Abstract
Consistent with its proposed status as an early branching eukaryote, Giardia has the most divergent actin of any eukaryote and lacks core actin regulators. Although conserved actin-binding proteins are missing from Giardia, its actin is utilized similarly to that of other eukaryotes and functions in core cellular processes such as cellular organization, endocytosis, and cytokinesis. We set out to identify actin-binding proteins in Giardia using affinity purification coupled with mass spectroscopy (multidimensional protein identification technology [MudPIT]) and have identified >80 putative actin-binding proteins. Several of these have homology to conserved proteins known to complex with actin for functions in the nucleus and flagella. We validated localization and interaction for seven of these proteins, including 14-3-3, a known cytoskeletal regulator with a controversial relationship to actin. Our results indicate that although Giardia lacks canonical actin-binding proteins, there is a conserved set of actin-interacting proteins that are evolutionarily indispensable and perhaps represent some of the earliest functions of the actin cytoskeleton.
INTRODUCTION
In addition to being a major parasite infecting more than 280 million people each year, Giardia lamblia (synonymous with Giardia intestinalis and Giardia duodenalis) belongs to one of the earliest diverging groups of eukaryotes (1–4). Therefore, investigation of Giardia biology has the potential to uncover evolutionarily deep cellular mechanisms. However, the placement of Giardia near the root of the eukaryotic tree, in addition to the placement of the root itself, is contentious (5). Nevertheless, Giardia is the most divergent eukaryote that can be manipulated in the laboratory with molecular and genetic tools (4, 6–10). In addition, many pathways in Giardia have fewer components than in the well-studied model eukaryotes (4). Thus, the combination of Giardia's highly divergent and minimalistic genome provides a unique perspective through which cellular processes may be examined. This perspective may potentially define both the minimal requirements for function and the portions of cellular mechanisms constrained throughout evolution.
A major point of divergence between Giardia and other eukaryotes is the cytoskeleton (11). Giardia lacks the canonical actin-binding proteins, once thought common to all extant eukaryotes, which perform critical functions in other eukaryotes (12). Their absence may indicate a split from the last eukaryotic common ancestor before the canonical set of actin-binding proteins was established. Alternatively, Giardia may have evolved a novel set of actin-interacting proteins that allowed for the gradual loss of the canonical set of actin-binding proteins (11, 13). Our previous work has shown that despite the lack of canonical actin-binding proteins, Giardia actin (giActin) is required for conserved cellular functions, including membrane trafficking, cytokinesis, polarity, and control of cellular morphology (13). The Giardia cytoskeleton is also quite elaborate, suggesting the presence of cytoskeletal regulators (Fig. 1). That giActin performs similar functions to actin in other eukaryotes suggests these processes were already associated with actin at the time Giardia split from the other eukaryotes (13). We have also shown that giRac, the sole Rho family GTPase in Giardia, regulates actin despite the absence of all actin-binding proteins known to link G-protein signaling to the actin cytoskeleton (Arp2/3, formin, wave, myosin, and cofilin) (13). Therefore, Giardia must contain a novel set of actin-interacting proteins comprised of ancient yet undiscovered and/or Giardia-specific actin regulators. We sought here to identify actin-binding proteins using actin affinity chromatography coupled with multidimensional protein identification technology (MudPIT) (14). The discovery of Giardia-specific actin-binding proteins with essential functions would open an avenue to potential therapeutic targets, while the discovery of conserved proteins would highlight an ancient relationship between actin and the identified protein, worthy of further exploration.
FIG 1.
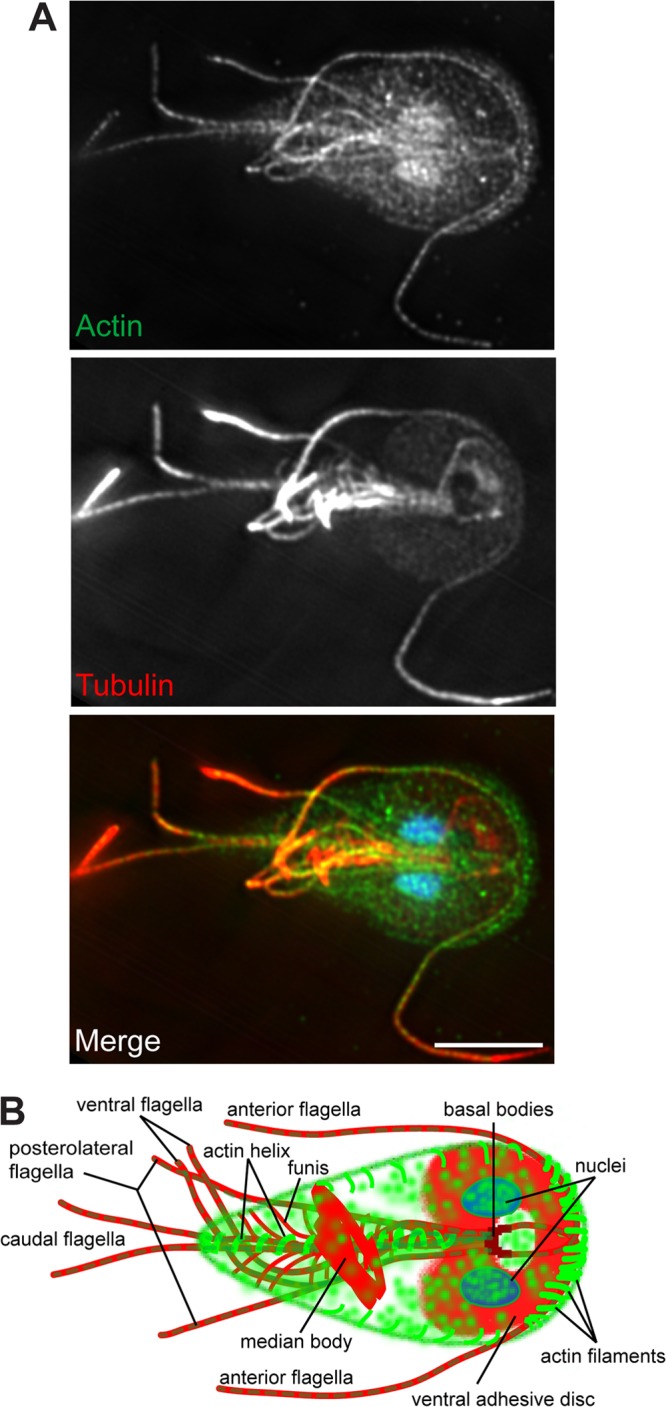
Giardia cytoskeletal organization. (A) Maximum projection of a Z-stack. Actin is green, tubulin is red, and DNA is blue. (B) Diagram of Giardia trophozoite with all of the prominent cytoskeletal structures labeled.
MATERIALS AND METHODS
Strain and culture conditions.
Giardia lamblia, strain WBC6 was cultured as described previously (15). Morpholino knockdown experiments and quantitative Western blotting were performed as described previously (9, 13). Large volume high-yield cultures required a method to increase the surface area. We filled standard wide-mouth media bottles with cut-to-length “jumbo drinking straws” and autoclaved them before filling with media (see Fig. S1 in the supplemental material). Giardia cell counts increased by ∼30% in straw-filled 500-ml bottles versus those without. After 3 days of growth, we did not observe unattached cells at the bottom of straw-filled culture vessels that are typical of overgrown cultures, while bottles without straws had a layer of cell sediment. After 72 h of growth 1-liter cultures regularly reach 2.5 × 106 cells/ml, exceeding maximum trophozoite concentrations obtained with Farthing's roller bottles, without needing specialized equipment (16).
Constructs.
The TS-Actin vector was constructed by modifying pGFPapac (17). A BamHI site was first introduced between BsrGI and NotI of enhanced green fluorescent protein (eGFP) using an oligonucleotide adapter; all primer sequences can be found in Table S1 in the supplemental material. The glutamate dehydrogenase (GDH) promoter was exchanged for the actin promoter by excising GDH with HindIII and NcoI, the actin promoter was subsequently ligated into the same position. Next. eGFP was excised with NcoI and BamHI, allowing for the TwinStrep tag to be ligated into the same position. Finally, the vector was digested with BamHI and NotI so that the actin gene could be ligated into the vector. All PCR amplification steps were performed with iProof high-fidelity polymerase (Bio-Rad), and the resulting vectors were verified by sequencing. The putative actin-interacting proteins were PCR amplified from genomic DNA and inserted into the pKS 3HA.NEO vector (10) using the restriction sites indicated in Table S1 in the supplemental material. All resulting constructs except for TS-Actin, GL50803_6744, and GL50803_13273 were linearized and integrated into the genome by homologous recombination to generate endogenously tagged proteins.
Actin affinity chromatography.
One-liter straw-packed and sterilized bottles were filled with medium and inoculated with two 13-ml confluent cultures containing wild-type (WT) or TwinStrep-giActin cell lines. After 3 days the cultures were incubated in an ice water bath for 1 h to detach cells. The media and unattached cells were transferred to centrifuge bottles and pelleted at 750 × g for 15 min. The resulting cell pellet was washed in 10 ml of cold HEPES-buffered saline, transferred to 15-ml conical tubes, and pelleted again. The cell pellet was resuspended in an equal volume (∼2 ml) of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 7.5% glycerol, 0.25 mM CaCl2, 0.25 mM ATP, 0.05 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 2× Halt protease inhibitors [Pierce]). The pellet was stored overnight at −80°C and, after thawing, the cells were sonicated, and the extract cleared at 10,000 × g for 10 min. The lysate was added to 200 μl of Streptactin-Sepharose beads (IBA) previously equilibrated with lysis buffer. Binding was performed for 1.5 h with end-over-end mixing at 4°C. The beads were washed once in batch (100 mM Tris, pH 8.0, 150 mM NaCl, 7.5% glycerol, 0.25 mM CaCl2, 0.25 mM ATP, 0.5 mM DTT) and then moved into a chromatography column (Bio-Rad) and washed four additional times with one column bed volume of wash buffer. Protein was eluted with 6 half-column bed volumes with elution buffer (100 mM Tris, pH 8.0, 150 mM NaCl, 7.5% glycerol, 0.25 mM CaCl2, 0.25 mM ATP, 0.5 mM DTT, 2 mM d-biotin).
Actin pelleting assay.
TwinStrep-Actin was purified as described above and then dialyzed for 2 h in G buffer (5 mM Tris, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM dithiothreitol). After a buffer exchange, the actin was dialyzed overnight. The dialyzed actin was cleared by centrifugation at 100,000 × g for 30 min to remove aggregates. A 1/10 volume of 10× KMEI80 (800 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole [pH 7.0]) was added to the cleared actin, followed by incubation for 30 min at room temperature. The KMEI80-actin mixture was then centrifuged at 100,000 × g for 30 min.
Mass spectroscopy.
Mass spectrometry was performed by the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory at UC Berkeley. The protein solution was adjusted to 8 M urea, subjected to carboxyamidomethylation of cysteines, and digested with trypsin. The sample was then desalted using a c18 spec tip (Varian). A nano-LC column was packed in a 100-μm-inner-diameter glass capillary with an emitter tip. The column consisted of 10 cm of Polaris c18 5-μm packing material (Varian), followed by 4 cm of Partisphere 5 SCX (Whatman). The column was loaded by using a pressure bomb and washed extensively with buffer A (see below). The column was then directly coupled to an electrospray ionization source mounted on a Thermo-Fisher LTQ XL linear ion trap mass spectrometer. An Agilent 1200 high-pressure liquid chromatograph equipped with a split line so as to deliver a flow rate of 300 nl/min was used for chromatography. Peptides were eluted using an eight-step MudPIT procedure (14). Buffer A was 5% acetonitrile–0.02% heptafluorobutyric acid (HBFA); buffer B was 80% acetonitrile–0.02% HBFA. Buffer C was 250 mM ammonium acetate–5% acetonitrile–0.02% HBFA; buffer D was the same as buffer C, but with 500 mM ammonium acetate. The programs SEQUEST and DTASelect were used to identify peptides and proteins from the Giardia database (18, 19).
Immunoprecipitation and Western blotting.
Immunoprecipitation began with a single confluent 13-ml tube per cell line. After detachment, cells were pelleted at 900 × g and washed once in HBS. The cells were resuspended in 300 μl of lysis buffer (50 mM Tris 7.5, 150 mM NaCl, 7.5% glycerol, 0.25 mM CaCl2, 0.25 mM ATP, 0.5 mM DTT, 0.5 mM PMSF, 0.1% Triton X-100, 2× Halt protease inhibitors [Pierce]) and sonicated. The lysate was cleared by centrifugation at 10,000 × g for 10 min at 4°C and then added to 30 μl of anti-HA beads (Sigma). After 1.5 h of binding, the beads were washed four times with wash buffer (25 mM Tris 7.5, 150 mM NaCl, 0.25 mM CaCl2, 0.25 mM ATP, 5% glycerol, 0.05% Tween 20) and then boiled in 50 μl of sample buffer. Western blotting was performed as described previously (13). Rabbit anti-giActin polyclonal (13) and anti-HA mouse monoclonal HA7 antibody (Sigma-Aldrich) were both used at 1:3,000. Fluorescent secondary antibodies (Li-Cor) were used at 1:15,000, horseradish peroxidase-linked anti-rabbit antibodies (Bio-Rad) were used at 1:7,000.
Microscopy.
Fixations were performed as described previously (13). Anti-HA mouse monoclonal HA7 antibody (Sigma-Aldrich) was used at 1:125, and anti-mouse and anti-rabbit secondary antibodies were used at 1:200 (Molecular Probes). Images were acquired on a DeltaVision Elite microscope using a 100× 1.4 NA objective and a CoolSnap HQ2 camera. Deconvolution was performed with SoftWorx (API, Issaquah, WA). Maximal projections were made with ImageJ (20), and figures were assembled using the Adobe Creative Suite (Mountain View, CA).
RESULTS
We set out to identify giActin interactors via an affinity chromatography approach utilizing the TwinStrep Tag (21–23). Actin is notoriously sensitive to chimeric fusions, because epitope or fluorescent protein fusions may cause steric interference or otherwise affect filament formation and dynamics (24). Thus, we devised a strategy to test whether our TwinStrep-Actin fusion (TS-Actin) was functional in vivo. Previous work demonstrated that actin can be effectively depleted with translation-blocking morpholinos (13). These antisense morpholinos bind to the start of the transcript and block translation initiation machinery from recognizing the start codon (9, 13). Therefore, by fusing TwinStrep to the N terminus of giActin, we generated a morpholino-insensitive version of giActin. In this case, morpholino treatment is expected to block translation of endogenous actin, whereas it should have no effect on the transgenic version. We also sought to maintain actin levels near endogenous levels by driving expression of our TS-Actin fusion with the native actin promoter.
Quantitative Western blotting with an anti-giActin polyclonal antibody (13) indicated that although we used the native promoter, there was roughly a 4-fold increase in total actin levels compared to nontransgenic controls; ca. 75% of this was TS-Actin (Fig. 2A). The higher levels of transgenic actin are presumably due to the copy number of our episomally maintained construct exceeding the number of endogenous actin genes. Morpholino treatment of the TS-Actin-expressing cell line behaved as predicted; the N-terminal epitope tag protected TS-Actin from being depleted by anti-actin morpholinos, while the endogenous actin was depleted to ∼20% of control levels (Fig. 2A). Further, we examined the morpholino-treated cells for morphological defects associated with actin depletion such as abnormal cell shape, out-of-position flagella, and multiple or out-of-position nuclei (13). In the control-treated cell line we observed a slight increase in the number of abnormal cells: 6.6% for TS-Actin (n = 600) versus 1.9% for wild-type (n = 400), indicating that the increased actin levels and/or the epitope tag mildly interfered with normal actin function (Fig. 2B). The transgenic line was, however, resistant to morpholino depletion since the proportion of abnormal cells remained at 6.8% (n = 600) after morpholino treatment. In contrast, 35.2% of the WT cells (n = 500) treated with the anti-actin morpholinos had abnormal morphology. Therefore, we conclude that TS-Actin can partially rescue endogenous actin depletion, indicating that TS-Actin is functional in vivo.
FIG 2.
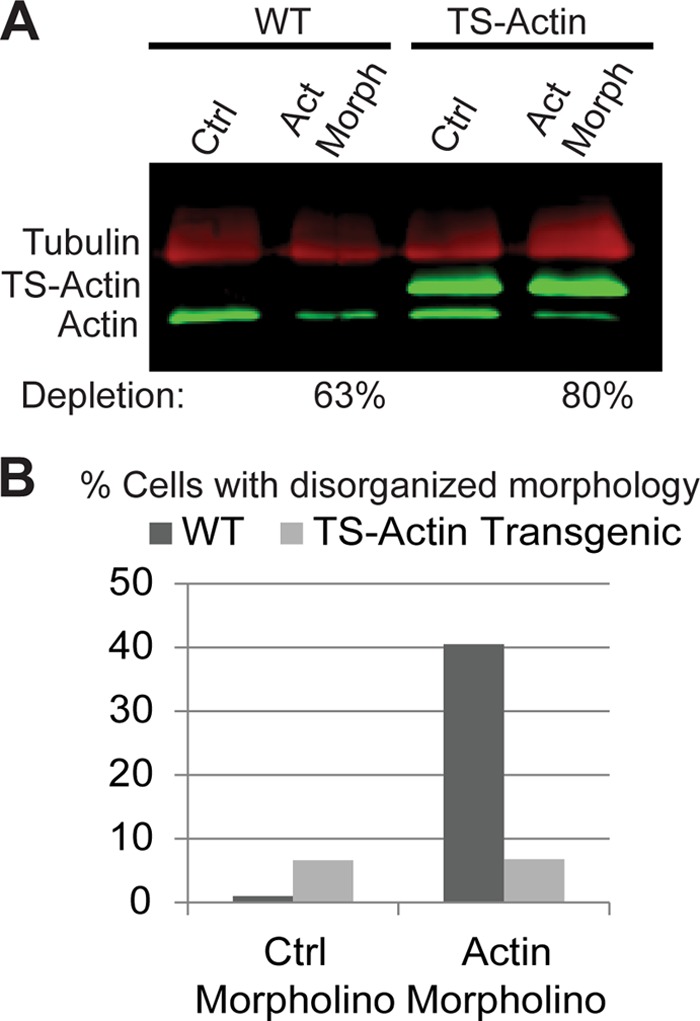
TS-Actin is functional in vivo. (A) Multiplex Western blot (actin, green; tubulin, red) showing that TS-Actin is morpholino resistant, while endogenous actin is significantly reduced. (B) Reducing endogenous actin results in cellular disorganization, morpholino-resistant TS-Actin can substitute for endogenous actin.
A particular challenge of producing large-scale Giardia cultures, sufficient for biochemical analysis, is the need to provide surface area for adherent growth. Giardia is an extracellular parasite that colonizes the host intestine by attaching via its “suction cup” organelle, the ventral disc (25, 26). Likewise, in the laboratory Giardia trophozoites grow attached to the sides of the culture tubes. Cultures cease to proliferate after the culture tubes are confluent with cells. Free-floating cells are often observed to have an aberrant morphology, indicating the importance of surface attachment, possibly because Giardia divides by an adhesion-dependent mechanism (27, 28). Custom “inside-out” roller bottles have been used by others to grow high-yield Giardia cultures, but these are not commercially available (16). We developed a low-cost high-yield method of growing Giardia by inserting common polypropylene drinking straws into wide mouth bottles (see Fig. S1 in the supplemental material and see Materials and Methods). Using our high-surface-area culture system, 1-liter cultures of WT and the TS-Actin transgenic cell lines produced ∼2-ml cell pellets. Extracts from these cell pellets were affinity purified in parallel. The elutions from a pilot experiment were concentrated before sodium dodecyl sulfate (SDS) analysis so that ∼50% of the eluted protein could be analyzed by SDS-PAGE. Many unique bands are apparent in the TS-Actin sample (Fig. 3A). The purification was repeated for mass spectroscopy analysis; Fig. 3B represents 5% of the elutions that were analyzed by mass spectroscopy. Table 1 lists 57 proteins that were unique to the TS-Actin sample and had a minimum of five detected peptides. The complete list, including low-abundance hits and proteins also identified in our mock control, is given in Table S2 in the supplemental material. Bioinformatics analysis was utilized to place these hits into six categories (Table 1; see Table S2 in the supplemental material).
FIG 3.
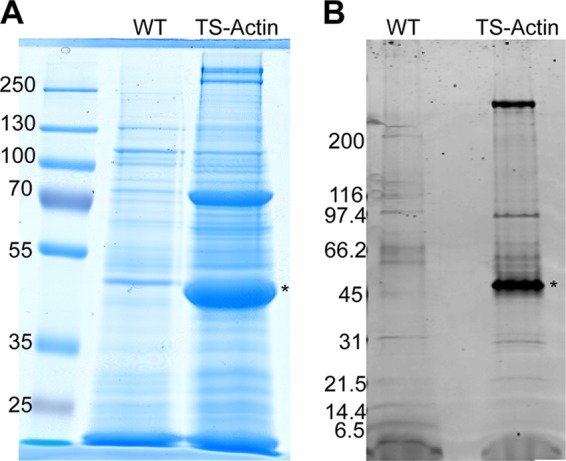
Isolation of TS-Actin and interacting proteins. (A) Elutions from streptactin columns for both WT and TS-Actin purifications were concentrated and then analyzed by SDS-PAGE. Note that several bands are unique to the TS-Actin cell line. Actin is marked with an asterisk. (B) Five percent of the TS-Actin purification used in the MudPIT analysis was loaded onto a 4 to 16% gradient gel and stained with SYPRO Ruby.
TABLE 1.
Identified interactors
| Protein identification no.a | Name and/or description | Mol wt | No. of peptides | Interactorb | Reference(s) | S. salmonicida GenBank no.c |
|---|---|---|---|---|---|---|
| Axonemal/cytoskeleton | ||||||
| GL50803_111950 | Axonemal dynein heavy chain | 570,319 | 900 | Precedents | 32, 39 | EST41976, 0.0 |
| GL50803_101138 | Axonemal dynein heavy chain | 578,219 | 849 | Precedents | 32, 39 | EST46166.1, 0.0 |
| GL50803_40496 | Axonemal dynein heavy chain | 553,424 | 778 | Precedents | 32, 39 | EST44588.1, 0.0 |
| GL50803_13273* | P28 axonemal dynein light chain | 26,895 | 294 | IP+++ | 32, 39 | EST43975.1, 6E–138 |
| GL50803_42285 | Axonemal dynein heavy chain 11 | 834,759 | 48 | Precedents | 32, 39 | EST41750.1, 2E–61 |
| GL50803_6744* | Centrin | 18,687 | 46 | IP+ | 32, 39 | EST41812.1, 2E–98 |
| GL50803_37985 | Dynein heavy chain | 118,679 | 26 | Precedents | 32, 39 | EST48250.1, 0.0 |
| GL50803_137716 | Axoneme-associated protein GASP-180 | 174,782 | 24 | No | ||
| GL50803_16424* | Myeloid leukemia factor like | 29,702 | 14 | IP− | 35 | EST47319.1, 9E–17 |
| GL50803_14242 | Dynein heavy chain-cytoplasmic | 633,335 | 11 | EST46283.1, 1E–27 | ||
| Chaperone | ||||||
| GL50803_11992 | TCP-1 chaperonin subunit epsilon | 61,193 | 223 | Precedents | 29 | EST46493.1, 0.0 |
| GL50803_11397 | TCP-1 chaperonin subunit beta | 56,604 | 204 | Precedents | 29 | EST41446.1, 0.0 |
| GL50803_16124 | TCP-1 chaperonin subunit eta | 64,752 | 196 | Precedents | 29 | EST48572.1, 0.0 |
| GL50803_13500 | TCP-1 chaperonin subunit theta | 60,646 | 187 | Precedents | 29 | EST46649.1, 0.0 |
| GL50803_10231 | TCP-1 chaperonin subunit zeta | 60,941 | 118 | Precedents | 29 | EST48976.1, 0.0 |
| GL50803_17482 | TCP-1 chaperonin subunit delta | 56,324 | 86 | Precedents | 29 | EST41630.1, 4E–173 |
| GL50803_91919 | TCP-1 chaperonin subunit alpha | 59,281 | 82 | Precedents | 29 | EST47029.1, 0.0 |
| GL50803_17411 | TCP-1 chaperonin subunit gamma | 61,559 | 71 | Precedents | 29 | EST41533.1, 0.0 |
| GL50803_88765* | Cytosolic HSP70 | 71,633 | 22 | IP+ | 52 | EST45839, 0.0 |
| GL50803_17121 | Bip | 74,360 | 12 | 53 | EST46254.1, 0.0 | |
| Nuclear | ||||||
| GL50803_9825* | TBP-interacting protein TIP49 | 51,418 | 276 | IP+++ | 54–56 | EST49365.1, 0.0 |
| GL50803_17565 | TBP-interacting protein TIP49 | 52,616 | 162 | Precedents | 54–56 | EST48144.1, 0.0 |
| GL50803_15113* | ARP7-like | 51,465 | 90 | IP+ | 54–56 | EST44310.1, 6E–31 |
| GL50803_9705 | Hypothetical/YEATS domain | 30,449 | 23 | EST47502.1, 1E–27 | ||
| GL50803_8125 | SMARCC1 | 47,004 | 17 | Precedents | 54–56 | No |
| GL50803_2851 | Histone acetyltransferase MYST2 | 49,916 | 12 | Precedents | 54–56 | EST45122.1, 9E–78 |
| GL50803_6886 | Prokaryotic SMC domain protein | 102,836 | 11 | No | ||
| GL50803_17461 | SWIRM domain protein | 124,241 | 7 | EST47570.1, 3E–51 | ||
| Signaling | ||||||
| GL50803_6317 | Putative DUB | 150,389 | 16 | EST43894.1,1E–14 | ||
| GL50803_22850* | ERK7-like/giERK2 | 41,096 | 15 | IP+ | EST48827.1, 0.0 | |
| GL50803_6430* | 14-3-3 protein | 28,576 | 8 | IP++ | EST46224.1, 6E–115 | |
| GL50803_12795 | Phosducin-like | 26,919 | 6 | EST41453.1, 1E–14 | ||
| GL50803_9413 | Protein disulfide isomerase PDI2 | 50,408 | 6 | Precedents | 57 | EST43183.1, 5E–43 |
| Trafficking | ||||||
| GL50803_14373* | Dynamin | 79,513 | 13 | IP− | 36 | EST46023.1, 0.0 |
| Unknown/Giardia specific | ||||||
| GL50803_15264 | Hypothetical protein | 404,245 | 153 | No | ||
| GL50803_39938 | Hypothetical protein | 716,474 | 95 | EST41505.1, 4E–25 | ||
| GL50803_17532 | Hypothetical protein | 66,334 | 57 | EST45241.1, 3E–19 | ||
| GL50803_15485 | Hypothetical protein | 751,987 | 55 | EST41949.1, 9E–20 | ||
| GL50803_13942 | Hypothetical Protein | 56,506 | 53 | No | ||
| GL50803_7807 | WD-40 repeat protein | 59,785 | 51 | EST44362.1, 2E–65 | ||
| GL50803_17266 | Ankyrin and WD repeat protein | 14,462 | 47 | No | ||
| GL50803_137711 | Hypothetical protein | 719,629 | 41 | EST41949.1, 2E–49 | ||
| GL50803_17596 | Zinc finger domain protein | 90,538 | 29 | EST4938.1, 2E–09 | ||
| GL50803_23897 | Hypothetical protein | 92,642 | 28 | No | ||
| GL50803_99726 | Hypothetical protein | 11,183 | 25 | No | ||
| GL50803_113592 | Hypothetical protein | 489,165 | 24 | No | ||
| GL50803_5859 | Hypothetical Protein | 20,141 | 22 | No | ||
| GL50803_20601 | Hypothetical protein | 11,136 | 22 | No | ||
| GL50803_3559 | Hypothetical protein | 24,801 | 18 | No | ||
| GL50803_14492 | Hypothetical protein | 54,020 | 15 | No | ||
| GL50803_35487 | Hypothetical protein | 884,517 | 14 | No | ||
| GL50803_8725 | Hypothetical protein | 51,753 | 11 | No | ||
| GL50803_37350 | Hypothetical protein | 796,898 | 11 | EST43354.1, 0.0 | ||
| GL50803_35341 | Hypothetical protein | 776,894 | 11 | EST45092.1, 1E–176 | ||
| GL50803_15442 | KIF binding domain | 108,189 | 8 | No | ||
| GL50803_137739 | Hypothetical protein | 211,474 | 8 | No | ||
| GL50803_92602 | Hypothetical protein | 342,328 | 6 | EST44629.1, 6E–73 |
*, This protein was chosen for testing interaction with actin, as described in Results.
+++, strong interactor; ++, moderate interactor; +, weak interactor; −, no interaction detected.
The exponential values are BLAST Expect values, indicating the level of conservation between the homologs.
We identified several hits that support the quality and relevance of this data set. For example, we identified all eight subunits of the TCP-1 chaperonin complex, which has an important role in folding actin (29). In addition, two proteins, p28 dynein light chain (p28 DLC) and centrin, were found in the giActin interactome, which we had previously hypothesized to be conserved actin-interacting proteins (13). Genetic and biochemical analysis of flagellar components has demonstrated that actin has an important role in flagella, where it functions in the inner dynein arm complexes (30–32). Within the inner dynein arms, p28 DLC and centrin, have been demonstrated to directly interact with actin (32). In Giardia, actin is readily detectable within all eight flagella, and both p28 DLC and centrin are conserved (13). In terms of peptides per molecular weight, p28 DLC was the most abundant interactor identified in our analysis. In addition to these examples, homologs of several other proteins that have been reported to complex with actin in other eukaryotes were identified and are indicated in Table 1.
The genome of Spironucleus salmonicida, another diplomonad and close relative of Giardia, was recently released (33). As part of our analysis, we compared our list of putative actin interactors to the S. salmonicida genome (Table 1) (33). Although most of the identified proteins are present in S. salmonicida, several appear to be specific to Giardia. We also searched the S. salmonicida genome for the presence of canonical actin-binding proteins. Intriguingly, we found that S. salmonicida contains several actin-binding proteins not found in Giardia; these include formin, cofilin, and coronin (see Table S3 in the supplemental material). S. salmonicida, however, lacks many canonical actin-binding proteins, including the Arp2/3 complex, nucleation-promoting factors, dynactin, capping protein, and myosin. Nevertheless, the subset of canonical actin-binding proteins in S. salmonicida suggests the loss of such proteins from Giardia. Without additional genomes, we can only speculate whether the diplomonads ever had the full complement of actin-binding proteins; however, the absence of myosin in Giardia, S. salmonicida, and Trichomonas vaginalis (a nondiplimonad excavate) remains consistent with the idea that a subset of excavates may have split from the other eukaryotes before the full complement of actin-binding proteins was established (4, 34). This possibility could help explain how Giardia could have lost proteins that are essential in the model eukaryotes.
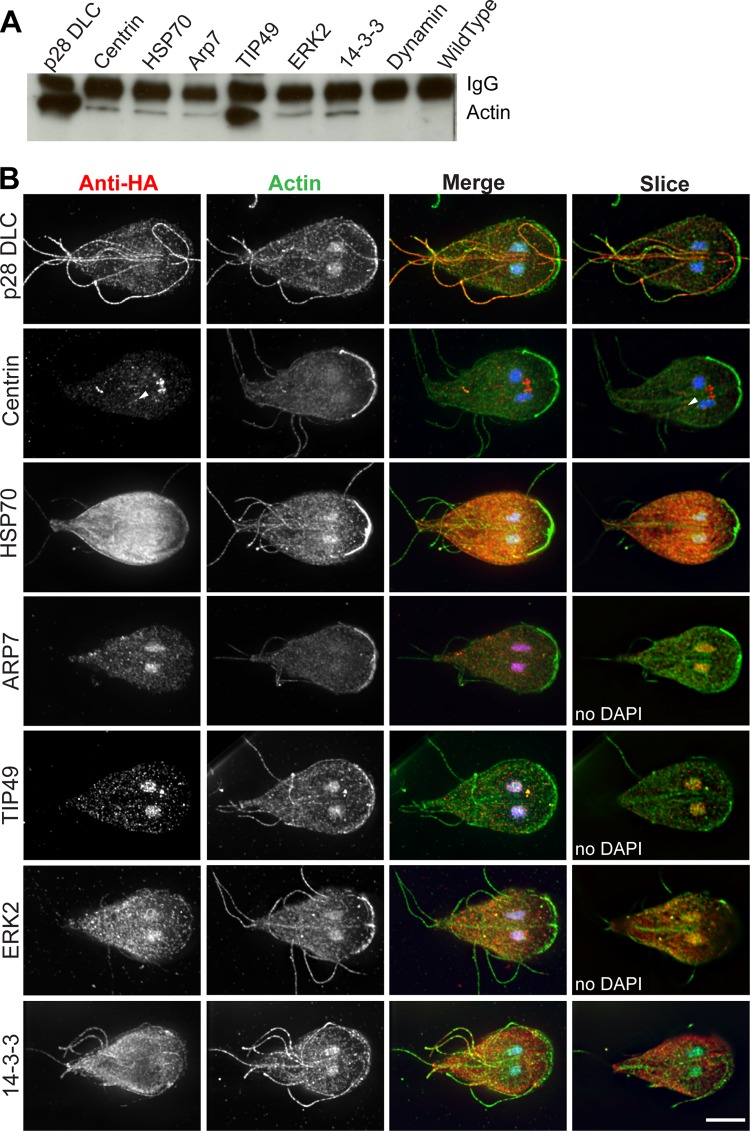
Next, we sought to validate a subset of the conserved interactions through reciprocal immunoprecipitations. We selected nine representative proteins, at least one from each of the conserved categories; these are indicated by an asterisk in Table 1. In each case, we tagged the identified protein with a C-terminal triple HA tag. We were able to verify complex formation with giActin for p28 DLC, centrin, HSP70, ARP7, TIP49, ERK2, and 14-3-3 (Fig. 4A). Attempts to validate dynamin (Fig. 4A) and myeloid leukemia factor (MLF; data not shown) were unsuccessful. Both dynamin and MLF have been shown to interact with actin and alter filament organization in other eukaryotes (35, 36). Although these hits may be false positives, it is also possible that the C-terminal tag disrupted interaction or that the lower concentration of cell extracts in our immunoprecipitation experiments versus large-scale affinity chromatography failed to maintain integrity of the complex.
FIG 4.
Validation of identified interacting proteins. (A) Immunoprecipitation from Giardia extracts of C-terminally HA-tagged interactors, followed by anti-actin Western blotting demonstrates that these proteins interact with actin. (B) Colocalization of actin and HA-tagged interacting proteins in Giardia trophozoites. Actin is green, HA tagged proteins are red, DNA is blue. The first three columns are maximal projections, and the last column is a single slice through the middle of the cell. Arrowhead marks centrin localization associated with the posterolateral flagella. Scale bar, 5 μm.
To better understand the relationship between these conserved interactors and actin, we colocalized actin and the tagged interactors (Fig. 4B). Each protein displayed a localization pattern consistent with its proposed function. p28 DLC localized to flagella. Centrin localized to the basal bodies and around a portion of the internal axonemes of the posterolateral flagella. ARP7, TIP49, and ERK2 localized to the nuclei with various amounts of non-nuclear localization. HSP70 and 14-3-3 were found throughout the cell with slight enrichment at the cell anterior. None of these conserved proteins colocalized with prominent filamentous actin structures (see Fig. 1), which is consistent with the idea that they complex with G-actin (discussed below). It should be noted that standard tools such as fluorescent phalloidin and DNase I typically used to distinguish between monomeric and filamentous actin do not work in Giardia (13).
DISCUSSION
In this study, we undertook a biochemical approach to identify actin interactors in Giardia. Our easily adopted method for growing large-scale cultures and the use of the TwinStrep tag have the potential to make the process of defining interactomes routine in Giardia. During the course of our study, Svard and coworkers published a TAP-tagging approach for proteomics in Giardia (37). Similar to our approach, these researchers used two tandem Strep II tags but also included a Flag tag, the entirety of which is known as the SF-TAP tag. They overcame the surface area issues by distributing 2 liters of medium among 40 50-ml conical tubes. Our straw method simplifies cell culture, and our ability to identify actin-interacting proteins in a single purification step suggests that tandem purification is not generally required. This is significant because single-step purifications are able to isolate weaker interactors commonly lost in two-step purifications (22). Indeed, our laboratories have already performed proteomic analysis on two additional proteins using this approach. In all cases, 1 liter of medium was sufficient for isolating protein-protein interactors. Although analysis is still under way, in each case a unique set of high-frequency hits were identified. Conversely, several low-frequency hits are common to our data sets. One data set is for Polo-like kinase (S. Gourguechon and W. Z. Cande, unpublished data); because we did not find Polo in the actin data set, nor did we find actin in the Polo data set, we believe the low-abundance hits are likely false positives. The identity of these low-abundance hits may be useful for others using our same approach; therefore, we have identified the overlapping hits in Table S2 in the supplemental material.
Although once controversial, it is now clear that actin is part of the nucleoskeleton responsible for many nuclear processes, including transcriptional regulation, chromatin remodeling, and general maintenance of genome organization and integrity (reviewed in reference 38). In contrast to localization studies performed in model eukaryotes, where it is difficult to detect actin in the nucleus, giActin is readily detectable in the nuclei, suggesting that it has an important role in nuclear function (see Fig. 3B). Although we validated complex formation with actin for ARP7 and TIP49 (the second most abundant hit in terms of peptides/molecular weight), we identified six other proteins containing domains that are consistent with a role in actin-based chromatin remodeling. It has been put forth that several proteins known to function in the cytoskeleton have roles in the nucleus; thus, they may have originally evolved to serve the genome (38). Our identification of conserved nuclear proteins and the lack of core cytoskeletal regulators are consistent with this notion.
Actin's role in the flagella is well established but largely ignored. Biochemical fractionation of flagella has shown that six of the seven inner dynein arm complexes are associated with actin (39). A conventional actin mutant of Chlamydomonas, ida5, lacks four of the inner-arm dynein complexes and, in this mutant, an actin-like protein, NAP, is upregulated to substitute for actin in the remaining two inner-arm dynein complexes, thus demonstrating the importance of actin to axonemal structure and function (39). Within the inner-arm dynein complexes, actin is thought to exist as a monomer in a complex with either a dimer of p28 or a monomer of centrin (32). Using super-resolution microscopy, we observed a regular repeating pattern for actin within all eight of the Giardia flagella (13). The precise molecular role of actin in the inner-arm dynein complexes remains enigmatic, but if actin simply acts as an adapter, it is perplexing to consider that Giardia may have lost proteins such as myosin, formin, and cofilin while maintaining actin's role in the flagella. Alternatively, some of the earliest functions of actin may include flagellar and nuclear processes.
Of the conserved proteins identified, 14-3-3 and ERK2 may be the most intriguing since they are likely regulators of actin dynamics or actin-related processes. 14-3-3 is known to play an important role in cytoskeletal regulation in other eukaryotes (40); however, the relationship between 14-3-3 and actin is complicated by multiple isoforms of 14-3-3 and conflicting results about 14-3-3 interactions with actin (reviewed in reference 41). In addition to our identification of 14-3-3 as an actin interactor, several efforts to define the 14-3-3 interactome in both Giardia and mammalian cells corroborate an actin–14-3-3 interaction (42–44). However, the current view is that 14-3-3 regulates actin through phosphodependent interaction with the actin-depolymerizing protein cofilin and does not bind to actin directly (45, 46). Notably, Giardia lacks both cofilin and its regulatory kinase LIM. Perhaps a more direct regulation of actin by 14-3-3 underlies the well-characterized cofilin–14-3-3 interaction. Our analysis of 14-3-3's role in actin regulation is in preparation (J. Krtková, J. W. Xu, and A. R. Paredez, unpublished data).
Giardial ERK2 is a potential regulator of actin-related processes. Giardia contains two ERK homologs: a prototypical ERK, giERK1, and the ERK7-like protein giERK2 (47, 48). Although ERK stands for extracellular signal-regulated kinase, ERK7, unlike other mitogen-activated proteins, is atypical in that it is thought to be auto-activated rather than responding to external signals (49, 50). giERK2 lacks the extended C-terminal domain found in ERK7 and may not be regulated in the same manner, and yet in vitro kinase assays have shown that giERK2 is much more active than giERK1 (48). In other eukaryotes, ERK7 kinases have been shown to regulate protein secretion and cell proliferation (50, 51). Our identification of an actin-ERK2 complex in Giardia and the localization of this kinase in the nucleus and throughout the cell do not exclude potentially conserved function.
This initial characterization of actin-associated proteins in Giardia has focused on validating interaction with conserved proteins (see above), both because of the evolutionary implications and because these conserved proteins serve as proof of principle for our proteomic strategy. The largest group of identified proteins is, however, the novel/Giardia-specific category (see Table 1). We have begun to analyze the Giardia-specific interactors with the same endogenous-tagging approach used to study the conserved actin-interacting proteins. We have tagged 13 of the Giardia-specific proteins listed in Table 1 and have been able to immunoprecipitate giActin with 10 of these proteins (in preparation). Notably, five of these proteins localize to the nuclei, one localizes to the flagella, and the remaining proteins show a punctate pattern. None of the proteins identified thus far appear to colocalize with filamentous actin structures. Most canonical actin-binding proteins exclusively recognize globular or filamentous actin. The buffer conditions used in our affinity purification approach contained Ca2+ and ATP but lacked the Mg2+ and KCl typically found in buffers that support actin filament formation. Therefore, additional interactors remain to be discovered, and the set of interactors described here likely represents the globular giActin interactome. We did, however, test the ability of TS-Actin to polymerize, as assayed by ultracentrifugation (see Fig. S2 in the supplemental material). We observed that TS-Actin has some ability to pellet under filament-forming conditions that are consistent with the partial rescue we observed in our morpholino-knockdown experiments (Fig. 2). Future work will explore the identification of actin interactors in buffer conditions that support actin filament formation.
In this first exploration of the giActin interactome, we found both conserved and Giardia-specific interactors. The subset of canonical actin-binding proteins in S. salmonicida suggests loss of actin-binding proteins from Giardia. Therefore, the retention of actin-interacting proteins in the nucleus and flagella suggest these processes are the most constrained of any actin processes. In any case, the role of actin in the nucleoskeleton and flagella are likely some of actin's most ancient functions and remain relatively unexplored compared to the role of actin in the cytoskeleton. The set of novel/Giardia-specific proteins remain intriguing. Many of these proteins have no recognizable domains; therefore, elucidation of their function will be a challenge. Simultaneously, if functional experiments demonstrate these proteins to be essential, they will become potential therapeutic targets to treat giardiasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Kohlstaedt and the QB3 P/MSL for their assistance with the MudPIT analysis. We thank S. Gourguechon for technical assistance, for sharing his proteomic data, and for critical reading of the manuscript. We thank M. Steele-Ogus, W. Hardin, and J. J. Vicente for critical reading of the manuscript.
This research was sponsored by National Institutes of Health grant A1054693 to W.Z.C., National Science Foundation Postdoctoral Fellowship 0705351, and UW Biology Startup funds to A.R.P.
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00041-14.
REFERENCES
- 1.Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int. J. Syst. Evol. Microbiol. 52:297–354. 10.1099/ijs.0.02058-0 [DOI] [PubMed] [Google Scholar]
- 2.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 3.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. 2009. Phylogenomic analyses support the monophyly of excavata and resolve relationships among eukaryotic “supergroups.” Proc. Natl. Acad. Sci. U. S. A. 106:3859–3864. 10.1073/pnas.0807880106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JE, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926. 10.1126/science.1143837 [DOI] [PubMed] [Google Scholar]
- 5.Koonin E. 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11:209. 10.1186/gb-2010-11-5-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keister DB. 1983. Axenic culture of Giardia lamblia in Tyi-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 77:487–488. 10.1016/0035-9203(83)90120-7 [DOI] [PubMed] [Google Scholar]
- 7.Gillin FD, Reiner DS, Gault MJ, Douglas H, Das S, Wunderlich A, Sauch JF. 1987. Encystation and expression of cyst antigens by Giardia lamblia in vitro. Science 235:1040–1043. 10.1126/science.3547646 [DOI] [PubMed] [Google Scholar]
- 8.Sun CH, Tai JH. 2000. Development of a tetracycline controlled gene expression system in the parasitic protozoan Giardia lamblia. Mol. Biochem. Parasitol. 105:51–60. 10.1016/S0166-6851(99)00163-2 [DOI] [PubMed] [Google Scholar]
- 9.Carpenter ML, Cande WZ. 2009. Using morpholinos for gene knockdown in Giardia intestinalis. Eukaryot. Cell 8:916–919. 10.1128/EC.00041-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gourguechon S, Cande WZ. 2011. Rapid tagging and integration of genes in Giardia intestinalis. Eukaryot. Cell 10:142–145. 10.1128/EC.00190-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SC, Paredez AR. 2013. Alternative cytoskeletal landscapes: cytoskeletal novelty and evolution in basal excavate protists. Curr. Opin. Cell Biol. 25:134–141. 10.1016/j.ceb.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollard TD. 2003. The cytoskeleton, cellular motility, and the reductionist agenda. Nature 422:741–745. 10.1038/nature01598 [DOI] [PubMed] [Google Scholar]
- 13.Paredez AR, Assaf ZJ, Sept D, Timofejeva L, Dawson SC, Wang CJ, Cande WZ. 2011. An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 108:6151–6156. 10.1073/pnas.1018593108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn MP, Wolters D, Yates JR., III 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- 15.Sagolla MS, Dawson SC, Mancuso JJ, Cande WZ. 2006. Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J. Cell Sci. 119:4889–4900. 10.1242/jcs.03276 [DOI] [PubMed] [Google Scholar]
- 16.Farthing MJG, Pereira MEA, Keusch GT. 1982. Giardia lamblia: evaluation of roller bottle cultivation. Exp. Parasitol. 54:410–415. 10.1016/0014-4894(82)90050-9 [DOI] [PubMed] [Google Scholar]
- 17.Singer SM, Yee J, Nash TE. 1998. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem. Parasitol. 92:59–69. 10.1016/S0166-6851(97)00225-9 [DOI] [PubMed] [Google Scholar]
- 18.Eng JK, McCormack AL, Yates JR. 1994. an approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989. 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 19.Tabb DL, McDonald WH, Yates JR., III 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26. 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt TG, Batz L, Bonet L, Carl U, Holzapfel G, Kiem K, Matulewicz K, Niermeier D, Schuchardt I, Stanar K. 2013. Development of the Twin-Strep-Tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 92:54–61. 10.1016/j.pep.2013.08.021 [DOI] [PubMed] [Google Scholar]
- 22.Witte CP, Noel LD, Gielbert J, Parker JE, Romeis T. 2004. Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55:135–147. 10.1007/s11103-004-0501-y [DOI] [PubMed] [Google Scholar]
- 23.Junttila MR, Saarinen S, Schmidt T, Kast J, Westermarck J. 2005. Single-step Strep-Tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics 5:1199–1203. 10.1002/pmic.200400991 [DOI] [PubMed] [Google Scholar]
- 24.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam RD. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447–475. 10.1128/CMR.14.3.447-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen WR, Tulyathan O, Dawson SC, Cande WZ, Fletcher DA. 2006. Giardia lamblia attachment force is insensitive to surface treatments. Eukaryot. Cell 5:781–783. 10.1128/EC.5.4.781-783.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benchimol M. 2004. Mitosis in Giardia lamblia: multiple modes of cytokinesis. Protist 155:33–44. 10.1078/1434461000162 [DOI] [PubMed] [Google Scholar]
- 28.Tumova P, Kulda J, Nohynkova E. 2007. Cell division of Giardia intestinalis: assembly and disassembly of the adhesive disc, and the cytokinesis. Cell Motil. Cytoskel. 64:288–298. 10.1002/cm.20183 [DOI] [PubMed] [Google Scholar]
- 29.Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. 1993. The T-complex polypeptide-1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl. Acad. Sci. U. S. A. 90:9422–9426. 10.1073/pnas.90.20.9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 183:923–932. 10.1083/jcb.200808050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piperno G, Luck DJL. 1979. Actin-like protein is a component of axonemes from Chlamydomonas flagella. J. Biol. Chem. 254:2187–2190 [PubMed] [Google Scholar]
- 32.Yanagisawa H, Kamiya R. 2001. Association between actin and light chains in Chlamydomonas flagellar inner-arm dyneins. Biochem. Biophys. Res. Commun. 288:443–447. 10.1006/bbrc.2001.5776 [DOI] [PubMed] [Google Scholar]
- 33.Xu F, Jerlstrom-Hultqvist J, Einarsson E, Astvaldsson A, Svard SG, Andersson JO. 2014. The genome of spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet. 10:e1004053. 10.1371/journal.pgen.1004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212. 10.1126/science.1132894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata K, Takagi K, Hashida T, Ichikawa Y. 1985. A monovalent cation-sensitive actin-binding factor in a myeloid leukemia cell line. Cell Struct. Funct 10:105–120. 10.1247/csf.10.105 [DOI] [PubMed] [Google Scholar]
- 36.Yamada H, Abe T, Satoh A, Okazaki N, Tago S, Kobayashi K, Yoshida Y, Oda Y, Watanabe M, Tomizawa K, Matsui H, Takei K. 2013. Stabilization of actin bundles by a dynamin 1/cortactin ring complex is necessary for growth cone filopodia. J. Neurosci. 33:4514–4526. 10.1523/jneurosci.2762-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jerlstrom-Hultqvist J, Stadelmann B, Birkestedt S, Hellman U, Svard SG. 2012. Plasmid vectors for proteomic analyses in Giardia: purification of virulence factors and analysis of the proteasome. Eukaryot. Cell 11:864–873. 10.1128/ec.00092-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon DN, Wilson KL. 2011. The nucleoskeleton as a genome-associated dynamic “network of networks.” Nat. Rev. Mol. Cell Biol. 12:695–708. 10.1038/nrm3207 [DOI] [PubMed] [Google Scholar]
- 39.KatoMinoura T, Hirono M, Kamiya R. 1997. Chlamydomonas inner-arm dynein mutant, Ida5, has a mutation in an actin-encoding gene. J. Cell Biol. 137:649–656. 10.1083/jcb.137.3.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth D, Birkenfeld J, Betz H. 1999. Dominant-negative alleles of 14-3-3 proteins cause defects in actin organization and vesicle targeting in the yeast Saccharomyces cerevisiae. FEBS Lett. 460:411–416. 10.1016/S0014-5793(99)01383-6 [DOI] [PubMed] [Google Scholar]
- 41.Sluchanko NN, Gusev NB. 2010. 14-3-3 proteins and regulation of cytoskeleton. Biochemistry (Mosc.) 75:1528–1546 [DOI] [PubMed] [Google Scholar]
- 42.Lalle M, Camerini S, Cecchetti S, Sayadi A, Crescenzi M, Pozio E. 2012. Interaction network of the 14-3-3 protein in the ancient protozoan parasite Giardia duodenalis. J. Proteome Res. 11:2666–2683. 10.1021/pr3000199 [DOI] [PubMed] [Google Scholar]
- 43.Liang S, Yu Y, Yang P, Gu S, Xue Y, Chen X. 2009. Analysis of the protein complex associated with 14-3-3 epsilon by a deuterated-leucine labeling quantitative proteomics strategy. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 877:627–634. 10.1016/j.jchromb.2009.01.023 [DOI] [PubMed] [Google Scholar]
- 44.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 2004. 14-3-3 affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation, and trafficking. Biochem. J. 379:395–408. 10.1042/BJ20031797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkenfeld J, Betz H, Roth D. 2003. Identification of cofilin and Lim-domain-containing protein kinase 1 as novel interaction partners of 14-3-3 zeta. Biochem. J. 369:45–54. 10.1042/BJ20021152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gohla A, Bokoch GM. 2002. 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr. Biol. 12:1704–1710. 10.1016/S0960-9822(02)01184-3 [DOI] [PubMed] [Google Scholar]
- 47.Manning G, Reiner DS, Lauwaet T, Dacre M, Smith A, Zhai Y, Svard S, Gillin FD. 2011. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 12:R66. 10.1186/gb-2011-12-7-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis JG, Davila M, Chakrabarti R. 2003. Potential involvement of extracellular signal-regulated kinase 1 and 2 in encystation of a primitive eukaryote, Giardia lamblia: stage-specific activation and intracellular localization. J. Biol. Chem. 278:1936–1945. 10.1074/jbc.M209274200 [DOI] [PubMed] [Google Scholar]
- 49.Abe MK, Kahle KT, Saelzler MP, Orth K, Dixon JE, Rosner MR. 2001. Erk7 is an autoactivated member of the MAPK family. J. Biol. Chem. 276:21272–21279. 10.1074/jbc.M100026200 [DOI] [PubMed] [Google Scholar]
- 50.Abe MK, Kuo WL, Hershenson MB, Rosner MR. 1999. Extracellular signal-regulated kinase 7 (Erk7), a novel Erk with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol. Cell. Biol. 19:1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C. 2011. Erk7 is a negative regulator of protein secretion in response to amino acid starvation by modulating Sec16 membrane association. EMBO J. 30:3684–3700. 10.1038/emboj.2011.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turturici G, Geraci F, Candela ME, Giudice G, Gonzalez F, Sconzo G. 2008. Hsp70 localizes differently from chaperone Hsc70 in mouse mesoangioblasts under physiological growth conditions. J. Mol. Histol. 39:571–578. 10.1007/s10735-008-9197-7 [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z, Liu H, Wang X, Li Z. 2011. Separation and identification of HSP-associated protein complexes from pancreatic cancer cell lines using 2D CN/SDS-PAGE coupled with mass spectrometry. J. Biomed. Biotechnol. 2011:193052. 10.1155/2011/193052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rando OJ, Zhao KJ, Janmey P, Crabtree GR. 2002. Phosphatidylinositol-dependent actin filament binding by the Swi/Snf-like Baf chromatin remodeling complex. Proc. Natl. Acad. Sci. U. S. A. 99:2824–2829. 10.1073/pnas.032662899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J, Wood MA, Cole MD. 2002. Baf53 forms distinct nuclear complexes and functions as a critical c-myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 22:1307–1316. 10.1128/MCB.22.5.1307-1316.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi J, Heo K, An WJ. 2009. Cooperative action of Tip48 and Tip49 in H2a.Z exchange catalyzed by acetylation of nucleosomal H2a. Nucleic Acids Res. 37:5993–6007. 10.1093/nar/gkp660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Safran M, Farwell AP, Leonard JL. 1992. Thyroid hormone-dependent redistribution of the 55-kilodalton monomer of protein disulfide isomerase in cultured glial cells. Endocrinology 131:2413–2418. 10.1210/endo.131.5.1425439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.