Abstract
Flippases are key regulators of membrane asymmetry and secretory mechanisms. Vesicular polysaccharide secretion is essential for the pathogenic mechanisms of Cryptococcus neoformans. On the basis of the observations that flippases are required for polysaccharide secretion in plants and the putative Apt1 flippase is required for cryptococcal virulence, we analyzed the role of this enzyme in polysaccharide release by C. neoformans, using a previously characterized apt1Δ mutant. Mutant and wild-type (WT) cells shared important phenotypic characteristics, including capsule morphology and dimensions, glucuronoxylomannan (GXM) composition, molecular size, and serological properties. The apt1Δ mutant, however, produced extracellular vesicles (EVs) with a lower GXM content and different size distribution in comparison with those of WT cells. Our data also suggested a defective intracellular GXM synthesis in mutant cells, in addition to changes in the architecture of the Golgi apparatus. These findings were correlated with diminished GXM production during in vitro growth, macrophage infection, and lung colonization. This phenotype was associated with decreased survival of the mutant in the lungs of infected mice, reduced induction of interleukin-6 (IL-6) cytokine levels, and inefficacy in colonization of the brain. Taken together, our results indicate that the lack of APT1 caused defects in both GXM synthesis and vesicular export to the extracellular milieu by C. neoformans via processes that are apparently related to the pathogenic mechanisms used by this fungus during animal infection.
INTRODUCTION
The mechanisms by which eukaryotic cells secrete molecules to the cell surface and/or to the extracellular space include both conventional and nonconventional pathways (1, 2). Conventional secretion requires the sequential traffic of molecules from the endoplasmic reticulum to the Golgi apparatus, from where eukaryotic molecules are transported to the cell surface (2). Proteins that engage this secretion pathway contain a signal peptide that is a marker for conventional export (3). Proteins lacking the signal peptide can use numerous alternative routes of export consisting of the unconventional secretory pathways (4). Most of the mechanisms involved in unconventional secretory routes require formation of extracellular vesicles (EVs) (1, 4).
Fungal cells export a wide variety of molecules to the extracellular space. Remarkably, most of the molecules trafficked by fungi to the extracellular milieu lack secretion signals (5–7). Extracellular fungal molecules include numerous proteins (8–11) but also pigments (12) and polysaccharides (13, 14). It is now well accepted that these molecules are at least partially exported to the outer space in EVs (15). It has been proposed that fungal EVs are derived from plasma membrane reshaping resulting in cytoplasmic subtractions (6), but the molecular regulators of formation of these compartments are unknown.
Lipid asymmetry is essential for the architecture of biological membranes (16, 17). This property is dependent on compositional differences between inner and outer leaflets in membrane bilayers. Phospholipids in the outer membrane leaflet, preferentially phosphatidylserine and phosphatidylethanolamine, are enzymatically transferred to the inner leaflet by type 4 P-type ATPase subfamily members (P4-ATPases) known as aminophospholipid translocases (APTs) or flippases (16–18). These enzymes, therefore, play key physiological roles as transmembrane lipid transporters responsible for maintaining membrane phospholipid asymmetry. Flippases are responsible for a number of other essential physiological steps in eukaryotic cells (16, 17), including membrane fusion events during vesicle biogenesis both at the plasma membrane (19) and in the trans-Golgi network (20–22). Recently, flippases have been linked to extracellular vesicle formation in Caenorhabditis elegans embryos (23, 24).
Cryptococcus neoformans is an encapsulated fungal pathogen that causes cryptococcosis, which kills approximately 500,000 people each year (25). The pathogenicity of C. neoformans is largely dependent on secretory mechanisms, which result in the transport of important virulence factors to the extracellular space, including fungal melanin, hydrolases, and immunomodulatory polysaccharides (26). Cryptococcal extracellular polysaccharides, which are considered to be the most important regulators of pathogenicity (27), are also required for capsule formation, which protects the fungus against a number of antifungal mechanisms used by host cells (reviewed in reference 28). C. neoformans polysaccharides are transported to the outer milieu in EVs (14).
In C. neoformans, the APT1 gene, which encodes a putative flippase, is required for protein secretion and fungal pathogenicity (29). However, key virulence factors of C. neoformans, including melanin production and capsule formation, are apparently not affected by APT1 deletion. In this study, we investigated polysaccharide traffic in C. neoformans mutant cells lacking Apt1. We observed that the Apt1 flippase was required for maintenance of the Golgi morphology and for polysaccharide export in C. neoformans both in vitro and in vivo, which resulted in important alterations in pathogenic steps and in the host response.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The C. neoformans strains mainly used in this study were the wild-type (WT) isolate H99, which was the background strain for the APT1 gene deletion (29), and the apt1Δ-40 mutant, which lacks expression of Apt1. Morphological analyses (nucleus and Golgi apparatus staining, observation of vacuoles, and capsular architecture tests) also included the complemented apt1+APT1-25 strain. Stock cultures were maintained on YPD solid medium (1% yeast extract, 2% peptone, 2% dextrose, and 2% agar) supplemented with Geneticin (G418) (200 μg/ml) for selection of C. neoformans APT1 deletion transformants. For most experiments, C. neoformans H99 and apt1Δ strains were grown in a minimal medium composed of glucose (15 mM), MgSO4 (10 mM), KH2PO4 (29.4 mM), glycine (13 mM), and thiamine-HCl (3 μM), pH 5.5, for 48 h at 25°C with shaking. For routine morphological analyses, C. neoformans cells (10-μl suspensions) were placed onto glass slides and mixed with India ink. The suspensions were covered with glass coverslips and analyzed with an Axioplan 2 (Zeiss, Germany) microscope. Image processing required a Color View SX digital camera and the analySIS (Soft Image System) software. Capsular dimensions were determined using the ImageJ software (version 1.45s).
Effects of APT1 deletion on phenotypic traits of C. neoformans.
Analyses of C. neoformans cells by fluorescence microscopy were performed for determination of capsular and Golgi morphologies. Staining of the Golgi apparatus was based on a previously described protocol (30). The Golgi staining reagent was C6-NBD-ceramide, which accumulates at the Golgi apparatus of either living or fixed cells (31). Yeast cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), followed by washing with the same buffer and incubation with C6-NBD-ceramide (20 mM) for 16 h at 4°C. The cells were then incubated with bovine serum albumin (BSA) (1%) at 4°C for 1 h to remove the excess C6-NBD-ceramide (31). For staining of the cell wall, the cells were then extensively washed and incubated for 15 min with Uvitex 2B (0.1 mg/ml) (Polysciences, Warrington, PA) at room temperature, followed by washing with PBS and analysis by fluorescence microscopy. Different staining patterns were determined in approximately 300 cells of each strain using the ImageJ software. For capsule staining, yeast cells (106) were fixed in 4% paraformaldehyde in PBS. Fixed yeast cells were washed twice with PBS and blocked with 1% BSA in PBS (PBS-BSA) for 1 h. For cell wall chitin staining, the cells were suspended in 100 μl calcofluor white (Invitrogen, Carlsbad, CA) (5 μg/ml) and incubated for 30 min at 37°C. For staining of glucuronoxylomannan (GXM), the cells were washed with PBS and incubated for 1 h in the presence of the 18B7 (1 μg/ml) monoclonal antibody (MAb), a mouse anti-GXM IgG1 that has been extensively used in a number of protocols aiming at determining the morphology and functions of capsular components (32). The cells were finally incubated with a fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG (Fc-specific) antibody (Sigma-Aldrich Corp., St. Louis, MO, USA). For a negative control, we used an isotype-matched irrelevant IgG at the same concentration used for MAb 18B7. Cell suspensions were mounted over glass slides as described above and analyzed under an Axioplan 2 (Zeiss, Germany) fluorescence microscope. The morphological aspects of the fungal cell surface were analyzed by regular protocols of scanning electron microscopy, as previously described by our group (33). Morphological analysis also included staining of the nucleus. Yeast cells were fixed in 4% paraformaldehyde in PBS for 30 min, washed three times with the same buffer, and incubated with 10 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at room temperature. After washing with PBS, the cells were analyzed microscopically as described above. On the basis of the demonstration that apt1Δ cells manifest defective kinetics for vacuolar formation (29), the number of large (>1-μm) intracellular vacuolar compartments in WT and mutant strains (100 cells of each strain) was also determined. For this analysis, fungal cells were analyzed microscopically as described above and photographed under the differential interferential contrast (DIC) model. Images were analyzed using ImageJ software (version 1.45s).
Indirect measurement of flippase activity.
In eukaryotes, phosphatidylserine (PS) is usually maintained in the cytosolic side of cell membranes by flippases. Therefore, PS is accessible to annexin V only if associated with the outer leaflet of the plasma membrane, which might result from reduced flippase activity. PS exposure in WT and apt1Δ cells was analyzed using the annexin V-FITC detection kit (APOAF; Sigma-Aldrich, St. Louis, MO, USA) following the manufacture's instructions. Flippase activity was also assessed based on the ability of C. neoformans to translocate a PS analog (34). Briefly, WT and mutant cells were cultivated for 24 h at 30°C in yeast extract-peptone-dextrose (YPD) medium. The cells were washed three times in PBS, and 2 × 107 yeast cells were suspended in a 50 μM solution (in PBS) of the fluorescent PS analog 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine (NBD-PS) (Avanti Polar Lipids, Alabaster, AL, USA) for further incubation at 37°C for 60 min. The cells were then transferred to ice, and 5 ml of ice-cold PBS-BSA was added to the suspension, resulting in extraction of NBD-PS from the outer leaflet of the plasma membrane. Alternatively, ice-cold PBS was used to wash the cells. After 15 min, the cells were pelleted, suspended in PBS, and then analyzed by flow cytometry in a Becton Dickinson LSRII cytometer. The index of lipid translocation was determined by dividing the mean fluorescence of fungal populations containing inner membrane leaflet-associated NBD-PS (obtained after extraction with BSA) by the mean fluorescence of similar samples that were not treated with PBS-BSA (total membrane-associated NBD-PS). The residual NBD-PS present in washing supernatants was measured in a Victor multilabel plate reader fluorimeter, using excitation and emission wavelengths of 485 and 535 nm, respectively, and a 0.1-s exposure.
Isolation of EVs.
Fungal extracellular vesicles (EVs) were isolated from culture supernatants as described in a number of studies by sequential centrifugation steps (8, 9, 11, 13, 14, 35). Briefly, the cells and debris were removed from culture fluids by centrifugation at 5,000 and 15,000 × g (15 min, 4°C). The remaining supernatants were filtered through 0.8-μm membranes and ultracentrifuged at 100,000 × g for 1 h at 4°C. The resulting pellets were washed three times with PBS under the same conditions. Extravesicular GXM was removed from vesicle preparations by immunoprecipitation. In these assays, pellets from the 100,000 × g centrifugation were suspended in 50 μl PBS and added to the wells of a 96-well enzyme-linked immunosorbent assay (ELISA) plate previously coated with MAb 18B7 (10 μg/ml, 1 h) and blocked with PBS–1% BSA (1 h at 37°C). The plates were incubated for 1 h at room temperature, and the unbound fraction, containing vesicles free of soluble GXM, was collected and again filtered through 0.8-μm membranes to remove any potential aggregate or contaminating cells. The resulting suspensions containing intact vesicles were used for diameter determination by dynamic light scattering (DLS), as described by Eisenman and colleagues (12). Alternatively, the suspensions were vacuum dried and suspended in chloroform-methanol (9:1, vol/vol) mixtures (14). After immediate formation of a precipitate, the suspension was centrifuged, and the resulting sediment was solubilized in PBS for quantitative ELISA for GXM determination (36) and analysis of polysaccharide dimensions by DLS (37). The chloroform-methanol supernatant was dried under a N2 stream and analyzed by thin-layer chromatography for quantitative sterol analysis (35). Sterols have been previously described as components of the membranes of fungal EVs that indirectly reflect the amount of isolated vesicles (14, 35). Therefore, all quantitative analysis of polysaccharides in cryptococcal extracellular vesicles included normalizations as follows: (polysaccharide mass)/(vesicular sterol concentration × cell number in the original culture−1).
Composition, size, and serological analyses of polysaccharide fractions.
Cryptococcal GXM fractions were obtained from culture supernatants and cell pellets. GXM was obtained from culture supernatants by ultrafiltration, as described by our group previously (38). Cellular polysaccharides were extracted with dimethyl sulfoxide (DMSO), following protocols that were established for efficient removal of GXM from C. neoformans cells (39). To ensure that intracellular GXM was extracted from fungal cells, we monitored membrane permeabilization with DMSO by propidium iodide staining, which revealed that the solvent permeabilized virtually 100% of the C. neoformans cells (data not shown). Polysaccharides were quantified by ELISA for specific GXM detection (36) and by the phenol-sulfuric acid method for total carbohydrate determination (40). Polysaccharide dimensions were determined by DLS, as previously described by Frases and colleagues (37). The monosaccharide composition of each polysaccharide fraction was determined by gas chromatography-mass spectrometry (GC-MS), following methanolysis and derivatization with trimethylsilane (TMS) (41, 42). GC was performed with a SE-54 column (30 m by 0.25 mm by 0.5 mm; Thomas Scientific) on a Trace GC (Thermo Fisher, Austin, TX) with the following running conditions: 140°C (2 min), 5°C/min gradient, 250°C (10 min) intermediate temperature, and 15°C/min gradient II with a final temperature of 265°C/min (5 min). The carrier gas was helium, with a constant flow rate of 1.5 ml/min. The molecules were ionized by electron impact at 70 eV. MS acquisition was performed with a linear scanning mode at the 40 to 650 m/z range (Polaris Q; Thermo Fisher). scyllo-Inositol added as an internal standard before methanolysis was used for normalization and quantification. Each sample was run at least three times, and the sugar residues were identified with the help of a monosaccharide mix (43). The reactivities of polysaccharide fractions from WT and mutant cells with different mouse MAbs to GXM (18B7 [IgG1] and 13A1, 12F1, and 2D10 [IgM]) (44–46) were analyzed by dot blotting and ELISA, as previously described (36, 38).
Determination of GXM during interaction of C. neoformans with host cells in vitro.
The effects of APT1 deletion during the interaction of C. neoformans with mammalian cells was assessed in experimental models using the murine macrophage-like lineage J774.A1, obtained from the American Type Culture Collection (ATCC). Cultures were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) at 37°C under a 5% CO2 atmosphere in the wells of 96-well culture plates. C. neoformans cells were suspended in fresh DMEM to form fungal suspensions at a density of 10 yeast cells per macrophage. This suspension was used to replace the culture medium of animal cells, and then the systems were incubated at 37°C with 5% CO2 for 24 h. Free yeast cells were removed by washing with PBS, and then the infected macrophages were lysed with cold water. The resulting suspension was plated onto YPD solid agar for counting of CFU. Alternatively, the suspension obtained by macrophage lysis was assessed for GXM quantification by ELISA (36). The CFU values were used as a normalization factor for the determination of GXM production during macrophage infection.
In vivo studies.
In vivo infection studies were conducted according to a previously described intranasal inhalation infection model (47) using three female C57BL/6 mice (approximately 12 to 14 weeks old) for each cage. Mice were anesthetized with ketamine (82.25 mg/kg of body weight) and xylazine (5.5 mg/kg) by intraperitoneal injection and then were suspended by their incisors on a thread to fully extend their necks. Yeast cell suspensions (5.0 × 104 cells in 50 μl) were slowly pipetted into the nares of each mouse. After 14 days of infection, the animals were euthanized by CO2 inhalation, and lungs and brains were removed. The organs were weighed, macerated, homogenized in PBS, and plated on Sabouraud agar for fungal growth analysis by counting CFU. Alternatively, lung macerates were prepared for GXM determination. For this purpose, the suspensions were supplemented with proteinase K (0.2 mg/ml, final concentration). After overnight incubation at 37°C, the samples were heated for 20 min at 100°C, placed on ice and centrifuged at 10,000 × g. The resulting supernatants were then used for GXM determination. Due to the high background levels usually observed in in vivo tests of GXM determination, we used dot blot analysis for densitometric quantification of polysaccharide production (30). Determination of the histopathological aspects of infected lungs followed previously described protocols (47). At day 14 postinfection, the lungs were fixed in 10% neutral buffered formalin. The tissue was then embedded in paraffin, cut into 5-μm-thick sections, stained with Mayer's mucicarmine (MM) to visualize the cryptococcal capsule, and then fixed on slides. Slides were examined by light microscopy. Capsule sizes were measured in at least 50 fungal cells from randomly chosen micrographs for each sample. The in vivo experiments were also used for cytokine determination in lung homogenates (47) with the BD cytometric bead array (CBA) mouse inflammation kit (Becton Dickinson, San Jose, CA) according to the manufacturer's instructions. The protocols for the experiments with mice (protocol A13–0093) were approved by the University of British Columbia Committee on Animal Care.
Statistics.
Statistical differences in paired systems were analyzed using the Student t test. Multiple comparisons were performed by analysis of variance (ANOVA). All statistical analyses were done using GraphPad Prism 6.0 software (GraphPad Software, Inc.). All values are reported as means with standard deviations (SD).
RESULTS
Deletion of ATP1 does not affect global flippase activity in C. neoformans.
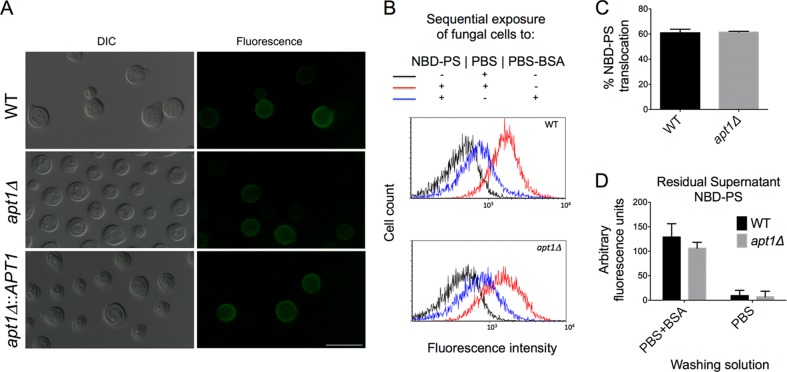
The C. neoformans APT1 gene encodes a predicted integral membrane type IV flippase. Although APT1 deletion causes important changes in the physiology of C. neoformans (29), the enzymatic activity of Apt1 has not been demonstrated. We first addressed whether the apt1Δ mutant had altered flippase activity by measuring PS exposure in annexin V-stained C. neoformans cells. The WT, mutant, and complemented strains had similar in levels of PS exposure (Fig. 1A). In all cases, the percentage of fluorescent cells was low (about 10% for all strains), as concluded by microscopic determination and confirmed by flow cytometry analysis (data not shown). The WT and mutant strains were also similar in their ability to translocate the fluorescent PS analog NBD-PS. Similar levels of fluorescence resulting from NBD-PS membrane binding were observed after WT and apt1Δ cells were incubated with the lipid and washed with PBS (Fig. 1B). Treatment of fungal cells with PBS-BSA for removal of NBD-PS from the outer membrane layer, as expected, caused a severe reduction in the fluorescence levels of yeast cells, which resulted in similar profiles of staining in both WT and apt1Δ cells. Quantification of NBD-PS translocation (Fig. 1C) confirmed the flow cytometry data. We also evaluated the remaining levels of fluorescence in the supernatants of fungal cells incubated with NBD-PS, and similar values were again obtained for WT and mutant cells. Altogether, these results indicate that the deletion of APT1 does not have a detectable influence on global PS translocation activity in C. neoformans cells.
FIG 1.
APT1 deletion does not affect the global flippase activity of C. neoformans. (A) Determination of phosphatidylserine (PS) exposure after treatment of fungal cells with FITC-annexin V reveals similar profiles of partial staining in wild-type (WT), mutant (apt1Δ), and complemented (apt1Δ::APT1) strains. Scale bar, 10 μm. (B) Analysis of the uptake of NBD-PS, a fluorescent analog of PS, by WT and mutant cells reveals that both strains were similarly efficient in incorporating the phospholipid derivative (red histograms) in comparison to unstained cells (black histograms). Treatment of C. neoformans cells with PBS-BSA for sequestration of NBD-PS molecules distributed into the external phospholipid layer of the plasma membrane resulted in cells with similar levels of fluorescence (blue histograms). (C and D) Accordingly, the levels of PS translocation were similar in WT and apt1Δ cells (C), as were the residual amounts of NBD-PS in the supernatants of cells that were washed with PBS alone or with PBS-BSA (D). P values resulting from the statistical comparison between WT and apt1Δ cells were higher than 0.5 in all cases.
Lack of Apt1 affects Golgi morphology and GXM synthesis.
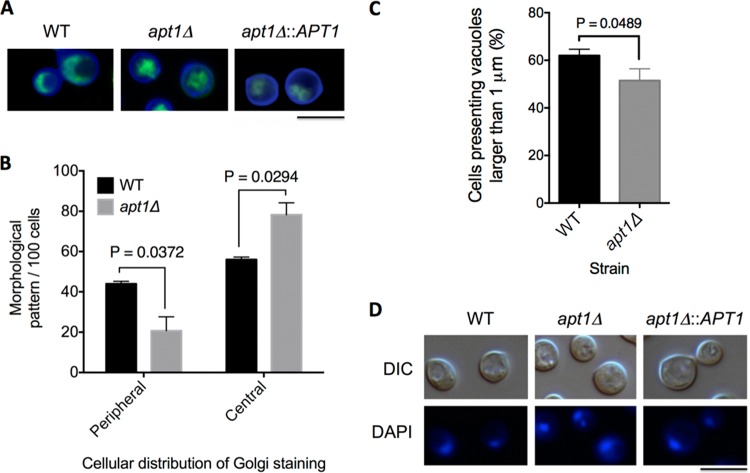
Apt1 orthologs were characterized as components of the Golgi apparatus in Saccharomyces cerevisiae (48). Due to the membranous nature of the Golgi apparatus and to the role of flippases in membrane architecture (16), we compared morphological aspects of this organelle in WT, apt1Δ, and complemented cells after staining yeast cells with Uvitex 2B (cell wall) and C6-NBD-ceramide (Golgi apparatus) (Fig. 2A). The typical peripheral distribution of the Golgi apparatus described for S. cerevisiae (49) and C. neoformans (30) predominated in WT and complemented cells. In apt1Δ cells, the cellular structures stained by C6-NBD-ceramide were mostly concentrated in the center of the cell. These morphological patterns were quantified in WT cells and in the apt1Δ mutant (Fig. 2B). The vast majority of the mutant population (85%) had C6-NBD-ceramide-stained structures that were concentrated in the central area of the cell, whereas only about 15% of the cells had the peripheral pattern of Golgi staining. The peripheral Golgi distribution was significantly more abundant in WT cells than in the mutant (P = 0.0372), while the central pattern of staining of this organelle prevailed in apt1Δ cells (P = 0.0294).
FIG 2.
Involvement of APT1 in morphological aspects of the Golgi apparatus in C. neoformans. (A) The Golgi apparatus of wild-type (WT), mutant (apt1Δ), and complemented (apt1Δ::APT1) cells was stained with C6-NBD-ceramide (green fluorescence), and the cell wall was stained with Uvitex 2B (blue fluorescence). (B) Quantitative analysis of the morphological profiles that predominated in WT and apt1Δ cells. (C) Quantification of intracellular vacuoles exceeding 1 μm in diameter in WT and mutant cells. (D) Analysis of nuclear morphology in WT, mutant, and complemented strains. C. neoformans cells are shown under differential interferential contrast (DIC) and fluorescence modes. Scale bar, 5 μm.
The morphological pattern of Golgi staining in WT cells suggested that the usually high vacuolization of C. neoformans cells could affect the morphology of this organelle. We therefore quantified the number of vacuole-like structures exceeding 1 μm in diameter in WT and mutant cells. The 1-μm cutoff was chosen based on the fluorescence microscopy analysis of the cellular segments usually associated with the Golgi apparatus, which were always greater than this value in diameter. The apt1Δ mutant had an approximately 10% reduction in the number of large vacuoles, in comparison to WT cells (Fig. 2C) (P = 0.0489). Considering that the number of WT cells manifesting the Golgi peripheral distribution was at least 2-fold higher than that of the apt1Δ mutant, we assumed that the Golgi morphology was not affected by cytoplasmic vacuoles. As a control, C. neoformans cells were stained with DAPI to evaluate whether vacuolization would affect the morphology of the nucleus. This analysis indicated similar nuclear morphologies in WT, mutant, and complemented cells (Fig. 2D). Altogether, these results indicate that the differences in the cellular distribution of the Golgi apparatus in C. neoformans cells lacking APT1 are not influenced by the morphology of intracellular vacuoles.
Structural organization is essential for the biological activity of the Golgi apparatus (50). Considering this observation and the fact that GXM synthesis was reported to occur at the Golgi apparatus for further vesicular export (51), we evaluated a number of properties of the polysaccharide in WT and mutant cells.
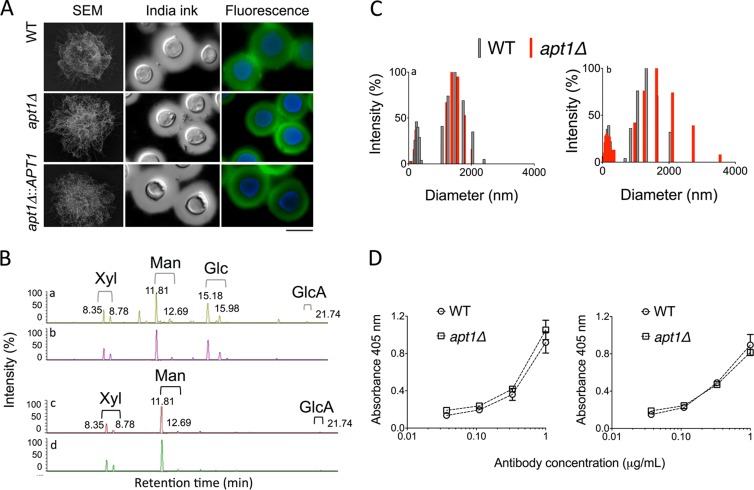
A previous report demonstrated that capsular dimensions were not affected by deletion of APT1 in C. neoformans (29). The capsular size is unquestionably important for cryptococcal pathogenicity (28), but other biological and physical chemical properties of capsular components have also been linked to the mechanisms by which C. neoformans interacts with the host (52). Therefore, we evaluated the relevance of APT1 to general properties of extracellular polysaccharides and capsular components of C. neoformans. By a combination of scanning electron microscopy, India ink counterstaining, and fluorescence microscopy with MAb 18B7, we observed that morphological aspects, serological properties, and dimensions of capsular components were similar in WT, mutant, and complemented cells (Fig. 3A).
FIG 3.
Morphological, structural, and serological analyses of capsular components in WT and Δapt1 cells of C. neoformans. (A) Morphological aspects of the capsule in WT, apt1Δ mutant, and complemented cells were visualized by scanning electron microscopy (SEM), India ink counterstaining, and fluorescence microscopy (green fluorescence, GXM; blue fluorescence, cell wall chitin). (B) GXM was isolated from C. neoformans WT (a) or mutant (b) cells or culture supernatants (c, WT cells; d, apt1Δ mutant) and analyzed by GC-MS. The chromatographic separation of monosaccharide components revealed no differences between fractions from WT and mutant cells. (C) Determination of molecular dimensions of cellular (a) or extracellular (b) polysaccharide fractions obtained from WT and apt1Δ cells reveals polysaccharide distributions in similar size ranges. (D) Serological tests with MAb 18B7 reveal that cellular (left) and extracellular (right) GXM fractions from WT and apt1Δ cells are similarly recognized by the antibody.
Compositional analyses of surface-associated and secreted GXM by GC-MS confirmed the notion that a lack of APT1 did not affect GXM structure. Chromatograms of the monosaccharide components of GXM after methanolysis of the polysaccharide revealed very similar profiles in fractions obtained from both WT and apt1Δ cells (Fig. 3B). Cellular fractions had the typical peaks of xylose (Xyl), mannose (Man), and glucuronic acid (GlcA) isomers, as well as the well-reported contamination of cellular extracts with glucose (Glc) (53). The typical GXM components were also similarly detected in extracellular fractions obtained from cultures of both WT and mutant cells. Analysis of each polysaccharide peak by MS fragmentation revealed the presence of the typical fragments of monosaccharide units, including m/z 204, 133, and 73 (Man, Glc, and Xyl) and m/z 217, 204, 133, and 73 (GlcA) (data not shown). Polysaccharides produced by both strains were analyzed by DLS, which revealed that GXM fractions obtained from either cell extracts or culture supernatants were distributed into similar ranges of effective diameter in both WT and mutant cells (Fig. 3C).
Extracellular and cell-associated GXM fractions were also tested for their reactivity with MAb 18B7, and no differences were observed between WT and mutant cells (Fig. 3D). Similar results were observed when MAb 18B7 was replaced with the anti-GXM IgMs 12A1, 2D10, and 13F1 (data not shown), validating the use of MAbs as quantitative tools for polysaccharide detection in further experiments. Altogether, these results indicated that capsule formation and the essential structural aspects of GXM were not affected by a lack of Apt1.
Cell-associated and extracellular polysaccharides were also quantified in WT and mutant cells. The GXM concentration in culture fluids of mutant cells lacking Apt1 was significantly lower than that found in WT cells supernatants (P = 0.0005) (Fig. 4A). These results suggested a defective export of GXM in the mutant. However, the possibility that the lack of Apt1 caused a general defect in GXM synthesis could not be ruled out. To address this hypothesis, we prepared GXM extracts from both WT and mutant cells for carbohydrate quantification. The total amount of GXM was again diminished in cells lacking Apt1 (P = 0.0028) (Fig. 4B). This observation was apparently specifically linked to GXM synthesis, since the total carbohydrate concentrations in WT and mutant cells were similar (P > 0.05) (Fig. 4C).
FIG 4.
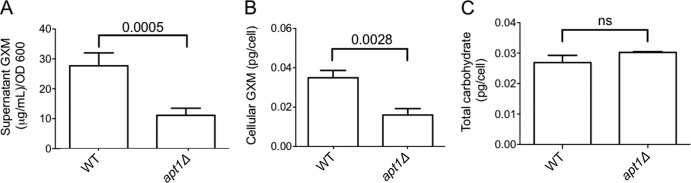
Lack of Apt1 results in attenuated GXM synthesis. (A) Quantification of supernatant GXM in cultures of WT and apt1Δ cells reveals a significantly decreased concentration of the polysaccharide in cultures of the mutant in comparison with the parental strain. (B) Similar results were obtained when cellular extracts were analyzed. (C) The defect in polysaccharide synthesis manifested by the mutant is apparently specific for GXM, since the total carbohydrate contents in both cells are similar. ns, not statistically significant (P > 0.05).
The apt1Δ mutant manifests defects in the vesicular export of GXM.
The connections between flippases, polysaccharide synthesis, and membrane architecture led us to evaluate the vesicular export of GXM in both WT and apt1Δ mutant cells. First, EVs were isolated from culture supernatants, and the distribution of their effective diameters was analyzed. Vesicles of variable dimensions were similarly observed in both WT and apt1Δ cells (Fig. 5A), which was consistent with the probable coisolation of fungal EVs of different origins in fungi, as extensively proposed in the literature (reviewed in reference 15). The size distribution of vesicles produced by WT cells included a narrow range of 10 to 150 nm and a wider span that included vesicular particles ranging from 400 to 1000 nm. Vesicles produced by mutant cells were distributed into two well-defined ranges of diameter. One of the distribution regions (10 to 150 nm) was similar to that found in WT vesicles. The larger population, however, was restricted to a range of 400 to 600 nm. GXM determination in vesicular fractions from both WT and apt1Δ cells revealed a significantly lower content of this polysaccharide in EVs produced by the mutant (P < 0.0001, Fig. 5B). The dimensions of polysaccharide fibers obtained from both strains, however, were similar (Fig. 5C). These results suggest that lack of Apt1 results in modified extracellular vesicle fractions, which contain smaller amounts of GXM.
FIG 5.
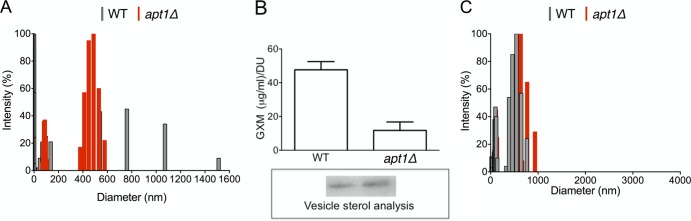
Lack of Apt1 affects C. neoformans EVs. (A) Diameter distribution of cryptococcal EVs. (B) Quantitative determination of vesicular GXM after sterol analysis (boxed area) of EVs produced by WT and mutant strains. DU, densitometry units used for normalization of GXM content to sterol concentration. The GXM concentration was significantly higher (P < 0.0001) in vesicles produced by WT cells. (C) Size determination of GXM fibers extracted from vesicles produced by WT and apt1Δ cells.
Apt1 is required for GXM secretion during interaction with the host.
GXM secretion is essential for the progression of cryptococcosis (reviewed in reference 28), and Apt1-related polysaccharide secretion might therefore represent an important aspect of pathogenesis. Our results suggest that Apt1 was involved in polysaccharide export during regular growth, which led us to evaluate whether similar findings would be observed during interaction with host cells. We therefore quantified the production of GXM by WT and apt1Δ mutant cells during macrophage infection in vitro and lung colonization in vivo. Quantification of GXM in extracts of macrophages infected with C. neoformans revealed a significantly decreased concentration of the polysaccharide when the apt1Δ strain was used to infect the phagocytes (P = 0.0087) (Fig. 6A). This difference was more accentuated when the production of GXM during lung infection was analyzed. For quantitative normalization in systems infected with WT or mutant cells, we first determined fungal CFU counts in mouse lungs. Determination of viable fungal cells in pulmonary tissue showed an approximate 1,000-fold reduction in CFU counts of mutant cells in comparison to the parental strain (Fig. 7B). The GXM concentration was then normalized to the number of CFU in lung macerates. The polysaccharide was abundantly detected in tissue samples from animals infected with the WT strain, but we were unable to detect GXM in lung preparations after infection with Δapt1 cells (Fig. 6B). We also attempted to determine GXM production in the brains of infected animals. However, we did not observe any viable C. neoformans cells in the brains of animals that were infected with the apt1Δ mutant (data not shown). The impossibility of quantitative normalization of GXM production to the number of living cells, therefore, led us to focus our analysis on infected lungs.
FIG 6.
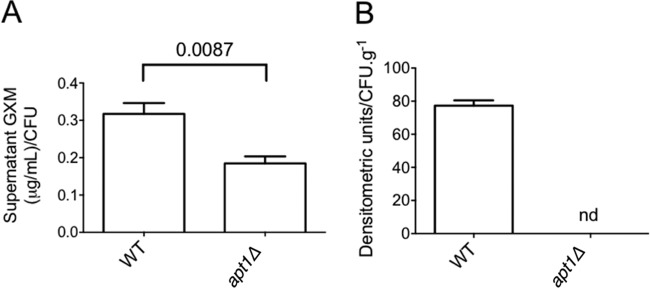
Apt1 is required for GXM secretion during infection of host cells. (A) Quantification of GXM in macrophage cultures by ELISA after infection with C. neoformans reveals a significantly decreased concentration of the polysaccharide when the phagocytes interact with apt1Δ mutant cells in comparison with the parental strain. (B) Soluble GXM is abundantly detected by dot blotting in lung macerates when mice are infected with WT C. neoformans cells. In systems where animals are infected with the apt1Δ mutant, GXM was not detected (nd).
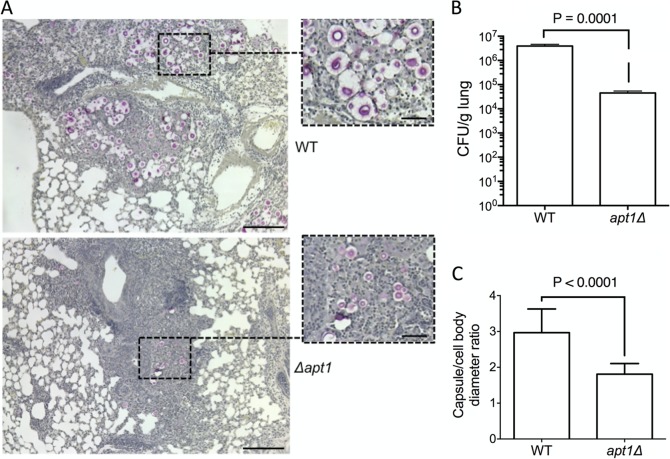
FIG 7.
Apt1 affects lung colonization, capsule formation, and host response during murine infection by C. neoformans. (A and B) The histopathology of mouse lungs after infection with WT or apt1Δ mutant cells (A) suggests a lower fungal burden when the Apt1-lacking cells are used for in vivo experimentation, which was confirmed by CFU determination (B). (C) Microscopic determination of capsule size (C) confirmed the supposition, based on visual analysis of higher-magnification fields, of reduced capsule formation in the mutant. Scale bars correspond to 200 μm (large panels) and 50 μm (insets). Data are representative of two experiments with similar results.
GXM has been reported to modulate many parameters of the host defense in favor of C. neoformans (28). The lack of GXM detection in macerates of lungs infected with the apt1Δ mutant led us to evaluate different pathogenic aspects of pulmonary cryptococcosis in animals that were infected with WT or apt1Δ cells. The histopathological analysis revealed key differences in the profile of pulmonary cellularity (Fig. 7A). In animals infected with WT cells, encapsulated cryptococci were abundantly observed. Large cellular infiltrates were only occasionally found and apparently were unable to contain fungal proliferation. A comparative analysis of these histopathological findings with those observed in lung sections infected with the apt1Δ mutant revealed fewer C. neoformans cells when the mutant was used to infect mice, as demonstrated by determination of CFU in lung macerates (Fig. 7B) (P = 0.0001). The cells of the mutant also appeared to be hypocapsular and efficiently contained within lung granulomas. Microscopic determination of capsule dimensions in histological sections confirmed that capsule formation was defective in the apt1Δ mutant in comparison with WT cells (Fig. 7C) (P < 0.0001).
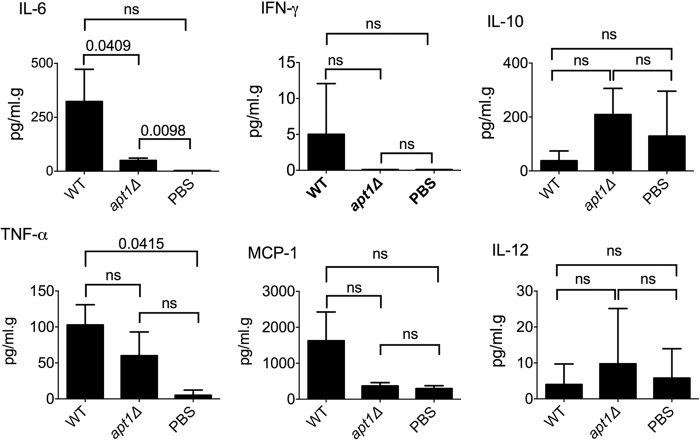
The differences in the cellular response in the lung and in capsule formation during infection by WT or apt1Δ cells were suggestive of alterations in orchestrators of the immune response. To evaluate this hypothesis, we quantified the variation of five different lung cytokines (interleukin-6 [IL-6], IL-10, IL-12, gamma interferon [IFN-γ], and tumor necrosis factor alpha [TNF-α]) and one chemokine (monocyte chemoattractant protein 1 [MCP-1]) produced in response to infection with the different strains of C. neoformans used in this study (Fig. 8). Both WT and apt1Δ cells were capable of inducing lung IL-6, but the parental strain was significantly more effective than the mutant (P = 0.0409) in this process. The levels of TNF-α, MCP-1, and IFN-γ tended to be augmented in lungs infected with WT cells. On the other hand, IL-10 and IL-12 showed a trend of increase in response to infection with the apt1Δ strain. However, the alteration in IL-6 concentration was the only statistically significant difference observed when WT and apt1Δ cells were compared. Therefore, we conclude that IL-6 was the only cytokine notably affected by lack of Apt1, among those investigated.
FIG 8.
Cytokine (IL-6, IL-10 IL-12, IFN-γ, and TNF-α) and chemokine (MCP-1) determination in the lungs of mice infected with WT or apt1Δ mutant cells versus mice receiving PBS as controls. Statistical comparisons between the values obtained from the lungs of mice infected with WT or apt1Δ mutant cells revealed that only IL-6 was differentially induced in the two systems. ns, not significant (P > 0.05).
DISCUSSION
Flippases are important regulators of secretion in a number of eukaryotes (16–18). In C. neoformans, mutants lacking the gene coding for Apt1 were recently characterized and demonstrated to be hypovirulent (29). The mutant showed an altered actin distribution and increased susceptibility to stress conditions and trafficking inhibitors (29). Notably, the lack of Apt1 resulted in a reduced export of acid phosphatase activity, confirming a link between secretion and flippase function in C. neoformans. Apt1 mutants, however, had apparently normal capsules upon gross morphological analysis (29). These findings agreed with those of Yoneda and Doering (51), who demonstrated that the C. neoformans sav1Δ mutant, which encodes a homolog of the Sec4/Rab8 subfamily GTPases, had altered GXM trafficking and diminished phosphatase activity but normal capsular dimensions. In the same study, GXM synthesis was demonstrated to occur at the Golgi apparatus, which had been characterized as one of the cellular locations of Apt1-like flippases in S. cerevisiae (48). The results described in our current study reveal that the lack of Apt1 results in altered Golgi morphology and reduced GXM synthesis. Therefore, based on these findings and on the above-mentioned reports, we speculate that Apt1 could function in the Golgi apparatus by exerting a primary role in controlling membrane asymmetry and an additional role in regulating the synthesis and/or export of cryptococcal molecules. Flippases have been reported to regulate Golgi morphology and polysaccharide synthesis in plants (54), which supports this hypothesis.
In this study, we observed that Apt1 was not required for the global flippase activity of C. neoformans, thus raising the possibility that this protein is not an active flippase. Other possibilities, however, cannot be ruled out. For instance, an analysis of the C. neoformans genome (H99 strain) (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html, November 2013) indicated the existence of at least four candidate phospholipid-translocating ATPases. Considering that the current methods for determination of flippase activity would not discriminate between the activities of these potentially different enzymes, it is possible that C. neoformans could compensate for APT1 deletion by upregulating the expression of other flippase genes, as suggested for other eukaryotic enzymes (55). It is also possible that the contribution of Apt1p to the overall flippase activity of C. neoformans is relatively low and below the sensitivity of the methods used in this study. In fact, determining the activity of flippases in fungal cells might be experimentally challenging, considering that the plasma membrane is the principal cellular site where flippases are active. In this context, tests using intact cells require externally added compounds, including lipids to be translocated and detection probes, to traverse the cell wall. Therefore, there may be challenges with kinetics and the availability of substrates, in comparison to the case for mammalian cells, where a cell wall is not present. This scenario suggests that assays of flippase activity in fungal cells might be particularly challenging and give underestimated results. Future studies to evaluate the functions of the other genes encoding potential flippases in C. neoformans may shed light on the relative contribution of Apt1p. The phenotypic traits of the apt1Δ mutant, however, were clear, and a direct relationship between fungal pathogenesis and APT1 expression was established in the current study and in a previous report characterizing apt1Δ mutants (29).
The reduced production of extracellular GXM in vivo was associated with changes in the host cytokine response. We observed a significant reduction in tissue levels of IL-6 in animals infected with the apt1Δ mutant, in comparison to the concentration of this cytokine in the lungs of animals infected with the parental strain. Previous reports demonstrated that GXM induced the production of IL-6 by human monocytes in a dose-dependent manner (56). Accordingly, IL-6 secretion by human neutrophils during interaction with C. neoformans was higher when encapsulated isolates were tested than with acapsular cells (57). On the basis of these observations, it seems plausible to assume that lower concentrations of GXM produced by the apt1Δ mutant resulted in reduced production of IL-6.
The efficacy of capsule formation is generally associated with the ability of C. neoformans to export polysaccharides (28). However, a number of secretory mutants of C. neoformans showed normal capsular dimensions but reduced concentrations of extracellular GXM (35, 51, 58, 59). This information might suggest that the efficiency of C. neoformans in synthesizing and exporting GXM may exceed the minimum quantitative requirements for full capsule formation. The characterization of C. neoformans mutants with normal capsules and reduced extracellular GXM also supports the existence of separate machineries for the synthesis and export of exopolysaccharides and capsular polysaccharides, as inferred from the observation that soluble and capsule-associated GXMs differ in biological functions and physical chemical properties (53). Interestingly, in our study, capsule formation was not significantly affected in vitro, but histopathological findings suggested a reduced capsule in pulmonary tissues. Models of capsule enlargement proposed so far include the export of GXM to the extracellular space for further incorporation into the growing capsule (60). It is reasonable to assume, therefore, that capsule enlargement is directly influenced by the fluidity of the extracellular environment, as suggested in previous studies (38). We therefore speculate that the impaired ability of the apt1Δ mutant to secrete GXM results in a reduced capsule in vivo due to the likely diminished fluidity in the tissue environment. This change might impact the ability of soluble molecules to dynamically interact with surface-associated GXM to promote capsule enlargement.
Flippases have been linked to extracellular vesicle formation (23, 24), which is in agreement with their proposed role in controlling lipid asymmetry, regulating membrane curvature, and consequently influencing the budding of vesicles (16–18). The mechanism of formation of fungal EVs is still unknown, but experimental evidence supports the requirement of membrane reshaping (6), Golgi functionality and morphology (30, 35), and endosome maturation with consequent exosome formation (10). Flippase activity has been in fact associated with each of these processes in eukaryotic cells (16–18), supporting a role for Apts in vesicle biogenesis. In our study, the loss of Apt1 resulted in EVs with a reduced GXM concentration. This observation could be a consequence of a general attenuation of GXM synthesis observed in the apt1Δ mutant. In this context, we cannot rule out the possibility that the expression of genes required for GXM synthesis is altered in the apt1Δ mutant. Nevertheless, analyses of the correlation between expression of APT1 and capsule-related genes would probably be highly intricate, given the number of genes involved in GXM synthesis and the potential for complex regulation (61). Reduced GXM secretion could also result from an altered cargo of secretory vesicles, which is agreement with the roles of flippases in the traffic of membranous compartments (18). However, the fact that the properties of EVs produced by the mutant were different from those of EVs produced by WT cells suggests additional functional attributes. Vesicles with diameters higher than 600 nm were absent in the apt1Δ mutant. This observation might implicate fungal flippases in the biogenesis of specific types of EVs. One example may be microvesicles, which are eukaryotic EVs in the diameter range of 1,000 nm (62). Our results are also consistent with the general conclusion that the currently used methods for isolation of fungal vesicles do not discriminate between membranous compartments of different cellular natures. Optimization of methods of fractionation of fungal EVs in the C. neoformans model has been proven to be complex and difficult due to the low yields of vesicle recovery in centrifugation gradients (14, 63). Genetic approaches have also been unsuccessful in turning off vesicle production (35), pointing to the need for the development of efficient biochemical separation methods, which are available for mammalian exosomes. Therefore, the development of new methods allowing the establishment of a direct relationship between flippase activity and vesicle formation in C. neoformans might be promising.
It is generally accepted that secretory mechanisms are fundamental for the pathogenicity of C. neoformans (15, 26). In this study, we demonstrated a previously unknown function of flippases in physiological and pathogenic secretion-related events used by C. neoformans. To the best of our knowledge, cryptococcal regulators of secretion described so far include the products of the SEC (51, 58, 64) and CAP genes (65–70), Golgi reassembly and stacking protein (30), protein kinase A (71), vacuolar Ca2+ transporters (35, 59), and the vacuolar protein Vps23 (72). Mutants with altered expression of the genes coding for each of these secretory regulators are hypovirulent or avirulent in mice. These observations and the fact that most of the virulence factors of C. neoformans are extracellular (6) support the notion that secretory regulators are important as components of the physiology of C. neoformans and as potential drug targets. Based on our findings, we propose the Apt1 flippase as an additional regulator of secretion and a potential drug target in C. neoformans.
ACKNOWLEDGMENTS
This project was supported by grants from the Canadian Institutes of Health Research (J.W.K.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (M.L.R. and S.F.), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (M.L.R. and S.F.), Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças Negligenciadas (INCT-IDN) (M.L.R.), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (M.L.R. and S.F.). I.C.A. is supported by NIH grant 8G12MD007592 and is special visiting-researcher fellow of the Science Without Borders program, CNPq-Brazil. We are grateful to the Biomolecule Analysis Core Facility (BACF) at BBRC/UTEP (NIH/NIMHD/RCMI grant 8G12MD007592) for the access to the LC-MS instrument used in this study. J.W.K. also gratefully acknowledges a Scholar Award in Molecular Pathogenic Mycology from the Burroughs Wellcome Fund. D.L.O. is a fellow of the Science Without Borders program of the National Council for Scientific and Technological Development (CNPq-Brazil). J.R. is a Ph.D. student affiliated with the Programa de Pós-Graduação em Química Biológica, IBqM/UFRJ, Brazil, that was supported in part by an Interhemispheric Research Training Grant in Infectious Diseases, Fogarty International Center at the Nosanchuk Laboratory (Albert Einstein College of Medicine, NY).
We are grateful to Arturo Casadevall for the gift of monoclonal antibodies to GXM, to Leonardo Nimrichter for numerous suggestions, to Vitor Cabral for help with flow cytometry, and to Joshua Nosanchuk for the use of labware and equipment.
Footnotes
Published ahead of print 13 December 2013
REFERENCES
- 1.Ding Y, Wang J, Wang J, Stierhof YD, Robinson DG, Jiang L. 2012. Unconventional protein secretion. Trends Plant Sci. 17:606–615. 10.1016/j.tplants.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 2.Schekman RW. 1994. Regulation of membrane traffic in the secretory pathway. Harvey Lect. 90:41–57 [PubMed] [Google Scholar]
- 3.Lyman SK, Schekman R. 1996. Polypeptide translocation machinery of the yeast endoplasmic reticulum. Experientia 52:1042–1049. 10.1007/BF01952100 [DOI] [PubMed] [Google Scholar]
- 4.Nickel W. 2010. Pathways of unconventional protein secretion. Curr. Opin. Biotechnol. 21:621–626. 10.1016/j.copbio.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues ML, Nakayasu ES, Almeida IC, Nimrichter L. 2014. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J. Proteomics 97:177–186. 10.1016/j.jprot.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigues ML, Franzen AJ, Nimrichter L, Miranda K. 2013. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr. Opin. Microbiol. 16:414–420. 10.1016/j.mib.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Oliveira DL, Rizzo J, Joffe LS, Godinho RM, Rodrigues ML. 2013. Where do they come from and where do they go: candidates for regulating extracellular vesicle formation in fungi. Int. J. Mol. Sci. 14:9581–9603. 10.3390/ijms14059581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJ, Longo LV, Ganiko L, Almeida IC, Puccia R. 2012. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J. Proteome Res. 11:1676–1685. 10.1021/pr200872s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallejo MC, Matsuo AL, Ganiko L, Medeiros LC, Miranda K, Silva LS, Freymuller-Haapalainen E, Sinigaglia-Coimbra R, Almeida IC, Puccia R. 2011. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-Galactosyl epitopes. Eukaryot. Cell 10:343–351. 10.1128/EC.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7:58–67. 10.1128/EC.00370-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. 2008. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 10:1695–1710. 10.1111/j.1462-5822.2008.01160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. 2009. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155:3860–3867. 10.1099/mic.0.032854-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albuquerque PC, Cordero RJ, Fonseca FL, Peres da Silva R, Ramos CL, Miranda KR, Casadevall A, Puccia R, Nosanchuk JD, Nimrichter L, Guimaraes AJ, Rodrigues ML. 2012. A Paracoccidioides brasiliensis glycan shares serologic and functional properties with cryptococcal glucuronoxylomannan. Fungal Genet. Biol. 49:943–954. 10.1016/j.fgb.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48–59. 10.1128/EC.00318-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues ML, Nosanchuk JD, Schrank A, Vainstein MH, Casadevall A, Nimrichter L. 2011. Vesicular transport systems in fungi. Future Microbiol. 6:1371–1381. 10.2217/fmb.11.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka K, Fujimura-Kamada K, Yamamoto T. 2011. Functions of phospholipid flippases. J. Biochem. 149:131–143. 10.1093/jb/mvq140 [DOI] [PubMed] [Google Scholar]
- 17.Pomorski T, Menon AK. 2006. Lipid flippases and their biological functions. Cell. Mol. Life Sci. 63:2908–2921. 10.1007/s00018-006-6167-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muthusamy BP, Natarajan P, Zhou X, Graham TR. 2009. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim. Biophys. Acta 1791:612–619. 10.1016/j.bbalip.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC. 2003. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14:1240–1254. 10.1091/mbc.E02-08-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CY, Ingram MF, Rosal PH, Graham TR. 1999. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147:1223–1236. 10.1083/jcb.147.6.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. 2002. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12:1623–1627. 10.1016/S0960-9822(02)01148-X [DOI] [PubMed] [Google Scholar]
- 22.Hua Z, Fatheddin P, Graham TR. 2002. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13:3162–3177. 10.1091/mbc.E02-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuck S. 2011. Extracellular vesicles: budding regulated by a phosphatidylethanolamine translocase. Curr. Biol. 21:R988–990. 10.1016/j.cub.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 24.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. 2011. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21:1951–1959. 10.1016/j.cub.2011.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues ML, Djordjevic JT. 2012. Unravelling secretion in Cryptococcus neoformans: more than one way to skin a cat. Mycopathologia 173:407–418. 10.1007/s11046-011-9468-9 [DOI] [PubMed] [Google Scholar]
- 27.McClelland EE, Bernhardt P, Casadevall A. 2006. Estimating the relative contributions of virulence factors for pathogenic microbes. Infect. Immun. 74:1500–1504. 10.1128/IAI.74.3.1500-1504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68:133–216. 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu G, Kronstad JW. 2010. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot. Cell 9:74–83. 10.1128/EC.00289-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kmetzsch L, Joffe LS, Staats CC, de Oliveira DL, Fonseca FL, Cordero RJ, Casadevall A, Nimrichter L, Schrank A, Vainstein MH, Rodrigues ML. 2011. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol. Microbiol. 81:206–218. 10.1111/j.1365-2958.2011.07686.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagano RE. 1989. A fluorescent derivative of ceramide: physical properties and use in studying the Golgi apparatus of animal cells. Methods Cell Biol. 29:75–85 [DOI] [PubMed] [Google Scholar]
- 32.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, Kozel TR, Lendvai N, Mukherjee J, Pirofski LA, Rivera J, Rosas AL, Scharff MD, Valadon P, Westin K, Zhong Z. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos CL, Fonseca FL, Rodrigues J, Guimaraes AJ, Cinelli LP, Miranda K, Nimrichter L, Casadevall A, Travassos LR, Rodrigues ML. 2012. Chitin-like molecules associate with Cryptococcus neoformans glucuronoxylomannan to form a glycan complex with previously unknown properties. Eukaryot. Cell 11:1086–1094. 10.1128/EC.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sipkens JA, Hahn NE, van Nieuw-Amerongen GP, Stehouwer CD, Rauwerda JA, van Hinsbergh VW, Niessen HW, Krijnen PA. 2011. Homocysteine induces phosphatidylserine exposure in cardiomyocytes through inhibition of Rho kinase and flippase activity. Cell. Physiol. Biochem. 28:53–62. 10.1159/000331713 [DOI] [PubMed] [Google Scholar]
- 35.Kmetzsch L, Staats CC, Simon E, Fonseca FL, de Oliveira DL, Sobrino L, Rodrigues J, Leal AL, Nimrichter L, Rodrigues ML, Schrank A, Vainstein MH. 2010. The vacuolar Ca(2)(+) exchanger Vcx1 is involved in calcineurin-dependent Ca(2)(+) tolerance and virulence in Cryptococcus neoformans. Eukaryot. Cell 9:1798–1805. 10.1128/EC.00114-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadevall A, Mukherjee J, Scharff MD. 1992. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 154:27–35. 10.1016/0022-1759(92)90209-C [DOI] [PubMed] [Google Scholar]
- 37.Frases S, Pontes B, Nimrichter L, Viana NB, Rodrigues ML, Casadevall A. 2009. Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc. Natl. Acad. Sci. U. S. A. 106:1228–1233. 10.1073/pnas.0808995106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimrichter L, Frases S, Cinelli LP, Viana NB, Nakouzi A, Travassos LR, Casadevall A, Rodrigues ML. 2007. Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot. Cell 6:1400–1410. 10.1128/EC.00122-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan RA, Zaragoza O, Zhang T, Ortiz G, Casadevall A, Dadachova E. 2005. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 4:465–475. 10.1128/EC.4.2.465-475.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167. 10.1038/168167a0 Error: More than 1 reference returned by Crossref [DOI] [PubMed] [Google Scholar]
- 41.Ferguson MA, Homans SW, Dwek RA, Rademacher TW. 1988. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239:753–759. 10.1126/science.3340856 [DOI] [PubMed] [Google Scholar]
- 42.Medeiros MM, Peixoto JR, Oliveira AC, Cardilo-Reis L, Koatz VL, Van Kaer L, Previato JO, Mendonca-Previato L, Nobrega A, Bellio M. 2007. Toll-like receptor 4 (TLR4)-dependent proinflammatory and immunomodulatory properties of the glycoinositolphospholipid (GIPL) from Trypanosoma cruzi. J. Leukoc. Biol. 82:488–496. 10.1189/jlb.0706478 [DOI] [PubMed] [Google Scholar]
- 43.Ruiz-Matute AI, Hernandez-Hernandez O, Rodriguez-Sanchez S, Sanz ML, Martinez-Castro I. 2011. Derivatization of carbohydrates for GC and GC-MS analyses. J. Chromatogr. B 879:1226–1240. 10.1016/j.jchromb.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 44.Feldmesser M, Rivera J, Kress Y, Kozel TR, Casadevall A. 2000. Antibody interactions with the capsule of Cryptococcus neoformans. Infect. Immun. 68:3642–3650. 10.1128/IAI.68.6.3642-3650.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleare W, Casadevall A. 1998. The different binding patterns of two immunoglobulin M monoclonal antibodies to Cryptococcus neoformans serotype A and D strains correlate with serotype classification and differences in functional assays. Clin. Diagn. Lab. Immunol. 5:125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nussbaum G, Cleare W, Casadevall A, Scharff MD, Valadon P. 1997. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J. Exp. Med. 185:685–694. 10.1084/jem.185.4.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng PY, Sham A, Kronstad JW. 2009. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect. Immun. 77:4284–4294. 10.1128/IAI.00628-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natarajan P, Wang J, Hua Z, Graham TR. 2004. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. U. S. A. 101:10614–10619. 10.1073/pnas.0404146101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine TP, Wiggins CA, Munro S. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 11:2267–2281. 10.1091/mbc.11.7.2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe M. 2011. Structural organization of the Golgi apparatus. Curr. Opin. Cell Biol. 23:85–93. 10.1016/j.ceb.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 51.Yoneda A, Doering TL. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17:5131–5140. 10.1091/mbc.E06-08-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues ML, Fonseca FL, Frases S, Casadevall A, Nimrichter L. 2009. The still obscure attributes of cryptococcal glucuronoxylomannan. Med. Mycol. 47:783–788. 10.3109/13693780902788621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frases S, Nimrichter L, Viana NB, Nakouzi A, Casadevall A. 2008. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot. Cell 7:319–327. 10.1128/EC.00378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poulsen LR, Lopez-Marques RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG. 2008. The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20:658–676. 10.1105/tpc.107.054767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bollen E, Prickaerts J. 2012. Phosphodiesterases in neurodegenerative disorders. IUBMB Life 64:965–970. 10.1002/iub.1104 [DOI] [PubMed] [Google Scholar]
- 56.Delfino D, Cianci L, Lupis E, Celeste A, Petrelli ML, Curro F, Cusumano V, Teti G. 1997. Interleukin-6 production by human monocytes stimulated with Cryptococcus neoformans components. Infect. Immun. 65:2454–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Retini C, Vecchiarelli A, Monari C, Tascini C, Bistoni F, Kozel TR. 1996. Capsular polysaccharide of Cryptococcus neoformans induces proinflammatory cytokine release by human neutrophils. Infect. Immun. 64:2897–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panepinto J, Komperda K, Frases S, Park YD, Djordjevic JT, Casadevall A, Williamson PR. 2009. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 71:1165–1176. 10.1111/j.1365-2958.2008.06588.x [DOI] [PubMed] [Google Scholar]
- 59.Kmetzsch L, Staats CC, Cupertino JB, Fonseca FL, Rodrigues ML, Schrank A, Vainstein MH. 2013. The calcium transporter Pmc1 provides Ca tolerance and influences the progression of murine cryptococcal infection. FEBS J. 280:4853–4864. 10.1111/febs.12458 [DOI] [PubMed] [Google Scholar]
- 60.Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 59:67–83. 10.1111/j.1365-2958.2005.04928.x [DOI] [PubMed] [Google Scholar]
- 61.Klutts JS, Yoneda A, Reilly MC, Bose I, Doering TL. 2006. Glycosyltransferases and their products: cryptococcal variations on fungal themes. FEMS Yeast Res. 6:499–512. 10.1111/j.1567-1364.2006.00054.x [DOI] [PubMed] [Google Scholar]
- 62.Schneider A, Simons M. 2013. Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 352:33–47. 10.1007/s00441-012-1428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliveira DL, Nimrichter L, Miranda K, Frases S, Faull KF, Casadevall A, Rodrigues ML. 2009. Cryptococcus neoformans cryoultramicrotomy and vesicle fractionation reveals an intimate association between membrane lipids and glucuronoxylomannan. Fungal Genet. Biol. 46:956–963. 10.1016/j.fgb.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chayakulkeeree M, Johnston SA, Oei JB, Lev S, Williamson PR, Wilson CF, Zuo X, Leal AL, Vainstein MH, Meyer W, Sorrell TC, May RC, Djordjevic JT. 2011. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol. Microbiol. 80:1088–1101. 10.1111/j.1365-2958.2011.07632.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Rivera J, Chang YC, Kwon-Chung KJ, Casadevall A. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385–392. 10.1128/EC.3.2.385-392.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang YC, Kwon-Chung KJ. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang YC, Kwon-Chung KJ. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang YC, Cherniak R, Kozel TR, Granger DL, Morris LC, Weinhold LC, Kwon-Chung KJ. 1997. Structure and biological activities of acapsular Cryptococcus neoformans 602 complemented with the CAP64 gene. Infect. Immun. 65:1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang YC, Penoyer LA, Kwon-Chung KJ. 1996. The second capsule gene of cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang YC, Wickes BL, Kwon-Chung KJ. 1995. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene 167:179–183. 10.1016/0378-1119(95)00640-0 [DOI] [PubMed] [Google Scholar]
- 71.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. 2013. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 81:292–302. 10.1128/IAI.01037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]