.1073/pnas.1303047110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrescia NG, Bamford DH, Grimes JM, Stuart DI. 2012. Structure unifies the viral universe. Annu. Rev. Biochem. 81:795–822. 10.1146/annurev-biochem-060910-095130 [DOI] [PubMed] [Google Scholar]
- 13.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 1:118–124. 10.1016/j.coviro.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 14.Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv. Virus Res. 82:33–62. 10.1016/B978-0-12-394621-8.00012-1 [DOI] [PubMed] [Google Scholar]
- 15.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 67:565–585. 10.1146/annurev-micro-092412-155633 [DOI] [PubMed] [Google Scholar]
- 16.Pietila MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72:307–319. 10.1111/j.1365-2958.2009.06642.x [DOI] [PubMed] [Google Scholar]
- 17.Roine E, Kukkaro P, Paulin L, Laurinavicius S, Domanska A, Somerharju P, Bamford DH. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 84:3682–3689. 10.1128/JVI.01879-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. 10.1126/science.1239181 [DOI] [PubMed] [Google Scholar]
- 19.Koonin EV. 2010. The two empires and three domains of life in the postgenomic age. Nat. Educ. 3:27 [Google Scholar]
- 20.Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. 2013. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J. Bacteriol. 195:941–950. 10.1128/JB.01801-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223. 10.1186/1743-422X-6-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol. Direct 1:29. 10.1186/1745-6150-1-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939. 10.1038/nrmicro2030 [DOI] [PubMed] [Google Scholar]
- 24.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anantharaman V, Iyer LM, Aravind L. 2010. Presence of a classical RRM-fold palm domain in Thg1-type 3′-5′ nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol. Direct 5:43. 10.1186/1745-6150-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875–3896. 10.1093/nar/gki702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbalenya AE, Koonin EV. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413–8440. 10.1093/nar/17.21.8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146:11–31. 10.1016/j.jsb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- 30.Krupovic M. 2013. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 3:578–586. 10.1016/j.coviro.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 31.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 75:610–635. 10.1128/MMBR.00011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yutin N, Raoult D, Koonin EV. 2013. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 10:158. 10.1186/1743-422X-10-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baltimore D. 1971. Expression of animal virus genomes. Bacteriol. Rev. 35:235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV. 1991. Genome replication/expression strategies of positive-strand RNA viruses: a simple version of a combinatorial classification and prediction of new strategies. Virus Genes 5:273–281. 10.1007/BF00568977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolja VV, Koonin EV. 2011. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 1:322–331. 10.1016/j.coviro.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375–430. 10.3109/10409239309078440 [DOI] [PubMed] [Google Scholar]
- 37.Dolja VV, Koonin EV. 2012. Capsid-less RNA viruses. Encyclopedia of life sciences; John Wiley & Sons, Ltd, Chichester, United Kingdom. 10.1002/9780470015902.a0023269 [DOI] [Google Scholar]
- 38.Cole TE, Hong Y, Brasier CM, Buck KW. 2000. Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch elm disease fungus, Ophiostoma novo-ulmi. Virology 268:239–243. 10.1006/viro.1999.0097 [DOI] [PubMed] [Google Scholar]
- 39.Fujimura T, Esteban R. 2007. Interactions of the RNA polymerase with the viral genome at the 5′- and 3′-ends contribute to 20S RNA narnavirus persistence in yeast. J. Biol. Chem. 282:19011–19019. 10.1074/jbc.M702432200 [DOI] [PubMed] [Google Scholar]
- 40.Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv. Virus Res. 86:149–176. 10.1016/B978-0-12-394315-6.00006-4 [DOI] [PubMed] [Google Scholar]
- 41.Hillman BI, Esteban R. 2012. Family Narnaviridae, p 1055–1060 In King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, 9th ed. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 42.Solorzano A, Rodriguez-Cousino N, Esteban R, Fujimura T. 2000. Persistent yeast single-stranded RNA viruses exist in vivo as genomic RNA.RNA polymerase complexes in 1:1 stoichiometry. J. Biol. Chem. 275:26428–26435. 10.1074/jbc.M002281200 [DOI] [PubMed] [Google Scholar]
- 43.Cai G, Myers K, Fry WE, Hillman BI. 2012. A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch. Virol. 157:165–169. 10.1007/s00705-011-1126-5 [DOI] [PubMed] [Google Scholar]
- 44.Hong Y, Cole TE, Brasier CM, Buck KW. 1998. Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246:158–169. 10.1006/viro.1998.9178 [DOI] [PubMed] [Google Scholar]
- 45.Rastgou M, Habibi MK, Izadpanah K, Masenga V, Milne RG, Wolf YI, Koonin EV, Turina M. 2009. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90:2525–2535. 10.1099/vir.0.013086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47:353–384. 10.1146/annurev-phyto-080508-081932 [DOI] [PubMed] [Google Scholar]
- 47.Nuss DL. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632–642. 10.1038/nrmicro1206 [DOI] [PubMed] [Google Scholar]
- 48.Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630. 10.1038/nature04546 [DOI] [PubMed] [Google Scholar]
- 49.Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467:929–934. 10.1038/nature09486 [DOI] [PubMed] [Google Scholar]
- 50.Adams MJ, Zerbini FM, French R, Rabenstein F, Stenger DC, Valconen JPT. 2012. Family Potyviridae, p 1069–1089 In King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, 9th ed. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 51.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. 2005. The tree of eukaryotes. Trends Ecol. Evol. 20:670–676. 10.1016/j.tree.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 52.Koonin EV. 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11:209. 10.1186/gb-2010-11-5-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nuss DL. 2011. Mycoviruses, RNA silencing, and viral RNA recombination. Adv. Virus Res. 80:25–48. 10.1016/B978-0-12-385987-7.00002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin EV, Choi GH, Nuss DL, Shapira R, Carrington JC. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U. S. A. 88:10647–10651. 10.1073/pnas.88.23.10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. 10.1016/S0092-8674(00)81614-1 [DOI] [PubMed] [Google Scholar]
- 56.Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5:896–904. 10.1128/EC.00373-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukuhara T, Gibbs MJ. 2012. Family Endornaviridae, p 519–521 In King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, 9th ed. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 58.Roossinck MJ, Sabanadzovic S, Okada R, Valverde RA. 2011. The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 92:2674–2678. 10.1099/vir.0.034702-0 [DOI] [PubMed] [Google Scholar]
- 59.Gibbs MJ, Koga R, Moriyama H, Pfeiffer P, Fukuhara T. 2000. Phylogenetic analysis of some large double-stranded RNA replicons from plants suggests they evolved from a defective single-stranded RNA virus. J. Gen. Virol. 81:227–233 [DOI] [PubMed] [Google Scholar]
- 60.Liu H, Fu Y, Jiang D, Li G, Xie J, Peng Y, Yi X, Ghabrial SA. 2009. A novel mycovirus that is related to the human pathogen hepatitis E virus and rubi-like viruses. J. Virol. 83:1981–1991. 10.1128/JVI.01897-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tuomivirta TT, Kaitera J, Hantula J. 2009. A novel putative virus of Gremmeniella abietina type B (Ascomycota: Helotiaceae) has a composite genome with endornavirus affinities. J. Gen. Virol. 90:2299–2305. 10.1099/vir.0.011973-0 [DOI] [PubMed] [Google Scholar]
- 62.Hacker CV, Brasier CM, Buck KW. 2005. A double-stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J. Gen. Virol. 86:1561–1570. 10.1099/vir.0.80808-0 [DOI] [PubMed] [Google Scholar]
- 63.Martelli GP, Adams MJ, Kreuze JF, Dolja VV. 2007. Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45:73–100. 10.1146/annurev.phyto.45.062806.094401 [DOI] [PubMed] [Google Scholar]
- 64.Xie J, Wei D, Jiang D, Fu Y, Li G, Ghabrial S, Peng Y. 2006. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 87:241–249. 10.1099/vir.0.81522-0 [DOI] [PubMed] [Google Scholar]
- 65.Howitt RL, Beever RE, Pearson MN, Forster RL. 2006. Genome characterization of a flexuous rod-shaped mycovirus, Botrytis virus X, reveals high amino acid identity to genes from plant ‘potex-like' viruses. Arch. Virol. 151:563–579. 10.1007/s00705-005-0621-y [DOI] [PubMed] [Google Scholar]
- 66.Howitt RL, Beever RE, Pearson MN, Forster RL. 2001. Genome characterization of Botrytis virus F, a flexuous rod-shaped mycovirus resembling plant ‘potex-like' viruses. J. Gen. Virol. 82:67–78 [DOI] [PubMed] [Google Scholar]
- 67.Koga R, Fukuhara T, Nitta T. 1998. Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol. Biol. 36:717–724. 10.1023/A:1005907310553 [DOI] [PubMed] [Google Scholar]
- 68.Koga R, Horiuchi H, Fukuhara T. 2003. Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol. Biol. 51:991–999. 10.1023/A:1023003412859 [DOI] [PubMed] [Google Scholar]
- 69.Ghabrial SA, Nibert ML, Maiss E, Lesker T, Baker TS, Tao YJ. 2012. Family Partitiviridae, p 523–534 In King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 70.Ryabov EV, Taliansky ME, Robinson DJ, Waterhouse PM, Murant AF, de Zoeten GAFBW, Vetten HJ, Gibbs MJ. 2012. Genus umbravirus, p 1191–1195 In King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 71.King MQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Virus taxonomy, 9th ed. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 72.Koonin EV, Dolja VV. 2013. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 3:546–557. 10.1016/j.coviro.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goremykin VV, Salamini F, Velasco R, Viola R. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 26:99–110. 10.1093/molbev/msn226 [DOI] [PubMed] [Google Scholar]
- 74.Eickbush TH, Jamburuthugoda VK. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134:221–234. 10.1016/j.virusres.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gladyshev EA, Arkhipova IR. 2011. A widespread class of reverse transcriptase-related cellular genes. Proc. Natl. Acad. Sci. U. S. A. 108:20311–20316. 10.1073/pnas.1100266108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beauregard A, Curcio MJ, Belfort M. 2008. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42:587–617. 10.1146/annurev.genet.42.110807.091549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koonin EV. 2007. The Biological Big Bang model for the major transitions in evolution. Biol. Direct 2:21. 10.1186/1745-6150-2-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Puigbo P, Wolf YI, Koonin EV. 2009. Search for a tree of life in the thicket of the phylogenetic forest. J. Biol. 8:59. 10.1186/jbiol159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rokas A, Carroll SB. 2006. Bushes in the tree of life. PLoS Biol. 4:e352. 10.1371/journal.pbio.0040352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rokas A, Kruger D, Carroll SB. 2005. Animal evolution and the molecular signature of radiations compressed in time. Science 310:1933–1938. 10.1126/science.1116759 [DOI] [PubMed] [Google Scholar]
- 81.Lambowitz AM, Zimmerly S. 2011. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 3:a003616. 10.1101/cshperspect.a003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon DM, Kelchner SA, Zimmerly S. 2009. A broadscale phylogenetic analysis of group II intron RNAs and intron-encoded reverse transcriptases. Mol. Biol. Evol. 26:2795–2808. 10.1093/molbev/msp193 [DOI] [PubMed] [Google Scholar]
- 83.Chiang CC, Lambowitz AM. 1997. The Mauriceville retroplasmid reverse transcriptase initiates cDNA synthesis de novo at the 3′ end of tRNAs. Mol. Cell. Biol. 17:4526–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Griffiths AJ. 1995. Natural plasmids of filamentous fungi. Microbiol. Rev. 59:673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simon DM, Zimmerly S. 2008. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 36:7219–7229. 10.1093/nar/gkn867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Callinan PA, Batzer MA. 2006. Retrotransposable elements and human disease. Genome Dyn. 1:104–115. 10.1159/000092503 [DOI] [PubMed] [Google Scholar]
- 87.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J. 2010. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 63:584–598. 10.1111/j.1365-313X.2010.04263.x [DOI] [PubMed] [Google Scholar]
- 89.Lisch D. 2012. Regulation of transposable elements in maize. Curr. Opin. Plant Biol. 15:511–516. 10.1016/j.pbi.2012.07.001 [DOI] [PubMed] [Google Scholar]
- 90.Clayton C. 2010. Repetitive elements in parasitic protozoa. BMC Biol. 8:64. 10.1186/1741-7007-8-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evgen'ev MB, Arkhipova IR. 2005. Penelope-like elements—a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet. Genome Res. 110:510–521. 10.1159/000084984 [DOI] [PubMed] [Google Scholar]
- 92.Gladyshev EA, Arkhipova IR. 2007. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 104:9352–9357. 10.1073/pnas.0702741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Babushok DV, Kazazian HH., Jr 2007. Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28:527–539. 10.1002/humu.20486 [DOI] [PubMed] [Google Scholar]
- 94.Ding W, Lin L, Chen B, Dai J. 2006. L1 elements, processed pseudogenes and retrogenes in mammalian genomes. IUBMB Life 58:677–685. 10.1080/15216540601034856 [DOI] [PubMed] [Google Scholar]
- 95.Barzilay G, Hickson ID. 1995. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays 17:713–719. 10.1002/bies.950170808 [DOI] [PubMed] [Google Scholar]
- 96.Barzilay G, Walker LJ, Robson CN, Hickson ID. 1995. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 23:1544–1550. 10.1093/nar/23.9.1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malik HS. 2005. Ribonuclease H evolution in retrotransposable elements. Cytogenet. Genome Res. 110:392–401. 10.1159/000084971 [DOI] [PubMed] [Google Scholar]
- 98.Coffin JM. 1992. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 176:143–164 [DOI] [PubMed] [Google Scholar]
- 99.Doolittle RF, Feng DF, Johnson MS, McClure MA. 1989. Origins and evolutionary relationships of retroviruses. Q. Rev. Biol. 64:1–30 [DOI] [PubMed] [Google Scholar]
- 100.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10:395–406. 10.1038/nrmicro2783 [DOI] [PubMed] [Google Scholar]
- 101.Weiss RA. 2013. On the concept and elucidation of endogenous retroviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120494. 10.1098/rstb.2012.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beck J, Nassal M. 2007. Hepatitis B virus replication. World J. Gastroenterol. 13:48–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bousalem M, Douzery EJ, Seal SE. 2008. Taxonomy, molecular phylogeny and evolution of plant reverse transcribing viruses (family Caulimoviridae) inferred from full-length genome and reverse transcriptase sequences. Arch. Virol. 153:1085–1102. 10.1007/s00705-008-0095-9 [DOI] [PubMed] [Google Scholar]
- 104.Glebe D, Bremer CM. 2013. The molecular virology of hepatitis B virus. Semin. Liver Dis. 33:103–112. 10.1055/s-0033-1345717 [DOI] [PubMed] [Google Scholar]
- 105.Geering AD, Scharaschkin T, Teycheney PY. 2010. The classification and nomenclature of endogenous viruses of the family Caulimoviridae. Arch. Virol. 155:123–131. 10.1007/s00705-009-0488-4 [DOI] [PubMed] [Google Scholar]
- 106.Staginnus C, Richert-Poggeler KR. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci. 11:485–491. 10.1016/j.tplants.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 107.Flavell AJ, Pearce SR, Heslop-Harrison P, Kumar A. 1997. The evolution of Ty1-copia group retrotransposons in eukaryote genomes. Genetica 100:185–195. 10.1023/A:1018385713293 [DOI] [PubMed] [Google Scholar]
- 108.Peterson-Burch BD, Voytas DF. 2002. Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol. Biol. Evol. 19:1832–1845. 10.1093/oxfordjournals.molbev.a004008 [DOI] [PubMed] [Google Scholar]
- 109.Piednoel M, Donnart T, Esnault C, Graca P, Higuet D, Bonnivard E. 2013. LTR-retrotransposons in R. exoculata and other crustaceans: the outstanding success of GalEa-like copia elements. PLoS One 8:e57675. 10.1371/journal.pone.0057675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik HS, Eickbush TH. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73:5186–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Malik HS, Eickbush TH. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11:1187–1197. 10.1101/gr.185101 [DOI] [PubMed] [Google Scholar]
- 112.Kojima KK, Jurka J. 2013. A superfamily of DNA transposons targeting multicopy small RNA genes. PLoS One 8:e68260. 10.1371/journal.pone.0068260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pal C, Papp B. 2013. From passengers to drivers: impact of bacterial transposable elements on evolvability. Mob. Genet. Elements 3:e23617. 10.4161/mge.23617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steiniger-White M, Rayment I, Reznikoff WS. 2004. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 14:50–57. 10.1016/j.sbi.2004.01.008 [DOI] [PubMed] [Google Scholar]
- 115.Bao W, Kapitonov VV, Jurka J. 2010. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob. DNA 1:3. 10.1186/1759-8753-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Capy P, Maisonhaute C. 2002. Acquisition and loss of modules: the construction set of transposable elements. Genetika 38:719–726 [PubMed] [Google Scholar]
- 117.Krylov DM, Koonin EV. 2001. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr. Biol. 11:R584–R587. 10.1016/S0960-9822(01)00357-8 [DOI] [PubMed] [Google Scholar]
- 118.Sirkis R, Gerst JE, Fass D. 2006. Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 364:376–387. 10.1016/j.jmb.2006.08.086 [DOI] [PubMed] [Google Scholar]
- 119.Ivanov D, Stone JR, Maki JL, Collins T, Wagner G. 2005. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell 17:137–143. 10.1016/j.molcel.2004.12.015 [DOI] [PubMed] [Google Scholar]
- 120.Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. 2003. The SCAN domain defines a large family of zinc finger transcription factors. Gene 310:29–38. 10.1016/S0378-1119(03)00509-2 [DOI] [PubMed] [Google Scholar]
- 121.Llorens C, Fares MA, Moya A. 2008. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 8:276. 10.1186/1471-2148-8-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Volff JN. 2009. Cellular genes derived from Gypsy/Ty3 retrotransposons in mammalian genomes. Ann. N. Y. Acad. Sci. 1178:233–243. 10.1111/j.1749-6632.2009.05005.x [DOI] [PubMed] [Google Scholar]
- 123.Lerat E, Capy P. 1999. Retrotransposons and retroviruses: analysis of the envelope gene. Mol. Biol. Evol. 16:1198–1207. 10.1093/oxfordjournals.molbev.a026210 [DOI] [PubMed] [Google Scholar]
- 124.Covey SN. 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14:623–633. 10.1093/nar/14.2.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leipe DD, Aravind L, Koonin EV. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389–3401. 10.1093/nar/27.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. 10.1159/000312913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Khan SA. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126–136. 10.1016/j.plasmid.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 128.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. 2013. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 11:525–538. 10.1038/nrmicro3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279–3285. 10.1093/nar/20.13.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koonin EV, Ilyina TV. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241–268. 10.1016/0303-2647(93)90074-M [DOI] [PubMed] [Google Scholar]
- 131.Koonin EV, Ilyina TV. 1992. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 73:2763–2766. 10.1099/0022-1317-73-10-2763 [DOI] [PubMed] [Google Scholar]
- 132.Krupovic M, Ravantti JJ, Bamford DH. 2009. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol. Biol. 9:112. 10.1186/1471-2148-9-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saccardo F, Cettul E, Palmano S, Noris E, Firrao G. 2011. On the alleged origin of geminiviruses from extrachromosomal DNAs of phytoplasmas. BMC Evol. Biol. 11:185. 10.1186/1471-2148-11-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roux S, Krupovic M, Poulet A, Debroas D, Enault F. 2012. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7:e40418. 10.1371/journal.pone.0040418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Delwart E, Li L. 2012. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 164:114–121. 10.1016/j.virusres.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosario K, Duffy S, Breitbart M. 2012. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch. Virol. 157:1851–1871. 10.1007/s00705-012-1391-y [DOI] [PubMed] [Google Scholar]
- 137.Diemer GS, Stedman KM. 2012. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct 7:13. 10.1186/1745-6150-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Roux S, Enault F, Bronner G, Vaulot D, Forterre P, Krupovic M. 2013. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 4:2700. [DOI] [PubMed] [Google Scholar]
- 139.Gibbs MJ, Smeianov VV, Steele JL, Upcroft P, Efimov BA. 2006. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 23:1097–1100. 10.1093/molbev/msj122 [DOI] [PubMed] [Google Scholar]
- 140.Stedman K. 2013. Mechanisms for RNA capture by ssDNA viruses: grand theft RNA. J. Mol. Evol. 76:359–364. 10.1007/s00239-013-9569-9 [DOI] [PubMed] [Google Scholar]
- 141.Sencilo A, Paulin L, Kellner S, Helm M, Roine E. 2012. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res. 40:5523–5534. 10.1093/nar/gks215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soler N, Gaudin M, Marguet E, Forterre P. 2011. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem. Soc. Trans. 39:36–44. 10.1042/BST0390036 [DOI] [PubMed] [Google Scholar]
- 143.Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66–84. 10.1006/plas.1999.1460 [DOI] [PubMed] [Google Scholar]
- 144.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41:331–368. 10.1146/annurev.genet.40.110405.090448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kapitonov VV, Jurka J. 2007. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 23:521–529. 10.1016/j.tig.2007.08.004 [DOI] [PubMed] [Google Scholar]
- 147.Kapitonov VV, Jurka J. 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 98:8714–8719. 10.1073/pnas.151269298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu H, Fu Y, Li B, Yu X, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. 2011. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol. Biol. 11:276. 10.1186/1471-2148-11-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oke M, Kerou M, Liu H, Peng X, Garrett RA, Prangishvili D, Naismith JH, White MF. 2011. A dimeric Rep protein initiates replication of a linear archaeal virus genome: implications for the Rep mechanism and viral replication. J. Virol. 85:925–931. 10.1128/JVI.01467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krupovic M, Bamford DH. 2007. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics 8:236. 10.1186/1471-2164-8-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Odegrip R, Haggard-Ljungquist E. 2001. The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage p2. J. Mol. Biol. 308:147–163. 10.1006/jmbi.2001.4607 [DOI] [PubMed] [Google Scholar]
- 152.Rohwer F. 2003. Global phage diversity. Cell 113:141. 10.1016/S0092-8674(03)00276-9 [DOI] [PubMed] [Google Scholar]
- 153.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812. 10.1038/nrmicro1750 [DOI] [PubMed] [Google Scholar]
- 154.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 158:2517–2521. 10.1007/s00705-013-1768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jurka J, Kapitonov VV, Kohany O, Jurka MV. 2007. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 8:241–259. 10.1146/annurev.genom.8.080706.092416 [DOI] [PubMed] [Google Scholar]
- 156.Kapitonov VV, Jurka J. 2006. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 103:4540–4545. 10.1073/pnas.0600833103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pritham EJ, Putliwala T, Feschotte C. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390:3–17. 10.1016/j.gene.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 158.Desnues C, Boyer M, Raoult D. 2012. Sputnik, a virophage infecting the viral domain of life. Adv. Virus Res. 82:63–89. 10.1016/B978-0-12-394621-8.00013-3 [DOI] [PubMed] [Google Scholar]
- 159.Fischer MG, Allen MJ, Wilson WH, Suttle CA. 2010. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. U. S. A. 107:19508–19513. 10.1073/pnas.1007615107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fischer MG, Suttle CA. 2011. A virophage at the origin of large DNA transposons. Science 332:231–234. 10.1126/science.1199412 [DOI] [PubMed] [Google Scholar]
- 161.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100–104. 10.1038/nature07218 [DOI] [PubMed] [Google Scholar]
- 162.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Gibson JA, Cavicchioli R. 2011. Virophage control of Antarctic algal host-virus dynamics. Proc. Natl. Acad. Sci. U. S. A. 108:6163–6168. 10.1073/pnas.1018221108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yutin N, Colson P, Raoult D, Koonin EV. 2013. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 10:106. 10.1186/1743-422X-10-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou J, Zhang W, Yan S, Xiao J, Zhang Y, Li B, Pan Y, Wang Y. 2013. Diversity of virophages in metagenomic data sets. J. Virol. 87:4225–4236. 10.1128/JVI.03398-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Colson P, de Lamballerie X, Fournous G, Raoult D. 2012. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology 55:321–332. 10.1159/000336562 [DOI] [PubMed] [Google Scholar]
- 166.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734. 10.1128/JVI.75.23.11720-11734.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV, Raoult D. 2012. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. U. S. A. 109:18078–18083. 10.1073/pnas.1208835109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, Kristjansson JK, Garrett R, Zillig W. 1999. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 34:217–226. 10.1046/j.1365-2958.1999.01573.x [DOI] [PubMed] [Google Scholar]
- 169.Wang Y, Duan Z, Zhu H, Guo X, Wang Z, Zhou J, She Q, Huang L. 2007. A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363:124–133. 10.1016/j.virol.2007.01.035 [DOI] [PubMed] [Google Scholar]
- 170.Handa H. 2008. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion 8:15–25. 10.1016/j.mito.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 171.Braithwaite DK, Ito J. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787–802. 10.1093/nar/21.4.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jeske S, Meinhardt F. 2006. Autonomous cytoplasmic linear plasmid pPac1-1 of Pichia acaciae: molecular structure and expression studies. Yeast 23:479–486. 10.1002/yea.1367 [DOI] [PubMed] [Google Scholar]
- 173.Shuman S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1–40 [DOI] [PubMed] [Google Scholar]
- 174.Yanez RJ, Rodriguez JM, Boursnell M, Rodriguez JF, Vinuela E. 1993. Two putative African swine fever virus helicases similar to yeast ‘DEAH' pre-mRNA processing proteins and vaccinia virus ATPases D11L and D6R. Gene 134:161–174. 10.1016/0378-1119(93)90090-P [DOI] [PubMed] [Google Scholar]
- 175.Magnoni F, Sala C, Forti F, Deho G, Ghisotti D. 2006. DNA replication in phage P4: characterization of replicon II. Plasmid 56:216–222. 10.1016/j.plasmid.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 176.Ravin NV. 2011. N15: the linear phage-plasmid. Plasmid 65:102–109. 10.1016/j.plasmid.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 177.Zhang Z, Liu Y, Wang S, Yang D, Cheng Y, Hu J, Chen J, Mei Y, Shen P, Bamford DH, Chen X. 2012. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology 434:233–241. 10.1016/j.virol.2012.05.036 [DOI] [PubMed] [Google Scholar]
- 178.Collins CM, Medveczky PG. 2002. Genetic requirements for the episomal maintenance of oncogenic herpesvirus genomes. Adv. Cancer Res. 84:155–174. 10.1016/S0065-230X(02)84005-2 [DOI] [PubMed] [Google Scholar]
- 179.Forterre P, Prangishvili D. 2009. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N. Y. Acad. Sci. 1178:65–77. 10.1111/j.1749-6632.2009.04993.x [DOI] [PubMed] [Google Scholar]
- 180.Szathmary E, Maynard Smith J. 1997. From replicators to reproducers: the first major transitions leading to life. J. Theor. Biol. 187:555–571. 10.1006/jtbi.1996.0389 [DOI] [PubMed] [Google Scholar]
- 181.Takeuchi N, Hogeweg P. 2008. Evolution of complexity in RNA-like replicator systems. Biol. Direct 3:11. 10.1186/1745-6150-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Takeuchi N, Hogeweg P. 2012. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys. Life Rev. 9:219–263. 10.1016/j.plrev.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Takeuchi N, Hogeweg P, Koonin EV. 2011. On the origin of DNA genomes: evolution of the division of labor between template and catalyst in model replicator systems. PLoS Comput. Biol. 7:e1002024. 10.1371/journal.pcbi.1002024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Koonin EV, Martin W. 2005. On the origin of genomes and cells within inorganic compartments. Trends Genet. 21:647–654. 10.1016/j.tig.2005.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Joyce GF. 2007. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 46:6420–6436. 10.1002/anie.200701369 [DOI] [PubMed] [Google Scholar]
- 186.Mills DR, Kramer FR, Spiegelman S. 1973. Complete nucleotide sequence of a replicating RNA molecule. Science 180:916–927. 10.1126/science.180.4089.916 [DOI] [PubMed] [Google Scholar]
- 187.Oehlenschlager F, Eigen M. 1997. 30 years later—a new approach to Sol Spiegelman's and Leslie Orgel's in vitro evolutionary studies. Dedicated to Leslie Orgel on the occasion of his 70th birthday. Orig. Life Evol. Biosph. 27:437–457. 10.1023/A:1006501326129 [DOI] [PubMed] [Google Scholar]
- 188.Spiegelman S. 1971. An approach to the experimental analysis of precellular evolution. Q. Rev. Biophys. 4:213–253. 10.1017/S0033583500000639 [DOI] [PubMed] [Google Scholar]
- 189.Diener TO. 1991. Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J. 5:2808–2813 [DOI] [PubMed] [Google Scholar]
- 190.Flores R, Hernandez C, Martinez de Alba AE, Daros JA, Di Serio F. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117–139. 10.1146/annurev.phyto.43.040204.140243 [DOI] [PubMed] [Google Scholar]
- 191.Flores R, Ruiz-Ruiz S, Serra P. 2012. Viroids and hepatitis delta virus. Semin. Liver Dis. 32:201–210. 10.1055/s-0032-1323624 [DOI] [PubMed] [Google Scholar]
- 192.Taylor J, Pelchat M. 2010. Origin of hepatitis delta virus. Future Microbiol. 5:393–402. 10.2217/fmb.10.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Koonin EV, Wolf YI. 2012. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front. Cell Infect. Microbiol. 2:119. 10.3389/fcimb.2012.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Valegard K, Liljas L, Fridborg K, Unge T. 1990. The three-dimensional structure of the bacterial virus MS2. Nature 345:36–41. 10.1038/345036a0 [DOI] [PubMed] [Google Scholar]
- 195.Forterre P. 2002. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 5:525–532. 10.1016/S1369-5274(02)00360-0 [DOI] [PubMed] [Google Scholar]
- 196.Forterre P. 2005. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie 87:793–803. 10.1016/j.biochi.2005.03.015 [DOI] [PubMed] [Google Scholar]
- 197.Koonin EV. 2011. The logic of chance: the nature and origin of biological evolution. FT Press, Upper Saddle River, NJ [Google Scholar]
- 198.Moran NA. 2003. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6:512–518. 10.1016/j.mib.2003.08.001 [DOI] [PubMed] [Google Scholar]
- 199.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 200.van der Giezen M. 2009. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 56:221–231. 10.1111/j.1550-7408.2009.00407.x [DOI] [PubMed] [Google Scholar]
- 201.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18:11–19. 10.1016/j.tim.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110:462–467. 10.1159/000084979 [DOI] [PubMed] [Google Scholar]