Abstract
The RASSF1A gene is one of the most frequently inactivated genes in over 30 different types of cancers (H. Donninger, M. D. Vos, and G. J. Clark, J. Cell Sci. 120:3163–3172, 2007, http://dx.doi.org/10.1242/jcs.010389). Despite the prevalence of RASSF1A silencing in human cancer, the mechanism by which RASSF1A functions as a tumor suppressor is not well understood. Characterization of the consequences of RASSF1A loss on epithelial cell proliferation revealed that RASSF1A expression suppresses both microRNA 21 (miR-21) expression and extracellular signal-regulated kinase 1/2 (ERK1/2) activation. The mechanism of the former is through restraint of SCFβTrCP-dependent destruction of the repressor element 1 silencing transcription factor (REST) tumor suppressor and consequent inhibition of miR-21 promoter activation. The mechanism of the latter is through physical sequestration of MST2, which results in accumulation of inactivating S259 phosphorylation of RAF1. Whether or not inactivation of these RASSF1A regulatory relationships can unleash enhanced proliferative capacity is dependent upon the coupling of SCFβTrCP and miR-21 to suppression of SKP2 protein translation and stability. Airway epithelial cultures retain this coupling and therefore respond to RASSF1A inactivation by p27-dependent cell cycle arrest. In contrast, colonic crypt-derived epithelial cells have uncoupled SCFβTrCP from SKP2 and respond to RASSF1A inactivation by enhanced proliferation rates. These observations help account for context-specific molecular etiology of oncogenic transformation and suggest intervention strategies for recently developed SKP2 inhibitors.
INTRODUCTION
RASSF1 has received significant scrutiny as a candidate tumor suppressor locus since its discovery within the minimal region of common loss of heterozygosity on chromosome 3 (3p21.3) in solid tumors (1–6). The locus produces at least 2 major gene products, RASSF1A and RASSF1C, which differ only in their amino termini due to distinct exon 1 selection that is specified by two independent promoter regions (7). Notably, selective inactivation of the RASSF1A promoter is one of the most common epigenetic lesions found in human tumors (1). The functional relevance of RASSF1A inactivation to disease etiology can be inferred from RASSF1A−/− mice, which display increased spontaneous and carcinogen-induced tumor susceptibility relative to control littermates (8, 9). In addition, ectopic expression of RASSF1A in a broad variety of human cancer cell lines impairs cell autonomous proliferation and survival and is sufficient to inhibit tumorigenicity (3, 7, 10–12).
A firm mechanistic account of the contribution of RASSF1A to cell and tissue homeostasis has been slow to crystalize, in part because of the seemingly bewildering array of cell biological phenotypes associated with gain and loss of function analyses. These include altered regulation of apoptosis, DNA damage, mitosis, and cell cycle control (1). Though absent any recognizable functional domains, RASSF1A-interacting proteins have directly implicated RASSF1A in Hippo pathway signaling, tumor necrosis factor receptor 1 (TNFR1) signaling, microtubule dynamics, cyclin-dependent kinase activation, and proteostasis (13).
Through efforts to characterize the contribution of RASSF1A inactivation to human epithelial cell transformation, we identified derepression of SCFβTrCP activity as a key mechanistic event that may account for many of the altered cell regulatory states observed upon loss of RASSF1A expression. Importantly, we find that RASSF1A inactivation leads to SCFβTrCP-dependent destruction of the repressor element 1 silencing transcription factor (REST) tumor suppressor and consequent derepression of onco-microRNA 21 (miR-21) expression. This tumor suppressor/oncogene cascade instructs enhanced proliferative potential that can either promote cell replication or cell cycle arrest according to context-specific activation of p27 and p53 checkpoint controls.
MATERIALS AND METHODS
Cell culture.
HeLa cells and BJ normal foreskin fibroblasts were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). U2OS and HCT116 cells were purchased from ATCC and cultured in 10% McCoy's medium. Human bronchial epithelial cells (HBEC30) were cultured in keratinocyte serum-free medium (SFM) plus supplements. Human colonic epithelial cells (HCECs) were a ind gift from the Shay/Wright lab and cultured as described previously (14). 293FT cells were purchased from Invitrogen and cultured in DMEM supplemented with 10% FBS.
Constructs.
pRK5-myc, pRK5-RASSF1A-myc, and pRK5-RASSF1C-myc constructs were generated previously as described in reference 15. pLenti-Skp2 3′UTR-luc vector (where UTR stands for untranslated region) was purchased from Applied Biological Materials Inc. A point mutation was detected at nucleotide (nt) 1925 and was mutated to wild-type Skp2 mRNA according to the NCBI sequence database. Celplor (Raleigh, NC) was used to construct point mutations within pLENTi-Skp2 3′UTR-luc vector. Using wild-type Skp2 mRNA as a template, two or three point mutations were made within pLENTi-Skp2 3′UTR-luc at potential miR-21 binding sites, designated site 1 and site 2. Mutant contructs were validated by sequencing by Celplor.
siRNAs.
Small interfering RNA (siRNA) sequences were as follows: RASSF1A-1, GACCUCUGUGGCGACUUCAdTdT; RASSF1A-2, CACGUGGUGCGACCUCUGU; Emi1, GAUGCUCAAACCAAGUUAU. For βTrCP silencing, the SMARTpool was obtained from Dharmacon (Lafayette, CO). The following siRNAs were custom ordered from Dharmacon: RASSF1A-3, 5′-UGUGGAGUGGGAGACACCUUU-3′; RASSF1A-4 (16), 5′-UCUUCUGCUCAAUCUCAGC-3′; REST single (17), 5′-GGGCCUAAACCUCUUAAUU-3′; MST2 (27), 5′-UUGCGACAACUUGACCGGAUU-3′. SiGenome pools for the following genes were ordered from Dharmacon: RASSF1, CDH1, p27 pools, p21 pools, p53 pools, REST pools. For negative-control siRNA, we used nontargeting pool 2 (catalog no. D-001206-14-20) from Dharmacon. MicroRNA sequences were purchased from Dharmacon: mimic negative control 1, catalog no. CN-001000-01-05; miR-21 mimic, 5′-UAGCUUAUCAGACUGAUGUUGA–3′; hairpin inhibitor negative control 1, catalog IN-001005-01-05; miR-21 hairpin inhibitor, 5′-UAGCUUAUCAGACUGAUGUUGA-3′. Lentiviral short hairpin RNA (shRNA) expression constructs were purchased from Open Biosystems: pLKO.1-shRASSF1-1 (clone identifier [ID] TRCN0000077854), pLKO.1-shRASSF1-2 (clone ID TRCN0000077857), pLKO.1-shp27-1 (clone ID TRCN0000009856), pLKO.1-shp27-2 (clone ID TRCN0000009857), and pLKO.1-shGFP (used as described in reference 18).
Reagents.
MG132 (C2211) was purchased from Sigma. Propidium iodide (catalog no. 550825) was purchased from BD Biosciences. Nocodazole (catalog no. M1404) was purchased from Sigma. Mouse anti-RASSF1A was purchased from Abcam (ab23950) and eBioscience (catalog no. 14-6888-82). Antibodies for cyclin A, cyclin B, and p53 (sc-126) were from Santa Cruz. Anti-CDH1 was from LabVision Corporation. Mouse anti-βTrCP (catalog no. 37-3400) was purchased from Zymed. Mouse anti-SKP2 (catalog no. 32-3300) and rabbit anti-Emi1 (catalog no. 38-5000) were purchased from Zymed (Invitrogen) for Western blotting. Rabbit anti-REST (catalog no. 07-579) was purchased from Millipore. Mouse antiactin (A1978) was from Sigma. Mouse anti-Ras was purchased from BD Transduction Labs. Rabbit anti-MST2, rabbit anti-p27 (catalog no. 3686), rabbit anti-p21 (catalog no. 2947), rabbit anti-p16 (catalog no. 4824), mouse anti-cyclin E (catalog no. 4129), mouse anti-cyclin D1 (catalog no. 2926), mouse anti-cyclin B (catalog no. 4135), rabbit anti-phospho-extracellular signal-regulated kinase (anti-phospho-ERK; catalog no. 9101), and rabbit anti-ERK (catalog no. 9102) were purchased from Cell Signaling. For immunofluorescence, antibromodeoxyuridine (anti-BrdU)-Alexa Fluor 488 (B35130) was purchased from Invitrogen.
Transfection.
Cells were transfected with RNAiMAX (Invitrogen) according to the manufacturer's protocol and harvested 72 h later.
3′UTR luciferase assays.
For reporter assays, 293FT cells were cotransfected with wild-type or mutant reporter plasmid (20 ng) and miRNA (25 nM) with Lipofectamine 2000 in 96-well plates. Reporter assays were performed 48 h after transfection, using the dual-luciferase assay system (Promega), and normalized for transfection efficiency by cotransfected renilla luciferase (2.5 ng).
Immunoblotting.
Cells were lysed in RIPA buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, protease inhibitors, 2-mercaptoethanol) or SDS buffer (100 mM Tris [pH 6.8], 4% SDS, 20% glycerol, protease inhibitors, 2-mercaptoethanol). Samples were separated by SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes. Immunoblot analysis was performed with the indicated antibodies and visualized with SuperSignal West Pico chemiluminescent substrate (Pierce Chemical).
Immunoprecipitation.
For immunoprecipitation of RASSF1A and βTrCP, HeLa cells were lysed in nondenaturing Triton-DOC buffer (20 mM Tris-HCl [pH 7.4], 137 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM MgCl2, 2 mM EGTA) plus protease and phosphatase inhibitors (Roche EDTA-free protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF, 1 mM NaVO4, 80 mM β-glycerophosphate). Cells were lysed for 15 min and then cleared at 20,000 × g for 10 min at 4°C. A total of 800 μg of lysate was diluted with lysis buffer to a concentration of 1 μg/μl. Complexes were immunoprecipitated with 2 μg of the indicated antibody for 1 h. Antibody-antigen complexes were precipitated with protein A/G-agarose beads for 1 h. Complexes were washed in lysis buffer 3 or 4 times for 5 min at 4°C.
Immunofluorescence.
For BrdU visualization, cells were treated with 30 μM BrdU for 24 h and then fixed in 3.7% formaldehyde. Cells were permeabilized with MeOH for 10 min at −20°C and then blocked in phosphate-buffered saline (PBS), 5% bovine serum albumin (BSA), and 1% Tween for a minimum of 15 min. Anti-BrdU was used at a dilution of 1:20. Cells were visualized on an Axiovert upright microscope (Zeiss) equipped with a Hamamatsu black and white camera.
Fluorescence-activated cell sorting (FACS) analysis.
Cells were trypsinized at 72 h posttransfection and resuspended in a 50:50 mixture of EtOH and PBS. Following fixation for 30 min, cells were washed and labeled with propidium iodide (Sigma) at 40 μg/ml for 30 min at 37°C. For each analysis, 10,000 cells were collected by FACScan and analyzed with the CellQuest program (Becton, Dickinson).
qPCR.
HeLa cells were transfected in 35-mm2 dishes with 100 nM siRNA. RNA was extracted from cells with TRIzol (Invitrogen) at 72 h posttransfection according to the manufacturer's protocol. cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. For cDNA synthesis, 1 μg of RNA and oligo(dT)12-18 primers was used. One-fifteenth of the cDNA reaction was used with the Roche Light Cycler system and the Light Cycler FastStart DNA master SYBR green I (Roche Applied Systems). Primers were chosen to flank at least two siRNA target sequences and lie on separate exons. Primers for SKP2 are 5′TGAGCTGAACCTCTCCTGGT 3′ (forward) and 5′CTGGCACGATTCCAAAAACT 3′ (reverse). Values were normalized using GAPDH and analyzed using the relative quantification mathematical model (Pfaffl). Quantitative PCR (qPCR) on miR-21 was performed using the TaqMan miRNA assay kit (Applied Biosystems) according to the manufacturer's protocol. GAPDH was used as a loading control.
Ras activity assay.
Ras activity assay was performed using Raf1–glutathione S-transferase (GST) beads according to the manufacturer's protocol (Upstate).
Soft agar colony formation.
Soft agar assays were performed as described elsewhere (14).
Image quantification.
Fluorescence micrographs were analyzed using Image J software. Error was estimated using the Student unpaired t test.
RESULTS
RASSF1A promotes accumulation of the REST tumor suppressor.
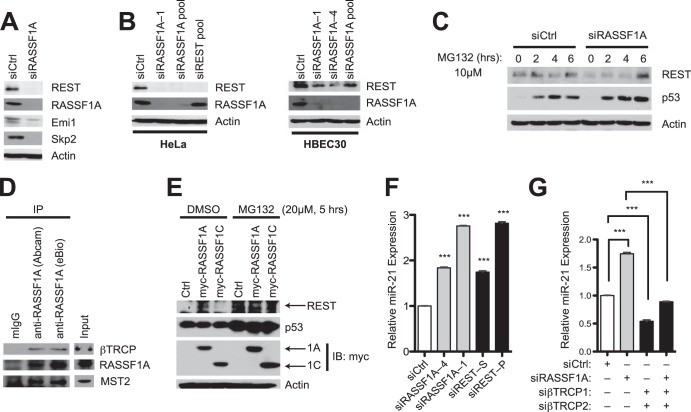
Although commonly lost in cancer, cell-based investigations indicate that the tumor suppressor RASSF1A makes a positive contribution to cell cycle progression, in CKI-responsive cells, by restraining promiscuous SCFβTrCP-dependent destruction of the APC/C inhibitor EMI1 (19). We therefore asked if additional effects of unrestrained SCFβTrCP activity might account for the tumor-promoting consequence of RASSF1A inactivation (20). Of note, we found that accumulation of the SCFβTrCP client protein REST was impaired upon RASSF1A depletion with multiple small interfering RNA (siRNA) oligonucleotides in the RASSF1A-positive cell lines HeLa and HBEC30 (Fig. 1A and B). REST is an enigmatic molecule that maintains neuronal pluripotency but shows context-dependent tumor suppressor activity (21) and is inactivated in colon cancer and small cell lung cancers (22, 23). REST protein accumulation was rescued by proteasome inhibition (Fig. 1C), consistent with ubiquitination-dependent REST degradation (22), in an RASSF1A-depleted background. Though not demonstrated here, the mechanism may be relatively direct, as endogenous RASSF1A immunoprecipitates βTrCP (Fig. 1D). In addition, expression of RASSF1A but not RASSF1C was sufficient to enhance REST protein accumulation (Fig. 1E).
FIG 1.
RASSF1A supports accumulation of the REST tumor suppressor. (A) HeLa cells were transfected with the indicated siRNA. Lysates were immunoblotted for detection of the indicated proteins at 72 h posttransfection. (B) HeLa and HBEC30 cells were treated as described for panel A; (C) HeLa cells were treated as described for panel A, followed by exposure to MG132 (10 μM) for the indicated times prior to lysis; (D) HeLa cells were lysed and mouse IgG (control) or RASSF1A antibodies were used for coimmunoprecipitation assays. Immunoprecipitates were blotted for detection of the indicated proteins. (E) The indicated cDNAs were transiently expressed in HeLa cells. Whole-cell lysates were then collected following a 5-h exposure to dimethyl sulfoxide (DMSO) or MG132 (20 μM) and immunoblotted for detection of the indicated proteins. (F and G) HeLa cells were transfected with indicated siRNAs, and consequences on endogenous miR-21 concentrations were measured by qPCR. Bars indicate mean relative expression ± standard errors from the means (SEM). ***, P < 0.001, Student's t test.
A key REST target in mice that can account for both maintenance of embryonic stem cell self-renewal and modulation of tumorigenicity is miR-21 (17). We found that REST-dependent suppression of miR-21 expression is conserved in human epithelial cells and is modulated by RASSF1A. Depletion of either RASSF1A or REST resulted in significant accumulation of endogenous miR-21 (Fig. 1F), while depletion of βTrCP was sufficient to inhibit miR-21 accumulation and to reverse the consequence of RASSF1A depletion on miR-21 activation (Fig. 1G). Thus, elevated expression of the miR-21 oncomir appears to be a collateral consequence of RASSF1A loss due to a cascade effect on the REST tumor suppressor via promiscuous SCFβTrCP activity.
SKP2 is an miR-21 target gene.
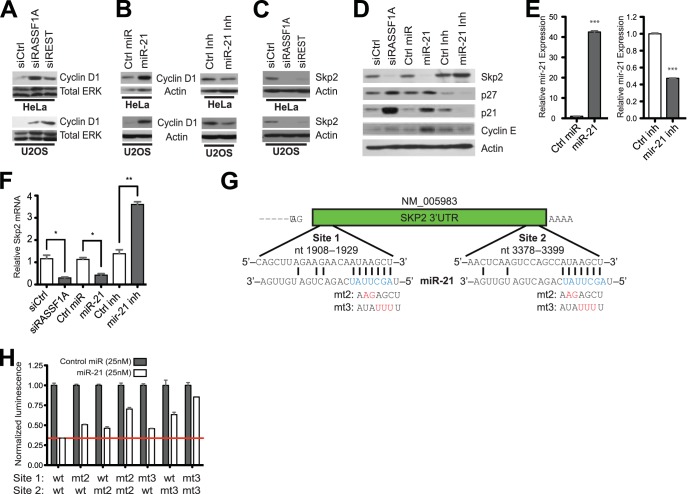
We next asked if REST depletion and/or miR-21 induction correlated with RASSF1A-dependent consequences on cell cycle progression machinery. Previous work indicated that an immediate consequence of RASSF1A loss is accumulation of cyclin D1 together with loss of SKP2 and consequent accumulation of SKP2 substrates, including the cyclin-dependent kinase inhibitor p27 (15, 19). Similarly, siRNA-mediated REST depletion or transfection of a nucleic acid mimic of miR-21 also resulted in accumulation of cyclin D1 (Fig. 2A and B) and loss of SKP2 (Fig. 2C and D). Importantly, transfection of an miR-21 inhibitor reduced endogenous miR-21 expression (Fig. 2E) and resulted in accumulation of SKP2, loss of SKP2 substrates p27, p21, and cyclin E, and loss of cyclin D1, suggesting native miR-21 regulates these proteins in an RASSF1A-positive background (Fig. 2D). miR-21-dependent alteration of SKP2 accumulation was associated with concomitant changes in SKP2 mRNA (Fig. 2F). This, together with the presence of two putative miR-21 seed sequence matches in the SKP2 3′UTR (Fig. 2G), suggested the possibility that SKP2 is an miR-21 target gene. Consistent with this, miR-21 suppressed expression of a luciferase reporter gene fused to the SKP2 3′UTR. This was partially rescued by introduction of two (mt2) or three (mt3) point mutations within either target site and completely reversed by abolishing miR-21 seed sequence pairing in both target sites (Fig. 2H).
FIG 2.
RASSF1A/REST-dependent restraint of mir-21 expression promotes SKP2 protein accumulation. (A to D) HeLa and U2OS cells were transfected with the indicated siRNAs, synthetic miRNA mimics, or synthetic miRNA inhibitors, and whole-cell lysates were immunoblotted for detection of the indicated proteins. In panel B, cyclin D1 exposure times were optimized for detection of the consequence of the miR-21 inhibitor on suppression of baseline cyclin D1 accumulation. (E) A synthetic miR-21 mimic (left) or inhibitor (right) was transfected into HeLa cells. miR-21 accumulation was measured by qPCR at 72 h posttransfection. Bars indicate mean relative expression ± SEM. ***, P < 0.001, Student's t test. (F) As described for panel E, except Skp2 mRNA accumulation was measured. **, P < 0.01; *, P < 0.05, Student's t test. (G) Seed sequence complementarity analysis of miR-21 within the SKP2 3′UTR indicates two 7-mer seed sequence matches. Predicted base pairing between miR-21 seed sequence and SKP2 3′UTR is shown. Engineered sequence alterations in the SKP2 3′UTR are indicated in red. (H) pLenti-SKP2–3′UTR-luc and renilla luciferase plasmids were cotransfected in 293FT cells with either control or the miR-21 mimetic. Luminescence was measured using the dual-luciferase reporter assay system. Wild-type SKP2 3′UTR seed sequence matches were mutated at a two (mt2) or three (mt3) position at site 1, site 2, or both. Luminescence values were normalized to control miRNA and renilla luciferase activity. Bars indicate mean relative reporter activity ± SEM.
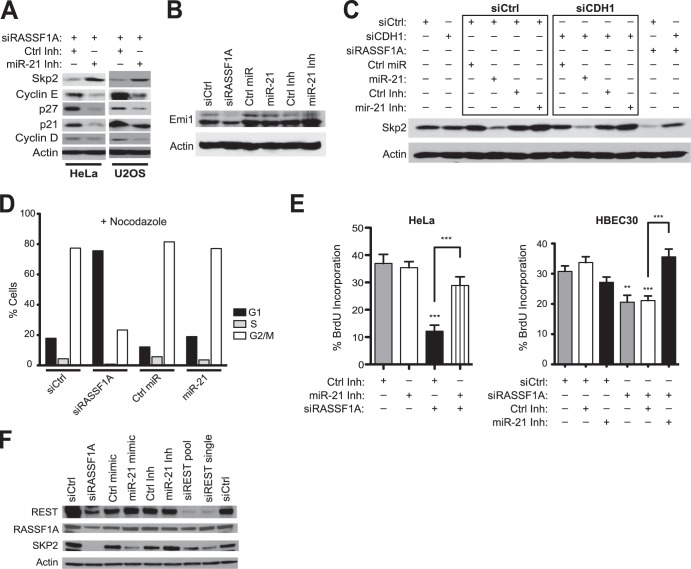
We had previously determined that RASSF1A loss results in APC/C-dependent destruction of SKP2 protein due to promiscuous degradation of the APC/C inhibitor EMI1 by SCFβTrCP (19). The capacity of miR-21 to modulate SKP2 suggests RASSF1A loss may have a combinatorial consequence on SKP2 protein accumulation via enhanced protein turnover together with inhibition of protein expression. Consistent with this, we found that RASSF1A depletion is associated with reduced accumulation of SKP2 mRNA (Fig. 2F). Furthermore, the miR-21 inhibitor rescued accumulation of SKP2 protein in RASSF1A-depleted cells and reversed accumulation of the SKP2 substrates p27 and p21 and the candidate substrate cyclin E (Fig. 3A). In contrast to RASSF1A depletion, and consistent with direct modulation of SKP2 mRNA by miR-21, miR-21 expression did not affect Emi1 accumulation (Fig. 3B). Likewise, miR-21-dependent inhibition of SKP2 accumulation was independent of the APC/C cofactor CDH1 (Fig. 3C). Ectopic miR-21 was insufficient to induce cell cycle arrest (Fig. 3D); however, depletion of endogenous miR-21 rescued proliferation of RASSF1A-depleted cells (Fig. 3E). Neither expression nor inhibition of miR-21 affected accumulation of REST or RASSF1 protein (Fig. 3F). These cumulative observations indicate that RASSF1A depletion results in SCFβTrCP-dependent REST degradation and derepression of expression of the oncomir miR-21. However, in checkpoint-responsive cells, this promiscuous miR-21 activity converges on inhibition of SKP2 accumulation and therefore contributes to G1 checkpoint activation upon RASSF1A inactivation.
FIG 3.
mir-21 partially mediates RASSF1A regulation of SKP2 and cell cycle progression. (A to C) HeLa and U2OS cells were cotransfected with the indicated oligonucleotides. Whole-cell lysates were immunoblotted for detection of the indicated proteins. (B and C) HeLa cells were treated as described for panel A. (D) HeLa cells were transfected with the indicated oligonucleotides and treated with nocodazole (100 ng/ml) for 16 h prior to staining with propidium iodide and FACS analysis. Bars indicate cell populations gated for 2N (G1), >2N and <4N (S), and 4N (G2/M) DNA content. (E) HeLa (left) and HBEC30 (right) cells were transfected as indicated followed by incubation with BrdU for 5 h (HeLa) or 20 h (HBEC30). Bars indicate mean relative BrdU incorporation ± SEM. ***, P < 0.001; **, P < 0.01, Student's t test. (F) HeLa cells were transfected with indicated siRNAs, and whole-cell lysates were immunoblotted for detection of the indicated proteins.
MST2 mediates RASSF1A suppression of ERK activation.
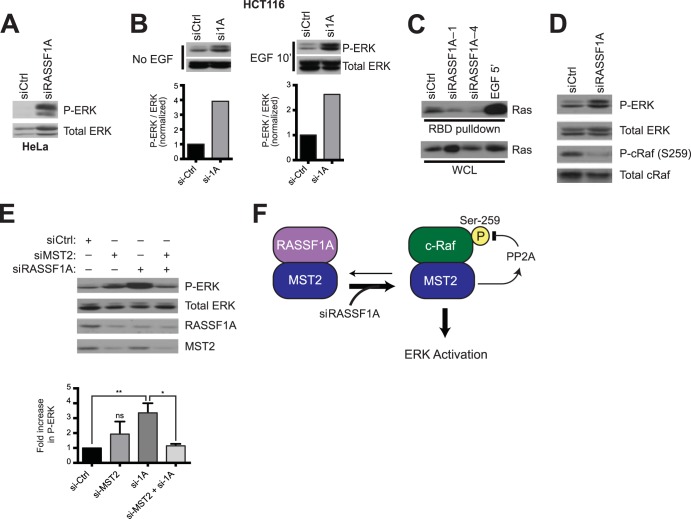
In the course of screening candidate oncogenic pathways reported to be inhibited by RASSF1A and induced by miR-21 (24, 25), we found that RASSF1A depletion resulted in significant elevation of baseline ERK1/2 activation and hyperresponsiveness of ERK1/2 to mitogenic signaling even in the background of oncogenic K-Ras (Fig. 4A and B). While this is consistent with miR-21 inhibition of Sprouty1/2 expression, thought to be a major factor mediating miR-21 oncogenic activity (25), we were unable to generate definitive evidence that ERK responsiveness to RASSF1A depletion was miR-21 dependent. RASSF1A depletion-induced ERK activation was associated with inhibition of Ras-GTP loading, suggesting the pathway is engaged downstream of Ras activation (Fig. 4C). Notably, the RASSF1A-interacting protein MST2 also interacts with Raf1 complexes (26). When associated with Raf1, MST2 participates in ERK pathway activation by stabilization of the PP2A phosphatase with consequent reversal of inhibitory Raf1 S259 phosphorylation (27). It was reported that disrupting Raf1 and MST2 complexes increased the interaction between MST2 and RASSF1A (28). Therefore, we asked if the opposite could be true, by which blocking RASSF1A and MST2 complex formation could promote a Raf1/MST2 interaction to enhance ERK activation. Indeed, RASSF1A depletion decreased levels of inhibitory phospho-cRaf (S259) (Fig. 4D) and increased activating phosphorylation of ERK in an MST2-dependent manner (Fig. 4E). This suggests the presence of latent mitogenic pathway activation upon RASSf1A loss (Fig. 4F) that is restrained due to collateral inhibition of SKP2 accumulation and concomitant accumulation of the cell cycle inhibitors p21 and p27 (19).
FIG 4.
MST2 mediates RASSF1A suppression of ERK activation. (A) HeLa cells were transfected with the indicated siRNA and immunoblotted for detection of phosphorylated ERK1/2 and total ERK1/2 as indicated. (B) HCT116 cells were transfected with the indicated siRNAs and stimulated with epidermal growth factor (EGF) for 10 min prior to lysis. Lysates were immunoblotted for detection of the indicated proteins (top). Under each condition (no EGF and EGF 10′), exposures were optimized for visualization of phospho-ERK within the linear range of detection. Quantification of P-ERK normalized to total ERK is indicated in the bottom panels. (C) Relative Ras-GTP accumulation was detected using affinity capture with Raf1-RBD-GST beads. Affinity-purified Ras (top) and total Ras (bottom) were detected with pan-Ras antibodies. Cells were stimulated with EGF for 5 min, as indicated, as a positive control. (D) HeLa cells were transfected with the indicated siRNAs and immunoblotted for detection of the indicated proteins and phosphor-proteins. (E) As described for panel D. The bar graph indicates relative accumulation of phospho-ERK normalized to total ERK ± SEM. **, P < 0.01; *, P < 0.05; ns, not significant, Student's t test. (F) Model of MST2 interaction with RASSF1A and c-Raf.
Context-specific modulation of cell cycle progression by the RASSF1A/SCFβTrCP/REST/miR-21 cascade.
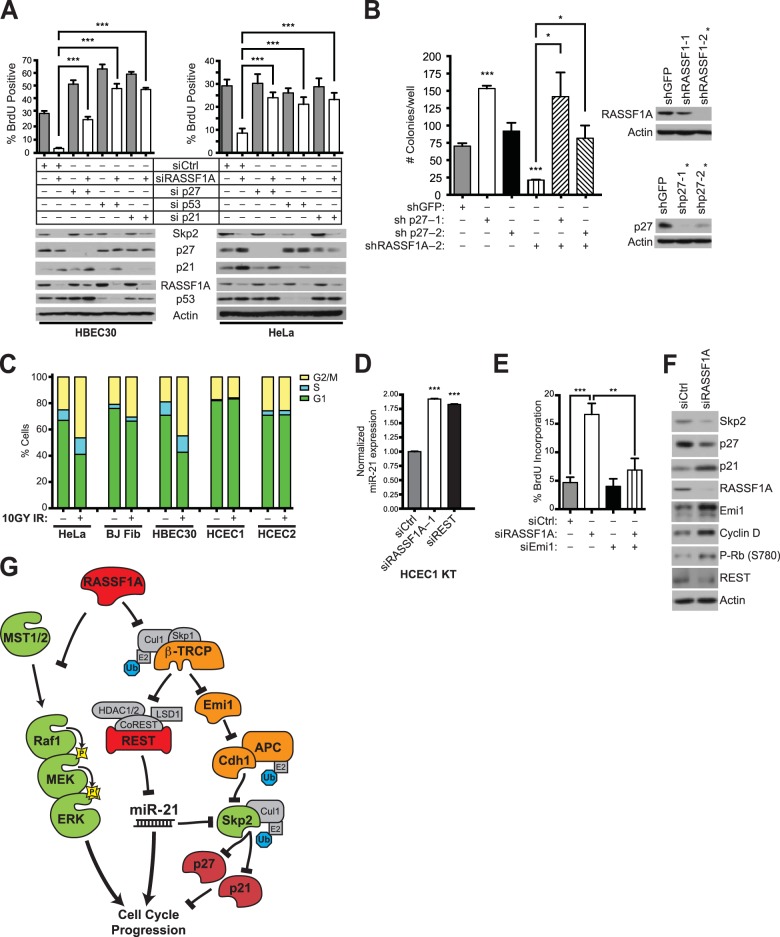
The coupling of cyclin-dependent kinase (CDK) inhibitor accumulation to RASSF1A loss may represent a fragile checkpoint against the otherwise tumorigenic consequences of REST degradation and miR-21 activation. Therefore, we asked if p27 and/or p21 accumulation was required for inhibition of cell cycle progression upon inhibition of RASSF1A expression. Consistent with previous work (19), RASSF1A depletion inhibited proliferation of telomerase-immortalized bronchiole epithelial cells. However, depletion of p27, p21, or the p53 tumor suppressor released additional proliferative potential that was largely independent of RASSF1A expression (Fig. 5A). Similarly, shRNA-mediated persistent suppression of RASSF1A expression in HeLa cells impaired soft-agar colony formation, which was rescued by codepletion of p27 (Fig. 5B).
FIG 5.
Context-specific stabilization of EMI1 determines the response of cell cycle progression to RASSF1A inactivation. (A) HBEC30 and HeLa cells were transfected with the indicated siRNAs. BrdU was added 5 h (HeLa) and 20 h (HBEC30) prior to fixation (top). Bars indicate mean BrdU incorporation ± SEM. ***, P < 0.001, Student's t test. Whole-cell lysates from parallel transfections were collected and immunoblotted for detection of the indicated proteins (bottom). (B) HeLa cells were infected with the indicated shRNAs and suspended in soft agar for 2 weeks. Bars indicate mean colonies per well ± SEM. ***, P < 0.001; *, P < 0.05, Student's t test. Lysates from parallel transfections were collected to assess knockdown efficiency by immunoblotting (right). The asterisk indicates shRNAs used for soft agar colony formation assay. (C) The indicated cell lines were treated (+IR) or not treated (−IR) with ionizing radiation (10 gy). After 8 h, cells were collected and stained with propidium iodide for FACS analysis of DNA content. Bars indicate cell population distributions in G1, S, and G2 phases or mitosis. (D) Telomerase-immortalized human colonic epithelial cells (HCEC1 KT) were transfected with the indicated siRNAs, and miR-21 expression was measured by qPCR. ***, P < 0.001, Student's t test. (E) HCECs were transfected with indicated siRNAs, and BrdU was added for 5 h prior to harvest. Bars indicate mean BrdU incorporation ± SEM. ***, P < 0.001, Student's t test. (F) HCEC lysates were collected following transfected with the indicated siRNAs and immunoblotted for detection of the indicated proteins. (G) Summary: RASSF1A restrains opposing cell cycle regulatory cascades.
These cumulative observations led us to consider the consequence of RASSF1A inactivation in cell biological contexts associated with tumor initiation, such as tissue progenitor cells which tend to possess a shorter G1 and may have weaker G1 checkpoints than lineage-restricted cells (29, 30). We therefore examined colonic crypt-derived human epithelial cells (HCECs) that express stem cell markers and display multilineage differentiation capability in culture (31). Notably, these cells are resistant to ionizing radiation-induced cell cycle arrest at doses that produce significant responses in HeLa cells, primary human fibroblasts, and HBEC30 (Fig. 5C). As in other RASSF1A-positive cell lines examined, HCECs respond to RASSF1A depletion and REST depletion by upregulation of miR-21 expression (Fig. 5D). However, in marked contrast to all other RASSF1A-positive cell lines examined (HeLa, HBEC, and MCF10A) (19), HCECs respond to RASSF1A depletion with greatly enhanced proliferative potential as measured by frequency of BrdU incorporation (Fig. 5E). Immunoblotting of whole-cell lysates indicated a discriminating feature was the resistance of EMI1 accumulation to RASSF1A loss, which correlated with SKP2 stability and the absence of p27 induction (Fig. 5F). The mechanistic relevance of this was indicated by the reversal of enhanced proliferation by codepletion of EMI1 together with RASSF1A (Fig. 5E). These cumulative observations suggest that the nature of the cell of origin and/or collaborating lesions in cell cycle checkpoint machinery can have significant consequences on the tumorigenic potential associated with RASSF1A inactivation.
DISCUSSION
Epigenetic inactivation of RASSF1A expression has been observed in nonmalignant neoplastic tissue associated with renal cell carcinoma (32), breast cancer (33), melanoma (24), and testicular germ cell tumors (34). This suggests that RASSF1A loss may be an early event in oncogenesis. In apparent contrast, shRNA-mediated RASSF1 depletion decreased proliferation of oncogenic Ras-transformed cells and reduced tumor burden when these cells were transplanted into mice (35). Similarly, independent studies have reported that siRNA-mediated selective depletion of the RASSF1A isoform impaired cell cycle progression in nontumorigenic epithelium-derived cell cultures (24). Here, we find that the cell autonomous response to RASSF1A depletion is directed by the presence or absence of SCFβTrCP-dependent destruction of the APC/C inhibitor EMI1. This relationship was most evident in the contrasting responses of lung and colonic epithelial cell behavior. We suggest that the context-dependent consequence of RASSF1A inactivation is an iteration of the mechanistic coupling of tumor suppressors to checkpoint controls that has been elaborated for proteins like VHL and PTEN. Mutations in VHL cause renal cell carcinoma, but VHL inactivation in primary fibroblasts results in p53 and HIF-independent senescence (36). This correlated with reduced expression of SKP2 and concomitant accumulation of p27 and Rb hypophosphorylation (36). Similarly, loss of PTEN can result in both p53-dependent and p53-independent cell cycle exit (37). Like RASSF1A inactivation, the latter is coupled to SKP2 and is p27 dependent (38). Similar to VHL and PTEN, the capacity of RASSF1A loss to promote oncogenic transformation is likely strongly linked to either the nature of the tumor cell of origin or the presence of collaborating lesions in cell cycle checkpoint control machinery.
RASSF1A support of SKP2 accumulation through both enhanced translation and protein stability may serve as a component of mechanisms that harness cell cycle progression to appropriate mitogenic signals. However, RASSF1A inactivation also appears to unleash substantial proliferative potential due to a cascading deregulation of tumor suppressor and oncogenic signaling networks. Evidence presented here suggests at least two major mechanisms account for this: MST2-dependent Raf kinase activation and SCFβTrCP-dependent destruction of the REST tumor suppressor. The former suggests RASSF1A concentrations may normally limit Raf kinase activation by competitive sequestration of MST2 protein, which would otherwise recruit PP2A to reverse inhibitory Raf1S259 phosphorylation. This implies that regulation of RASSF1A concentration is a mechanism to modulate the available pool of RAF1 that is responsive to upstream activators. Epigenetic inactivation of RASSF1 expression would ablate this regulation, sensitizing the Raf/Map kinase pathway for maximal activation by instructive signals.
Loss of RASSF1A restraint of SCFβTrCP had the unexpected consequence of inducing miR-21 expression. This is notable given the pervasive contribution of miR-21 to tumorigenic transformation (39, 40). The latter has stimulated much interest in development of strategies for miR-21 inhibition as a therapeutic intervention (25, 39). The observation that baseline miR-21 expression, in nontumorigenic cells, directly suppresses SKP2 translation suggests that such strategies should be considered with caution, as they could have unintended consequences on cell cycle checkpoint control. This is illustrated by our observation that SKP2 protein accumulation, upon treatment of cells with miR-21 inhibitors, destabilizes p27, which in turn enhances the proliferative capacity of epithelial cells in culture. The recent development of chemical inhibitors of SKP2 (41, 42) may provide an effective combinatorial intervention strategy if these mechanistic relationships are conserved in the tumor microenvironment.
ACKNOWLEDGMENTS
We thank J. Shay, W. Wright, and J. Minna for some of the cell lines used in these studies.
This work was supported by grants from the NCI (CA71443) and the Welch Foundation (I-1414). R.R.R. received support from T32-GM008203.
Footnotes
Published ahead of print 14 April 2014
REFERENCES
- 1.Donninger H, Vos MD, Clark GJ. 2007. The RASSF1A tumor suppressor. J. Cell Sci. 120:3163–3172. 10.1242/jcs.010389 [DOI] [PubMed] [Google Scholar]
- 2.Pan ZG, Kashuba VI, Liu XQ, Shao JY, Zhang RH, Jiang JH, Guo C, Zabarovsky E, Ernberg I, Zeng YX. 2005. High frequency somatic mutations in RASSF1A in nasopharyngeal carcinoma. Cancer Biol. Ther. 4:1116–1122. 10.4161/cbt.4.10.2023 [DOI] [PubMed] [Google Scholar]
- 3.Agathanggelou A, Cooper WN, Latif F. 2005. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 65:3497–3508. 10.1158/0008-5472.CAN-04-4088 [DOI] [PubMed] [Google Scholar]
- 4.Chan MW, Chan LW, Tang NL, Lo KW, Tong JH, Chan AW, Cheung HY, Wong WS, Chan PS, Lai FM, To KF. 2003. Frequent hypermethylation of promoter region of RASSF1A in tumor tissues and voided urine of urinary bladder cancer patients. Int. J. Cancer 104:611–616. 10.1002/ijc.10971 [DOI] [PubMed] [Google Scholar]
- 5.Tomizawa Y, Kohno T, Kondo H, Otsuka A, Nishioka M, Niki T, Yamada T, Maeshima A, Yoshimura K, Saito R, Minna JD, Yokota J. 2002. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin. Cancer Res. 8:2362–2368 [PubMed] [Google Scholar]
- 6.Yu MY, Tong JH, Chan PK, Lee TL, Chan MW, Chan AW, Lo KW, To KF. 2003. Hypermethylation of the tumor suppressor gene RASSFIA and frequent concomitant loss of heterozygosity at 3p21 in cervical cancers. Int. J. Cancer 105:204–209. 10.1002/ijc.11051 [DOI] [PubMed] [Google Scholar]
- 7.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J. Natl. Cancer Inst. 93:691–699. 10.1093/jnci/93.9.691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. 2005. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 65:92–98 [PubMed] [Google Scholar]
- 9.van der Weyden L, Tachibana KK, Gonzalez MA, Adams DJ, Ng BL, Petty R, Venkitaraman AR, Arends MJ, Bradley A. 2005. The RASSF1A isoform of RASSF1 promotes microtubule stability and suppresses tumorigenesis. Mol. Cell. Biol. 25:8356–8367. 10.1128/MCB.25.18.8356-8367.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 25:315–319. 10.1038/77083 [DOI] [PubMed] [Google Scholar]
- 11.Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER. 2001. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 98:7504–7509. 10.1073/pnas.131216298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzmin I, Gillespie JW, Protopopov A, Geil L, Dreijerink K, Yang Y, Vocke CD, Duh FM, Zabarovsky E, Minna JD, Rhim JS, Emmert-Buck MR, Linehan WM, Lerman MI. 2002. The RASSF1A tumor suppressor gene is inactivated in prostate tumors and suppresses growth of prostate carcinoma cells. Cancer Res. 62:3498–3502 [PubMed] [Google Scholar]
- 13.Richter AM, Pfeifer GP, Dammann RH. 2009. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim. Biophys. Acta 1796:114–128. 10.1016/j.bbcan.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 14.Eskiocak U, Kim SB, Ly P, Roig AI, Biglione S, Komurov K, Cornelius C, Wright WE, White MA, Shay JW. 2011. Functional parsing of driver mutations in the colorectal cancer genome reveals numerous suppressors of anchorage-independent growth. Cancer Res. 71:4359–4365. 10.1158/0008-5472.CAN-11-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. 2002. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell. Biol. 22:4309–4318. 10.1128/MCB.22.12.4309-4318.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrabaud E, Lassot I, Blot G, Le Rouzic E, Tanchou V, Quemeneur E, Daviet L, Margottin-Goguet F, Benarous R. 2007. RASSF1C, an isoform of the tumor suppressor RASSF1A, promotes the accumulation of beta-catenin by interacting with betaTrCP. Cancer Res. 67:1054–1061. 10.1158/0008-5472.CAN-06-2530 [DOI] [PubMed] [Google Scholar]
- 17.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. 2008. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453:223–227. 10.1038/nature06863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, Brekken R, Wurz R, Tasker A, Polverino T, Tan SL, White MA. 2011. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell 41:458–470. 10.1016/j.molcel.2011.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehurst AW, Ram R, Shivakumar L, Gao B, Minna JD, White MA. 2008. The RASSF1A tumor suppressor restrains anaphase-promoting complex/cyclosome activity during the G1/S phase transition to promote cell cycle progression in human epithelial cells. Mol. Cell. Biol. 28:3190–3197. 10.1128/MCB.02291-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frescas D, Pagano M. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8:438–449. 10.1038/nrc2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ. 2005. A genetic screen for candidate tumor suppressors identifies REST. Cell 121:837–848. 10.1016/j.cell.2005.03.033 [DOI] [PubMed] [Google Scholar]
- 22.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ. 2008. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452:370–374. 10.1038/nature06780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. 2010. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 29:5828–5838. 10.1038/onc.2010.321 [DOI] [PubMed] [Google Scholar]
- 24.Calipel A, Abonnet V, Nicole O, Mascarelli F, Coupland SE, Damato B, Mouriaux F. 2011. Status of RASSF1A in uveal melanocytes and melanoma cells. Mol. Cancer Res. 9:1187–1198. 10.1158/1541-7786.MCR-10-0437 [DOI] [PubMed] [Google Scholar]
- 25.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. 2010. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 18:282–293. 10.1016/j.ccr.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill E, Rushworth L, Baccarini M, Kolch W. 2004. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 306:2267–2270. 10.1126/science.1103233 [DOI] [PubMed] [Google Scholar]
- 27.Kilili GK, Kyriakis JM. 2010. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J. Biol. Chem. 285:15076–15087. 10.1074/jbc.M109.078915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'Neill E. 2007. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell 27:962–975. 10.1016/j.molcel.2007.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. 2006. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell Physiol. 209:883–893. 10.1002/jcp.20776 [DOI] [PubMed] [Google Scholar]
- 30.Hong Y, Stambrook PJ. 2004. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 101:14443–14448. 10.1073/pnas.0401346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roig AI, Eskiocak U, Hight SK, Kim SB, Delgado O, Souza RF, Spechler SJ, Wright WE, Shay JW. 2010. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology 138:1012–1021. 10.1053/j.gastro.2009.11.052 [DOI] [PubMed] [Google Scholar]
- 32.Peters I, Rehmet K, Wilke N, Kuczyk MA, Hennenlotter J, Eilers T, Machtens S, Jonas U, Serth J. 2007. RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis. Mol. Cancer 6:49. 10.1186/1476-4598-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo W, Wong WL, Wong N, Law BK, Tse GM, Zhong S. 2005. High frequency of promoter hypermethylation of RASSF1A in tumorous and non-tumourous tissue of breast cancer. Pathology 37:125–130. 10.1080/00313020500058623 [DOI] [PubMed] [Google Scholar]
- 34.Honorio S, Agathanggelou A, Wernert N, Rothe M, Maher ER, Latif F. 2003. Frequent epigenetic inactivation of the RASSF1A tumour suppressor gene in testicular tumours and distinct methylation profiles of seminoma and nonseminoma testicular germ cell tumours. Oncogene 22:461–466. 10.1038/sj.onc.1206119 [DOI] [PubMed] [Google Scholar]
- 35.Vicent S, Chen R, Sayles LC, Lin C, Walker RG, Gillespie AK, Subramanian A, Hinkle G, Yang X, Saif S, Root DE, Huff V, Hahn WC, Sweet-Cordero EA. 2010. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J. Clin. Invest. 120:3940–3952. 10.1172/JCI44165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young AP, Schlisio S, Minamishima YA, Zhang Q, Li L, Grisanzio C, Signoretti S, Kaelin WG., Jr 2008. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat. Cell Biol. 10:361–369. 10.1038/ncb1699 [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:725–730. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. 2010. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature 464:374–379. 10.1038/nature08815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X, Wang ZX, Wang R. 2010. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 10:1224–1232. 10.4161/cbt.10.12.14252 [DOI] [PubMed] [Google Scholar]
- 40.Selcuklu SD, Donoghue MT, Spillane C. 2009. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 37:918–925. 10.1042/BST0370918 [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. 2012. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem. Biol. 19:1515–1524. 10.1016/j.chembiol.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, Logothetis CJ, Hung MC, Zhang S, Lin HK. 2013. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 154:556–568. 10.1016/j.cell.2013.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]