Abstract
The ability to acquire nutrients during infections is an important attribute in microbial pathogenesis. Amino acids are a valuable source of nitrogen if they can be degraded by the infecting organism. In this work, we analyzed histidine utilization in the fungal pathogen of humans Candida glabrata. Hemiascomycete fungi, like C. glabrata or Saccharomyces cerevisiae, possess no gene coding for a histidine ammonia-lyase, which catalyzes the first step of a major histidine degradation pathway in most other organisms. We show that C. glabrata instead initializes histidine degradation via the aromatic amino acid aminotransferase Aro8. Although ARO8 is also present in S. cerevisiae and is induced by extracellular histidine, the yeast cannot use histidine as its sole nitrogen source, possibly due to growth inhibition by a downstream degradation product. Furthermore, C. glabrata relies only on Aro8 for phenylalanine and tryptophan utilization, since ARO8, but not its homologue ARO9, was transcriptionally activated in the presence of these amino acids. Accordingly, an ARO9 deletion had no effect on growth with aromatic amino acids. In contrast, in S. cerevisiae, ARO9 is strongly induced by tryptophan and is known to support growth on aromatic amino acids. Differences in the genomic structure of the ARO9 gene between C. glabrata and S. cerevisiae indicate a possible disruption in the regulatory upstream region. Thus, we show that, in contrast to S. cerevisiae, C. glabrata has adapted to use histidine as a sole source of nitrogen and that the aromatic amino acid aminotransferase Aro8, but not Aro9, is the enzyme required for this process.
INTRODUCTION
As an opportunistic fungal pathogen, Candida glabrata is one of the leading causes of candidiasis (1, 2) and is responsible for several thousand systemic infections and deaths worldwide each year (3), yet C. glabrata is much more closely related to the nonpathogenic yeast Saccharomyces cerevisiae than to other pathogenic fungi, like Candida albicans. Identifying differences in the genetic, phenotypic, and metabolic setups of S. cerevisiae and C. glabrata is therefore highly relevant for understanding the basis of C. glabrata virulence (4, 5).
C. glabrata can cause several types of infections, ranging from superficial to systemic. During the course of these infections, C. glabrata cells face many different and rapidly changing host environments and have to adapt to each of these microniches. The ability to use as many nutrient sources as possible is vital for this kind of flexibility. Thus, a detailed knowledge of the fungal metabolic pathways involved can help in understanding, and consequently in fighting, fungal infections (6). Examples of the relevance of nutrient uptake in the host are trace elements, like iron or zinc (7–10), but also carbon and nitrogen compounds (11, 12). For the nitrogen supply of infecting fungi, amino acids are especially important (13). They are present in free form in low millimolar concentration at different host sites, like blood or urine (14, 15), or can be liberated from proteins and peptides by the actions of fungal hydrolases (16). For their uptake, fungi like S. cerevisiae and C. glabrata possess large families of different small and broad-spectrum permeases (17). In C. albicans and some of its pathogenic close relatives, these permease families are significantly expanded compared to other related, nonpathogenic species (18), indicating a role for flexibility of nitrogen utilization in pathogenicity. This flexibility includes the ability to use proteins, peptides, amino acids, and other low-molecular-weight nitrogen sources.
For humans, histidine is an essential amino acid. In the body, it can be converted to histamine by a histidine decarboxylase or to urocanic acid and ammonia by a histidine ammonia-lyase (HAL), also known as histidinase (19). In this work, we investigate the initiation of histidine degradation in C. glabrata and show that it uses an aromatic aminotransferase instead of a histidinase for histidine utilization.
MATERIALS AND METHODS
Strains and growth conditions.
The following strains were used in the experiments: C. glabrata ATCC 2001, S. cerevisiae ATCC 9763, C. albicans SC5314, and Saccharomyces kluyveri CBS3082. The fungi were maintained on solid YPD medium (2% glucose, 1% yeast extract, 1% peptone, 2% agar) and grown at 30°C. The aro8Δ deletion mutant has been described previously (20).
Growth curve experiments were performed in 1× YNB medium (BD Biosciences, CA, USA) with 2% glucose (Roth, Germany), 100 mM phosphate buffer (pH 5.8), and 10 mM the respective amino acid (Roth) or 0.5% NH4SO4 (Roth). For competitive-inhibition experiments, 10 mM (aminooxy)acetate (AOA) (Sigma) was added to these media. In selected experiments, 5 mM histidine was combined with 5 mM other amino acids as indicated. Fungi were inoculated from a YPD overnight culture at an optical density at 600 nm (OD600) of 0.2. Growth was measured every 15 min in a 1-ml volume in a 24-well plate (TPP, Switzerland) using a Tecan Infinite M200 microplate reader at 30°C with intermittent shaking.
In vitro transaminase assays.
The C. glabrata Aro8 (CgAro8) protein was expressed heterologously in Escherichia coli and purified via a His tag, as described previously (20). The transaminase activity of the protein was measured spectrophotometrically in an assay solution consisting of 100 mM Tris buffer, pH 7.5, 1 μM pyridoxal phosphate (Sigma), 5 μg purified protein, and different amounts of the amino acid to be tested (0.75 to 36 mM) (all from Roth, Germany). To start the reaction, the amino acceptor α-ketoglutarate (10 mM) was added. The formation of indolepyruvate (from tryptophan; molar extinction coefficient [ε] = 9,300 M−1 cm−1 at 338 nm), phenylpyruvate (from phenylalanine; ε = 5,900 M−1 cm−1 at 300 nm), and imidazol-5-yl-pyruvate (from histidine; ε = 11,000 M−1 cm−1 at 293 nm) was followed in an Amersham Ultrospec 3100pro spectrophotometer (21). As a negative control for intrinsic transaminase activity of the expression system, LacZ protein expressed and purified under the same conditions was used. Km and Vmax values were calculated from the data using GraphPad Prism software version 5.03 (GraphPad Software, San Diego, CA, USA).
Histidine uptake assay.
Yeast cells were cultivated in 1× YNB medium supplemented with 10 mM histidine as the sole nitrogen source at 30°C. Samples of cultivation broths were harvested after 1, 2, 4, and 24 h of cultivation and kept at −80°C until further analysis. For histidine quantification, samples were diluted 20-fold in high-performance liquid chromatography (HPLC)-grade water before analysis on a Biochrom 30+ amino acid analyzer (Biochrom) according to the manufacturer's protocol for standard amino acid determination in physiological fluids. Briefly, the diluted samples were deproteinized with sulfosalicylic acid, and amino acids were HPLC separated, followed by postcolumn derivatization using ninhydrin and photometrical detection at 570 nm.
Transcription analyses.
Yeast cells were grown in media containing 10 mM the appropriate amino acid, as described previously. Samples were taken at 0 min (YPD preculture) and after 1, 4, and 8 h of growth. RNA was isolated using an RNeasy kit (Qiagen) after glass bead disruption in a Precellys homogenizer (PeqLab). RNA quality was ensured with the Bioanalyzer instrument (Agilent). Expression of ARO8, ARO9, and the reference ACT1 genes was detected in C. glabrata and S. cerevisiae using an EvaGreen quantitative-PCR (qPCR) mix (Bio&Sell) with appropriate primers in a CFX96 real-time detection system (Bio-Rad). The expression levels of three biological replicates were normalized to ACT1 mRNA levels.
Alignment of the genomic locus.
The nucleotide alignment of selected regions of the C. glabrata and S. cerevisiae genomes was performed with the GATAligner tool, version 1.0, and displayed using GATAPlotter version 0.7 (22). The parameters used were as follows: window size, 7; cutoff score, 5 bits; and standard BLASTN parameters. Genome sequence data were downloaded from the Candida and yeast genome databases (www.candidagenome.org and www.yeastgenome.org, respectively).
RESULTS
C. glabrata and C. albicans can use histidine as the sole nitrogen source.
We measured the ability of the hemiascomycetes C. glabrata, C. albicans, S. cerevisiae, and S. kluyveri to use different amino acids as their sole nitrogen source (Fig. 1). When grown in minimal medium with glucose as a carbon source and 10 mM single amino acids as a nitrogen source, C. glabrata and S. cerevisiae were able to use most of the standard proteinogenic amino acids. Only cysteine and lysine cannot support the growth of either yeast. Glycine and histidine enable good growth of C. glabrata, but not of S. cerevisiae. The yeast S. kluyveri is similar to S. cerevisiae in its growth profile, with the exception of lysine, which can be used by S. kluyveri but not by baker's yeast. In contrast, C. albicans was able to use all 20 classical proteinogenic amino acids as its sole source of nitrogen. As expected, all fungi grew best in YPD complex medium or in minimal medium with ammonium sulfate as the nitrogen source.
FIG 1.
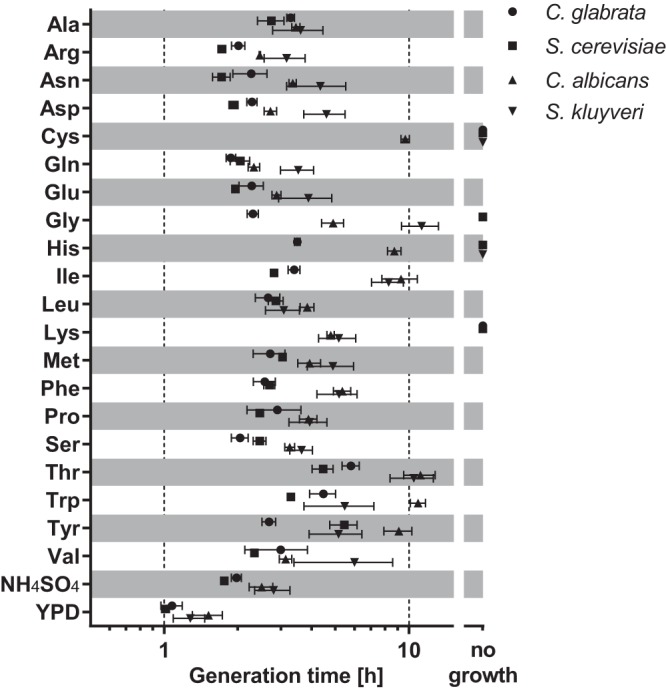
Growth of selected hemiascomycetes at 30°C with amino acids as sole nitrogen sources. Generation times of >12 h were generally considered absence of growth. Both C. glabrata and C. albicans were able to grow with histidine as the sole nitrogen source. Only C. albicans was able to use any amino acid for growth, while C. glabrata and S. cerevisiae generally grew faster than C. albicans or S. kluyveri under the conditions tested. Ammonium sulfate (0.5% NH4SO4) and YPD complex medium served as controls. All data are means ± standard deviations (SD).
Furthermore, the overall growth pattern of C. glabrata resembles that of S. cerevisiae more than that of C. albicans, with the exceptions noted above. Compared to the other species, C. albicans grows much more slowly with aromatic amino acids (phenylalanine, tryptophan, tyrosine, and, for both Candida species, histidine). Importantly, histidine supports the growth of C. glabrata, with a generation time of 3.5 h, but both S. cerevisiae and the more basal S. kluyveri cannot grow, while the more distantly related C. albicans multiplies with a generation time of 8.7 h.
The HAL gene is absent in the hemiascomycetes.
Histidine ammonia-lyases (HALs), or histidinases (EC 4.3.1.3), catalyze the conversion of histidine into ammonium and urocanic acid, enabling organisms to use histidine as a nitrogen source. Histidine ammonia-lyases are ubiquitous enzymes and can be found in archaea, bacteria, and eukaryota: a pfam search (23) for the conserved phenylalanine and histidine ammonia-lyase family (PF00221) detected HALs in 573 species from all three kingdoms (data not shown). Among the fungi, HALs can be found in many of the ascomycota and the basidiomycota. However, no conserved HAL sequences can be found in the hemiascomycete branch of the ascomycota, which contains (among other fungi) the fungal species investigated here, S. cerevisiae, C. albicans, C. glabrata, and S. kluyveri. In contrast, HALs can be found in the Pezizomycotina, as a related euascomycete phylum, for example, in Aspergillus and Penicillium spp. Evidently, the histidine ammonia-lyase gene was lost in the evolution of the Saccharomycotina. A TBLASTN search of fungal DNA sequences with the Aspergillus clavatus phenylalanine and histidine ammonia-lyase protein sequences as the query further supported the pfam results (data not shown). While this absence of a histidine ammonia-lyase would explain the lack of growth of S. cerevisiae and S. kluyveri with histidine as the sole source of nitrogen, both C. albicans and C. glabrata must have other means available to obtain the nitrogen from histidine. Thus, we propose that these fungi have evolved alternative metabolic pathways in order to use histidine as a potential nitrogen source.
Deletion of ARO8 in C. glabrata.
Recently, we created a mutant of C. glabrata that lacks the homologue of the yeast S. cerevisiae aromatic aminotransferase I gene, ScARO8 (20). In S. cerevisiae, aromatic aminotransferases are known to catalyze the transfer of the amino group from phenylalanine, tyrosine, and tryptophan to amino acceptors like 2-oxoglutarate (24). As anticipated, growth with the three aromatic amino acids was strongly reduced in the C. glabrata aro8Δ mutant (20), highlighting the broad substrate spectrum of the aminotransferase CgAro8. Unexpectedly, we found that the mutant was also nearly unable to use histidine as a nitrogen source, a phenotype resembling that of wild-type S. cerevisiae (Fig. 2). Therefore, is seems that the CgAro8 aminotransferase contributes significantly to the growth of C. glabrata on histidine as the sole nitrogen source.
FIG 2.
Growth curves of different mutants of C. glabrata and the S. cerevisiae wild type, used for comparison. Deletion of ARO8 nearly abolished growth of C. glabrata with histidine, tryptophan, or tyrosine and strongly reduced growth with phenylalanine as the sole nitrogen source. Deleting the aromatic aminotransferase gene ARO9 or the decarboxylase gene ARO10 had much less effect on growth. All mutant strains grew nearly identically to the C. glabrata wild type in YPD complex medium.
In contrast, deleting the C. glabrata homologue of the yeast aromatic aminotransferase II gene, ScARO9, or of the downstream decarboxylase gene ScARO10, had only a negligible effect on growth with histidine as the sole nitrogen source (Fig. 2). Growth with the aromatic amino acid tryptophan, phenylalanine, or tyrosine was moderately reduced in the C. glabrata aro9Δ and aro10Δ mutants. In YPD complex medium, none of the deletion mutants displayed a reduced growth rate, while a minor growth defect was observed with ammonium sulfate as the nitrogen source.
Addition of the competitive aromatic aminotransferase inhibitor AOA to C. glabrata cultures produced an effect similar to that of deletion of CgARO8. AOA is a broad-spectrum inhibitor of pyridoxal phosphate-dependent reactions like transaminations (25) and has been shown to inhibit, among others, fungal transaminases (26). In our hands, it strongly inhibited the growth of C. glabrata on histidine and all other aromatic amino acids at a concentration of 10 mM (Fig. 3). Growth with ammonium sulfate in the presence of the inhibitor was also reduced (as in the aro8Δ deletion mutant). There was no general growth reduction by addition of up to 10 mM AOA to YPD complex medium, showing that the reduced growth is likely not due to a general toxicity of AOA at these concentrations. Stronger growth inhibition was also observed for C. albicans. After a long lag phase, growth mostly resumed for C. glabrata. For C. albicans, which grew significantly more slowly even in the absence of the inhibitor (Fig. 1), no significant growth with any single aromatic amino acid was detected even after 36 h in the presence of AOA (Fig. 3). Overall, these results, in combination with the data from the deletion mutants, demonstrate that, like the utilization of other aromatic amino acids, histidine utilization is dependent on the aminotransferase activity of CgAro8 in C. glabrata.
FIG 3.
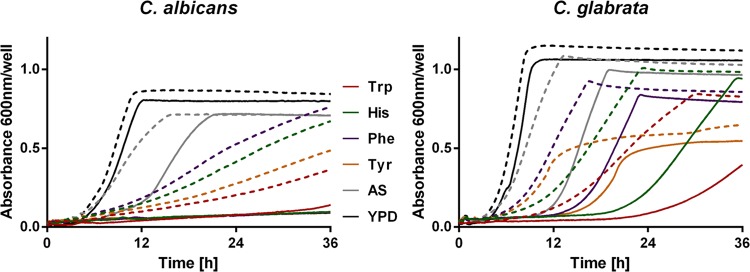
Growth of C. albicans and C. glabrata with 10 mM the aromatic aminotransferase inhibitor AOA. The presence of AOA reduces growth with all tested aromatic amino acids and ammonium sulfate (AS). Growth in YPD complex medium is nearly unaffected. Dashed lines, without inhibitor; solid lines, with the inhibitor AOA.
Finally, uptake of histidine was measured by HPLC analyses of the supernatants from growing yeast cultures (Fig. 4). After 24 h, complete removal (a concentration below the detection limit of 34 μM) of the initial 10 mM histidine was observed from the supernatant of C. glabrata wild-type cultures. In contrast, nearly all of the histidine (9.8 mM) remained in the supernatant of the aro8Δ mutant. The remaining histidine level of the S. cerevisiae supernatant (7.4 mM) showed that the yeast can take up histidine, albeit at significantly lower levels than C. glabrata. However, since growth of S. cerevisiae is not supported by histidine as a sole nitrogen source, the exact fate of histidine in S. cerevisiae remains to be elucidated.
FIG 4.

Residual histidine in the medium after incubation with the C. glabrata wild type, the S. cerevisiae wild type, or the C. glabrata aro8Δ mutant. The C. glabrata wild type used up the available histidine to levels below the detection threshold (nd), while most histidine remained in the medium in cultures with S. cerevisiae or the aro8Δ mutant.
Aro8 and Aro9 regulation in C. glabrata and S. cerevisiae.
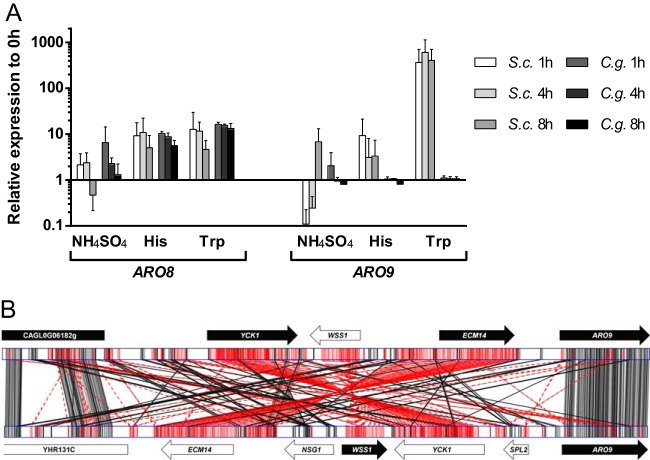
Previous microarray analyses indicated differences in regulation of ARO8 and ARO9, the genes for aromatic aminotransferases I and II, respectively, between C. glabrata and S. cerevisiae (20). We investigated this regulation on the transcriptional level in more detail using qRT-PCR on RNA isolated from liquid cultures of S. cerevisiae and C. glabrata cells exposed to histidine, tryptophan, or ammonium sulfate as the sole nitrogen source (Fig. 5A).
FIG 5.
(A) Relative expression of ARO8 and ARO9 after transfer from complex medium to minimal medium containing ammonium sulfate, histidine, or tryptophan as the sole source of nitrogen. In both S. cerevisiae (S.c.) and C. glabrata (C.g.), ARO8 expression was upregulated compared to its expression in the preculture at all time points when the amino acids were offered. There was no regulation observable for ARO9 in C. glabrata, while S. cerevisiae very strongly upregulated ARO9 when tryptophan was present. The error bars indicate standard deviations. (B) Genomic-locus alignment of the ARO9 gene and 10-kbp upstream regions in C. glabrata (top) and S. cerevisiae (bottom). An inversion event was detected encompassing the upstream genes YCK1, WSS1, and ECM14. The genes CAGL0G06182 and YHR131C are homologues with identical orientations with respect to the ARO9 genes. The solid arrows indicate ORFs with the same orientation as ARO9, and the open arrows indicate opposite orientations. Alignment was performed with GATA (22). Black lines indicate alignments in the same orientation (+/+), and red lines indicate those in inverse orientation (+/−). Darker shadings indicate better alignment scores.
ScARO8 was nearly 10-fold upregulated already at 1 h when histidine or tryptophan was offered as a nitrogen source. An early, lower upregulation was also observed with ammonium sulfate in the medium, but the transcript levels of ARO8 decreased over time again in both C. glabrata and S. cerevisiae. This likely reflects the catabolic and anabolic activities of the enzymes, respectively, in the different media.
As described previously (27), the main aromatic aminotransferase gene, ScARO9, was strongly upregulated in S. cerevisiae when tryptophan was provided as the source of nitrogen but downregulated with ammonium sulfate initially at 1 h compared to the preculture (as opposed to ScARO8). With histidine, an increase of ScARO9 mRNA levels was observed, but to a much lesser extent.
For C. glabrata, the most striking difference from S. cerevisiae was the complete lack of induction of the CgARO9 gene under any of our experimental conditions. Even with tryptophan, the strongest inducer of ScARO9 transcription in S. cerevisiae, there was no detectable difference from the preculture RNA levels in C. glabrata. In contrast, the transcription of CgARO8 was upregulated in histidine- and in tryptophan-containing media, in agreement with our previous data (20). With these data, we confirmed the differences in ARO gene regulation between S. cerevisiae and C. glabrata and found that the CgARO9 gene seems to be largely nonresponsive to external aromatic amino acids or histidine.
Genomic differences between the ARO8 and ARO9 loci in C. glabrata and S. cerevisiae.
To find an explanation for the differences in the regulation of these aromatic aminotransferase genes, we examined the structures of their respective genomic loci. Using YGOB, the yeast gene order browser (28), we found that CgARO8 and the next gene located upstream, CAGL0G01232g (an ScKEX1 homologue), form a pair with synteny to its counterpart in S. cerevisiae and the predicted common ancestor. Downstream of CgARO8 and upstream of the KEX1 homologue, the synteny is broken.
For CgARO9, there is an inversion upstream of the open reading frame (ORF) compared to the genome of S. cerevisiae and all other available genomes of the YGOB. Sequence analysis showed that this inversion encompasses several kilobases and includes the neighboring predicted genes YCK1, WSS1, and ECM14 (Fig. 5B). In addition, the inversion may also include one or more transcription factor binding sites of the immediate CgARO9 upstream region. Such a removal of the binding sites from CgARO9 gives a possible explanation for its lack of regulation in the presence of aromatic amino acids.
In vitro activity of the Aro8 proteins.
For further confirmation of the role of Aro8 in histidine degradation, we heterologously expressed a histidine-tagged CgAro8 protein in E. coli and affinity purified the enzyme. As we have shown before (20), the CgAro8 protein converted tryptophan into indole pyruvate. The Km of this reaction was 14.9 ± 2.5 mM, and the Vmax was 593.3 ± 48.5 nmol · min−1 · mg−1 under our in vitro assay conditions (with α-ketoglutarate as the amino acceptor). In addition, the protein was also able to convert phenylalanine into phenylpyruvate under the same conditions, with a Km of 1.39 ± 0.16 mM and a Vmax of 196.7 ± 3.9 nmol · min−1 · mg−1. This reaction is similar to that of the S. cerevisiae Aro8 protein (29). However, most interestingly, a test with l-histidine as the amino donor revealed that CgAro8 is also able to transfer the amino group of this amino acid to α-ketoglutarate, producing imidazol-5-yl-pyruvate (Fig. 6A), with a Km of 8.80 ± 1.41 mM and a Vmax of 274.9 ± 19.8 nmol · min−1 · mg−1. Interestingly, a heterologously expressed Aro8 protein from S. cerevisiae had a similar ability to catalyze the aminotransferase reaction with l-histidine as a donor (data not shown), implying a principal ability of S. cerevisiae for histidine transamination.
FIG 6.

(A) Proposed aromatic amino acid aminotransferase reaction by Aro8 with α-ketoglutarate and histidine as substrates, producing glutamate and imidazol-5-yl-pyruvate. The pathway from histidine to urocanic acid via the HAL does not take place in C. glabrata or S. cerevisiae due to the absence of the HAL gene. The transferred amine group is marked in boldface. (B) Effects of histidine supplementation on S. cerevisiae growth with different nitrogen sources. Addition of 5 mM histidine abolishes growth with the aromatic amino acids phenylalanine and tryptophan and reduces growth with alanine or ammonium sulfate as the nitrogen source. Addition of the aromatic amino acid aminotransferase inhibitor AOA slightly increases the growth rate when alanine is supplemented with histidine.
Interaction of histidine with other nitrogen sources in S. cerevisiae.
The Aro8 proteins of both species were able to catalyze the histidine aminotransferase reaction in vitro, and in both species, the ARO8 gene was upregulated in response to histidine. We therefore investigated the effect of adding histidine to the medium on the growth of S. cerevisiae. To this end, we mixed histidine 1:1 (5 mM each) with different nitrogen sources: alanine as an amino acid with an independent degradation pathway, phenylalanine as an aromatic amino acid that induces ARO8 expression, and ammonium sulfate as a non-amino-acid nitrogen source (Fig. 6B).
As before, S. cerevisiae was unable to grow with histidine as the sole nitrogen source. Strikingly, the comparatively good growth with 5 mM phenylalanine or tryptophan (generation times, 2.8 h and 4.9 h, respectively) was completely abolished when 5 mM histidine was added. Growth with 5 mM alanine (3.2 h) in the presence of additional histidine still took place but was slowed down significantly (9.9 h). Interestingly, addition of the inhibitor AOA partly rescued the growth in medium with alanine and histidine (to 7.2 h), while it reduced the growth rate in medium with alanine as a sole nitrogen source (to 4.2 h). Finally, growth with ammonium sulfate (1.7 h) was only slightly reduced (2.5 h) by addition of histidine. This indicates that histidine is indeed taken up by S. cerevisiae and transaminated under conditions of ScARO8 or ScARO9 induction. However, while histidine degradation can be initiated, downstream processing of the transaminated product seems to be disturbed or interferes with other metabolic pathways.
DISCUSSION
The comparatively close phylogenetic relationship between the normally nonpathogenic yeast S. cerevisiae and the pathogenic yeast C. glabrata allows the detection of specific factors and traits that are associated with fungal pathogenesis (4). These include, for example, the ability of C. glabrata to grow efficiently at body temperature, but also its ability to use nutrients from particular host niches, which likely differ significantly from S. cerevisiae's normal habitats.
Both S. cerevisiae and C. glabrata were able to grow with most of the proteinogenic amino acids as sole nitrogen sources. For baker's yeast, the only exceptions were cysteine, glycine, histidine, and lysine, in accordance with previous observations (30). The pathogenic yeast C. glabrata, on the other hand, was additionally able to grow with either glycine or histidine as the sole nitrogen source. Evidently, this pathogen is even more versatile than S. cerevisiae in its ability to use different nitrogen sources, though not as flexible as C. albicans, which can utilize all 20 proteinogenic amino acids.
We hypothesized that the inability of S. cerevisiae to use histidine as a nitrogen source was due to the loss of the histidine ammonia-lyase gene. Since this loss is common to all hemiascomycetes, it should also affect C. albicans and C. glabrata, yet these two species can utilize histidine as their sole nitrogen source. Comparison with S. kluyveri, which also cannot use histidine for a nitrogen supply, indicates that at least C. glabrata, and probably C. albicans, has independently gained this ability in a lineage that is unable to use histidine. As the pathogenic yeasts in this study, C. albicans and C. glabrata, were most flexible with regard to the utilization of amino acids, it is tempting to speculate that a correlation may exist between the ability to use a broad range of nitrogen sources and the pathogenic potential of these fungi.
Having a broad set of catabolic pathways for the specific nutrient sources in the host is clearly advantageous for pathogenic fungi (6). On the other hand, unused metabolic pathways are quickly lost. One striking example of this is the absence of the kynurenine pathway in C. glabrata (31). In baker's yeast, the aromatic amino acid tryptophan can be degraded by two distinct pathways, via the kynurenine pathway to NAD or via the Ehrlich pathway to indole-3-ethanol (32). C. glabrata has lost six of the seven BNA genes of the kynurenine pathway (31, 33) and is therefore auxotrophic for niacin, as opposed to S. cerevisiae and C. albicans. Due to these losses, the only remaining pathway for tryptophan degradation in C. glabrata is via the Ehrlich pathway, and therefore via the CgAro8 aromatic amino acid aminotransferase (20).
Here, we show that degradation of histidine in C. glabrata also occurs via the Ehrlich pathway. The aromatic amino acid aminotransferase Aro8 is both sufficient for in vitro aminotransferase activity and required for growth with histidine as the sole nitrogen source. The Km values of C. glabrata Aro8 for the transamination reactions of phenylalanine and tryptophan seem high for its biological function but are similar to those published for its S. cerevisiae counterpart (1.4 versus 0.3 mM and 14.9 versus 6 mM, respectively [24]). Thus, a similar biological role for CgAro8 in the degradation of external aromatic amino acids seems likely, and the histidine transamination observed in vitro should be sufficient to enable in vivo growth with histidine.
Like the deletion of CgARO8, inhibition of the aromatic aminotransferase with the substance AOA led to a severe growth defect of C. glabrata (and C. albicans) with tryptophan and histidine as the sole nitrogen sources. The growth defect with ammonium sulfate is likely due to the general inhibition of aminotransferase activities that are also required in amino acid anabolic processes. In S. cerevisiae, deletion of both ScARO8 and ScARO9 leads to phenylalanine and tryptophan auxotrophy of the double mutant (34). Thus, with only ammonium sulfate as a nitrogen source in the medium, AOA likely blocked the biosynthesis of aromatic and other amino acids by inhibition of the respective aminotransferases, which could lead to a limitation of building blocks for anabolic purposes. Alternatively, inhibition of aminotransferases could lead to accumulation of α-ketoacids that provoke general toxic effects and may thus cause reduced growth rates. However, the normal growth rate in YPD medium in the presence of AOA makes the latter scenario rather unlikely, although the possibility that differences in AOA uptake depending on the applied growth medium result in reduced toxicity cannot be excluded. Nevertheless, the strong growth-inhibitory effect of any of the aromatic amino acids used as a sole nitrogen source further indicates that, despite its low specificity, aromatic aminotransferases of C. glabrata were indeed inhibited by AOA treatment.
An aminotransferase reaction for histidine degradation has already been proposed to occur in other organisms. In the rat's liver, for example, this reaction has been reported to take place, but it is most likely not used for catabolic purposes. Instead, it serves a function during mitochondrial transport (35). No gene has been found so far to code for the enzyme. In E. coli and other bacteria, like Pseudomonas testosteroni (36), a similar histidine transaminase reaction has been described, but the well-investigated E. coli K-12 strain lacks this reaction and is unable to catabolize histidine. Other fungi, like Aspergillus spp., possess the standard histidine ammonia-lyase, and the protein has been shown to be active, for example, in A. nidulans when histidine is the sole nitrogen source (37). Therefore, to our knowledge, catabolism via an aromatic aminotransferase instead of a histidinase presents a novel route for fungal growth with histidine as a nitrogen source.
Interestingly, S. cerevisiae Aro8 also catalyzes the aminotransferase reaction from histidine, although the fungus is unable to grow with histidine as the sole nitrogen source. Our data indicate, however, that histidine degradation may lead to an inhibiting metabolic intermediate in yeast, while histidine did not support growth, it also abolished growth under conditions where the Ehrlich pathway was active (in the presence of phenylalanine or tryptophan as a nitrogen source). Under these conditions, histidine will likely be deaminated by the aromatic amino acid aminotransferases. Growth with ammonium sulfate was only slightly reduced in the presence of histidine, since ammonium sulfate is a preferred nitrogen source and does not lead to the activation of the Ehrlich pathway. Additionally, high expression of amino acid permeases like Gap1 for histidine uptake is unlikely (38). Finally, with alanine, the growth rate was reduced to an intermediate level. Alanine belongs to the amino acids with intermediate growth support (Fig. 1), and it appears likely that general amino acid permeases are expressed and allow uptake of histidine. Since inhibition of aromatic amino acid aminotransferases increased the growth rate under these conditions, Aro8 and Aro9 appear to be active and to produce an intermediate that causes the growth inhibition. In summary, these effects would explain why Aro8 transaminase activity is not sufficient for use of histidine as a nitrogen source in S. cerevisiae: at least one of the histidine degradation products itself likely inhibits growth.
Besides the observation that aromatic amino acid aminotransferases in S. cerevisiae are able to transaminate histidine but cannot support its utilization, it is interesting that the gene product of ScARO9 is the main aromatic aminotransferase in S. cerevisiae, and the gene is accordingly upregulated when tryptophan is offered as the sole source of nitrogen (27). In contrast, C. glabrata shows no upregulation of the corresponding gene; however, CgARO8 expression is increased in the presence of external aromatic amino acids (reference 20 and this work). A possible explanation for this phenomenon is an inversion event in the direct upstream region of the CgARO9 gene, which may have caused the loss of some or all regulatory genetic elements. Most of the aromatic amino acids, including histidine, will therefore likely be processed by CgAro8, since CgARO8 is the only aromatic amino acid transferase gene of C. glabrata upregulated in the presence of aromatic amino acids. Thus, the inversion event in the phylogenetic lineage of C. glabrata likely contributed to the observed differences between C. glabrata and S. cerevisiae.
The presence of an unusual nitrogen metabolic pathway has precedents in the hemiascomycetes. These yeasts catabolize urea via a urea amidolyase instead of the urease of other higher fungi (39). Deleting the urea amidolyase gene, DUR1,2, in C. albicans leads to a reduction in virulence and, importantly, of the fungal burden in organs with high urea content (40). This can, at least in part, be explained by the dependency of fungi on external nitrogen sources during infections (40). Hence, like the Dur1,2 pathway in C. albicans, the unusual histidine metabolism of C. glabrata may be considered a possible target for drugs that could reduce growth of the fungus in the host.
ACKNOWLEDGMENTS
We thank Markus Gressler for his valuable input.
This work was supported by the German Federal Ministry of Education and Health (BMBF), FKZ 01EO1002 from the Integrated Research and Treatment Center, Center for Sepsis Control and Care (CSCC), and the ERA-NET PathoGenoMics Program (Candicol; BMBF 0315 901 B). S.R. was supported by the graduate school, Jena School for Microbial Communication (JSMC) (www.jsmc.uni-jena.de).
Footnotes
Published ahead of print 11 April 2014
REFERENCES
- 1.Arendrup MC. 2010. Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 16:445–452. 10.1097/MCC.0b013e32833e84d2 [DOI] [PubMed] [Google Scholar]
- 2.Richardson M, Lass-Florl C. 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14(Suppl 4):5–24. 10.1111/j.1469-0691.2008.01978.x [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163. 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roetzer A, Gabaldon T, Schuller C. 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314:1–9. 10.1111/j.1574-6968.2010.02102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunke S, Hube B. 2013. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 15:701–708. 10.1111/cmi.12091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock M. 2009. Fungal metabolism in host niches. Curr. Opin. Microbiol. 12:371–376. 10.1016/j.mib.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 7.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217. 10.1371/journal.ppat.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog. 8:e1002777. 10.1371/journal.ppat.1002777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amich J, Vicentefranqueira R, Leal F, Calera JA. 2010. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot. Cell 9:424–437. 10.1128/EC.00348-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron—a metal at the host-pathogen interface. Cell. Microbiol. 12:1691–1702. 10.1111/j.1462-5822.2010.01529.x [DOI] [PubMed] [Google Scholar]
- 11.Rhodes JC. 2006. Aspergillus fumigatus: growth and virulence. Med. Mycol. 44(Suppl 1):S77–S81. 10.1080/13693780600779419 [DOI] [PubMed] [Google Scholar]
- 12.Ramachandra S, Linde J, Brock M, Guthke R, Hube B, Brunke S. 2014. Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS One 9:e92734. 10.1371/journal.pone.0092734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez P, Ljungdahl PO. 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25:9435–9446. 10.1128/MCB.25.21.9435-9446.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan IK, Gajra B. 2006. Plasma and urine amino acid profiles in a healthy adult population of Singapore. Ann. Acad. Med. Singapore 35:468–475 [PubMed] [Google Scholar]
- 15.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. 2011. The human serum metabolome. PLoS One 6:e16957. 10.1371/journal.pone.0016957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400–428. 10.1128/MMBR.67.3.400-428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317–328. 10.1007/s002940050506 [DOI] [PubMed] [Google Scholar]
- 18.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchi M, Sano H, Mizuno H, Wada Y. 1995. Molecular cloning and structural characterization of the human histidase gene (HAL). Genomics 29:98–104. 10.1006/geno.1995.1219 [DOI] [PubMed] [Google Scholar]
- 20.Brunke S, Seider K, Almeida RS, Heyken A, Fleck CB, Brock M, Barz D, Rupp S, Hube B. 2010. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol. Microbiol. 76:25–47. 10.1111/j.1365-2958.2010.07052.x [DOI] [PubMed] [Google Scholar]
- 21.Paris CG, Magasanik B. 1981. Tryptophan metabolism in Klebsiella aerogenes: regulation of the utilization of aromatic amino acids as sources of nitrogen. J. Bacteriol. 145:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nix DA, Eisen MB. 2005. GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinformatics 6:9. 10.1186/1471-2105-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kradolfer P, Niederberger P, Hutter R. 1982. Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases. Arch. Microbiol. 133:242–248. 10.1007/BF00415010 [DOI] [PubMed] [Google Scholar]
- 25.Wallach DP. 1961. Studies on the GABA pathway. II. The lack of effect of pyridoxal phosphate on GABA-KGA transaminase inhibition induced by amino-oxyacetic acid. Biochem. Pharmacol. 8:328–331 [DOI] [PubMed] [Google Scholar]
- 26.Preuss J, Hort W, Lang S, Netsch A, Rahlfs S, Lochnit G, Jortzik E, Becker K, Mayser PA. 2013. Characterization of tryptophan aminotransferase 1 of Malassezia furfur, the key enzyme in the production of indolic compounds by M. furfur. Exp. Dermatol. 22:736–741. 10.1111/exd.12260 [DOI] [PubMed] [Google Scholar]
- 27.Iraqui I, Vissers S, Andre B, Urrestarazu A. 1999. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3360–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15:1456–1461. 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. 1998. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238–248. 10.1007/s004380050644 [DOI] [PubMed] [Google Scholar]
- 30.Large P. 1986. Degradation of organic nitrogen compounds by yeasts. Yeast 2:1–34. 10.1002/yea.320020102 [DOI] [Google Scholar]
- 31.Li YF, Bao WG. 2007. Why do some yeast species require niacin for growth? Different modes of NAD synthesis. FEMS Yeast Res. 7:657–664. 10.1111/j.1567-1364.2007.00231.x [DOI] [PubMed] [Google Scholar]
- 32.Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74:2259–2266. 10.1128/AEM.02625-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wogulis M, Chew ER, Donohoue PD, Wilson DK. 2008. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry 47:1608–1621. 10.1021/bi701172v [DOI] [PubMed] [Google Scholar]
- 34.Urrestarazu A, Vissers S, Iraqui I, Grenson M. 1998. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol. Gen. Genet. 257:230–237. 10.1007/s004380050643 [DOI] [PubMed] [Google Scholar]
- 35.Emes AV, Hassall H. 1973. The degradation of l-histidine in the rat. The formation of imidazolylpyruvate, imidazolyl-lactate and imidazolylpropionate. Biochem. J. 136:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacking AJ, Hassall H. 1975. The purification and properties of l-histidine-2-oxoglutarate aminotransferase from Pseudomonas testosteroni. Biochem. J. 147:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polkinghorne MA, Hynes MJ. 1982. l-Histidine utilization in Aspergillus nidulans. J. Bacteriol. 149:931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jauniaux JC, Grenson M. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39–44 [DOI] [PubMed] [Google Scholar]
- 39.Navarathna DH, Harris SD, Roberts DD, Nickerson KW. 2010. Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res. 10:209–213. 10.1111/j.1567-1364.2009.00602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarathna DH, Lionakis MS, Lizak MJ, Munasinghe J, Nickerson KW, Roberts DD. 2012. Urea amidolyase (DUR1,2) contributes to virulence and kidney pathogenesis of Candida albicans. PLoS One 7:e48475. 10.1371/journal.pone.0048475 [DOI] [PMC free article] [PubMed] [Google Scholar]