Abstract
Vacuolar H+-ATPases (V-ATPases) are highly conserved ATP-driven proton pumps responsible for acidification of intracellular compartments. V-ATPase proton transport energizes secondary transport systems and is essential for lysosomal/vacuolar and endosomal functions. These dynamic molecular motors are composed of multiple subunits regulated in part by reversible disassembly, which reversibly inactivates them. Reversible disassembly is intertwined with glycolysis, the RAS/cyclic AMP (cAMP)/protein kinase A (PKA) pathway, and phosphoinositides, but the mechanisms involved are elusive. The atomic- and pseudo-atomic-resolution structures of the V-ATPases are shedding light on the molecular dynamics that regulate V-ATPase assembly. Although all eukaryotic V-ATPases may be built with an inherent capacity to reversibly disassemble, not all do so. V-ATPase subunit isoforms and their interactions with membrane lipids and a V-ATPase-exclusive chaperone influence V-ATPase assembly. This minireview reports on the mechanisms governing reversible disassembly in the yeast Saccharomyces cerevisiae, keeping in perspective our present understanding of the V-ATPase architecture and its alignment with the cellular processes and signals involved.
INTRODUCTION
Vacuolar H+-ATPases (V-ATPases) are ATP-driven proton pumps distributed throughout the endomembrane system of all eukaryotic cells (1, 2). V-ATPase proton transport acidifies organelles and energizes secondary transport systems. Zymogen activation, protein processing and trafficking, and receptor-mediated endocytosis are fundamental cellular processes that require V-ATPase activity. Cells specialized for active proton secretion express also V-ATPases at the plasma membrane. Proton transport by plasma membrane V-ATPases in osteoclasts, epididymal clear cells, and renal intercalated cells is necessary for bone resorption, sperm maturation, and maintenance of the systemic acid-base balance, respectively (3, 4). V-ATPase has been implicated in several pathological states, including osteopetrosis, distal renal tubular acidosis, male infertility, and cancers (2). Not surprisingly, studies of V-ATPase function and regulation are increasing, as is our knowledge of these dynamic proteins.
V-ATPase structure and function are highly conserved and well characterized in Saccharomyces cerevisiae (referred to here as yeast). Lack of V-ATPase function leads to a conditionally lethal phenotype that is characterized by pH sensitivity in yeast; complete lack of V-ATPase function is lethal in higher eukaryotes (5). Recent atomic- and pseudo-atomic-resolution structures of V-ATPase and its subunits have helped shed light on the molecular dynamics that regulate V-ATPase function (6, 7). V-ATPases are large multisubunit complexes structurally organized into two major domains, V1 and Vo (Fig. 1). Eight peripheral subunits (A to H) form the V1 domain, where ATP hydrolysis takes place. Six subunits (a, c, c′, c″, d, and e) comprise Vo, the membrane intrinsic domain that forms the path for proton transport. An important mechanism by which cells control organelle acidification is by disassembling and reassembling the V-ATPase complex (1, 2, 8, 9). Disassembly rapidly inactivates the pumps, resulting in three constituents: V1 subunit C, V1 (without subunit C), and Vo (Fig. 2). Disassembly is reversible, and reassociation of the three components rapidly restores ATP hydrolysis and proton transport across membranes. Catalytic inactivation and reactivation entail conformational changes in V1 subunit H (Fig. 2) (12, 13). This subunit is necessary to silence cytosolic V1 and activate V1Vo complexes (11, 15).
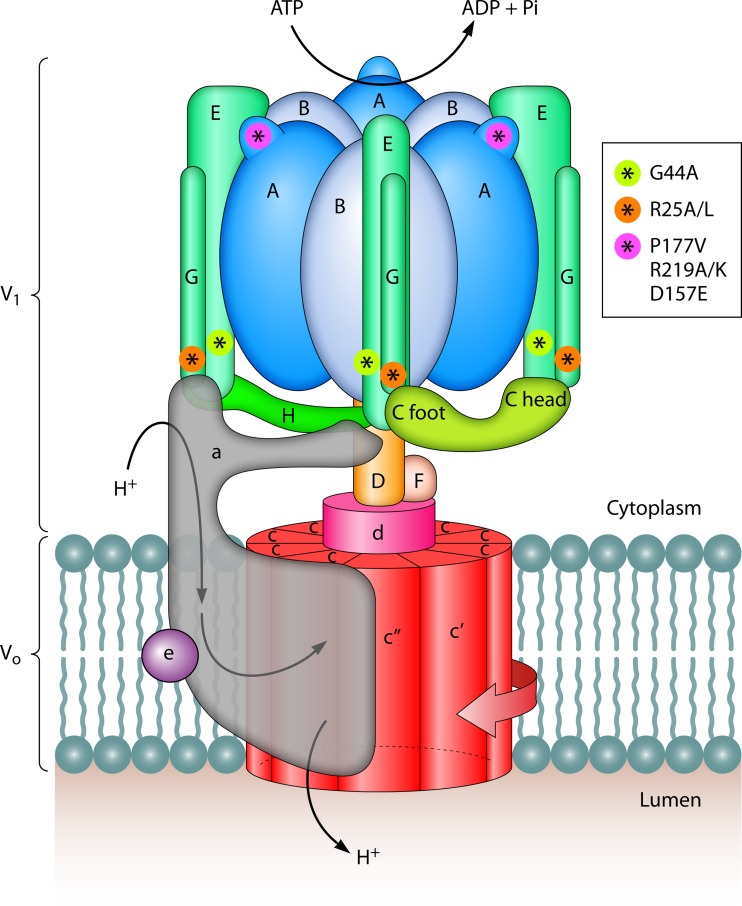
FIG 1.
The V-ATPase complex: subunit composition and organization. V-ATPase is composed of 14 different subunits, organized into two major domains: V1 is the catalytic ATPase domain and Vo is the proton translocation domain. Active transport of protons across the membrane entails rotation of a rotor (subunits F, D, d, c, c′, and c″) that is driven by ATP hydrolysis in V1 (subunits A). Three elongated peripheral stalks (subunits EG) connect the V1 and Vo domains and allow relative rotation of subunits during catalysis, by working as stators. Three stators are necessary for regulation of V-ATPase by disassembly and reassembly. Shown are mutations in the peripheral stalk subunits E (G44A) and G (R25A/L) and the catalytic subunit A at its nonhomologous domain (P177V and R219A/K). These mutations simultaneously alter V1Vo disassembly and catalysis, suggesting that disassembly requires wild-type catalytic activity (rotation). The mutation D157E in subunit A, which also prevents V1Vo disassembly, does not affect catalysis; it is proposed that D157E acts by stabilizing subunit-subunit interactions.
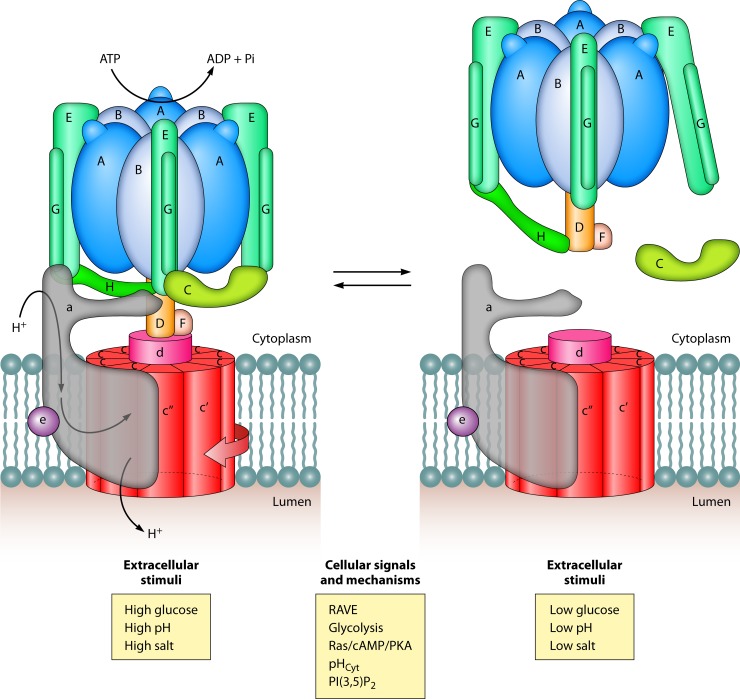
FIG 2.
Reversible disassembly of V-ATPase: extracellular stimuli and intracellular signals. V-ATPase disassembly breaks the complex apart, as V1, Vo, and the V1 subunit C separate. Disassembly is reversible, and reassembly of the three components restores ATP hydrolysis and proton transport. Yeast cells adjust the number of assembled V-ATPases in response to environmental stressors, including changes in glucose, pH, and salts. These extracellular cues are communicated to V-ATPases by several signals and unknown mechanisms that require an assembly factor (RAVE) and are intertwined with glycolysis and glycolytic enzymes, RAS/cAMP/PKA components, cytosolic pH (pHCyt) homeostasis, and PI(3,5)P2.
Yeast V-ATPase inactivation by disassembly is a response to glucose deficit (10). V1Vo disassembly prevents energy depletion (e.g., loss of ATP). Reassembly is a response to glucose readdition following a brief period of glucose deprivation; it rapidly restores vacuolar acidification. Because Vo is not an open proton pore and cytosolic V1 cannot hydrolyze MgATP (7, 11–14), protons do not leak across membranes and cellular ATP is not depleted. Thus, disassembly can be sustained for a long time. Long-term disassembly is also rapidly reversed by addition of glucose (8), indicating that the structural and functional integrity of the V1 and Vo domains is preserved in the midst of scarcity.
All eukaryotic V-ATPases may be built with the potential to reversibly disassemble. However, not all V-ATPases appear to disassemble and reassemble. V-ATPase subunit isoforms and V-ATPase interactions with an assembly factor (RAVE; discussed below) in the cytosol and phosphoinositides at the membrane can dictate which pumps reversibly disassemble in response to environmental cues (16, 17). Recent studies have begun to elucidate the mechanisms that allow cells to communicate extracellular signals to intracellular V-ATPases located at the vacuolar membrane. In yeast, V-ATPase assembly is regulated by glucose, pH, and osmotic stress, and it is intertwined with glycolysis, RAS/cyclic AMP (cAMP)/protein kinase A (PKA), and phosphatidylinositol-(3,5)-bisphosphate [PI(3,5)P2] (16, 18–21). In insects, starvation and hormone stimulation influence V1Vo assembly by mechanisms involving cAMP/PKA signaling (9, 22, 23). In higher eukaryotes, glucose (renal epithelial cells) and mechanical stimulation (dendritic cells) have been shown to modulate V-ATPase assembly by a process that requires PIP 3-kinase and mTOR activation (24–27).
This review reports on the mechanisms of reversible disassembly in yeast, particularly in regard to our present understanding of the V-ATPase architecture. Next, we summarize recent structural discoveries on the yeast V-ATPase, their interrelation with V-ATPase regulation by reversible disassembly, and our current understanding of the mechanisms and signals involved.
ARCHITECTURE OF EUKARYOTIC V-ATPase
ATPase rotary motors include F-ATP synthase, archaeal A-type ATP synthase, bacterial A/V-like ATPase, and eukaryotic V-ATPase (28). V-ATPase and other members in this family share common structural features essential for the mechanical rotation of protein subunits during ATP catalysis. They all have (i) a protuberant globular domain peripherally attached to the membrane that houses three catalytic sites, (ii) a membrane domain that forms the path for ion transport, (iii) a centrally located rotor that couples ATP hydrolysis and ion transport across membranes, and (iv) one or more peripheral stalks that connect the peripheral and membrane domains.
Rotation of rotor-forming subunits relative to the steady catalytic sites is driven by hydrolysis of ATP inside the globular structure of V1 (A3B3) (Fig. 1). ATP hydrolysis promotes rotation of the rotor's shaft (subunits D, F, and d) at the center of the A3B3 hexamer. The shaft is connected to a hydrophobic proteolipid ring inside the membrane (c-ring), which consists of subunits c, c′, and c″ and transfers the protons. Active transport requires entrance of cytosolic protons to the Vo subunit a in order to reach the c-ring. The protons bind to an acidic residue in the c-ring, and after a 360° rotation, protons exit the other side of the membrane, traveling through Vo subunit a. This general mechanism of rotational catalysis is shared with all rotary ATPases (28).
Eukaryotic V-ATPases distinguish themselves from other rotary ATPases in three ways. First, V-ATPases are dedicated proton pumps. Second, V-ATPases are regulated by reversible disassembly. Third, V-ATPases contain three peripheral stalks. In contrast, the A and bacterial A/V-ATPases have two peripheral stalks and F-ATPases have one (28). The V-ATPase peripheral stalks are made of a heterodimer of E and G subunits; reversible disassembly requires the third peripheral stalk (EG3) (Fig. 3) (6, 29). It also requires a soluble subunit that is absent in other rotary ATPases (subunit C). The yeast subunit C contains two globular domains, the head (Chead) and foot (Cfoot) (30). The Chead domain interacts with EG3 with high affinity (6, 31). Through its Cfoot domain, subunit C interacts with the second peripheral stalk (EG2) and the N terminus of the Vo subunit a (a-NT). These subunit interactions are broken and reformed when V-ATPases disassemble and reassemble.
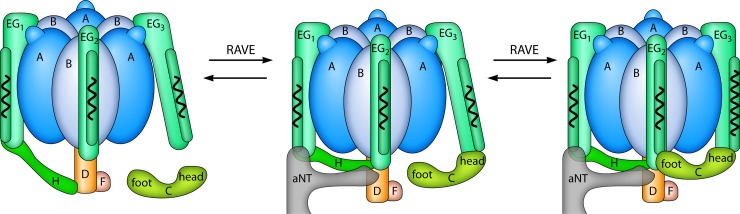
FIG 3.
Spring-loading: a model for disassembly and reassembly. The V1 domain and subunit C detach from Vo at the membrane and are released into the cytosol during disassembly. Reassembly requires reassociation of subunit C with the peripheral stalks EG3 and EG2 and the N terminus domain of the Vo subunit a (a-NT). Restoration of the native complex probably requires bending of EG3, like spring-loading, which is accomplished with the assistance of the chaperone complex RAVE. The tension contained in EG3 within the assembled V1Vo complex, is then released when V1Vo disassembles.
Subunit C is released to the cytosol during disassembly (8). Reassembly requires the subunit C to be rapidly reincorporated into the complex and its interactions with EG3, EG2, and a-NT to be restored. Reintroduction of subunit C into V1Vo requires significant bending of the third peripheral stalk (6, 29). This compression imposes physical stress in its coiled-coil structure, like “spring-loading.” The EG3 tension, which persists within assembled V1Vo complexes, is released when V1Vo disassembles. Thus, it is proposed that spring-loading requires energy for reassembly and primes V-ATPases to easily disassemble after glucose depletion, when ATP must be preserved.
These new structural discoveries hopefully will lead to a better understanding of how glucose and other cellular signals regulate V-ATPase function and assembly. The spring-loading mechanism of reversible disassembly is compatible with our current understanding of the structural architecture of the eukaryotic V-ATPase complex. It is also consistent with our knowledge of the major cellular processes associated with V1Vo disassembly and reassembly. Below, we discuss our view of the alignment of the V-ATPase architecture with these cellular processes and signals.
CONNECTING GLUCOSE METABOLISM TO V-ATPase ASSEMBLY
The concept of spring-loading requires energy to bend EG3 and reestablish proper binding of subunit C in V1Vo (6). Glucose, the primary energy source for most organisms, is an important driver of reassembly, suggesting that glucose oxidation could provide the necessary chemical energy (e.g., ATP). In addition to glucose, reassembly of V1Vo can be triggered by fructose and mannose, other rapidly fermentable sugars, suggesting that glycolysis itself may be necessary for V1Vo reassembly and spring-loading of EG3 (19). Further evidence that glucose metabolism is involved includes the facts that (i) conversion of glucose-6-phosphate to fructose-6 phosphate is necessary for reassembly and (ii) the intracellular pool of assembled V1Vo complexes is proportional to the concentration of glucose in the growth medium, demonstrating that V1Vo reassembly is not an all-or-none response (19).
The glycolytic enzymes aldolase (21), phosphofructokinase-1 (32), and glyceraldehyde-3-phosphate dehydrogenase (33) interact with V-ATPase. These enzymes coimmunoprecipitate with V-ATPases and can be detected in yeast vacuolar membrane fractions. Aldolase binding to V-ATPase is glucose dependent and necessary for stable V1Vo complex formation (21, 34). Lu et al. (21) were able to differentiate the function of aldolase in glycolysis from its function for V-ATPase assembly. The authors showed reduced V1Vo complex formation in an aldolase mutant that retained catalytic activity in vitro. These studies suggest that aldolase may play a direct role in V1Vo reassembly. Whether the same holds true for other glycolytic enzymes is not known. Glycolytic mutants cannot efficiently utilize glucose, which suppresses glycolysis and glucose-dependent signals, altering V1Vo assembly. This makes it challenging to study the interplay of V-ATPase with other glycolytic enzymes. Nonetheless, these studies merit additional examination because phosphofructokinase-1 can directly bind yeast and human V-ATPase subunits (24), suggesting that several aspects of this mechanism are conserved.
The interrelation between V-ATPase and glycolysis cannot be overlooked; it is conserved in yeast (1, 19, 35, 36), plants (37), and mammals (38, 39). Moreover, V-ATPase mutations that impair binding to phosphofructokinase-1 are associated with distal renal tubular acidosis (24), and V-ATPase regulation by glycolysis plays a role in viral infections (40) and the metabolic switch in cancers (41, 42). It has been proposed that glycolytic enzymes form a supercomplex with V-ATPase that funnels ATP directly to V-ATPase and propels proton transport (21, 24, 32, 34, 37, 43). A similar molecular machinery has been described at synaptic vesicle membranes where ATP synthesized by phosphoglycerate kinase supports glutamate uptake (44); this process is energized by V-ATPase proton pumps. A model of this kind will require glycolytic ATP production at the yeast vacuolar membrane, but functional interactions of phosphoglycerate kinase or pyruvate kinase (glycolytic enzymes that produce ATP) with V-ATPase have yet to be demonstrated. However, it is clear that ATP levels modulate V-ATPase coupling efficiency in vitro (45). ATP-dependent modifications of V-ATPase proton transport in vivo will probably need to work tightly coupled with glycolysis, the main source of ATP; glycolytic enzymes at the membrane could produce the ATP that fine-tunes the number of protons transported per ATP hydrolyzed. Collectively, these data suggest that V-ATPase reassembly and/or V-ATPase activity can be controlled by interactions with glycolytic enzymes and the ATP that they produce.
ONE “RAVE” PATH TOWARD V1Vo REASSEMBLY
The regulator of ATPase of vacuoles and endosomes (RAVE) complex is a V-ATPase-exclusive assembly factor. It is required for V1Vo assembly at steady state (biosynthetic assembly) and reassembly in response to glucose readdition to glucose-deprived cells (46–48). The RAVE complex chaperones loading of subunit C into V1Vo, a job that requires aligning Chead with the EG3 and EG2 peripheral stalks in addition to introducing structural stress in EG3 (Fig. 3) (6). In the absence of RAVE, V-ATPases at the vacuolar membrane are unstable and inactive, with V1 and subunit C loosely associated (48). Importantly, although several assembly factors are required for V-ATPase assembly (49–54), only RAVE appears to be involved in V-ATPase reversible disassembly.
The RAVE complex has three components, the adaptor protein Skp1p and its two subunits, Rav1p and Rav2p (46). Skp1p associates with other cellular complexes. Rav1p and Rav2p are solely found in the RAVE complex. Of the two subunits, Rav1p constitutes the central component; it binds Rav2p and Skp1p (47). Rav1p also forms the interface between RAVE and V-ATPase subunits. In the cytosol, Rav1p binds V-ATPase subunit C and the V1 peripheral stalk-forming subunits EG (48). At the membrane, Rav1p interacts with the N-terminal domain of Vo subunit a (17). Genetic and biochemical data have shown that binding of Rav1p to subunit C can occur independently of its binding to V1. Preloading RAVE with subunit C and V1 simultaneously in the cytosol may expedite reassembly, which is known to be a fast response completed within 3 to 5 min of glucose readdition (19, 55). Importantly, formation of RAVE-C and RAVE-V1 subcomplexes in the cytosol is not glucose dependent, indicating that RAVE binding is not the signal for V1Vo reassembly.
Deletion of the genes RAV1 and RAV2 leads to growth defects characteristic of V-ATPase mutants (46, 47); the vacuolar membrane ATPase (vma) growth phenotype displays growth sensitivity at pH 7.5 and in the presence of calcium (1). The rav1Δ and rav2Δ mutant cells also exhibit temperature sensitivity, but the vma traits are detected at 37°C. This phenotype is more substantial in rav1Δ than rav2Δ cells (46, 47), likely because Rav1p constitutes the functional subunit of the RAVE complex. The rav1Δ mutant has major V-ATPase assembly and functional defects in vivo, although its vma growth phenotype is fairly mild and considered “partial.”
The rav1Δ mutant resembles the yeast mutant strain vph1Δ, which lacks the isoform Vph1p of the Vo subunit a (56). The Vo subunit a is the only yeast V-ATPase subunit encoded by two functional homologs, VPH1 and STV1 (56, 57); VPH1 encodes the vacuolar isoform and STV1 has sorting information for the Golgi/endosomal compartments (58). Genome-level synthetic genetic analyses (17) showed that a synthetic vma growth phenotype can be generated after combining the rav1Δ mutation with class E mutants of endosomal and vacuolar transport (59), suggesting that the physiological basis for the rav1Δ partial vma phenotype is that RAVE is a Vph1p-specific chaperone. The discovery that RAVE assists in the assembly of Vph1p-containing V-ATPases but that Stv1p-containing complexes do not need RAVE to properly assemble is in agreement with prior studies showing that Vph1p-containing V-ATPases are more responsive to glucose than are Stv1p-containing pumps (60). Since Vph1p targets V-ATPase to the vacuole and Stv1p to the Golgi and endosomal compartments, these results also suggest that only vacuole-associated pumps reversibly disassemble.
The functions of the RAVE complex are likely conserved in other eukaryotes. The Rav1p sequence homologs, rabconnectins, are necessary for acidification of endosomes and synaptic vesicles (61–63). The human Vo subunit a exists in four different isoforms; mutations in particular isoforms cause osteopetrosis and renal tubular acidosis (64). Identifying the Vo subunit a isoform(s) recognized by rabconnectins could help in understanding these and other pathologies associated with V-ATPases. The RAVE subunit Rav2p is found only in fungi and does not have human homologs. Therefore, Rav2p offers unique opportunities to selectively disrupt RAVE complex functions in fungal human pathogens for which V-ATPase pumps are desirable targets (65–72).
IS ATP HYDROLYSIS NECESSARY FOR V-ATPase DISASSEMBLY AND REASSEMBLY?
How energy can be used to reassemble V1Vo is virtually unknown. ATP facilitates in vitro reconstitution of V1 and Vo (73–75), suggesting that ATP could promote reassembly in vivo. Genetic screens aimed at identifying V-ATPase mutants defective in V1Vo reassembly are challenging because V1Vo disassembly is not absolute. There is a cellular fraction of V-ATPase complexes that does not disassemble in response to glucose depletion; it probably yields basal V-ATPase activity necessary to support critical cellular functions (1, 10). These pumps constitute about 30% of the total V1Vo and likely include V-ATPases at nonvacuolar membranes (Stv1p-containing V-ATPase pumps) (19, 60). Coincidentally, 25 to 30% of V-ATPase activity is sufficient to support wild-type growth, which makes the growth phenotype of these types of mutants very subtle. A few mutants, primarily disassembly mutants, have been identified by site-directed mutagenesis experiments. Intriguingly, many of those mutations also alter V-ATPase catalysis. Those studies suggest that peripheral stalks may regulate rotational catalysis by influencing ATP binding, chemical reaction, or release of ADP/Pi (76–78).
As expected, site-directed mutations at conserved amino acids of the peripheral stalk subunits E and G can suppress glucose-dependent disassembly (Fig. 1) (76, 79). The mutations vma4-D44A and vma10-R25A/L in subunits E and G, respectively, suppress disassembly; they also stimulate the enzyme activity (76, 79). The vma4-T202A mutation near the C terminus enhances Vmax by about 2-fold without significantly affecting the Km of the enzyme (77), resembling the mutant vma4-D44A (76). Although the ability of vma4-T202A to reversibly disassemble has not been determined, these studies indicate that the peripheral stalks can communicate with the catalytic sites inside the A3B3 hexamer and they can affect disassembly and catalysis.
Paradoxically, the rate of ATP hydrolysis by V-ATPase can alter V1Vo disassembly (Fig. 1). Pharmacologic inhibition of V-ATPase pumps with the V-ATPase inhibitor concanamycin A reduces V1Vo disassembly, without affecting reassembly (19). Inactive V-ATPases carrying mutations at a Vo proton-binding subunit (vma11-E145L) of the c-ring (19) or the V1 catalytic subunit A (vma1-R219K) (78) are defective for disassembly. Likewise, vma1-P177V and vma1-R219A, which are partially inactive (by 30% to 50%), also are defective in disassembly. Thus, it appears that wild-type levels of activity are necessary to disassemble V1Vo; hyperactive, hypoactive, and inactive pumps cannot sufficiently disassemble (19, 45, 76). Obviously, not all disassembly mutants are catalytically impaired. The mutations vma1-G150 and vma1-D157E inhibit disassembly without affecting V-ATPase activity (78), perhaps by stabilizing subunit interactions.
From these studies, it becomes clear how little we still know about the intramolecular mechanisms that drive disassembly and reassembly. Although additional studies will be necessary to determine how intrinsic subunit interactions and differential conformations impact disassembly and reassembly, it appears that V-ATPases adopt conformations prone for disassembly during a catalytic cycle of rotation driven by ATP hydrolysis (19). Still, we cannot exclude the possibility that catalysis may also drive reassembly. In this context, ATP-driven subunit rotation in V1 may stimulate its own reassembly in the presence of RAVE. This process will require the inhibitory subunit of V1 (subunit H) to be released from its rotor-locking inhibitory position during reassembly (Fig. 3) (7, 11, 12, 80).
YEAST V-ATPase: AT THE CROSSROADS OF MULTIPLE INTRACELLULAR SIGNALS
In support of the spring-loading hypothesis, there are no known chaperones that aid in V-ATPase disassembly; V-ATPase may be primed to disassemble (29). In addition to glucose depletion, exposure to less preferred carbon sources (galactose, glycerol/ethanol, and raffinose) causes disassembly (8, 19). These data further argue that little or no glucose is the driving signal of disassembly. There is evidence indicating that the Ras/cAMP/PKA pathway probably mediates reversible disassembly (18, 20). Ras/cAMP/PKA signaling controls metabolism in response to sudden availability of rapidly fermentable sugars (81), compatible with a role for Ras/cAMP/PKA during V1Vo reassembly (Fig. 2). Constitutively active Ras and PKA suppress disassembly by glucose depletion (18). These studies suggest that the Ras/cAMP/PKA pathway acts upstream of V-ATPase. In an independent study linking V-ATPase reassembly to cAMP and PKA, reassembly appeared to be an upstream activator of PKA (20). That study suggested that alkalization of the cytosol after glucose readdition is the signal for reassembly. Although these results seem contradictory, the possibility that a positive feedback mechanism may regulate V-ATPase assembly cannot be excluded (Fig. 4). The reassembled V-ATPase may activate PKA signaling, which in turn enhances the V-ATPase assembly.
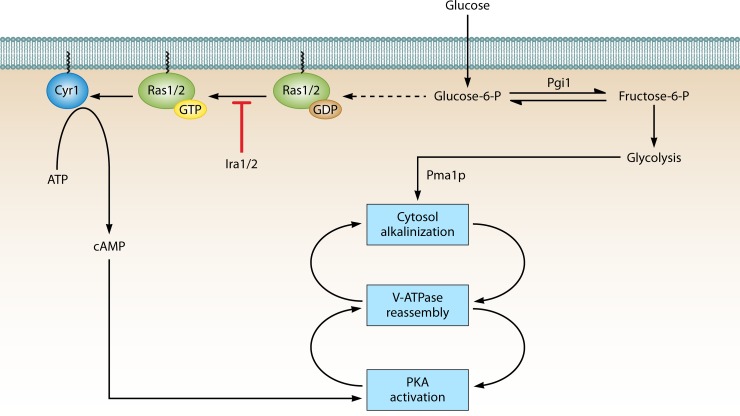
FIG 4.
Hypothetical positive feedback for the regulation of V-ATPase assembly by cytosolic pH and Ras/cAMP/PKA signaling. In this model, two positive feedback loops control glucose-dependent yeast V-ATPase assembly by promoting cytosol pH alkalinization and protein kinase A (PKA) activation. Glucose readdition after glucose depletion activates PKA and alkalinizes the cytosol, which promotes V-ATPase reassembly. More assembled V-ATPase helps maintain alkalinized cytosol pH. The reassembled V-ATPase may activate PKA signaling, which enhances V-ATPase assembly, upregulates glycolysis, and mediates the rapid transition from respiratory to fermentative growth. Arrows and bars represent positive and negative interactions, respectively. Dashed lines represent putative or indirect interactions.
Cytosolic pH is emerging as a key regulator for various cellular functions (82), and V-ATPase affects cytosolic pH homeostasis (83). In addition to activating V-ATPase catalysis and proton transport, readdition of glucose activates the plasma membrane ATPase, Pma1p, which is the main efflux pump responsible for yeast cytosolic pH regulation (83, 84). V-ATPases are necessary for cytosolic pH homeostasis because (i) active V-ATPases are necessary for normal Pma1p levels to be present at the plasma membrane, and (ii) cytosolic pH homeostasis is maintained by the coordinated action of V-ATPase and Pma1p (83). In the evaluation of the signals for reassembly, the contribution of cytosolic pH merits additional investigation. Addressing whether glucose-dependent Pma1p activation precedes glucose-dependent V-ATPase activation may help clarify the role of cytosolic pH for reassembly.
Fungi grow more rapidly at acidic than neutral pH (85). It should come as no surprise that V1Vo disassembly in response to glucose depletion is affected by environmental stress signals, such as elevated pH (86). At pH 7.0, V-ATPase disassembly is significantly suppressed compared to disassembly at pH 5.0, the optimal pH for yeast growth. Although the mechanisms involved in the prevention of disassembly by pH remain elusive, adaptation to high pH appears to have both PI(3,5)P2-dependent and -independent components (16). Knowing whether glucose and pH use common mechanisms to regulate V1Vo disassembly requires additional studies. Notably, it may help in the understanding of fungal pathogenicity; Candida albicans adaptation to neutral-to-alkaline pH environments in the host stimulates cellular signals that trigger its morphological change from the yeast form (nonpathogenic) to the hyphal form (pathogenic) (65, 68, 72, 87).
V-ATPase function is necessary for adaptation to stress conditions. Vacuoles are yeast storage compartments and an important mechanism of protection against toxic metals and drugs (88). By modulating V-ATPase disassembly, yeast protects the vacuolar luminal pH and maintains secondary transport systems across the membrane. Exposure of yeast to osmotic shock increases the total pool of vacuolar V1Vo assembled (89). This involves a mechanism that requires the signaling lipid PI(3,5)P2 interacting with the Vo subunit a isoform Vph1p (16). Interestingly, PI(3,5)P2 has little or no effect on glucose-dependent reversible disassembly of the V-ATPase, indicating that the cellular signals behind hyperosmotic stress- and glucose-induced V-ATPase reassembly are independent. High salts and high pH can increase V1Vo assembly levels at steady state in the presence of glucose, when cellular energy is abundant and most V-ATPase complexes are assembled. How this may work is not clear. It suggests that vacuolar membranes may contain subpopulations of V-ATPases specialized to respond to different signals, adding a layer of complexity to this intricate regulatory event.
CONCLUDING REMARKS
Structural data are beginning to support a collection of studies investigating how glucose signals are communicated to V-ATPases. The new concept is that V1Vo may be structurally built with an inherent facility to disassemble but that its reassembly imposes energetic constrains. This concept has reinforced our view of disassembly and reassembly as two independently controlled events. A variety of extracellular cues that control V-ATPase assembly and/or disassembly are emerging, although glucose is the main and strongest external stimulus. We do not know what is the glucose sensor or the mechanism involved in this communication. Our understanding of V-ATPase regulation by reversible disassembly is incomplete. The spring-loading hypothesis has not been experimentally tested. If all V-ATPases are structurally suited to reversibly disassemble, why do not all of them do so? Vph1p-containing V-ATPases disassemble and reassemble, but not Stv1p-containing V-ATPases. There are many questions that remain unanswered regarding the roles of glycolysis, RAS/cAMP/PKA, and V1 catalysis. Some of these questions include the following: (i) do glycolytic enzymes and/or glycolysis control V-ATPase at steady state and during glucose depletion/readdition; (ii) are glycolysis and RAS/cAMP/PKA elements of a common pathway or different pathways that work in parallel to control V-ATPase assembly and function; (iii) is V-ATPase upstream of PKA or downstream; (iv) what is the yeast V-ATPase subunit that is phosphorylated, if any; and (v) what phosphatase enzyme is involved.
ACKNOWLEDGMENTS
We gratefully acknowledge support, in whole or in part, from the NIH grant R01GM086495 (to K.J.P.), AHA grant 14PRE19020015 (to C.-Y.C.), and the UNM Health Sciences Center RAC Award (to J.C.).
We thank Colleen Fordyce and David Vander Jagt for the helpful discussions and revisions. We also thank Jessica “DJ” Binder for assistance with the illustrations.
Footnotes
Published ahead of print 4 April 2014
REFERENCES
- 1.Kane PM. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191. 10.1128/MMBR.70.1.177-191.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929. 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- 3.Breton S, Brown D. 2013. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 28:318–329. 10.1152/physiol.00007.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobacchi C, Frattini A, Orchard P, Porras O, Tezcan I, Andolina M, Babul-Hirji R, Baric I, Canham N, Chitayat D, Dupuis-Girod S, Ellis I, Etzioni A, Fasth A, Fisher A, Gerritsen B, Gulino V, Horwitz E, Klamroth V, Lanino E, Mirolo M, Musio A, Matthijs G, Nonomaya S, Notarangelo LD, Ochs HD, Superti Furga A, Valiaho J, van Hove JL, Vihinen M, Vujic D, Vezzoni P, Villa A. 2001. The mutational spectrum of human malignant autosomal recessive osteopetrosis. Hum. Mol. Genet. 10:1767–1773. 10.1093/hmg/10.17.1767 [DOI] [PubMed] [Google Scholar]
- 5.Kane PM. 2007. The long physiological reach of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 39:415–421. 10.1007/s10863-007-9112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oot RA, Huang LS, Berry EA, Wilkens S. 2012. Crystal structure of the yeast vacuolar ATPase heterotrimeric EGC(head) peripheral stalk complex. Structure 20:1881–1892. 10.1016/j.str.2012.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benlekbir S, Bueler SA, Rubinstein JL. 2012. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-A resolution. Nat. Struct. Mol. Biol. 19:1356–1362. 10.1038/nsmb.2422 [DOI] [PubMed] [Google Scholar]
- 8.Kane PM. 1995. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270:17025–17032 [PubMed] [Google Scholar]
- 9.Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H. 1995. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270:5649–5653. 10.1074/jbc.270.10.5649 [DOI] [PubMed] [Google Scholar]
- 10.Kane PM, Parra KJ. 2000. Assembly and regulation of the yeast vacuolar H(+)-ATPase. J. Exp. Biol. 203:81–87 [DOI] [PubMed] [Google Scholar]
- 11.Parra KJ, Keenan KL, Kane PM. 2000. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275:21761–21767. 10.1074/jbc.M002305200 [DOI] [PubMed] [Google Scholar]
- 12.Jefferies KC, Forgac M. 2008. Subunit H of the vacuolar (H+) ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F. J. Biol. Chem. 283:4512–4519. 10.1074/jbc.M707144200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diab H, Ohira M, Liu M, Cobb E, Kane PM. 2009. Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J. Biol. Chem. 284:13316–13325. 10.1074/jbc.M900475200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkens S, Forgac M. 2001. Three-dimensional structure of the vacuolar ATPase proton channel by electron microscopy. J. Biol. Chem. 276:44064–44068. 10.1074/jbc.M106579200 [DOI] [PubMed] [Google Scholar]
- 15.Ho MN, Hirata R, Umemoto N, Ohya Y, Takatsuki A, Stevens TH, Anraku Y. 1993. VMA13 encodes a 54-kDa vacuolar H(+)-ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae. J. Biol. Chem. 268:18286–18292 [PubMed] [Google Scholar]
- 16.Li SC, Diakov TT, Xu T, Tarsio M, Zhu W, Couoh-Cardel S, Weisman LS, Kane PM. 2014. The signaling lipid PI(3,5)P2 stabilizes V1-Vo sector interactions and activates the V-ATPase. Mol. Biol. Cell 25:1251–1262. 10.1091/mbc.E13-10-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smardon AM, Diab HI, Tarsio M, Diakov TT, Nasab ND, West RW, Kane PM. 2014. The RAVE complex is an isoform-specific V-ATPase assembly factor in yeast. Mol. Biol. Cell 25:356–367. 10.1091/mbc.E13-05-0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bond S, Forgac M. 2008. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J. Biol. Chem. 283:36513–36521. 10.1074/jbc.M805232200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra KJ, Kane PM. 1998. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 18:7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29:2515–2526. 10.1038/emboj.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M, Ammar D, Ives H, Albrecht F, Gluck SL. 2007. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 282:24495–24503. 10.1074/jbc.M702598200 [DOI] [PubMed] [Google Scholar]
- 22.Tiburcy F, Beyenbach KW, Wieczorek H. 2013. Protein kinase A-dependent and -independent activation of the V-ATPase in Malpighian tubules of Aedes aegypti. J. Exp. Biol. 216:881–891. 10.1242/jeb.078360 [DOI] [PubMed] [Google Scholar]
- 23.Voss M, Vitavska O, Walz B, Wieczorek H, Baumann O. 2007. Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A. J. Biol. Chem. 282:33735–33742. 10.1074/jbc.M703368200 [DOI] [PubMed] [Google Scholar]
- 24.Su Y, Blake-Palmer KG, Sorrell S, Javid B, Bowers K, Zhou A, Chang SH, Qamar S, Karet FE. 2008. Human H+ATPase a4 subunit mutations causing renal tubular acidosis reveal a role for interaction with phosphofructokinase-1. Am. J. Physiol. Renal Physiol. 295:F950–F958. 10.1152/ajprenal.90258.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. 2005. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell. Biol. 25:575–589. 10.1128/MCB.25.2.575-589.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. 2003. Activation of lysosomal function during dendritic cell maturation. Science 299:1400–1403. 10.1126/science.1080106 [DOI] [PubMed] [Google Scholar]
- 27.Liberman R, Bond S, Shainheit MG, Stadecker MJ, Forgac M. 2014. Regulated assembly of vacuolar ATPase is increased during cluster disruption-induced maturation of dendritic cells through a phosphatidylinositol 3-kinase/mTOR-dependent pathway. J. Biol. Chem. 289:1355–1363. 10.1074/jbc.M113.524561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muench SP, Trinick J, Harrison MA. 2011. Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44:311–356. 10.1017/S0033583510000338 [DOI] [PubMed] [Google Scholar]
- 29.Stewart AG, Stock D. 2012. Priming a molecular motor for disassembly. Structure 20:1799–1800. 10.1016/j.str.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 30.Drory O, Frolow F, Nelson N. 2004. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5:1148–1152. 10.1038/sj.embor.7400294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oot RA, Wilkens S. 2010. Domain characterization and interaction of the yeast vacuolar ATPase subunit C with the peripheral stator stalk subunits E and G. J. Biol. Chem. 285:24654–24664. 10.1074/jbc.M110.136960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Zhou A, Al-Lamki RS, Karet FE. 2003. The a-subunit of the V-type H+-ATPase interacts with phosphofructokinase-1 in humans. J. Biol. Chem. 278:20013–20018. 10.1074/jbc.M210077200 [DOI] [PubMed] [Google Scholar]
- 33.Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409:581–588. 10.1038/35054500 [DOI] [PubMed] [Google Scholar]
- 34.Lu M, Holliday LS, Zhang L, Dunn WA, Jr, Gluck SL. 2001. Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276:30407–30413. 10.1074/jbc.M008768200 [DOI] [PubMed] [Google Scholar]
- 35.Kane PM. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191. 10.1128/MMBR.70.1.177-191.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kane PM. 2012. Targeting reversible disassembly as a mechanism of controlling V-ATPase activity. Curr. Protein Peptide Sci. 13:117–123. 10.2174/138920312800493142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konishi H, Yamane H, Maeshima M, Komatsu S. 2004. Characterization of fructose-bisphosphate aldolase regulated by gibberellin in roots of rice seedling. Plant Mol. Biol. 56:839–848. 10.1007/s11103-004-5920-2 [DOI] [PubMed] [Google Scholar]
- 38.Merkulova M, Hurtado-Lorenzo A, Hosokawa H, Zhuang Z, Brown D, Ausiello DA, Marshansky V. 2011. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am. J. Physiol. Cell Physiol. 300:C1442–C1455. 10.1152/ajpcell.00076.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura S. 2004. Glucose activates H(+)-ATPase in kidney epithelial cells. Am. J. Physiol. Cell Physiol. 287:C97–C105. 10.1152/ajpcell.00469.2003 [DOI] [PubMed] [Google Scholar]
- 40.Kohio HP, Adamson AL. 2013. Glycolytic control of vacuolar-type ATPase activity: a mechanism to regulate influenza viral infection. Virology 444:301–309. 10.1016/j.virol.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 41.Fogarty FM, O'Keeffe J, Zhadanov A, Papkovsky D, Ayllon V, O'Connor R. 21 October 2013. HRG-1 enhances cancer cell invasive potential and couples glucose metabolism to cytosolic/extracellular pH gradient regulation by the vacuolar-H ATPase. Oncogene 10.1038/onc.2013.403 [DOI] [PubMed] [Google Scholar]
- 42.Avnet S, Di Pompo G, Lemma S, Salerno M, Perut F, Bonuccelli G, Granchi D, Zini N, Baldini N. 2013. V-ATPase is a candidate therapeutic target for Ewing sarcoma. Biochim. Biophys. Acta 1832:1105–1116. 10.1016/j.bbadis.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 43.Lu M, Sautin YY, Holliday LS, Gluck SL. 2004. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 279:8732–8739. 10.1074/jbc.M303871200 [DOI] [PubMed] [Google Scholar]
- 44.Ikemoto A, Bole DG, Ueda T. 2003. Glycolysis and glutamate accumulation into synaptic vesicles. Role of glyceraldehyde phosphate dehydrogenase and 3-phosphoglycerate kinase. J. Biol. Chem. 278:5929–5940. 10.1074/jbc.M211617200 [DOI] [PubMed] [Google Scholar]
- 45.Shao E, Forgac M. 2004. Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J. Biol. Chem. 279:48663–48670. 10.1074/jbc.M408278200 [DOI] [PubMed] [Google Scholar]
- 46.Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384–391. 10.1038/35070067 [DOI] [PubMed] [Google Scholar]
- 47.Smardon AM, Tarsio M, Kane PM. 2002. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277:13831–13839. 10.1074/jbc.M200682200 [DOI] [PubMed] [Google Scholar]
- 48.Smardon AM, Kane PM. 2007. RAVE is essential for the efficient assembly of the C subunit with the vacuolar H(+)-ATPase. J. Biol. Chem. 282:26185–26194. 10.1074/jbc.M703627200 [DOI] [PubMed] [Google Scholar]
- 49.Hirata R, Umemoto N, Ho MN, Ohya Y, Stevens TH, Anraku Y. 1993. VMA12 is essential for assembly of the vacuolar H(+)-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J. Biol. Chem. 268:961–967 [PubMed] [Google Scholar]
- 50.Hill KJ, Stevens TH. 1994. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol. Biol. Cell 5:1039–1050. 10.1091/mbc.5.9.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill KJ, Stevens TH. 1995. Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H(+)-ATPase complex. J. Biol. Chem. 270:22329–22336. 10.1074/jbc.270.38.22329 [DOI] [PubMed] [Google Scholar]
- 52.Ryan M, Graham LA, Stevens TH. 2008. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol. Biol. Cell 19:5131–5142. 10.1091/mbc.E08-06-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malkus P, Graham LA, Stevens TH, Schekman R. 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell 15:5075–5091. 10.1091/mbc.E04-06-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis-Kaplan SR, Compton MA, Flannery AR, Ward DM, Kaplan J, Stevens TH, Graham LA. 2006. PKR1 encodes an assembly factor for the yeast V-type ATPase. J. Biol. Chem. 281:32025–32035. 10.1074/jbc.M606451200 [DOI] [PubMed] [Google Scholar]
- 55.Kane PM, Tarsio M, Liu J. 1999. Early steps in assembly of the yeast vacuolar H+-ATPase. J. Biol. Chem. 274:17275–17283. 10.1074/jbc.274.24.17275 [DOI] [PubMed] [Google Scholar]
- 56.Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. 1992. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267:14294–14303 [PubMed] [Google Scholar]
- 57.Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. 1994. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J. Biol. Chem. 269:14064–14074 [PubMed] [Google Scholar]
- 58.Finnigan GC, Cronan GE, Park HJ, Srinivasan S, Quiocho FA, Stevens TH. 2012. Sorting of the yeast vacuolar-type, proton-translocating ATPase enzyme complex (V-ATPase): identification of a necessary and sufficient Golgi/endosomal retention signal in Stv1p. J. Biol. Chem. 287:19487–19500. 10.1074/jbc.M112.343814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coonrod EM, Stevens TH. 2010. The yeast vps class E mutants: the beginning of the molecular genetic analysis of multivesicular body biogenesis. Mol. Biol. Cell 21:4057–4060. 10.1091/mbc.E09-07-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawasaki-Nishi S, Nishi T, Forgac M. 2001. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276:17941–17948. 10.1074/jbc.M010790200 [DOI] [PubMed] [Google Scholar]
- 61.Yan Y, Denef N, Schupbach T. 2009. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17:387–402. 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Einhorn Z, Trapani JG, Liu Q, Nicolson T. 2012. Rabconnectin3alpha promotes stable activity of the H+ pump on synaptic vesicles in hair cells. J. Neurosci. 32:11144–11156. 10.1523/JNEUROSCI.1705-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sethi N, Yan Y, Quek D, Schupbach T, Kang Y. 2010. Rabconnectin-3 is a functional regulator of mammalian Notch signaling. J. Biol. Chem. 285:34757–34764. 10.1074/jbc.M110.158634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toei M, Saum R, Forgac M. 2010. Regulation and isoform function of the V-ATPases. Biochemistry 49:4715–4723. 10.1021/bi100397s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayek SR, Lee SA, Parra KJ. 2014. Advances in targeting the vacuolar proton-translocating ATPase (V-ATPase) for anti-fungal therapy. Front. Pharmacol. 5:4. 10.3389/fphar.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raines SM, Rane HS, Bernardo SM, Binder JL, Lee SA, Parra KJ. 2013. Deletion of vacuolar proton-translocating ATPase V(o)a isoforms clarifies the role of vacuolar pH as a determinant of virulence-associated traits in Candida albicans. J. Biol. Chem. 288:6190–6201. 10.1074/jbc.M112.426197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, Lee SA. 2013. Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation. Eukaryot. Cell 12:1369–1382. 10.1128/EC.00118-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patenaude C, Zhang Y, Cormack B, Kohler J, Rao R. 2013. Essential role for vacuolar acidification in Candida albicans virulence. J. Biol. Chem. 288:26256–26264. 10.1074/jbc.M113.494815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang YQ, Gamarra S, Garcia-Effron G, Park S, Perlin DS, Rao R. 2010. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6:e1000939. 10.1371/journal.ppat.1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang YQ, Rao R. 2010. Beyond ergosterol: linking pH to antifungal mechanisms. Virulence 1:551–554. 10.4161/viru.1.6.13802 [DOI] [PubMed] [Google Scholar]
- 71.Schachtschabel D, Arentshorst M, Lagendijk EL, Ram AF. 2012. Vacuolar H(+)-ATPase plays a key role in cell wall biosynthesis of Aspergillus niger. Fungal Genet. Biol. 49:284–293. 10.1016/j.fgb.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 72.Parra KJ. 2012. Vacuolar ATPase (V-ATPase): a model proton pump for antifungal drug discovery, p 89–100 In Tegos G, Mylonakis E. (ed), Antimicrobial drug discovery. Emerging strategies. Advances in molecular and cellular microbiology. CABI, Wallingford, Oxfordshire, United Kingdom [Google Scholar]
- 73.Parra KJ, Kane PM. 1996. Wild-type and mutant vacuolar membranes support pH-dependent reassembly of the yeast vacuolar H+-ATPase in vitro. J. Biol. Chem. 271:19592–19598. 10.1074/jbc.271.32.19592 [DOI] [PubMed] [Google Scholar]
- 74.Tomashek JJ, Garrison BS, Klionsky DJ. 1997. Reconstitution in vitro of the V1 complex from the yeast vacuolar proton-translocating ATPase. Assembly recapitulates mechanism. J. Biol. Chem. 272:16618–16623 [DOI] [PubMed] [Google Scholar]
- 75.Imamura H, Funamoto S, Yoshida M, Yokoyama K. 2006. Reconstitution in vitro of V1 complex of Thermus thermophilus V-ATPase revealed that ATP binding to the A subunit is crucial for V1 formation. J. Biol. Chem. 281:38582–38591. 10.1074/jbc.M608253200 [DOI] [PubMed] [Google Scholar]
- 76.Okamoto-Terry H, Umeki K, Nakanishi-Matsui M, Futai M. 2013. Glu-44 in the amino-terminal alpha-helix of yeast vacuolar ATPase E subunit (Vma4p) has a role for VoV1 assembly. J. Biol. Chem. 288:36236–36243. 10.1074/jbc.M113.506741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Owegi MA, Carenbauer AL, Wick NM, Brown JF, Terhune KL, Bilbo SA, Weaver RS, Shircliff R, Newcomb N, Parra-Belky KJ. 2005. Mutational analysis of the stator subunit E of the yeast V-ATPase. J. Biol. Chem. 280:18393–18402. 10.1074/jbc.M412567200 [DOI] [PubMed] [Google Scholar]
- 78.Shao E, Nishi T, Kawasaki-Nishi S, Forgac M. 2003. Mutational analysis of the non-homologous region of subunit A of the yeast V-ATPase. J. Biol. Chem. 278:12985–12991. 10.1074/jbc.M212096200 [DOI] [PubMed] [Google Scholar]
- 79.Charsky CM, Schumann NJ, Kane PM. 2000. Mutational analysis of subunit G (Vma10p) of the yeast vacuolar H+-ATPase. J. Biol. Chem. 275:37232–37239. 10.1074/jbc.M006640200 [DOI] [PubMed] [Google Scholar]
- 80.Rizzo JM, Tarsio M, Martinez-Munoz GA, Kane PM. 2007. Diploids heterozygous for a vma13Delta mutation in Saccharomyces cerevisiae highlight the importance of V-ATPase subunit balance in supporting vacuolar acidification and silencing cytosolic V1-ATPase activity. J. Biol. Chem. 282:8521–8532. 10.1074/jbc.M607092200 [DOI] [PubMed] [Google Scholar]
- 81.Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56:1–32. 10.1007/s00294-009-0287-1 [DOI] [PubMed] [Google Scholar]
- 82.Orij R, Urbanus ML, Vizeacoumar FJ, Giaever G, Boone C, Nislow C, Brul S, Smits GJ. 2012. Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pH(c) in Saccharomyces cerevisiae. Genome Biol. 13:R80. 10.1186/gb-2012-13-9-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez-Munoz GA, Kane P. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309–20319. 10.1074/jbc.M710470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rao R, Slayman CW. 1993. Mutagenesis of conserved residues in the phosphorylation domain of the yeast plasma membrane H(+)-ATPase. Effects on structure and function. J. Biol. Chem. 268:6708–6713 [PubMed] [Google Scholar]
- 85.Blank LM, Sauer U. 2004. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 150:1085–1093. 10.1099/mic.0.26845-0 [DOI] [PubMed] [Google Scholar]
- 86.Diakov TT, Kane PM. 2010. Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J. Biol. Chem. 285:23771–23778. 10.1074/jbc.M110.110122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Biswas S, Van Dijck P, Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376. 10.1128/MMBR.00009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li SC, Kane PM. 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793:650–663. 10.1016/j.bbamcr.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li SC, Diakov TT, Rizzo JM, Kane PM. 2012. Vacuolar H+-ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress. Eukaryot. Cell 11:282–291. 10.1128/EC.05198-11 [DOI] [PMC free article] [PubMed] [Google Scholar]