Abstract
The gene gating hypothesis put forth by Blobel in 1985 was an alluring proposal outlining functions for the nuclear pore complex (NPC) in transcription and nuclear architecture. Over the past several decades, collective studies have unveiled a full catalog of nucleoporins (Nups) that comprise the NPC, structural arrangements of Nups in the nuclear pore, and mechanisms of nucleocytoplasmic transport. With this foundation, investigations of the gene gating hypothesis have now become possible. Studies of several model organisms provide credence for Nup functions in transcription, mRNA export, and genome organization. Surprisingly, Nups are not only involved in transcriptional events that occur at the nuclear periphery, but there are also novel roles for dynamic Nups within the nucleoplasmic compartment. Several tenants of the original gene gating hypothesis have yet to be addressed. Knowledge of whether the NPC impacts the organization of the genome to control subsets of genes is limited, and the cooperating molecular machinery or specific genomic anchoring sequences are not fully resolved. This minireview summarizes the current evidence for gene gating in Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and mammalian model systems. These examples highlight new and unpredicted mechanisms for Nup impacts on transcription and questions that are left to be explored.
INTRODUCTION
It is often said that all good things take time. Early observations of eukaryotic nuclear structure sparked a long-standing curiosity regarding the mechanisms that coordinate three-dimensional genome organization and potentially impact gene expression. In 1985, a provocative “gene gating hypothesis” was put forth by Blobel (1). Key tenets focused on specific interactions between nuclear pore complexes (NPCs) and transcriptionally active gene loci, with the location of NPCs throughout the nuclear envelope reflecting the organization of active versus inactive chromatin (1). Such NPC-gene links were further speculated as inherent to directing the export of transcripts from a given gene through a given NPC. Furthermore, linking precise genomic regions to the NPC was suggested to impact the transcriptional regulation and targeting of subsets of genes to NPCs during development and in the establishment of cell polarity. Indeed, studies of chromatin organization identified interchromatin compartments (ICs), or nuclear subcompartments, where proteins necessary for RNA transcription and splicing along with the DNA replication and repair machinery reside (2, 3). These ICs were speculated to act as tracks to direct mRNAs transcribed within the nuclear interior to select NPCs. However, in the late 1980s, the protein composition of the NPC was only beginning to be dissected. Armed now with both the identification of NPC proteins (nucleoporins [Nups]) and technological innovations in microscopy and molecular biology, evidence for several aspects of the gene gating hypothesis has accumulated.
This minireview highlights recent progress made in testing the gene gating hypothesis as gained from the Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and mammalian model systems. Work with S. cerevisiae has yielded important evidence for NPC interactions with the genome as well as identifying additional factors that bridge and establish these interactions. Several aspects are conserved in metazoan systems, although distinct mechanisms have arisen likely due to the increasing complexity of the genome as well as the transcriptional programs required during development and differentiation. Overall, there is now a deep appreciation of functional roles for Nups in transcription and exciting opportunities for future discoveries.
DEFINING THE NPC COMPOSITION AND ROLES IN RNA EXPORT
Direct analysis of NPC roles in gene gating inherently required first revealing the catalog of Nup composition in different model organisms that would also allow robust transcriptional analysis. In addition, critical information has come from understanding the functional roles for Nups in nuclear import and export and the substructural location of Nups in the NPC (4). Embedded in nuclear envelope pores, the 60- to 100-MDa NPCs are assembled from ∼30 different Nups whose identities were revealed through combined biochemical and genetic strategies. A subset of the Nups are specifically pore membrane anchored and termed “Poms,” whereas the majority are soluble peripheral proteins. Based on the NPC apparent 8-fold rotational symmetry, each Nup is present in at least eight copies per NPC (5–9). However, some are exclusive to the cytoplasmic side of the NPC and contribute to formation of cytoplasmic filaments. Others reside only on the nuclear NPC face and form basket-like structures. In general, the majority of the Nups show structural conservation across species with noted exceptions among the Poms and in the asymmetrically localized cytoplasmic and nuclear Nups (10). In terms of gene gating, the distinct Nups in the nuclear basket are positioned for connections to intranuclear structures, transcriptional machinery, and chromatin. Moreover, analysis of Nup dynamics in living cells finds that some Nups transiently associate with the nuclear-envelope-embedded complex and thus also have potential intranuclear residence (11, 12).
The clearest link for NPCs in facilitating gene expression is via essential roles in RNA export. Transport from the nucleus to the cytoplasm is critical for all RNAs, including tRNA and pre-microRNA (pre-miRNA) molecules as well as RNA-protein (RNP) complexes for mRNA and the large- and small-ribosomal-subunit-associated RNAs (4, 13). In regard to gene gating, the factors mediating mRNA transcription and messenger RNP (mRNP) assembly are of prime importance (14–17). During the nuclear synthesis and maturation process, mRNP protein composition undergoes changes with association and subsequent dissociation of proteins that mediate pre-mRNA processing and the ultimate recruitment of export receptors. Quality control mechanisms target incompletely processed and incorrectly assembled mRNPs to the nuclear exosome for decay (18, 19). Association of the export receptor(s) in the mRNP with specific Nups is the key determinant for NPC targeting. A series of molecular events are required for facilitating mRNP translocation through NPCs and executing directional release of mRNPs into the cytoplasm. Detailed reviews have been published recently outlining the current understanding of these mechanisms (4, 20–22). However, additional work has been required to resolve whether Nup export functions are distinct from or connected with roles for Nups in transcription and genome organization.
EVIDENCE OF TRANSCRIPTIONAL REGULATION BY Nups IN S. CEREVISIAE
The nuclear periphery is both a repressive and permissive environment for mRNA transcription. In S. cerevisiae, silenced regions of the genome, including the rDNA locus, centrosome, and telomeres, are anchored to the periphery through associations with inner-nuclear-envelope-associated proteins. Alternating between these silent regions are the NPCs that are predicted to demarcate active zones of gene expression. Interestingly, Nup2 in the nuclear basket and Prp20 (the guanine nucleotide exchange factor [GEF] for Ran in nuclear import/export) both exhibit boundary activity and prevent the spread of silenced heterochromatin (23, 24). These functions link to the NPC nuclear basket and suggest a role for the NPC in establishing or distinguishing between alternating zones of silent heterochromatin and transcriptionally active euchromatin.
NPCs associate with transcriptionally active regions through interactions with chromatin and components of the mRNA export machinery. Examples of inducible genes, including GAL1, GAL2, HXK1, INO1, TSA2, HSP104, SUC2, and MFA2, relocalize from the nucleoplasmic interior where they are inactive to the periphery for proper expression (25–34). The integrity and function of the NPC are required for maximal transcriptional activity of these inducible genes, and multiple Nups are necessary for their proper positioning to the nuclear periphery. The peripheral positioning of GAL1, INO1, TSA2, and HSP104 also requires DNA elements within their respective promoters (27, 35). Such gene recruitment sequences (GRSs) are necessary and also sufficient for peripheral nuclear positioning when placed at the ectopic URA3 locus (35). Peripheral recruitment of GRS-containing genes requires the Snf1p-dependent SAGA (Spt-Ada-Gcn5-acetyltransferase) complex and mRNA export factors, indicating that the NPC interactions occur as the promoter transitions to a transcriptionally active state (26, 28, 29, 32). Most surprisingly, identical GRSs found within the INO2 and TSA2 gene loci are positioned in the same site at the nuclear periphery, thus occupying the same gene territory (36). This gene clustering requires a specialized transcription factor (36) which might target these GRS-containing genes to a designated NPC at the nuclear periphery (37). Alternatively, Nups with DNA or RNA binding affinity could facilitate peripheral gene positioning (38, 39). In addition to GRS elements, memory recruitment sequences (MRSs) maintain positioning at the periphery for several hours after the gene is inactivated, and this peripheral positioning allows the gene to be reactivated with faster kinetics (40). The transcriptional memory of these MRS-containing genes specifically requires the histone variant H2A.Z and Nup100 (25, 40). This identification of DNA sequence elements that confer three-dimensional positioning in the nucleus provides evidence for NPC functional roles in genome organization.
Multiple cellular inputs likely contribute to the dynamics of peripheral recruitment and release. The positioning of the INO1 and GAL1 loci changes in S phase to localization away from the periphery in a mechanism involving Cdk1 phosphorylation of Nup1 (41). It is currently unclear how environmental cues signal and result in genome organization changes. Many of the gene-positioning studies have also examined only one or two gene loci at a time. Future experiments utilizing chromosome conformation capture (3C)-derived techniques will help to resolve whether specific gene expression programs alter global nuclear architecture and/or coordinate expression of coclustering genes (42). It is also intriguing that a few Nups have reported roles in gene silencing. For example, Nup170 is required for peripheral tethering and silencing of subtelomeric regions through cooperation with the RSC chromatin-remodeling complex and Sir4 (43). Thus, the NPC and Nups are potentially multifaceted regulators of transcription, effectively extending the tenets of gene gating. Together, these studies of S. cerevisiae have identified Nup transcriptional roles that are exclusive to the nuclear periphery. With the wealth of knowledge of this model system, the field is now poised to address further outstanding questions, including whether mRNPs show enhanced export through the NPC or distinct cytoplasmic fates when transcribed from genes that are targeted to the nuclear periphery.
GENE POSITIONING TO THE C. ELEGANS NPC
Nuclear architecture throughout development is highly dynamic; however, converging evidence suggests that these dynamics are nonrandom and correlate with important cell fate decisions. The C. elegans model is well suited for visualizing differentiating cells within a living organism and provides an ideal system to track gene positioning throughout development. To visualize the positioning of gene promoters in C. elegans, the most robust strategy available is the LacO array/GFP-LacI system. The LacO arrays (based on repeats of LacO from bacteria) are inserted adjacent to transgenes containing cell-type-specific promoters. This, in combination, with the expression of a green fluorescent protein (GFP)-LacI repressor allows for visualization of promoter location in the embryo and throughout larval stages in development. Importantly, for tracking genes with the LacO repeats inserted, small arrays are used so as to not introduce potential chromatin structure artifacts (e.g., the array acting as a silencer itself) (44), and results are further validated by fluorescence in situ hybridization (FISH). With the LacO system, both gut (pha-4)- and muscle (myo-3)-specific promoters are inactive and sequestered at the nuclear periphery throughout early development; however, the promoters are transcriptionally active and found in the nuclear interior in differentiated gut and muscle cells, respectively (45). Based on these results, the peripheral sequestration of genes determining cell fate might limit expression until the cell reaches the correct stage of development, during which the gene is repositioned to a transcriptionally active nucleoplasmic compartment. In a C. elegans model for Emery-Dreifus muscular dystrophy, the loss of nuclear organization specifically links to the muscular defects observed in the diseased animals (46). A reduction in muscle-specific gene expression correlates with the loss of interior nuclear localization of the myo-3 promoter in differentiated muscle cells. A recent genetic screening strategy has also identified histone H3 lysine 9 (H3K9) methylation as a key determinant for peripheral tethering of silenced heterochromatin at the nuclear envelope (47). Within this developmental context, it is unclear whether the NPC and its stably associated Nups contribute to gene positioning and silencing of peripheral heterochromatin regions; however, dynamic Nups might play important roles in the activation of developmental genes in the nucleoplasm.
Interestingly, the stress-induced hsp-16.2 gene shows distinct positioning patterns upon transcriptional activation. Small integrated LacO arrays containing the hsp-16.2 promoter normally reside at the periphery in regions lacking NPCs, and under conditions of heat shock, they reposition so that they directly associate with NPCs (48). Superresolution microscopy studies confirm that the NPC/hsp-16.2 association occurs at the periphery similar to the events described above for S. cerevisiae when inducible genes reposition to the periphery coincident with active transcription. Overall, peripheral targeting of inducible genes in S. cerevisiae and C. elegans has similar requirements for cis-promoter elements, transcription factors, and factors associated with the SAGA histone acetyltransferase and THO/TREX mRNA export complex (48). These shared determinants might represent conserved gene gating mechanisms for stress-responsive genes in eukaryotes. Overall, in C. elegans, two classes of genes appear to exhibit distinct patterns of gene positioning upon transcriptional activation. Stress-induced genes operate by mechanisms similar to induced genes in S. cerevisiae, whereas developmentally induced genes behave oppositely and migrate from the periphery to the interior. Thus, these distinct modes of gene regulation may potentially reveal distinct mechanisms for gene gating.
DROSOPHILA Nup ASSOCIATION WITH THE GENOME
Studies of D. melanogaster have provided convincing evidence for specific, enriched interactions between chromatin regions and Nups. Chromatin immunoprecipitation with microarray technology (ChIP-chip) experiments show genomic regions spanning 5 to 500 kb that interact with nuclear basket Nups Nup153 and Megator (49). These Nup-associated regions (NARs) are enriched across the X chromosome in male flies, which require high levels of transcription for dosage compensation. The NARs within the X chromosome are positioned to the periphery and show decreased expression and loss of peripheral localization upon depletion of Nup153. Though other NARs do not preferentially position to the periphery, the expression also decreases in the absence of Nup153. Additional studies of D. melanogaster report the dynamic association of Nup50, Nup62, Sec13, Nup98, and MAb414-positive phenylalanine-glycine (FG)-containing Nups with both developmental and stress-induced genes (50, 51). This association occurs in both the nucleoplasmic compartment and at the nuclear periphery. These results suggest new roles for Nups in the organization of the Drosophila genomic NARs. Importantly, the dynamic association of Nups with transcriptionally active chromatin occurs throughout the nucleoplasm irrespective of the steady-state positioning of Nups within the NPC at the nuclear periphery. In 1985, Nup dynamics at the NPC and the existence of soluble Nups within the nucleoplasm remained unexplored. As such, these new studies provide evidence that effectively extends the gene gating hypothesis to include novel functions for Nups in gene expression within the nucleoplasmic compartment. Future studies are needed to continue to dissect which subsets of Nups have direct roles in the initiation and elongation phases of transcription and which subsets are involved in establishing connections with the genome to channel mRNPs from sites of transcription to NPCs for export.
DYNAMIC MAMMALIAN Nups IMPACT GENE EXPRESSION
The original gene gating hypothesis proposed that the NPC participates in organizing chromatin into interchromatin compartments (ICs) which would provide channels between NPCs and sites of transcription (1). Multiple lines of evidence suggest that mRNPs exhibit a trajectory from the nucleoplasm to the nuclear periphery and diffuse through ICs (52–55). Most recently, single particle tracking of mRNPs from sites of transcription observed several mRNPs traveling the same pathway through the nucleoplasm (55). Evidence in mice and in human cells supports a role for the transmembrane Nup, Nup210, in the proper induction of genes required for myogenesis and neurogenesis (56). The role of Nup210 in the positioning and regulation of these developmentally expressed genes is yet to be fully examined, though possibly Nup210 or other nuclear-envelope/NPC-anchored Nups further influence chromatin structure to establish a path for newly synthesized mRNPs in channeling to the NPC.
Alternatively, soluble and dynamic Nups might participate directly in the synthesis of mRNPs at sites of transcription and influence the trafficking of an mRNP to the NPC. Similar to studies of C. elegans, nuclear positioning of several developmentally regulated genes shifts from peripheral to nucleoplasmic in human cell culture systems. This includes the Mash1 locus during neurogenesis, the GFAB locus during astrocyte differentiation, and the β-globin locus during erythroid maturation (57–59). Similarly in myogenesis, the MyoD gene remains at the nuclear periphery until the myoblast transitions into a myotube upon which MyoD shifts to the interior and colocalizes with the TATA box-binding protein-associated factor 3 (TAF3) transcription factor IID (TFIID) subunit necessary for full expression (60). These examples indicate that the active compartment for developmentally induced genes is not the periphery, as it is in S. cerevisiae. However, this does not exclude possible functions for Nups in gene gating. In fact, accumulating evidence has also identified transcriptional roles for Nup98 in the nucleoplasm (61–64). The dynamic shuttling of Nup98 between the NPC and nucleoplasm requires active transcription (64). Moreover, Nup98 aids in preventing oncogenesis by regulating mRNA and protein levels of the p21 tumor suppressor. The association of Nup98 with the 3′ untranslated region (3′UTR) of p21 mRNA protects p21 mRNA from degradation by the exosome (63). Nup98 also performs functions in transcriptional memory and contributes to faster reactivation of gamma interferon (IFN-γ)-induced genes (65). This mechanism of transcriptional memory is partially conserved between Nup98 and its S. cerevisiae ortholog Nup100. Similarly, Nup98 associates with the promoters of recently activated promoters and contributes to epigenetic changes in H3K4 dimethylation. However, unlike in S. cerevisiae where Nup100 transcriptional memory is associated with the peripheral localization of the INO1 gene (40), IFN-γ-induced genes position to the nucleoplasmic interior and likely require a dynamic pool of Nup98 (65). Continuing work is necessary to resolve the mechanisms by which the dynamic Nup98 influences transcription and expression of developmentally regulated and stress-induced genes.
CONCLUSIONS
From elegant work in diverse eukaryotic model organisms, the mechanisms for the proposed gene gating functions of the NPC are continuing to come to light (as summarized in Fig. 1). Roles have been identified for Nups in enhancing transcription of genes induced under environmental stress and during development. Inducible genes in S. cerevisiae are positioned to the nuclear periphery and associate with Nups; thus, studies thus far strongly support gene expression roles for Nups at the NPC in S. cerevisiae. Several examples of peripheral roles for Nups in C. elegans and Drosophila have also been reported. What was most unexpected in the 1980s is the now known presence of Nups not only at NPCs but also within the nucleoplasm. Remarkably, in metazoans, dynamic Nups in the nucleoplasm function in transcription. In addition to enhancing the initial activation of gene transcription, the human Nup98 and S. cerevisiae Nup100 share conserved roles in subsequent rounds of reactivation and provide memory for previous transcriptional events (65). These dynamic Nups with clear nucleoplasmic activities in transcription give an important additional twist to the original model.
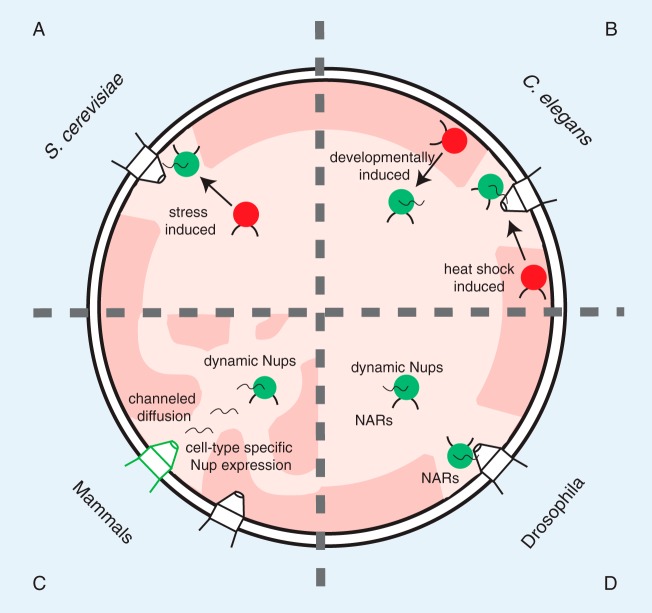
FIG 1.
Shared and distinct roles for Nups in gene gating across different model systems. (A) In S. cerevisiae, Nups are required to recruit multiple genes to the nuclear periphery for optimal gene expression. This mechanism requires specific DNA sequences in the promoters termed DNA zip codes. (B) The developmentally induced myo-3 and pha-4 promoters in C. elegans are positioned in the nucleoplasmic interior when transcriptionally active, whereas the heat shock-induced promoter hsp-16.2 is juxtaposed to the NPC, implying distinct gene gating mechanisms for different contexts of gene expression. (C) Drosophila Nups, including Nup98, Sec13, Nup50, and Nup62, impact transcriptional events in nucleoplasm and at the NPC. NARs are defined regions of the genome that interact with the Nup153 and Megator Nups. (D) The dynamic Nup98 regulates transcriptional memory of gamma interferon (IFN-γ)-induced genes in human cells. In mice, cell type-specific expression of the NE-anchored Nup210 regulates gene expression during myogenesis. Channeled diffusion of mRNPs from the nuclear interior to NPCs occurs through interchromatin compartments. Active genes (green circles), inactive genes (red circles), euchromatin and interchromatin compartments (light pink), and heterochromatin (dark pink) are indicated. The figure is not drawn to scale.
Blobel's gene gating hypothesis envisioned “a circumscribed space subjacent to the nuclear pore complex and extending into the interior of the nucleus in the form of channels” to serve as a transcriptional and posttranscriptional compartment of the nucleus (1). As noted above, single mRNP tracking from sites of transcription to the NPC indicates that the mRNPs travel by channeled diffusion through the interchromatin space. Whether Nups or additional factors influence the particular route of travel to select NPCs or whether all transcripts from a given gene target the same NPC remains unknown.
In electron microscopy images of vertebrate cell nuclei, the NPC is visibly associated with regions of active chromatin at the nuclear periphery, whereas inactive heterochromatin occupies the non-NPC regions (Fig. 1). A NPC-associated protein Tpr plays roles in establishing the heterochromatin exclusion zones at NPCs (66). In S. cerevisiae and C. elegans, the localization of an active gene to the periphery suggests that the gene is positioned to a nuclear envelope (NE) region affiliated with NPCs, whereas an inactive gene at the periphery is proposed to associate with silenced heterochromatin regions at the NE (or C. elegans nuclear lamina). These conclusions remain controversial, and additional work is required to test whether gene positions that correlate with NE regions (NPC and lamina) are functionally important. New superresolution microscopy methods and approaches to biochemically probe DNA-Nup interactions that occur exclusively at the NPC are now available to determine whether this partitioning of the nuclear periphery exists. Ultimately, these studies will aid in the further understanding of the mechanisms required to establish and maintain regions of active chromatin at the nuclear periphery.
Several implications of the gene gating hypothesis still extend beyond our current understanding, and there are potential paradigms to be further analyzed for NPC regulation of gene expression. One proposed function of the NPC is in regulating cell-specific three-dimensional organizations of the genome. As discussed above, cell-specific expression of Nups contributes to differentiation of muscle cells (56). It is possible that distinct Nups bind distinct regions of the genome and participate in specification of additional cell types. Technological advances in global 3C methods and single-cell approaches to track individual genes can now begin to test which Nups influence genomic arrangements required to specify cell fate (67–69). It will also be important to tease out a detailed map of Nup-chromatin associations and whether gene recruitment sequences (GRSs) identified in S. cerevisiae exist in metazoans. If so, a key goal will be to define the factors that bridge the NPC-chromatin interactions and understand how these interactions are affected by modifications to chromatin and/or cellular signaling events.
Tremendous attention has been paid to mRNA gene gating. It is well documented that tRNA and pre-miRNA nuclear processing steps are coupled to export (13, 70). Moreover, the clustering of rDNA loci and ribosomal biogenesis in the nucleolus is a hallmark of genome organization. In S. cerevisiae, the nucleolus is juxtaposed to a region of the nuclear envelope wherein the NPCs lack the Mlp1 and Mlp2 Nups (71). Given the roles of Mlp1 and Mlp2 in monitoring mRNA export (72), this has suggested that some NPCs are specialized for mRNA export, whereas others are committed to ribosome/rRNA export. The field is positioned to investigate whether these other classes of exporting RNA also obey the same gene gating mechanisms.
Finally, another postulated function for the NPC is in gating mRNAs to specific sites within the cytoplasm to promote cellular asymmetry. Both cis and trans elements are known to target mRNPs to distinct cellular locales, but whether NPCs directly influence cytoplasmic mRNA localization is unknown. The single-celled alga Chlamydomonas reinhardtii provides an extreme example for NPC positioning dictating cellular locale of a transcript during flagellar generation (73). The C. reinhardtii NPCs are asymmetrically positioned when transcription required for flagellar biogenesis is dramatically upregulated. Export through these NPCs effectively results in the localization of β2-tubulin mRNA to cellular compartments enriched in polysomes (73). In S. cerevisiae, the nuclear basket Nup60 is required for the export and proper localization of ASH1 mRNA to the bud tip (74). If select Nups are required for the export of differentially localized mRNPs, then the composition of NPC may dictate whether a transcript is targeted to a given cytoplasmic compartment. NPCs may additionally determine whether transcripts are rapidly translated, stored for later translation, or degraded. Further analysis of single transcripts is needed to determine whether NPC composition distinguishes which “gate” a given transcript exits though and its resulting cytoplasmic fate. If so, via gene gating, the NPC and Nups will be involved in both nuclear and cytoplasmic events that encompass the full cycle of gene expression.
ACKNOWLEDGMENTS
We thank members of the Wente laboratory for critical discussions.
This work was funded by the NIH (R37GM51219 to S.R.W.).
Footnotes
Published ahead of print 10 March 2014
REFERENCES
- 1.Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. U. S. A. 82:8527–8529. 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. 1993. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1:93–106. 10.1007/BF00710032 [DOI] [PubMed] [Google Scholar]
- 3.Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, Scherthan H, Ried T, Cremer C, Lichter P. 1993. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp. Quant. Biol. 58:777–792. 10.1101/SQB.1993.058.01.085 [DOI] [PubMed] [Google Scholar]
- 4.Aitchison JD, Rout MP. 2012. The yeast nuclear pore complex and transport through it. Genetics 190:855–883. 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg MW, Allen TD. 1992. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 119:1429–1440. 10.1083/jcb.119.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. 2007. The molecular architecture of the nuclear pore complex. Nature 450:695–701. 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- 8.Bilokapic S, Schwartz TU. 2012. 3D ultrastructure of the nuclear pore complex. Curr. Opin. Cell Biol. 24:86–91. 10.1016/j.ceb.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Martinez J, Rout MP. 2012. A jumbo problem: mapping the structure and functions of the nuclear pore complex. Curr. Opin. Cell Biol. 24:92–99. 10.1016/j.ceb.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothballer A, Kutay U. 2012. SnapShot: the nuclear envelope II. Cell 150:1084–1084.e1081. 10.1016/j.cell.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 11.Rabut G, Doye V, Ellenberg J. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- 12.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. 10.1083/jcb.153.7.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- 14.Natalizio BJ, Wente SR. 2013. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 23:365–373. 10.1016/j.tcb.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Navarro S, Hurt E. 2011. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 23:302–309. 10.1016/j.ceb.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Babour A, Dargemont C, Stutz F. 2012. Ubiquitin and assembly of export competent mRNP. Biochim. Biophys. Acta 1819:521–530. 10.1016/j.bbagrm.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 17.Muller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14:275–287. 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- 18.Tutucci E, Stutz F. 2011. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 12:377–384. 10.1038/nrm3119 [DOI] [PubMed] [Google Scholar]
- 19.Schmid M, Jensen TH. 2013. Transcription-associated quality control of mRNP. Biochim. Biophys. Acta 1829:158–168. 10.1016/j.bbagrm.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 20.Folkmann AW, Noble KN, Cole CN, Wente SR. 2011. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus 2:540–548. 10.4161/nucl.2.6.17881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valkov E, Dean JC, Jani D, Kuhlmann SI, Stewart M. 2012. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim. Biophys. Acta 1819:578–592. 10.1016/j.bbagrm.2012.02.017 [DOI] [PubMed] [Google Scholar]
- 22.Nino CA, Herissant L, Babour A, Dargemont C. 2013. mRNA nuclear export in yeast. Chem. Rev. 113:8523–8545. 10.1021/cr400002g [DOI] [PubMed] [Google Scholar]
- 23.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171:955–965. 10.1083/jcb.200509061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109:551–562. 10.1016/S0092-8674(02)00756-0 [DOI] [PubMed] [Google Scholar]
- 25.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774–778. 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- 27.Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441:770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- 29.Dieppois G, Iglesias N, Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870. 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427–439. 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- 31.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21:379–391. 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- 32.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049. 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- 33.Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M. 2006. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 25:4253–4262. 10.1038/sj.emboj.7601305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. 2007. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 175:1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118. 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell 22:1234–1246. 10.1016/j.devcel.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns LT, Wente SR. 2012. Nuclear GPS for interchromosomal clustering. Dev. Cell 22:1119–1120. 10.1016/j.devcel.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo HS, Blus BJ, Jankovic NZ, Blobel G. 2013. Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc. Natl. Acad. Sci. U. S. A. 110:16450–16455. 10.1073/pnas.1316607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball JR, Dimaano C, Bilak A, Kurchan E, Zundel MT, Ullman KS. 2007. Sequence preference in RNA recognition by the nucleoporin Nup153. J. Biol. Chem. 282:8734–8740. 10.1074/jbc.M608477200 [DOI] [PubMed] [Google Scholar]
- 40.Light WH, Brickner DG, Brand VR, Brickner JH. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 40:112–125. 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brickner DG, Brickner JH. 2010. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol. Biol. Cell 21:3421–3432. 10.1091/mbc.E10-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hakim O, Misteli T. 2012. SnapShot: chromosome confirmation capture. Cell 148:1068.e1-2. 10.1016/j.cell.2012.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152:969–983. 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubarry M, Loiodice I, Chen CL, Thermes C, Taddei A. 2011. Tight protein-DNA interactions favor gene silencing. Genes Dev. 25:1365–1370. 10.1101/gad.611011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. 2010. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24:766–782. 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. 2011. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr. Biol. 21:1603–1614. 10.1016/j.cub.2011.08.030 [DOI] [PubMed] [Google Scholar]
- 47.Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150:934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- 48.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. 2013. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J. Cell Biol. 200:589–604. 10.1083/jcb.201207024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6:e1000846. 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140:360–371. 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 51.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140:372–383. 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence JB, Singer RH, Marselle LM. 1989. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell 57:493–502. 10.1016/0092-8674(89)90924-0 [DOI] [PubMed] [Google Scholar]
- 53.Huang S, Spector DL. 1991. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 5:2288–2302. 10.1101/gad.5.12a.2288 [DOI] [PubMed] [Google Scholar]
- 54.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. 2005. Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 102:17008–17013. 10.1073/pnas.0505580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12:543–552. 10.1038/ncb2056 [DOI] [PubMed] [Google Scholar]
- 56.D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. 2012. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 22:446–458. 10.1016/j.devcel.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, Fisher AG. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132–140. 10.1242/jcs.02727 [DOI] [PubMed] [Google Scholar]
- 58.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447–1457. 10.1101/gad.1419506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. 2008. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 22:489–498. 10.1101/gad.1634608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao J, Fetter RD, Hu P, Betzig E, Tjian R. 2011. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 25:569–580. 10.1101/gad.2021411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. 2013. Dynamic association of NUP98 with the human genome. PLoS Genet. 9:e1003308. 10.1371/journal.pgen.1003308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatel G, Desai SH, Mattheyses AL, Powers MA, Fahrenkrog B. 2012. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 177:81–89. 10.1016/j.jsb.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer S, Zhao R, Barsotti AM, Ouwehand A, Fazollahi M, Coutavas E, Breuhahn K, Neumann O, Longerich T, Pusterla T, Powers MA, Giles KM, Leedman PJ, Hess J, Grunwald D, Bussemaker HJ, Singer RH, Schirmacher P, Prives C. 2012. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol. Cell 48:799–810. 10.1016/j.molcel.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13:1282–1297. 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Light WH, Freaney J, Sood V, Thompson A, D'Urso A, Horvath CM, Brickner JH. 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 11:e1001524. 10.1371/journal.pbio.1001524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673. 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. 2013. Live visualization of chromatin dynamics with fluorescent TALEs. Nat. Struct. Mol. Biol. 20:1321–1324. 10.1038/nsmb.2680 [DOI] [PubMed] [Google Scholar]
- 68.Bickmore WA, van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152:1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 69.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194:43–67. 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao X, Wu CY, Blobel G. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605–611. 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24:813–823. 10.1038/sj.emboj.7600527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colon-Ramos DA, Salisbury JL, Sanders MA, Shenoy SM, Singer RH, Garcia-Blanco MA. 2003. Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of beta2-tubulin mRNA in Chlamydomonas reinhardtii. Dev. Cell 4:941–952. 10.1016/S1534-5807(03)00163-1 [DOI] [PubMed] [Google Scholar]
- 74.Powrie EA, Zenklusen D, Singer RH. 2011. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. RNA 17:134–144. 10.1261/rna.1210411 [DOI] [PMC free article] [PubMed] [Google Scholar]