Abstract
Bromodomains are highly conserved acetyl-lysine binding domains found mainly in proteins associated with chromatin and nuclear acetyltransferases. The Trypanosoma cruzi genome encodes at least four bromodomain factors (TcBDFs). We describe here bromodomain factor 3 (TcBDF3), a bromodomain-containing protein localized in the cytoplasm. TcBDF3 cytolocalization was determined, using purified antibodies, by Western blot and immunofluorescence analyses in all life cycle stages of T. cruzi. In epimastigotes and amastigotes, it was detected in the cytoplasm, the flagellum, and the flagellar pocket, and in trypomastigotes only in the flagellum. Subcellular localization of TcBDF3 was also determined by digitonin extraction, ultrastructural immunocytochemistry, and expression of TcBDF3 fused to cyan fluorescent protein (CFP). Tubulin can acquire different posttranslational modifications, which modulate microtubule functions. Acetylated α-tubulin has been found in the axonemes of flagella and cilia, as well as in the subpellicular microtubules of trypanosomatids. TcBDF3 and acetylated α-tubulin partially colocalized in isolated cytoskeletons and flagella from T. cruzi epimastigotes and trypomastigotes. Interaction between the two proteins was confirmed by coimmunoprecipitation and far-Western blot assays with synthetic acetylated α-tubulin peptides and recombinant TcBDF3.
INTRODUCTION
Trypanosoma cruzi is a protozoan parasite and the causative agent of Chagas disease. It has a complex life cycle that alternates between two hosts and at least four distinct developmental stages. Amastigotes and bloodstream trypomastigotes are present in the mammalian host, whereas epimastigotes and the infective metacyclic trypomastigotes are present in the insect vectors (from the families Triatominae and Reduviidae). The differentiation event from epimastigotes to metacyclic trypomastigotes, occurring inside the insect, is called metacyclogenesis. This process can be induced in vitro using artificial media that resemble the conditions inside the vector and occurs spontaneously in old epimastigote cultures (1).
Trypanosomatids are characterized by the presence of a particular cytoskeleton responsible for the modulation of cell shape between the different life cycle stages and for motility and attachment to the host cell surface (2). These parasites present a layer of microtubules (MTs), the subpellicular microtubules, located below the plasma membrane, and a flagellum with a typical 9-plus-2 pattern of axonemal microtubules. The flagellum emerges from a membrane invagination called the flagellar pocket (FP). Four cytoplasmic microtubules are nucleated close to the basal body and run around the FP and along the entire flagellum attachment zone (FAZ) to the anterior cellular pole. The FP lacks the layer of subpellicular microtubules and is the place where endocytosis and exocytosis occur (3).
Microtubules can acquire a variety of evolutionarily conserved posttranslational modifications (PTMs). It has been proposed that these modifications dictate the recruitment of protein complexes that might regulate microtubule-based functions in different cellular locations (4–7). Acetylation occurs on lysine 40 of α-tubulin (4, 5), and it was thought that MT stabilization was a consequence of this PTM (6, 7). However, it was recently demonstrated that acetylation of MTs does not necessarily affect their stability (8, 9).
In trypanosomatids, acetylated α-tubulin is found in the subpellicular microtubules and in the flagella of T. cruzi and Trypanosoma brucei (10, 11). This PTM is also present in the ephemeral microtubules of the mitotic spindle of T. brucei (11). The presence of acetylated α-tubulin in early mitotic spindles in several organisms and the fact that protozoa like T. brucei and T. cruzi exhibit global α-tubulin acetylation reinforce the idea that the PTM is not restricted to stable MTs (reviewed in reference 12).
Although tubulin acetylation was described a long time ago, the enzymes responsible for this reversible modification have recently started to emerge. The first tubulin deacetylase described in mammals was HDAC6 (class II histone deacetylase [HDAC]), which can also modify other nonnuclear proteins (13). SIRT2 is a NAD-dependent deacetylase (class III histone deacetylase, or sirtuin) capable of modifiying α-tubulin (14). In the T. cruzi genome (http://tritrypdb.org/tritrypdb/), there are two coding sequences (CDSs) for histone deacetylases homologous to HDAC6 and two CDSs for sirtuins. The Leishmania infantum sirtuin (LiSIR2RP1) is a NAD-dependent deacetylase and ADP-ribosyltransferase capable of deacetylating α-tubulin (15). Also, two tubulin acetyltransferases have been described. The ELP3 subunit of the Elongator complex is known to modify microtubules of the cortical neurons (16), and αTAT-1 has recently been proposed as the major α-tubulin K40 acetyltransferase in mammals and nematodes (17). The T. cruzi genome contains two sequences homologous to ELP3. In T. brucei, ELPa (TbELPa) and TbELPb have been reported (18). Also, a putative sequence homologous to αTAT-1 is present in trypanosomatids but has not yet been characterized.
Bromodomains are conserved protein modules capable of binding acetylated lysines and are found in proteins associated with chromatin and in nearly every nuclear histone acetyltransferase. They have an atypical left-handed four-helix bundle structure (helices αA, αB, αC, and αZ) connected by two loops (loop ZA and loop BC) that constitute the surface-accessible hydrophobic pocket, where the acetyl-lysine binding site is located (19). Bromodomains can interact with other proteins in an acetylation-dependent manner and form multisubunit complexes (20). The bromodomain is considered a nuclear domain, but a small number of bromodomain-containing proteins have a dual nuclear-cytosolic localization (21–23).
Genes coding for putative bromodomain-containing factors (BDFs) were found in the TriTryp genomes (T. brucei, T. cruzi, and Leishmania spp.) (24). We previously characterized T. cruzi BDF2 (TcBDF2), which binds histone H4 (25). Here, we describe bromodomain factor 3 from T. cruzi (TcBDF3), the first exclusively nonnuclear bromodomain-containing protein reported so far. TcBDF3 is expressed in all life cycle stages and interacts with acetylated α-tubulin, the major component of the flagellar and subpellicular microtubules. In both metacyclic and bloodstream trypomastigotes, TcBDF3 was found to be concentrated in the flagellum and in the flagellar pocket region. Even though the precise function of TcBDF3 remains unrevealed, the results presented here suggest the participation of this bromodomain factor in cytoskeleton dynamics.
MATERIALS AND METHODS
Ethics statement.
All experiments were approved by the Institutional Animal Care and Use Committee of the School of Biochemical and Pharmaceutical Sciences (National University of Rosario, Rosario, Argentina) and were conducted according to the specifications of the U.S. National Institutes of Health guidelines for the care and use of laboratory animals.
Cell culture.
The Vero cell line was routinely cultivated in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS), 0.15% (wt/vol) NaHCO3, 100 U ml−1 penicillin, and 100 mg ml−1 streptomycin at 37°C in a humid atmosphere containing 5% CO2.
Parasites.
Epimastigotes of T. cruzi strain Dm28c were cultured in liver infusion-tryptose (LIT) medium (26) supplemented with 10% FCS at 28°C. The parasites were kept in exponential growth phase by subculturing every 3 days. Intracellular forms and trypomastigotes were obtained by infecting Vero cells with trypomastigotes, as previously described (27, 28).
To obtain metacyclic trypomastigotes, epimastigotes were differentiated in vitro following the procedure described by Contreras and coworkers (1) under chemically defined conditions using triatomine artificial urine medium (TAU; 190 mM NaCl, 17 mM KCl, 2 mM MgCl, 2 mM CaCl2, 8 mM phosphate buffer, pH 6.0, 0.035% sodium bicarbonate). Culture supernatants were collected after 24, 48, and 72 h of incubation in TAU3AAG medium (TAU supplemented with 20 mM l-proline, 50 mM l-glutamate, 2 mM l-aspartate, and 10 mM glucose).
Plasmid construction and expression of TcBDF3.
The TcBDF3 gene was amplified by PCR using BDF3Fw (5′AAGGATCCATGGGCTCTACGGGTCGG) and BDF3Rv (5′AACTCGAGCCTCGTCCTCCACCGCC) oligonucleotides. The TcBDF3ΔC fragment was amplified using BDF3Fw and BDF3ΔCRv (5′AACTCGAGTGCTCTTCCGCAAGACGCTCC) oligonucleotides. The restriction sites BamHI and XhoI (underlined) were inserted in the oligonucleotides. Proofreading DNA polymerase was used, and DNA purified from cultured T. cruzi epimastigotes served as the template.
The PCR products were inserted into the pCR 2.1TOPO vector (Invitrogen) and sequenced. TcBDF3 and TcBDF3ΔC coding regions were inserted into a pENTR3C vector (Gateway System Invitrogen) and then transferred by recombination to pDEST17 (Gateway System Invitrogen) and pTcCFPN (29), using LR clonase II enzyme mix (Invitrogen), to generate histidine tag and cyan fluorescent protein (CFP) fusions. pDEST17-TcBDF3 was transformed into Escherichia coli BL21 pLysS, and the recombinant protein (fused to a His tag) was obtained by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. The protein was purified by affinity chromatography using an Ni-nitrilotriacetic acid (NTA) agarose resin (Qiagen), following the manufacturer's instructions. The secondary structure of recombinant TcBDF3 was measured by circular dicroism.
Polyclonal antibodies.
Rabbit and mouse polyclonal antisera against TcBDF3 were obtained by subcutaneously inoculating recombinant TcBDF3 three times, first using complete Freund's adjuvant and subsequently with incomplete adjuvant. Specific antibodies were purified from the antisera obtained by chromatography through an Ni-NTA agarose column containing cross-linked TcBDF3-His. Antibodies were eluted with 100 mM triethylamine, pH 11; neutralized to pH 7; and stored at 4°C or at −20°C with 50% glycerol. The specificity of the purified antibodies was tested by Western blot assays.
Protein extracts.
Exponentially growing epimastigotes were washed twice with cold phosphate-buffered saline (PBS), and the pellets were resuspended in lysis buffer (20 mM HEPES, 8 M urea) and incubated for 30 min at room temperature with gentle agitation. Insoluble debris was eliminated by centrifugation. The same procedure was applied to amastigote and trypomastigote cellular pellets. To obtain nuclear extracts, exponentially growing epimastigotes were washed with PBS and lysed in hypotonic buffer A (10 mM HEPES, pH 8, 50 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1% [vol/vol] Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], aprotinin, 0.25% Triton X-100), 5% (vol/vol) glycerol was added, and the pellet was collected by centrifugation. The pellets were washed with buffer B (10 mM HEPES, pH 8, 140 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 5% [vol/vol] glycerol, 1 mM PMSF, 10 μg ml−1 aprotinin) and incubated for 10 min on ice. Nuclei were collected by centrifugation and resuspended in buffer C (10 mM HEPES, pH 8, 400 mM NaCl, 0.1 mM EDTA, 0.5 mM dithiothreitol [DTT], 5% [vol/vol] glycerol, 1 mM PMSF, 10 μg ml−1 aprotinin), incubated for 1 h on ice, and sonicated. This extraction was repeated three times, and the supernatants were precipitated with 20% trichloroacetic acid (TCA) overnight at 4°C.
T. cruzi cytoskeleton-enriched extracts were prepared as previously described by Schneider and coworkers (11). Briefly, cells were harvested, washed twice with PBS, and incubated with EB buffer (50 mM HEPES, 5 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, 2 mM MgCl2, 0.1% Triton X-100, and protease inhibitor cocktail set I [Calbiochem]) on ice for 30 min. The lysate was centrifuged at 10,000 × g for 15 min, and the supernatant, which contained soluble proteins, was named Sn1. The remaining pellet was incubated with EB buffer supplemented with 1 M NaCl on ice for 5 min, sonicated, and centrifuged at 20,000 × g for 15 min. The supernatant obtained contained soluble flagellar and cytoskeletal proteins (Sn2). The remaining pellet contained insoluble flagellar and cytoskeletal proteins. The supernatants were precipitated with 20% TCA overnight at 4°C.
For immunoprecipitation assays, cells were incubated with 1 mM dithiobis(succinimidylpropionate) (DSP) (Thermo Scientific) for 2 h on ice. This reagent was used to cross-link protein complexes within the cell. After incubation, the reaction was stopped with 20 mM Tris, pH 7.5. Then, the cells were harvested and incubated with EB buffer supplemented with 1 M NaCl on ice for 30 min. The lysate was centrifuged at 10,000 × g for 15 min, and the supernatant obtained was used to perform coimmunoprecipitation experiments.
Western blotting and slot blotting.
Protein extracts (30 to 50 μg per well) were separated by SDS-PAGE and transferred to nitrocellulose membranes. The transferred proteins were visualized with Ponceau S. The membranes were treated with 10% nonfat milk in PBS for 2 h and then incubated with specific antibodies diluted in PBS for 3 h. The antibodies used were polyclonal rabbit and mouse anti-TcBDF3, monoclonal mouse anti-acetylated α-tubulin antibody clone 6-11B-1 (Sigma-Aldrich), monoclonal mouse anti-trypanosome α-tubulin clone TAT-1 (a gift from K. Gull, University of Oxford, Oxford, England, United Kingdom), mouse anti-paraflagellar rod protein 2 (a gift from Ariel Silber, University of São Pablo, São Pablo, Brazil), mouse anti-T. cruzi histone H4 (a gift from Sergio Shenkman, Universidade Federal de São Paulo, São Paulo, Brazil), rabbit anti-T. cruzi high-mobility group B (TcHMGB) (30), and anti-T. cruzi bromodomain factor 2 (TcBDF2) (25). Bound antibodies were detected using peroxidase-labeled anti-mouse or anti-rabbit IgGs (GE Healthcare) and ECL Prime (GE Healthcare) according to the manufacturer's protocol.
Slot blotting was performed by immobilizing 10 μg of synthetic peptides—α-tubulin (PDGAMPSDKTIGVEDDA; Genscript), α-tubulin acetylated (ac) on lysine 40 (PDGAMPSDKacTIGVEDDA; Genscript), and histone H4 acetylated on lysine 14 (AKGKKSGEAKGTQKacRQ; [31])—onto nitrocellulose membranes. The membranes were incubated with recombinant His-tagged TcBDF3 or TcBDF2 for 3 h (0.5 μg/ml), and bound proteins were visualized using anti-histidine antibodies (GE Healthcare) and detected as described above.
Subcellular localization of TcBDF3 by digitonin extraction.
Parasites in exponential phase were collected, washed, and resuspended in buffer A (20 mM Tris-HCl, pH 7.2, with 225 mM sucrose, 20 mM KCl, 10 mM KH2PO4, 5 mM MgCl2, 1 mM Na2EDTA, and 1 mM DTT) at a protein concentration of 1 mg ml−1 and supplemented with digitonin (0 to 1 mg; final volume, 1 ml for each digitonin concentration). The resuspended parasites were incubated at 28°C for 20 min before being centrifuged at 14,000 × g for 2 min at 4°C. The enzymatic activities of alpha-hydroxyacid dehydrogenase (αHAdH) (cytosolic marker) and malate dehydrogenase (MdH) (glycosomal and mitochondrial marker) were determined in the supernatant (S) and occasionally in the cell pellet (P) in the presence of 0.1% (vol/vol) Triton X-100 and 150 mM NaCl. To measure the enzymatic activity of αHAdH, the extracts were incubated with 4 μl of 0.5 mM NADH, 7 μl of 1 mM phenylpyruvate, 2 μl 20% (vol/vol) Tritón X-100, and 40 μl of the protein extract in 10 mM Tris-HCl, pH 8. The percentage of activity was determined spectrophotometrically by measuring the oxidation of NADH at 339 nm (NADH ε339 = 6,220 M−1 cm−1) at 340 nm. To measure the enzymatic activity of MdH, the extracts were incubated with 5 μl of 0.5 mM NADH, 10 μl of 1 mM oxalacetate, 2 μl 20% (vol/vol) Tritón X-100, and 20 μl of the protein extract in 10 mM Tris-HCl, pH 8. The percentage of activity was determined spectrophotometrically by measuring the oxidation of NADH at 339 nm (NADH ε339 = 6,220 M−1 cm−1) at 340 nm.
Equal volumes of selected S and P fractions were subjected to SDS-PAGE, blotted onto nitrocellulose membranes, and probed with specific antibodies. The antibodies used were anti-tyrosine amino transferase (TcTAT), anti-glycosomal malate dehydrogenase (TcMdHglyc), and anti-mitochondrial malate dehydrogenase (TcMdHmit) antibodies (all gifts from Cristina Nowicki, Universidad de Buenos Aires, Buenos Aires, Argentina) and anti-paraflagellar rod 2 (TcPAR2) and mouse monoclonal anti-trypanosome α-tubulin clone TAT-1 (α-tubulin) antibodies.
Ultrastructural immunocytochemistry.
Parasites were fixed in 0.3% glutaraldehyde, 4% formaldehyde, and 1% picric acid diluted in 0.1 M cacodylate buffer at pH 7.2 and then dehydrated at 20°C using a graded acetone series and progressively infiltrated with Unicryl Resin (Polysciences, Inc.) at lower temperatures. The polymerization of the resin was carried out in BEEM capsules (Ted Pella, Inc.) at 20°C for 5 days under UV light. Ultrathin sections were obtained in a Leica ultramicrotome (Reichert Ultracuts), and the grids containing the sections were incubated with 50 mM NH4Cl for 30 min. The grids were then incubated with blocking solution (3.5% bovine serum albumin [BSA], 0.5% teleostean gelatin, 0.02% Tween 20 diluted in PBS, pH 8.0) for 30 min and finally with goat serum diluted (1:200) in blocking solution. Grids containing ultrathin sections were incubated for 1 h with anti-TcBDF3 antibodies diluted in blocking solution (1:1) and washed with PBS. The grids were then incubated with gold-labeled goat anti-rabbit IgG diluted 1:200 for 45 min, washed with blocking solution, and stained with uranyl acetate and lead citrate for further observation using a Zeiss 900 transmission electron microscope. In control assays, sections were not incubated with the primary antiserum.
Immunocytolocalization.
Trypomastigotes and exponentially growing epimastigotes were centrifuged, washed twice with PBS, settled on polylysine-coated coverslips, and fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. The fixed parasites were washed with PBS and permeabilized with 0.2% Triton X-100 in PBS for 10 min. After washing with PBS, the parasites were incubated with the appropriate primary antibody diluted in 1% BSA in PBS for 3 h at room temperature. In colocalization experiments, the two antibodies were incubated together. Nonbound antibodies were washed with 0.01% Tween 20 in PBS, and then the slides were incubated with fluorescence-conjugated anti-rabbit (fluorescein; Jackson Immuno Research) or anti-mouse (rhodamine; Calbiochem) IgG antibodies and 2 μg ml−1 DAPI (4′,6-diamidino-2-phenylindole) for 1 h. Alternatively, DNA was stained with propidium iodide (Invitrogen) according to the manufacturer's instructions. The slides were washed with 0.01% Tween 20 in PBS and finally mounted with VectaShield (Vector Laboratories). To analyze intracellular amastigotes, Vero cell monolayers were grown on coverslips and infected with T. cruzi trypomastigotes as described above. Three days postinfection, the cultures were washed with PBS and fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. The same procedure described above was followed for immunodetection. Images were acquired with a confocal Nikon Eclipse TE-2000-E2 microscope using Nikon EZ-C1 software or an epifluorescence Nikon Eclipse Ni-U microscope. Adobe Photoshop CS and ImageJ software (32) were used to pseudocolor and process all images.
The isolated cytoskeletons and flagellar complexes were obtained for immunocytolocalization as previously described by Sasse and Gull (33) and prepared for immunofluorescence assay as described above.
Transfection of parasites.
Epimastigotes were grown at 28°C in LIT medium supplemented with 10% FCS to a density of approximately 3 × 107 cells ml−1. Parasites were then harvested by centrifugation at 4,000 × g for 5 min at room temperature, washed once with PBS, and resuspended in 0.35 ml of transfection buffer, pH 7.5 (0.5 mM MgCl2, 0.1 mM CaCl2 in PBS), to a density of 1 × 108 cells ml−1. The cells were then transferred to a 0.2-cm-gap cuvette (Bio-Rad), and 15 to 100 μg of DNA was added in a final volume of 40 μl. The mixture was placed on ice for 15 min and then subjected to a pulse of 450 V and 500 μF using a GenePulser II (Bio-Rad, Hercules, CA, USA). After electroporation, the cells were transferred into 5 ml of LIT medium containing 10% FCS and maintained at room temperature for 15 min. Then, the cells were incubated at 28°C. After 24 h, the antibiotic G418 (Genbiotech) was added to an initial concentration of 125 μg ml−1, and 72 to 96 h after electroporation, the cultures were diluted 1:5 and the antibiotic concentration was doubled. Stable resistant cells were obtained approximately 30 days after transfection.
Coimmunoprecipitation.
Epimastigotes were grown up to 107 parasites ml−1, and 1010 parasites were used per coimmunoprecipitation experiment. The antibodies anti-TcBDF3 and total IgGs purified from nonimmunized rabbits were immobilized to magnetic beads (Dynabeads; Invitrogen) following the manufacturer's instructions. The antibody-coupled beads were incubated with the protein extracts for 3 h at 4°C with gentle shaking. Then, the beads were washed three times with MME buffer (50 mM HEPES, 5 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, 2 mM MgCl2) and two times with last wash buffer (30 mM Tris, pH 7.5, 0.02% Tween 20). The protein complexes were eluted with 0.5 M HN4OH and neutralized. The eluted proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes for Western blot analysis.
Molecular exclusion chromatography.
Molecular exclusion chromatography was carried out using a Superdex 75 column (GE Healthcare) equilibrated with 10 mM sodium phosphate buffer, pH 8, at the recommended flow rate of 0.25 ml/min, and the absorbance at 280 nm was continuously monitored using an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare). The standard curve was constructed using lysozyme (14.7 kDa), glutathione S-transferase (GST) (27 kDa for the monomeric form and 54 kDa for the dimeric form), and bovine serum albumin (67 kDa). All buffers and samples were filtered (0.45 μm) before use.
RESULTS AND DISCUSSION
T. cruzi BDF3.
The TcBDF3 coding sequence has two almost identical variant haplotypes in the T. cruzi genome (TcCLB.510719.70 and TcCLB.509747.110 [http://tritrypdb.org/tritrypdb]) that encode a 223-amino-acid polypeptide. This coding sequence contains a bromodomain (pfam PF00439) in its N-terminal portion from R37 to L141 (see Fig. S1A in the supplemental material). TcBDF3 has a predicted molecular mass of 24.7 kDa and an isoelectric point of 5.2. The C-terminal portion of the sequence shows no similarity to any domain present in databases. However, it is enriched in acidic amino acids (30% glutamic acid plus aspartic acid), basic amino acids (37% lysine plus arginine), and serine (14%) (see Fig. S1A in the supplemental material). These highly charged low-complexity sequences are generally involved in protein-protein interactions.
Orthologous genes are present in other trypanosomatids; the T. brucei (Tb927.11.10070) and Leishmania major (LmjF.36.3360) proteins have sequence identities of 63% and 42% and similarities of 75.9% and 54.9%, respectively, to TcBDF3. The similarity between the four T. cruzi BDFs and bromodomains from other organisms is low, with sequence identities always below 20%. However, the amino acids involved in binding the acetyl-lysine are conserved or conservatively substituted, supporting the hypothesis that TcBDF3 has a functional bromodomain (see Fig. S1B in the supplemental material). The secondary structure of the TcBDF3 bromodomain region was modeled with the Phyre2 server (34), retrieving the characteristic four-α-helix pattern of these domains (see Fig. S1C in the supplemental material). When analyzed by molecular exclusion chromatography, most of the recombinant TcBDF3 was found to be a dimer in solution (see Fig. S1D in the supplemental material).
TcBDF3 is a cytoplasmic protein.
To study the expression of TcBDF3 in T. cruzi, antibodies raised against recombinant protein were purified by affinity chromatography. Western blot analysis with rabbit and mouse antibodies showed a single band of the expected molecular weight in total lysates of T. cruzi epimastigotes (see Fig. S2A in the supplemental material). To test the specificity of the anti-TcBDF3 antibodies, they were competed with recombinant TcBDF3 and then used in Western blot (see Fig. S2B in the supplemental material) and immunofluorescence (see Fig. S2C in the supplemental material) assays.
TcBDF3 subcellular localization was predicted using pSORTII (http://psort.hgc.jp/form2.html) and Wolf pSORT (http://wolfpsort.org/). Surprisingly, the highest score was assigned to a nonnuclear localization. Also, the TFPP server (a tool for recognizing flagellar proteins in T. brucei) (35) predicted a flagellar localization for TcBDF3 and its orthologue in T. brucei (Tb927.11.10070). Although Tb927.11.10070 has not been identified in the flagellar proteome of T. brucei (36–38), it is worth mentioning that, due to technical limitations, many flagellar proteins fail to be detected or cannot be assigned to the flagellum with certainty by proteomic studies.
Next, we evaluated the expression of TcBDF3 in nuclear and nonnuclear epimastigote extracts by Western blot analysis. The TcBDF3-specific immunoreactive band was observed only in the nonnuclear fraction. TcTAT (39) is a cytosolic protein, and TcHMGB (30), TcBDF2 (25), and histone H4 (40) are all nuclear proteins (Fig. 1A). We decided to confirm the nonnuclear localization of this protein through several approaches.
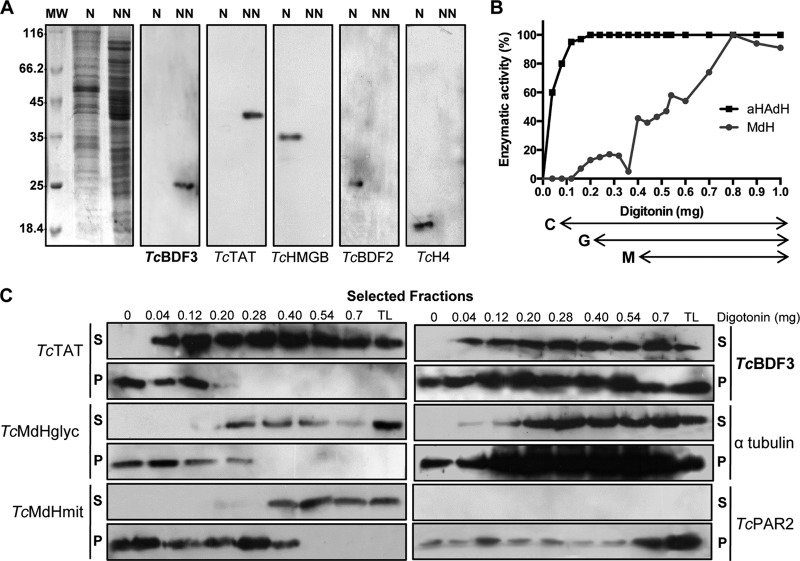
FIG 1.
TcBDF3 is a cytoplasmic bromodomain-containing protein in epimastigotes. (A) Nuclear (N) and nonnuclear (NN) protein extracts (30 μg per well) were subjected to Western blot analysis using rabbit anti-TcBDF3 antibodies, anti-TcTAT (cytosolic), anti-TcHMGB (nuclear), anti-TcBDF2 (nuclear), and anti-T. cruzi H4 histone (TcH4) (nuclear). On the left is the Coomassie-stained gel. (B) Enzymatic activity of αHAdH and MdH in epimastigotes treated with increasing concentrations of digitonin. Activities were measured and normalized to the protein concentration in the extracts. The arrows below the graph indicate the digitonin concentrations at which cytosolic (C), glycosomal (G), and mitochondrial (M) proteins are released. (C) Equal volumes of selected soluble (S) and insoluble (P) fractions obtained at different digitonin amounts (0 to 0.7 mg) were subjected to Western blot analysis using anti-TcBDF3 antibodies (TcBDF3) and known markers for different organelles. The antibodies used were anti-TcTAT (cytosolic), anti-TcMdHglyc (glycosomal), anti-TcMdHmit (mitochondrial), anti-TcPAR2 (flagellar), and anti-α-tubulin (α-tubulin) (cytoskeletal). TL, total lysate.
First, we performed Western blot analysis in subcellular fractions of epimastigotes obtained by digitonin extraction (39). By this approach, plasma membranes with high sterol content are specifically permeabilized at a low concentration of digitonin, whereas higher concentrations are required to permeabilize glycosomal and mitochondrial membranes (41). By measuring the activity of αHAdH (a cytosolic protein), we determined in which fractions the cytosolic content was released (starting at 0.04 mg of digitonin). MdH presents a glycosomal and a mitochondrial isoform. By measuring the activity of this enzyme, we determined when the glycosomal content (starting at 0.2 mg of digitonin) and the mitochondrial content (starting at 0.4 mg of digitonin) were released (Fig. 1B). The extraction pattern of TcBDF3 was monitored by Western blot analysis of selected soluble and insoluble fractions (Fig. 1C, 0 to 0.7 mg of digitonin). In agreement with our previous results, TcBDF3 was released at low digitonin concentrations. However, TcBDF3 was not completely extracted in the soluble fraction, and an immunoreactive band was observed in all the insoluble fractions, showing a pattern similar to that of PAR2 (a flagellar protein) and α-tubulin. These results suggest that TcBDF3 is a cytoplasmic protein that could also be associated with insoluble proteins in the membrane or cytoskeleton of T. cruzi.
Second, TcBDF3 was immunolocalized in thin sections of epimastigotes by transmission electron microscopy (TEM). TcBDF3 was not observed over DNA-containing structures, such as the nucleus or kinetoplast (Fig. 2A to C). TcBDF3 was immunolocalized at the flagellum (Fig. 2B, D, and F, white arrows), especially in its inner part, which is attached to the cell body. Labeling was also observed dispersed in the cytoplasm and close to the flagellar pocket (Fig. 2A to D, black arrows). In control assays with no primary antibody, no immunogold particles were detected (data not shown).
FIG 2.
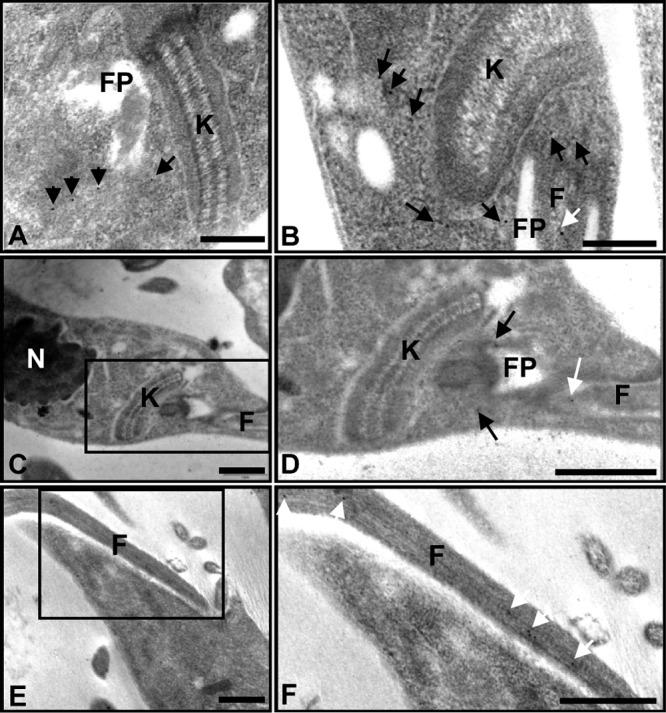
TcBDF3 is localized at the cytoplasm, the flagellum, and the flagellar pocket of epimastigotes. (A to C and E) Immunoelectron microscopy of TcBDF3 in T. cruzi epimastigotes using purified rabbit anti-TcBDF3 antibodies. The nucleus (N), kinetoplast (K), flagellar pocket (FP), and flagellum (F) are indicated. Gold particles are indicated with black and white arrows. The white arrows indicate flagellar labeling. (D) Enlarged image of the boxed area in panel C. (F) Enlarged image of the boxed area in panel E. Bars = 1 μm.
Next, TcBDF3 expression during the T. cruzi life cycle was addressed using epimastigote, trypomastigote, and amastigote total protein extracts. A single band of the expected molecular weight was observed for all developmental stages (Fig. 3A). The subcellular localization of TcBDF3 was assessed by immunofluorescence microscopy in different stages (Fig. 3B). In epimastigotes, TcBDF3 was present in the cytoplasm, the flagellum, and the flagellar pocket region, similar to the pattern obtained by TEM. In amastigotes, we observed an expression pattern similar to that in epimastigotes. However, the flagellar pocket region was more deeply marked and the plasma membrane appeared to be labeled. Surprisingly, in infective trypomastigotes, the TcBDF3 expression pattern changed and was localized exclusively in the flagellum.
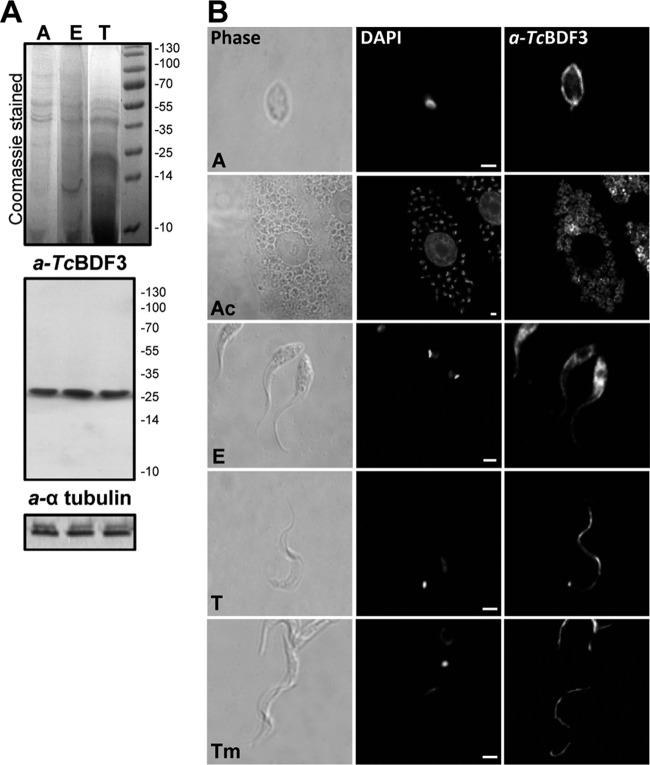
FIG 3.
TcBDF3 is expressed throughout the T. cruzi life cycle in vitro. (A) Western blot analysis using purified rabbit anti-TcBDF3 antibodies (a-TcBDF3) and mouse anti-α-tubulin (a-α-tubulin) as a loading control. A, amastigote total protein extracts; E, epimastigote total protein extracts; T, trypomastigote total protein extracts (30 μg per well). (B) Immunofluorescence assay using purified anti-TcBDF3 and parasites at different stages of the T. cruzi life cycle. A, free amastigote; Ac, amastigotes inside a Vero cell; E, epimastigote; T, trypomastigote from infected Vero cells; Tm, metacyclic trypomastigote obtained in vitro. Anti-rabbit IgG conjugated to fluorescein was used as a secondary antibody. Nuclei and kinetoplasts were labeled with DAPI. Bars = 2 μm.
This change in TcBDF3 localization during the epimastigote-trypomastigote transition was also analyzed using N-terminal fusions to CFP. A full-length form of TcBDF3 (pTcCFPN-TcBDF3) and a truncated form containing the first 131 amino acids (pTcCFPN-TcBDF3ΔC) were overexpressed in epimastigotes. Both fusion proteins were detected in the whole cell body of epimastigotes in a pattern similar to that observed for TcBDF3 by immunocytolocalization (see Fig. S3 in the supplemental material). When transfected parasites were differentiated in vitro to trypomastigotes, CFPN-TcBDF3 was found concentrated at the flagellum, as was wild-type TcBDF3 (see Fig. S3A in the supplemental material). However, truncated CFPN-TcBDF3ΔC remained dispersed in the cell body (see Fig. S3B in the supplemental material). Expression of CFPN-TcBDF3 and CFPN-TcBDF3ΔC was corroborated by Western blot analysis with anti-TcBDF3 antibodies (see Fig. S3C in the supplemental material). These results suggest that the C-terminal portion of TcBDF3 could be important for the flagellar localization of TcBDF3 in trypomastigotes.
The presence of a bromodomain-containing protein outside the nucleus has been described in other models, but always with a dual cytoplasmic-nuclear localization and restricted to some specific mammalian cells (21–23). The existence of a bromodomain factor in the flagellum, as described here, suggests a completely new role for these protein modules. Given that in the last few years several authors, using high-resolution mass spectrometry, identified thousands of acetylated proteins involved in a wide variety of cellular processes in different organisms (42–45), it is not unrealistic to think that bromodomains could be playing regulatory roles outside the nuclear compartment. In 2000, Kouzarides proposed that acetylation might rival phosphorylation as a regulator of cell function and that the bromodomain may be analogous to the phosphotyrosine-recognizing SH2 domain (46). The presence of TcBDF3 outside the nucleus opens new perspectives for a possible role of lysine acetylation as a regulatory switch in complex cellular processes, as proposed by many authors (47–50).
To study in further detail the differential localization of TcBDF3, in vitro metacyclic trypomastigotes were obtained using TAU3AAG medium. During the differentiation process between epimastigotes and trypomastigotes, intermediate stages were distinguished and classified based on the positions of the kinetoplast, the nucleus, and the flagellum, as described by Ferreira and coworkers (51) (a schematic representation is shown in Fig. 4, right). The immunocytolocalization of TcBDF3 and acetylated α-tubulin was analyzed in the intermediate differentiation stages (1a, b, and c). As is shown in Fig. 4, TcBDF3 concentrates in the flagellum during metacyclogenesis. As previously reported (10), acetylated α-tubulin was detected in the whole cell body in the intermediate stages, with no significant changes. However, we observed a partial colocalization of TcBDF3 and acetylated α-tubulin in the flagella of the intermediate stages (Fig. 4), which strongly suggested that TcBDF3 was associated with the flagellar microtubule structure.
FIG 4.
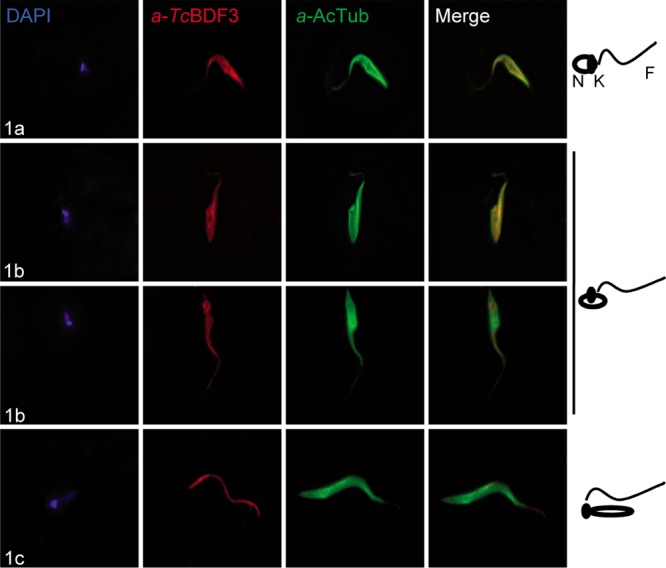
TcBDF3 changes its location during in vitro metacyclogenesis. Immunofluorescence assays used purified rabbit anti-TcBDF3 (α-TcBDF3) and mouse monoclonal anti-acetylated α-tubulin (α-AcTub) antibodies in intermediate stages 1a, 1b, and 1c, as defined by Ferreira et al. (51). On the right are schematic diagrams of the positions of the flagellum (F), nucleus (N), and kinetoplast (K) in the three selected intermediate differentiation stages. Anti-rabbit IgG conjugated to fluorescein (green) and anti-mouse IgG conjugated to rhodamine (red) were used as secondary antibodies. Nuclei and kinetoplasts were labeled with DAPI (blue).
TcBDF3 binds to acetylated α-tubulin.
The above-mentioned results, as well as the known ability of bromodomain-containing proteins to bind acetylated proteins, led us to think that TcBDF3 could bind acetylated proteins in the cytoskeleton and flagellum of T. cruzi.
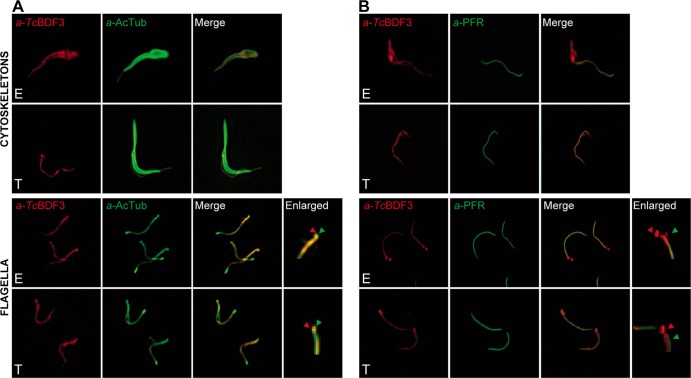
As mentioned above, microtubules are components of the subpellicular corset, the axoneme, the flagellar pocket, and the flagellum attachment zone in trypanosomatids (52). Multiple tubulin isotypes are present in microtubules, due to a series of posttranslational modifications. Among all isoforms of α-tubulin associated with the T. cruzi subpellicular and axonemal microtubules, the acetylated form seems to be predominant (10). Isolated subpellicular microtubules and flagellar complexes were obtained from epimastigotes and trypomastigotes in order to verify the presence of TcBDF3 in these cellular components. TcBDF3, as well as acetylated α-tubulin and PAR2 (α-PFR) (paraflagellar rod 2 protein) were detected by immunofluorescence (Fig. 5). In epimastigotes, TcBDF3 was present in discrete regions of the cytoskeleton with a stronger signal in the flagellar pocket region (Fig. 5A, top). Also, it was observed in the flagellum, where it partially colocalized with acetylated α-tubulin (Fig. 5A, bottom). The protocol performed to obtain isolated flagella, as Sasse and Gull stated in 1988, did not always lead to complete solubilization, and a group of subpellicular MTs attached to basal bodies and forming the flagellar pocket appeared to be resistant to treatment (33). In the enlarged images of the isolated flagella, we observed the presence of TcBDF3 and acetylated α-tubulin in these resistant structures (Fig. 5A, green and red arrowheads), which further corroborated the results obtained by TEM. These results suggest that TcBDF3 localization correlates with acetylated α-tubulin, although the opposite is not necessarily true. In trypomastigotes, TcBDF3 was present only in the flagellum, as was previously determined using intact parasites, where it colocalized partially with acetylated α-tubulin (Fig. 5A). Localizations of TcBDF3 and PAR2 were compared in detail to determine if TcBDF3 was present in the paraflagellar rod. The paraflagellar rod is present from the point where the flagellum exits the flagellar pocket and runs alongside the axoneme right to the distal tip. We observed that in the flagella of epimastigotes and trypomastigotes, the two proteins did not colocalize but seemed to run side by side, supporting the hypothesis that TcBDF3 is present in the flagellar axoneme of T. cruzi but not in the paraflagellar rod (Fig. 5B). The presence of TcBDF3, but not of PAR2, in the flagellar pocket region is clearly observed in the enlarged images of the isolated flagella (Fig. 5B, green and red arrowheads).
FIG 5.
TcBDF3 is detected in the cytoskeletons and flagella of epimastigotes (E) and only in the flagella of metacyclic trypomastigotes (T). Immunofluorescence assays used purified rabbit anti-TcBDF3 (α-TcBDF3) and mouse monoclonal anti-acetylated α-tubulin (α-AcTub) (A) and mouse anti-PAR2 (α-PFR) (B) antibodies on isolated cytoskeletons and flagella of epimastigotes and metacyclic trypomastigotes. Anti-rabbit IgG conjugated to fluorescein (green) and anti-mouse IgG conjugated to rhodamine (red) were used as secondary antibodies. The last right lanes are enlarged images of the detergent-resistant structures that correspond to MTs attached to basal bodies and forming the flagellar pocket. The green arrowheads indicate TcBDF3 localization, and the red arrowheads indicate acetylated α-tubulin (A) and PAR2 (B) localization in these structures.
Next, Western blot assays with anti-TcBDF3 and anti-acetylated α-tubulin were performed using protein extracts enriched in cytoskeletal and flagellar proteins (Fig. 6A). Three enriched fractions that corresponded to soluble proteins (Sn1), soluble cytoskeletal and flagellar proteins (Sn2), and insoluble cytoskeletal and flagellar proteins were obtained by differential extraction with detergent, as described by Schneider and coworkers (11). TcBDF3 and acetylated α-tubulin seemed to be fractionated together from soluble protein pools and from insoluble flagellar and cytoskeletal protein complexes. These results suggested that the interaction between these proteins is restricted to some specific cellular compartments.
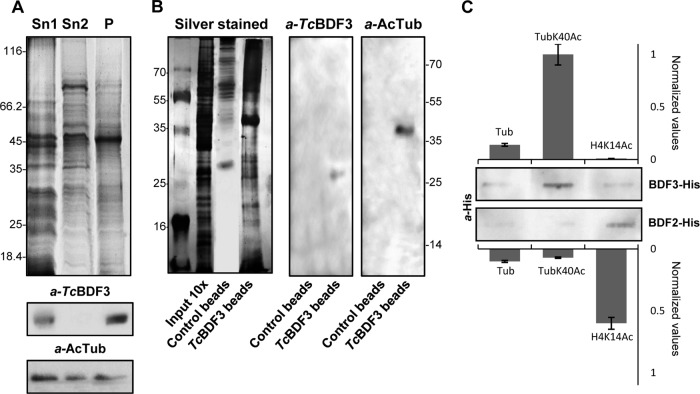
FIG 6.
TcBDF3 interacts with acetylated α-tubulin. (A) Coomassie-stained SDS-PAGE and Western blot analyses of epimastigote protein extracts enriched in cytoskeletal and flagellar proteins. Sn1, soluble protein extracts; Sn2, soluble cytoskeletal and flagellar protein extracts; P, insoluble cytoskeletal and flagellar protein extracts (50 μg per well). Rabbit anti-TcBDF3 antibodies (α-TcBDF3) and mouse anti-acetylated α-tubulin antibodies (α-AcTub) were used. (B) Coimmunoprecipitation assay using purified anti-TcBDF3 antibodies covalently coupled to magnetic beads (TcBDF3 beads). Magnetic beads coupled to IgGs (purified from antisera of nonimmunized rabbits) were used as a negative control (Control beads). On the left is a silver-stained SDS-PAGE gel of total cytoskeletal extracts and the elutions obtained after the immunoprecipitation experiment. On the right is a Western blot analysis of the eluted proteins after coimmunoprecipitation using purified rabbit anti-TcBDF3 antibodies (α-TcBDF3) and mouse monoclonal anti-acetylated α-tubulin antibodies (α-AcTub). (C) Slot far-Western blot assay. Acetylated tubulin (TubK40Ac), nonacetylated tubulin (Tub), and acetylated histone H4 (H4K14Ac) peptides were blotted onto a nitrocellulose membrane and incubated with His-tagged recombinant TcBDF3 (BDF3-His) or TcBDF2 (BDF2-His). Bound recombinant proteins were detected with anti-histidine antibodies (α-His). Signals were quantified by densitometry and normalized using the interaction with acetylated α-tubulin peptide as a reference (assigned the arbitrary value of 1). The bars and error bars indicate means ± standard deviations (SD) from the results of three independent experiments.
Immunoprecipitation assays were performed to study the interaction of TcBDF3 with acetylated α-tubulin (Fig. 6B). We used anti-TcBDF3 antibodies coupled to magnetic beads. Both TcBDF3 and acetylated α-tubulin were detected by Western blot analysis in the immunoprecipitated complexes. These results demonstrated that the two proteins interact in vivo. We did not detect any immunoreactive band using magnetic beads coupled to purified IgG (negative control).
To test TcBDF3 binding specificity for acetylated α-tubulin, we blotted acetylated and nonacetylated α-tubulin (Ac α-tubulin and α-tubulin) and histone H4 acetylated in lysine 14 (H4K14Ac) peptides onto a nitrocellulose membrane. Then, the membrane was incubated with recombinant TcBDF3 and TcBDF2 (fused to a histidine tag), and the bound proteins were visualized by Western blot analysis with anti-histidine antibodies (Fig. 6C). There was no cross-reactivity between bromodomain factors: recombinant TcBDF2 recognized only the H4K14Ac peptide (25), and TcBDF3 recognized only the acetylated α-tubulin peptide. This suggests that each BDF can recognize and bind to one (or a limited number of) specific acetylated lysine residues.
Although tubulin acetylation is a widespread modification present in all eukaryotic cells, its precise function in cytoskeleton dynamics has not yet been completely elucidated. Recent reports suggested a function for this PTM in axoneme-related cell structures. The acetylation of α-tubulin at K40 by the specific enzyme MEC-17 was associated with ciliogenesis and efficient mechanosensation in Caenorhabditis elegans (17, 53). Trypanosomes are evolutionarily early-branched species that have some unique characteristics. The participation of a bromodomain-containing protein complex during metacyclogenesis could also be a unique feature of these particular eukaryotic cells. It has been proposed that cilia and flagella emerged early in eukaryotic evolution and that in primitive eukaryotes, microtubule PTMs existed as a cilium- and flagellum-specific phenomenon, which was later adapted to other microtubule structures (6).
The results presented here show that TcBDB3 binds to acetylated α-tubulin both ex vivo and in vivo. This interaction seems to be associated with changes in the amount of TcBDF3 in the flagellum during metacyclogenesis. Although the exact meaning of this observation cannot be understood yet, it is important to mention that TcBDF3 is the first described bromodomain-containing protein that recognizes a modification in tubulin and, hence, the first candidate to be able to read α-tubulin PTM. At least two basic models can be proposed for its mode of action. Analogously to those already proposed for histone-binding bromodomains, a TcBDF3-containg complex could be carrying enzymatic activity to the flagellum to modify tubulin or any other cytoskeletal component. Another hypothesis is that TcBDF3 might carry acetylated α-tubulin from the cell body to the flagellum. However, we cannot rule out the possibility that TcBDF3 simply binds to the acetylated lysine residue to protect it from the actions of modifying enzymes.
A bromodomain-containing protein complex involved in the remodeling of the cytoskeleton in T. cruzi may also be a new chemotherapeutic target for Chagas' disease. Recently, two inhibitors that target bromodomains from the BET family have shown selective activity in a squamous cell carcinoma model (54). Many other inhibitors of the bromodomain–acetyl-lysine interaction were developed later, putting bromodomains alongside KATs (lysine acetyltransferases) and KDACs (lysine deacetylases) as interesting targets for diseases caused by aberrant acetylation of lysine residues (55). The presence of an exclusively cytoplasmic bromodomain like TcBDF3 could be another feature of trypanosomatids absent in mammalian host cells and hence could be considered a potential target for the development of new drugs against trypanosomiasis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Research Council (CONICET) grant PIP2010-0685 and National Agency of Scientific and Technological Promotion (ANPCyT) and Glaxo SmithKline joint grant PICTO2011-0046.
V.L.A. and C.R. are fellows and E.C.S., P.C., and G.V.V. are researchers of CONICET, Argentina.
We thank K. Gull for his generous gift of anti-T. brucei α-tubulin antibodies, C. Nowiki for the anti-T. cruzi tyrosine aminotransferase and anti-T. cruzi malate dehydrogenase antibodies, A. Silver for the anti-PAR2 antibodies, Sergio Schenkman for the anti-histone H4 antibodies and H4K14 acetylated peptides, and Lisvane Silva for helping to raise the mouse anti-TcBDF3 antisera.
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00341-13.
REFERENCES
- 1.Contreras VT, Araujo-Jorge TC, Bonaldo MC, Thomaz N, Barbosa HS, de Meirelles MN, Goldenberg S. 1988. Biological aspects of the Dm 28c clone of Trypanosoma cruzi after metacyclogenesis in chemically defined media. Mem. Inst. Oswaldo Cruz 83:123–133. 10.1590/S0074-02761988000100016 [DOI] [PubMed] [Google Scholar]
- 2.Hill KL. 2010. Parasites in motion: flagellum-driven cell motility in African trypanosomes. Curr. Opin. Microbiol. 13:459–465. 10.1016/j.mib.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field MC, Carrington M. 2009. The trypanosome flagellar pocket. Nat. Rev. Microbiol. 7:775–786. 10.1038/nrmicro2221 [DOI] [PubMed] [Google Scholar]
- 4.Hammond JW, Cai D, Verhey KJ. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20:71–76. 10.1016/j.ceb.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhey KJ, Gaertig J. 2007. The tubulin code. Cell Cycle 6:2152–2160. 10.4161/cc.6.17.4633 [DOI] [PubMed] [Google Scholar]
- 6.Janke C, Bulinski JC. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786. 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- 7.Cambray-Deakin MA, Burgoyne RD. 1987. Acetylated and detyrosinated alpha-tubulins are co-localized in stable microtubules in rat meningeal fibroblasts. Cell Motil. Cytoskeleton 8:284–291. 10.1002/cm.970080309 [DOI] [PubMed] [Google Scholar]
- 8.Belmadani S, Pous C, Fischmeister R, Mery PF. 2004. Post-translational modifications of tubulin and microtubule stability in adult rat ventricular myocytes and immortalized HL-1 cardiomyocytes. Mol. Cell. Biochem. 258:35–48. 10.1023/B:MCBI.0000012834.43990.b6 [DOI] [PubMed] [Google Scholar]
- 9.Janke C, Kneussel M. 2010. Tubulin post-translational modifications: encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 33:362–372. 10.1016/j.tins.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 10.Souto-Padron T, Cunha e Silva NL, de Souza W. 1993. Acetylated alpha-tubulin in Trypanosoma cruzi: immunocytochemical localization. Mem. Inst. Oswaldo Cruz 88:517–528. 10.1590/S0074-02761993000400004 [DOI] [PubMed] [Google Scholar]
- 11.Schneider A, Sherwin T, Sasse R, Russell DG, Gull K, Seebeck T. 1987. Subpellicular and flagellar microtubules of Trypanosoma brucei brucei contain the same alpha-tubulin isoforms. J. Cell Biol. 104:431–438. 10.1083/jcb.104.3.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alonso VL, Serra EC. 2012. Lysine acetylation: elucidating the components of an emerging global signaling pathway in trypanosomes. J. Biomed. Biotechnol. 2012:452934. 10.1155/2012/452934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- 14.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437–444. 10.1016/S1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
- 15.Tavares J, Ouaissi A, Santarem N, Sereno D, Vergnes B, Sampaio P, Cordeiro-da-Silva A. 2008. The Leishmania infantum cytosolic SIR2-related protein 1 (LiSIR2RP1) is an NAD+-dependent deacetylase and ADP-ribosyltransferase. Biochem. J. 415:377–386. 10.1042/BJ20080666 [DOI] [PubMed] [Google Scholar]
- 16.Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L. 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136:551–564. 10.1016/j.cell.2008.11.043 [DOI] [PubMed] [Google Scholar]
- 17.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. 2010. The major {alpha}-tubulin K40 acetyltransferase {alpha}TAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U. S. A. 107:21517–21522. 10.1073/pnas.1013728107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsford S, Horn D. 2011. Elongator protein 3b negatively regulates ribosomal DNA transcription in African trypanosomes. Mol. Cell. Biol. 31:1822–1832. 10.1128/MCB.01026-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng L, Zhou MM. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124–128. 10.1016/S0014-5793(01)03309-9 [DOI] [PubMed] [Google Scholar]
- 20.Yang XJ. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26:1076–1087. 10.1002/bies.20104 [DOI] [PubMed] [Google Scholar]
- 21.Crowley T, Brunori M, Rhee K, Wang X, Wolgemuth DJ. 2004. Change in nuclear-cytoplasmic localization of a double-bromodomain protein during proliferation and differentiation of mouse spinal cord and dorsal root ganglia. Brain Res. Dev. Brain Res. 149:93–101. 10.1016/j.devbrainres.2003.12.011 [DOI] [PubMed] [Google Scholar]
- 22.Crowley TE, Kaine EM, Yoshida M, Nandi A, Wolgemuth DJ. 2002. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 16:1727–1737. 10.1210/me.2001-0353 [DOI] [PubMed] [Google Scholar]
- 23.Trousdale RK, Wolgemuth DJ. 2004. Bromodomain containing 2 (Brd2) is expressed in distinct patterns during ovarian folliculogenesis independent of FSH or GDF9 action. Mol. Reprod. Dev. 68:261–268. 10.1002/mrd.20059 [DOI] [PubMed] [Google Scholar]
- 24.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. 10.1126/science.1112680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villanova GV, Nardelli SC, Cribb P, Magdaleno A, Silber AM, Motta MC, Schenkman S, Serra E. 2009. Trypanosoma cruzi bromodomain factor 2 (BDF2) binds to acetylated histones and is accumulated after UV irradiation. Int. J. Parasitol. 39:665–673. 10.1016/j.ijpara.2008.11.013 [DOI] [PubMed] [Google Scholar]
- 26.Camargo ME, Segura EL, Kagan IG, Souza JM, da Carvalheiro JR, Yanovsky JF, Guimaraes MC. 1986. Three years of collaboration on the standardization of Chagas' disease serodiagnosis in the Americas: an appraisal. Bull. Pan Am. Health Organ. 20:233–244 [PubMed] [Google Scholar]
- 27.Tonelli RR, Silber AM, Almeida-de-Faria M, Hirata IY, Colli W, Alves MJ. 2004. L-proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell Microbiol. 6:733–741. 10.1111/j.1462-5822.2004.00397.x [DOI] [PubMed] [Google Scholar]
- 28.Yoshida N, Mortara RA, Araguth MF, Gonzalez JC, Russo M. 1989. Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect. Immun. 57:1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batista M, Marchini FK, Celedon PA, Fragoso SP, Probst CM, Preti H, Ozaki LS, Buck GA, Goldenberg S, Krieger MA. 2010. A high-throughput cloning system for reverse genetics in Trypanosoma cruzi. BMC Microbiol. 10:259. 10.1186/1471-2180-10-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cribb P, Perozzi M, Villanova GV, Trochine A, Serra E. 2011. Characterization of TcHMGB, a high mobility group B family member protein from Trypanosoma cruzi. Int. J. Parasitol. 41:1149–1156. 10.1016/j.ijpara.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 31.da Cunha JP, Nakayasu ES, de Almeida IC, Schenkman S. 2006. Post-translational modifications of Trypanosoma cruzi histone H4. Mol. Biochem. Parasitol. 150:268–277. 10.1016/j.molbiopara.2006.08.012 [DOI] [PubMed] [Google Scholar]
- 32.Rasband WS. 2011. ImageJ software. U. S. National Institutes of Health, Bethesda, MD. http://imagej.nih.gov/ij/ [Google Scholar]
- 33.Sasse R, Gull K. 1988. Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 90:577–589 [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Shen Y, Ding G, Tian Y, Liu Z, Li B, Wang Y, Jiang C. 2013. TFPP: an SVM-based tool for recognizing flagellar proteins in Trypanosoma brucei. PLoS One 8:e54032. 10.1371/journal.pone.0054032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227. 10.1038/nature04541 [DOI] [PubMed] [Google Scholar]
- 37.Hart SR, Lau KW, Hao Z, Broadhead R, Portman N, Huhmer A, Gull K, McKean PG, Hubbard SJ, Gaskell SJ. 2009. Analysis of the trypanosome flagellar proteome using a combined electron transfer/collisionally activated dissociation strategy. J. Am. Soc. Mass Spectrom. 20:167–175. 10.1016/j.jasms.2008.08.014 [DOI] [PubMed] [Google Scholar]
- 38.Oberholzer M, Langousis G, Nguyen HT, Saada EA, Shimogawa MM, Jonsson ZO, Nguyen SM, Wohlschlegel JA, Hill KL. 2011. Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol. Cell. Proteomics 10:M111.010538. 10.1074/mcp.M111.010538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowicki C, Montemartini M, Duschak V, Santome JA, Cazzulo JJ. 1992. Presence and subcellular localization of tyrosine aminotransferase and p-hydroxyphenyllactate dehydrogenase in epimastigotes of Trypanosoma cruzi. FEMS Microbiol. Lett. 71:119–124 [DOI] [PubMed] [Google Scholar]
- 40.Marciano D, Maugeri DA, Cazzulo JJ, Nowicki C. 2009. Functional characterization of stage-specific aminotransferases from trypanosomatids. Mol. Biochem. Parasitol. 166:172–182. 10.1016/j.molbiopara.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 41.Marciano D, Llorente C, Maugeri DA, de la Fuente C, Opperdoes F, Cazzulo JJ, Nowicki C. 2008. Biochemical characterization of stage-specific isoforms of aspartate aminotransferases from Trypanosoma cruzi and Trypanosoma brucei. Mol. Biochem. Parasitol. 161:12–20. 10.1016/j.molbiopara.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 42.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 43.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23:607–618. 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 44.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18:1529–1536 [PubMed] [Google Scholar]
- 45.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327:1000–1004. 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176–1179. 10.1093/emboj/19.6.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polevoda B, Sherman F. 2002. The diversity of acetylated proteins. Genome Biol. 3:reviews0006. 10.1186/gb-2002-3-5-reviews0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norvell A, McMahon SB. 2010. Cell biology. Rise of the rival. Science 327:964–965. 10.1126/science.1187159 [DOI] [PubMed] [Google Scholar]
- 49.Close P, Creppe C, Gillard M, Ladang A, Chapelle JP, Nguyen L, Chariot A. 2010. The emerging role of lysine acetylation of non-nuclear proteins. Cell. Mol. Life Sci. 67:1255–1264. 10.1007/s00018-009-0252-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim GW, Yang XJ. 2011. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 36:211–220. 10.1016/j.tibs.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 51.Ferreira LR, de Dossin FM, Ramos TC, Freymuller E, Schenkman S. 2008. Active transcription and ultrastructural changes during Trypanosoma cruzi metacyclogenesis. An. Acad. Bras. Cienc. 80:157–166. 10.1590/S0001-37652008000100011 [DOI] [PubMed] [Google Scholar]
- 52.Kohl L, Gull K. 1998. Molecular architecture of the trypanosome cytoskeleton. Mol. Biochem. Parasitol. 93:1–9. 10.1016/S0166-6851(98)00014-0 [DOI] [PubMed] [Google Scholar]
- 53.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. 2010. MEC-17 is an alpha-tubulin acetyltransferase. Nature 467:218–222. 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller S, Filippakopoulos P, Knapp S. 2011. Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 13:e29. 10.1017/S1462399411001992 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.