Abstract
During adipocyte differentiation, significant alternative splicing changes occur in association with the adipogenic process. However, little is known about roles played by splicing factors in this process. We observed that mice deficient for the splicing factor SRSF10 exhibit severely impaired development of subcutaneous white adipose tissue (WAT) as a result of defects in adipogenic differentiation. To identify splicing events responsible for this, transcriptome sequencing (RNA-seq) analysis was performed using embryonic fibroblast cells. Several SRSF10-affected splicing events that are implicated in adipogenesis have been identified. Notably, lipin1, known as an important regulator during adipogenesis, was further investigated. While lipin1β is mainly involved in lipogenesis, its alternatively spliced isoform lipin1α, generated through the skipping of exon 7, is primarily required for initial adipocyte differentiation. Skipping of exon 7 is controlled by an SRSF10-regulated cis element located in the constitutive exon 8. The activity of this element depends on the binding of SRSF10 and correlates with the relative abundance of lipin1α mRNA. A series of experiments demonstrated that SRSF10 controls the production of lipin1α and thus promotes adipocyte differentiation. Indeed, lipin1α expression could rescue SRSF10-mediated adipogenic defects. Taken together, our results identify SRSF10 as an essential regulator for adipocyte differentiation and also provide new insights into splicing control by SRSF10 in lipin1 pre-mRNA splicing.
INTRODUCTION
Alternative splicing is a major mechanism that controls gene expression and increases protein diversity in mammalian cells. Increasing evidence indicates that precise regulation of alternative splicing is responsible for tissue type and developmental stage determination (1–4).
Adipose tissue is an important organ for energy storage and the endocrine system, which is comprised mostly of adipocytes (5, 6). During adipocyte differentiation, significant alternative splicing changes occur in association with the implementation of the adipogenic program. More importantly, different splice variants can exert different and even opposing effects on adipogenesis. For example, transcriptional corepressor NcoR has multiple splice variants, of which the NcoRω variant inhibits adipogenesis, but the NcoRδ variant promotes adipogenesis (7). However, examples of splicing factors that regulate alterative splicing during adipogenesis are rarely reported. Only a recent study suggested that splicing regulator Sam68 is involved in this process (9). Using genome-wide exon usage profiling in white adipose tissue (WAT), Huot and coworkers reported that Sam68 deficiency activates mTOR intron 5 retention, producing a noncoding mRNA isoform. This alteration leads to reduced mTOR protein and consequently reduced adipogenic capability.
Splicing factor SRSF10 (previously known as SRp38) is an atypical SR protein that can function as a sequence-dependent splicing activator (10). Consistent with this, SRSF10 was also shown to be a key regulator of alternative splicing, influencing selection of GluR-B mutually exclusive exons or activating triadin 3′ alternative exon inclusion (2, 10). Significantly, SRSF10 knockout (KO) mice died progressively from midgestation until birth due to multiple cardiac defects (2). These findings suggested that SRSF10-regulated splicing events play significant roles in certain physiologically important processes.
In this study, we observed that SRSF10 KO embryos display significantly impaired development of subcutaneous WAT compared to that in wild-type (WT) controls. We further observed severe adipogenic defects in induced differentiation of SRSF10-deficient mouse embryonic fibroblast (MEF) cells, and those defects can be reproduced in another cell line C3H10T1/2 by knockdown of SRSF10 expression. To investigate the molecular basis for this, we used a transcriptome sequencing (RNA-seq) approach to identify SRSF10-regulated splicing events in MEF cells. Several SRSF10-affected splicing events which have been implicated in adipogenesis have been identified, including axin1 (11), ACLY (12, 13), Upf1 (14), and lipin1 (15).
Next, we investigated whether deregulated splicing of lipin1 might contribute to the adipogenic defects caused by SRSF10 deletion. The two alternatively spliced isoforms of lipin1 differ only in whether exon 7 is included in the mRNA, with lipin1α lacking exon 7 and lipin1β containing it. By performing a sequential order of deletions within a lipin1 minigene construct, we identified an SRSF10-dependent cis element in the lipin1 exon 8 that directly controls the skipping of exon 7. SRSF10 specifically binds to the sequence element and governs its activity, as expression of SRSF10 is closely related to the relative abundance of lipin1α mRNA. A series of experiments demonstrated that SRSF10 controls the production of lipin1α and thus promotes adipocyte differentiation. Indeed, exogenous expression of the lipin1α isoform has a significantly greater enhancing effect in driving the differentiation of C3H10T1/2 cells than lipin1β overexpression. Taken together, our results identify SRSF10 as an essential regulator for adipocyte differentiation and also provide new insights into splicing control of lipin1 pre-mRNA by SRSF10.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
For the lipin1 minigene construct, a mouse genomic DNA fragment containing lipin1 exon 6 to exon 8 was amplified by high-fidelity PCR and inserted between the HindIII and XhoI sites of the pcDNA3.1(+) vector. Six deletion mutants, including ΔE6, ΔI6, ΔE7, ΔI7, and ΔE8, were generated by site-directed mutagenesis based on the lipin1 minigene plasmid. ΔE8-SRSF10 and ΔE8-random plasmids were constructed based on ΔE8. Two primer pairs (F1/R1 and F2/R2) were designed for in vivo splicing analysis, with each pair containing the forward or reverse primer that is complementary only to the vector sequences. Full-length lipin1α or lipin1β cDNA was cloned into p3xFLAG-CMV14 vector. All primer sequences are listed in Table S1 in the supplemental material.
Cell culture, transfection, and adipocyte differentiation.
Primary MEF cells were isolated from embryonic day 13.5 (E13.5) mouse embryos following a standard procedure as previously described (16). An immortalized culture of MEF cells was acquired by serial passage of primary MEF cells. Primary/immortalized MEF cells and C3H10T1/2 cells were cultured with Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Biochrom). 293GPG cells were maintained as previously described (17). All transfection assays were performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Two days after cells reached confluence, primary MEF cells and retrovirus-infected C3H10T1/2 cells were induced to adipogenic differentiation with medium containing a differentiation cocktail (0.5 mM 3-isobutyl-1-methylxanthine [IBMX], 10 μg/ml insulin, 1 μM dexamethasone, and 0.5 μg/ml troglitazone [all reagents purchased from Sigma]). Two days later, differentiation medium was switched to maintenance medium supplemented only with 10 μg/ml insulin, which was changed every 2 days. Differentiated adipocytes were stained with oil red O (ORO) following a standard protocol (18).
Western blotting, RNA extraction, RT-PCR, and qPCR analysis.
Western blotting, RNA extraction, and reverse transcription-PCR (RT-PCR) were performed as described previously (2). Primary antibody against tubulin was purchased from Santa Cruz Biotechnology; antibody against FLAG tag was purchased from Sigma. Anti-FABP4 antibody was ordered from Abcam. Anti-SRSF10 antibodies were kindly provided by J. L. Manley (Columbia University, NY) and applied as described previously (2). The primer sequences used in RT-PCR and quantitative PCR (qPCR) are listed in Table S1 in the supplemental material. Real-time PCR was performed with SYBR Premix (TaKaRa) on a StepOne Plus system (Applied Biosystems).
RNA-seq, mapping of reads, and alternative splicing analysis.
Total RNA from primary MEF cells was collected and extracted using TRIzol reagent (Invitrogen). High-purity mRNA was obtained after poly(A) bead enrichment, and then a cDNA library was built. RNA-seq was performed with the cDNA library and paired-end sequencing adaptors on a HiSeq 2000 system (Illumina). Reads were mapped to a reference mouse genome and transcriptome using TopHat (19). The Mixture-of-Isoforms (MISO v0.4.9) program was used to detect the exons differentially regulated between KO and WT SRSF10. The exon-centric analysis of MISO was performed according to the MISO documentation for paired-end reads. The BAM alignment files produced by TopHat and the files for alternative events (for mm10) provided by MISO were used as inputs. To identify SRSF10-regulated events, we used the following criteria: (i) the absolute value of the difference for percentage spliced in (PSI) between KO and WT is ≥0.2, (ii) the sum of inclusion and exclusion reads is greater than 10 with both inclusion and exclusion reads ≥1, and (iii) the Bayes factor is >10.
Recombinant protein purification and gel shift assays.
Recombinant human SRSF1 and SRSF10 proteins were purified as previously described (2, 10). A DNA fragment containing lipin1 exon 7 or exon 8 was PCR amplified and inserted into the pSP64 vector (Promega) for in vitro transcription. Gel shift assays were performed as previously described (20) with recombinant SRSF1/SRSF10 protein and 32P-labeled lipin1 exon 7/8 RNA.
Retrovirus package and infection.
Plasmids for short hairpin RNA (shRNA)-expressing retrovirus were constructed based on a pSIREN-retroQ vector (Clontech); shRNA-targeted sequences were designed by using Clontech RNAi Target Sequence Selector and are listed in Table S1 in the supplemental material. Plasmids for retrovirus-mediated overexpression were constructed by insertion of human SRSF10 cDNA into the pMSCV-hygro vector (Clontech). Retrovirus packaging was performed with 293GPG cells as previously described (17). Establishment of stable cell lines to knock down or overexpress a gene of interest was based on the manufacturer's recommendation (Clontech).
Mice and histological analysis.
SRSF10 heterozygous mice were kindly provided by J. L. Manley (Columbia University, NY) and mated as previously described (2). Embryos were removed from pregnant mice at E18.5 and quickly frozen. Transverse sections at the heart level were cut at 12 μm and transferred onto polylysine-coated slides. Sections were fixed with 4% formaldehyde and then subjected to ORO staining followed by hematoxylin counterstaining (21). Experiments involving the use of animals were approved by the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences.
Statistics analysis.
The data represent means ± standard deviations (SD) from at least three independent experiments except where indicated. Statistical analysis was performed with Student's t test with a P value of <0.05 considered significant. Quantification of RT-PCR was done by using ImageJ software, version 1.43u.
RNA-seq data accession number.
The raw sequence data and processed data have been submitted to Gene Expression Omnibus with accession number GSE54226.
RESULTS
Development of axillary subcutaneous WAT is greatly impaired in SRSF10 KO mice.
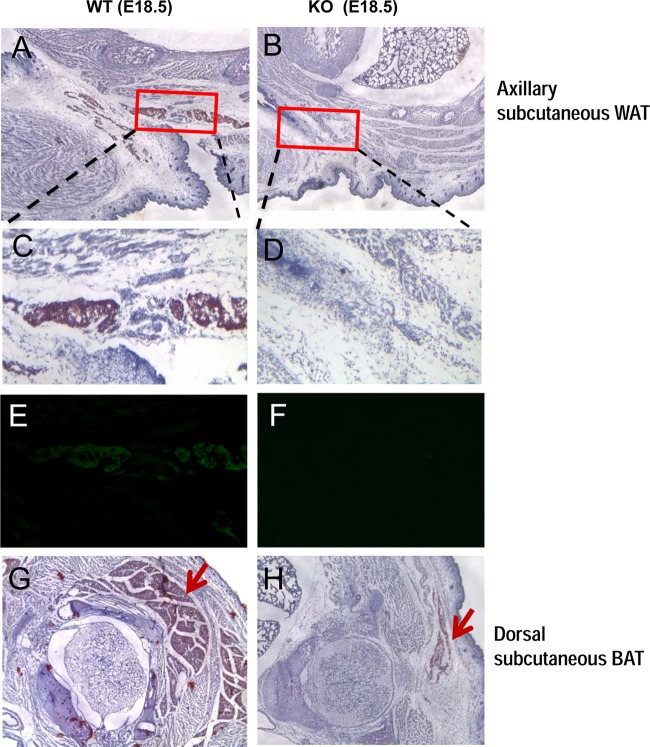
WAT mainly develops postnatally; however, lipid accumulation in axillary subcutaneous WAT can be detected at mouse embryonic day 18.5 (E18.5) (21). Our previous findings demonstrated that SRSF10 KO embryos die at various points between E13.5 and E18.5 as result of multiple cardiac defects (2). This allows us to examine the physiological impact of SRSF10 depletion on adipogenesis in vivo. To this end, transverse sections of the fat pad at heart level were stained with oil red O (ORO), followed by counterstaining with hematoxylin. As shown in Fig. 1A and B, severe impairment of axillary subcutaneous WAT development was observed in KO embryos compared to WT embryos, as indicated by red squares. High magnification further revealed an absence of lipid accumulation in KO sections, in contrast to apparent lipid deposition in WT embryos (Fig. 1C and D). Accordingly, the expression of adipocyte marker FABP4 was detected in WT sections but not in KO sections, as examined by immunostaining (Fig. 1E and F). These observations suggested that SRSF10 plays an essential role in the development of WAT. In addition, there was much less lipid accumulation observed in the dorsal subcutaneous brown adipose tissue (BAT) of KO embryos than in that of WT embryos (Fig. 1G and H).
FIG 1.
Development of axillary subcutaneous WAT is greatly impaired in SRSF10 KO mouse embryos at E18.5. Transverse sections of subcutaneous adipocyte tissues from WT and SRSF10 KO mouse embryos at E18.5 were stained with oil red O (ORO), followed by counterstaining with hematoxylin (A, B, C, D, G, and H). The adjacent sections were also subjected to immunostaining with antibodies to the adipocyte marker FABP4 (E and F). WAT, white adipocyte tissue; BAT, brown adipocyte tissue. The red squares indicate regions containing axillary subcutaneous WAT.
SRSF10 is required for hormone-induced adipogenesis.
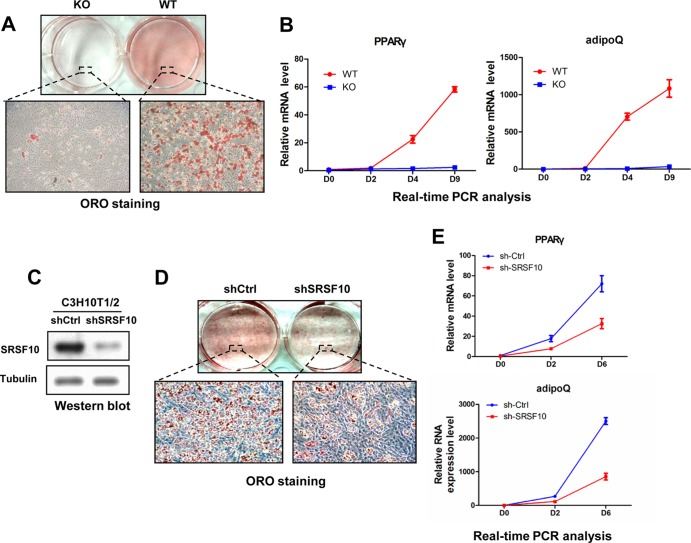
To rule out the possibility that this adipogenic defect is caused by the developmental delays observed in SRSF10 KO embryos, we further examined the adipogenic potential of primary MEF cells derived from KO embryos, along with differentiation of WT MEF cells for comparison. As shown in Fig. 2A, loss of SRSF10 resulted in a marked reduction in the adipogenic capability of KO MEF cells compared to control cells, which was observed consistently across multiple preparations. Consistent with the observed phenotype, mRNA expression levels of adipogenic markers such as peroxisome proliferator-activated receptor γ (PPARγ) and AdipoQ were significantly decreased during a time course of differentiation in KO MEF cells compared to WT cells, as analyzed by real-time PCR (Fig. 2B). These results indicate that SRSF10 is important for the adipogenic differentiation of MEF cells.
FIG 2.
SRSF10 is required for hormone-induced adipogenesis in different cell models. (A) WT and SRSF10 KO primary MEF cells were treated with insulin, IBMX, and dexamethasone plus troglitazone for induction of adipogenesis. At day 9 postinduction, adipocyte differentiation was assessed by ORO staining. (B) WT and KO primary MEF cells were induced to differentiate as described for panel A. RNAs were extracted from these cells at indicated induction days. Relative PPARγ or AdipoQ mRNA levels were measured by real-time PCR and normalized to 36b4 mRNA. Values shown are the mean ± SD (n = 3). The significance of each detected change was evaluated by Student's t test. (C) C3H10T1/2 cells were infected with retroviruses expressing either SRSF10-specific shRNA (shSRSF10) or control shRNA (shCtrl) and then selected for puromycin resistance. Western blotting was carried out to confirm SRSF10 knockdown efficiency in selected cells. Antitubulin blotting was used as a loading control. (D) shSRSF10 and shCtrl C3H10T1/2 stable cells were induced to differentiate as for panel A. Cells were stained with ORO at day 6 postinduction. (E) RNA was extracted from shSRSF10 or shCtrl C3H10T1/2 stable cells at the indicated days and the relative level of PPARγ or AdipoQ mRNA was measured as for panel B.
To provide more evidence that SRSF10 plays an essential role in adipogenesis, we used another hormone-induced adipogenesis model of C3H10T1/2 mesenchymal progenitor cells (22). A stable SRSF10-knockdown cell line was established by retrovirus-mediated shRNA delivery in C3H10T1/2 cells. A control cell line expressing negative-control oligonucleotides was generated in a similar manner. After selection with puromycin, the cells were cultured to confluence and treated with a hormone cocktail. SRSF10 protein in the shSRSF10 cells was reduced by approximately 80% compared with that in the control cell line (Fig. 2C). ORO staining revealed that shSRSF10 cells accumulated far fewer lipid droplets than the control cells (Fig. 2D). The results of real-time PCR confirmed the greatly attenuated expression of adipocyte markers in shSRSF10 cells during differentiation (Fig. 2E).
To exclude off-target effects of the shRNA, the human SRSF10 cDNA, which is resistant to shRNA specific for mouse SRSF10, was reintroduced into the shSRSF10 C3H10T1/2 cell line using retroviral transduction (see Fig. S1A in the supplemental material). Cells with the incorporated gene were selected with hygromycin and then induced to differentiate. Although human SRSF10 was modestly expressed in the shSRSF10 cells, the defective adipogenesis could be partially rescued, as assessed by ORO staining (see Fig. S1B in the supplemental material) and the expression of PPARγ and AdipoQ (see Fig. S1C in the supplemental material). Collectively, the experiments with both MEFs and C3H10T1/2 cells confirmed that SRSF10 is essential for hormone-induced adipogenesis.
In addition, we also observed that C3H10T1/2 cells with modest expression of SRSF10 seemed to differentiate into adipocytes as well as control cells (see Fig. S2A and B in the supplemental material). Since relative mRNA levels of SRSF10 remained largely unchanged during adipogenesis of both MEF and C3H10T1/2 cells (see Fig. S2C and D in the supplemental material) and SRSF10 is indeed detectable in abdominal WAT of adult mice (see Fig. S2E in the supplemental material), these findings likely indicate that the wild-type amount of SRSF10 is enough to drive the cells to differentiate.
RNA-seq and bioinformatic analysis of splicing targets of SRSF10.
Given that SRSF10 functions as a sequence-specific activator as well as an important regulator of alternative splicing, we performed RNA-seq analysis using RNA extracted from SRSF10 KO and WT MEF cells (see Materials and Methods) to identify alternative splicing events regulated by SRSF10. Briefly, WT and KO cDNA libraries were prepared and sequenced in a 2× 100-bp format using an Illumina HiSeq instrument. We then used TopHat (v2.0.6) to align clean reads against the mouse genome sequence (GRCm38.fa) and its corresponding annotated genes (GRCm38.71.gtf). After mapping, approximately 16,310,111 WT (31.5%) and 16,969,267 KO (32.7%) reads were mapped on splice junctions (Table 1).
TABLE 1.
Summary of RNA-seq data mapping results
| Reads | No. (%) in: |
|
|---|---|---|
| MEF control cells | MEF knockout cells | |
| Total clean | 53,331,490 (100) | 53,333,140 (100) |
| Mapped | 51,767,143 (97.07) | 518,34,710 (97.19) |
| Uniquea | 49,940,111 (96.47) | 50,183,039 (96.81) |
| Multimappedb | 1,827,032 (3.53) | 1,651,671 (3.19) |
| Junctionc | 16,310,111 (31.51) | 16,969,267 (32.74) |
| Unmapped | 1,564,347 (2.93) | 1,498,430 (2.81) |
Includes paired-end reads with an unalignable mate.
With multiple alignments.
All junction reads with ≥3 threshold alignments were considered to be a real junction.
We then performed transcriptome-wide screening to identify SRSF10-regulated alternative splicing events using the Mixture-of-Isoforms (MISO) software (see Materials and Methods) (23). As shown in Table 2 and in Table S2 in the supplemental material, 100 alternative splicing events were significantly altered in the KO samples compared to WT controls from all splicing events tested. A majority of the affected splicing events (71) belong to the category of cassette exon; the remaining events fall into other splicing modes.
TABLE 2.
Summary of SRSF10-affected splicing events in MEF cells
| Type of splicing event | No. of events |
|
|---|---|---|
| Total | Affected by SRSF10 | |
| Cassette exon | 7,939 | 71 |
| Alternative 5′ splice site | 2,341 | 10 |
| Alternative 3′ splice site | 3,744 | 10 |
| Mutually exclusive exon | 421 | 2 |
| Retained intron | 1,277 | 7 |
Validation of SRSF10-affected alternative splicing targets.
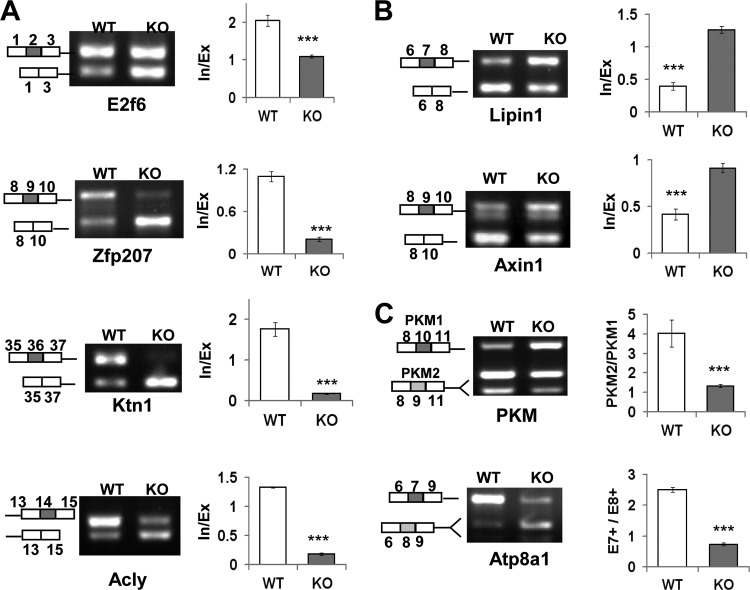
To validate potential alternative splicing events regulated by SRSF10, we designed primer pairs detecting alternative exons in MISO-predicated target genes and used them to analyze RNAs isolated from WT and KO cells by RT-PCR. Of 36 target genes analyzed, 16 revealed significant differences between WT and KO samples (Fig. 3; see Fig. S3 in the supplemental material). Specifically, cassette exon-containing mRNA isoforms were predominantly expressed in WT cells, such as for E2f6, Zfp207, Ktn1, and ACLY transcripts; SRSF10 depletion significantly decreased exon inclusion to a large extent for each target (Fig. 3A). These results thus confirm that SRSF10 functions as a splicing activator to promote exon inclusion in vivo.
FIG 3.
SRSF10 depletion causes both exon inclusion and exclusion in vivo. (A) Examples of SRSF10-dependent exon inclusion. The indicated transcripts containing alternatively spliced exons were analyzed by RT-PCR using RNAs extracted from WT and KO MEF cells. RNA products are indicated schematically on the left. Quantification of their RNA products measured as inclusion/exclusion (In/Ex) ratios is shown on the right bar graphs. Values shown are the mean ± SD (n = 3). Significance for each detected change was evaluated by Student's t test. (B) Examples of SRSF10-dependent exon exclusion were analyzed as described for panel A. (C) Examples of mutually exclusive exons regulated by SRSF10 were analyzed as described for panel A. Note that the PCR products were digested with either PstI (PKM) or BglII (Atp8a1) to distinguish their isoforms with the same size.
On the other hand, an increased inclusion ratio of lipin1 exon 7 or axin1 exon 9 was observed upon SRSF10 depletion (Fig. 3B), suggesting that SRSF10 can also repress exon inclusion in vivo. In addition, selection of mutually exclusive exons in the PKM and Atp8a1 pre-mRNAs was significantly affected by SRSF10 (Fig. 3C). In summary, of the 16 validated exons, 12 regulated exons were activated by SRSF10, while only three exons were repressed by SRSF10. These findings suggest that SRSF10 functions predominantly as a splicing activator to enhance exon inclusion.
Lipin1 alternative splicing is deregulated during differentiation of KO MEF cells.
Among the verified splicing targets, several SRSF10-affected splicing events have been implicated in adipogenesis, such as ACLY (13), axin1 (11, 12), Upf1 (14), and lipin1 (15). Significantly, two alternatively spliced isoforms of lipin1 have been shown to display distinct functions in adipocyte differentiation (8).
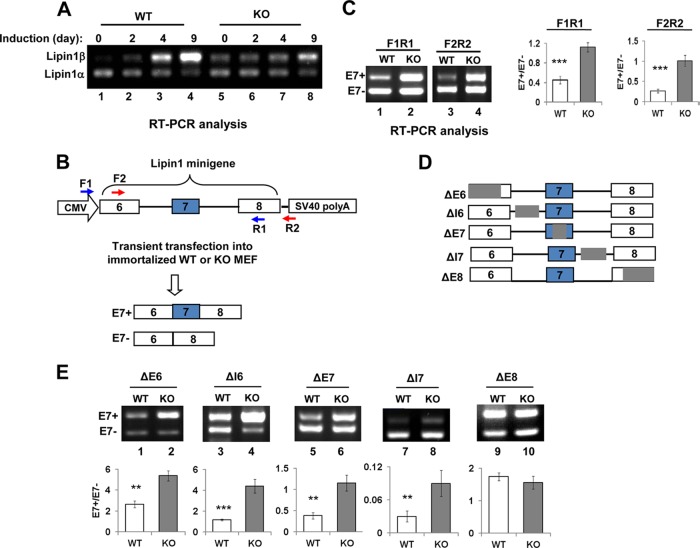
As an initial step to investigate whether deregulated splicing of lipin1 might contribute to the adipogenic defects caused by SRSF10 deletion, we first measured expression of two lipin1 isoforms during differentiation of WT and KO MEF cells by RT-PCR. As shown in Fig. 4A, lipin1α was most prominently expressed at the initial stages of differentiation of WT MEF cells, whereas lipin1β expression significantly increased with differentiation, and this isoform predominated in mature adipocytes (compare lanes 2 to 4 with lane 1). The expression patterns of the two lipin1 isoforms were similar to that observed during the differentiation of 3T3L1 preadipocytes (8). In contrast, the relative lipin1β levels were higher than those in WT cells due to SRSF10 depletion (Fig. 4A, compare lane 5 and lane 1), but expression of lipin1β showed only a slight increase during the differentiation of KO cells (Fig. 4A, compare lanes 5 to 8 with lanes 1 to 4). In addition, there were no significant changes observed for other SRSF10-regulated events during adipocyte differentiation (see Fig. S4 in the supplemental material). Together, these data suggested that SRSF10 accelerated differentiation probably through maintaining relatively high levels of the lipin1α isoform in MEF cells by repressing exon 7 inclusion.
FIG 4.
Mapping of SRSF10-regulated cis element using lipin1 minigene derivatives. (A) Lipin1α/β mRNA and PPARγ were measured by RT-PCR at the indicated differentiation days (top). (B) Diagram of lipin1 minigene plasmid and its two splicing isoforms containing or lacking exon 7. The intact lipin1 exon 6 to 8 genomic fragment was cloned into pcDNA3.1 vector. The alternative exon 7 is shown in the blue box, while constitutive exons 6 and 8 are shown in white boxes. The positions of two primer pairs for in vivo splicing analysis are indicated as numbered arrows. (C) RT-PCR was performed with RNAs extracted from immortalized WT and KO MEF cells transiently transfected with the lipin1 minigene plasmid, and products were analyzed directly on a 1.5% agarose gel and visualized with ethidium bromide (EtBr). Results with the F1/R1 and F2/R2 primer pairs are shown (left). Quantification of lipin1 minigene RNA products measured as the exon 7 inclusion/exclusion (E7+/E7−) ratio is shown on the right histogram. Values shown are the mean ± SD (n = 3). The significance of each detected change was evaluated by Student's t test. (D) Diagram of lipin1 minigene truncated plasmids. Note that the gray region was deleted from each of indicated mutants. (E) Each of the lipin1 minigene mutants was transfected into immortalized WT and KO MEF cells. RT-PCR analysis of each mutant and quantification were performed as described for panel C. Note that F1/R1 primer pairs were used for all truncates except ΔE8; F2/R2 primer pairs were used for ΔE8.
Mapping of SRSF10-dependent sequences for inhibiting lipin1 exon 7 inclusion.
Our results suggested that SRSF10 represses inclusion of exon 7 in lipin1 pre-mRNA splicing; this seems to contradict the proposed activation for the protein in splicing regulation (10). We next investigated the mechanisms by which SRSF10 represses splicing of exon 7 and thereby maintains relatively high levels of the lipin1α isoform in WT cells. To this end, we constructed a lipin1 minigene plasmid in which the genomic DNA fragment of lipin1 exons 6 to 8 containing the complete intervening intronic sequences was driven under the control of CMV promoter and followed by a poly(A) site. This plasmid was transfected into immortalized WT and KO MEF cells, and total RNAs were extracted and analyzed by RT-PCR with two primer pairs (Fig. 4B). As shown in Fig. 4C, the splicing pattern of the minigene precisely mirrored the lipin1α/β ratio of the endogenous mRNAs in WT cells, with a prevalence of products that did not contain exon 7. This pattern also drastically changed upon deletion of SRSF10, resulting in an approximately 1:1 ratio between exon skipping and exon inclusion (compare lane 1 with lane 2 and compare lane 3 with lane 4).
To identify functional cis-acting elements involved in SRSF10-repressed exon 7 inclusion, the lipin1 minigenomic sequences were scanned by performing a sequential order of deletions (Fig. 4D). Both the ΔI6 and ΔI7 constructs have large intronic sequences (approximately 2,000 nucleotides [nt]) deleted from the middle region of intron 6 or intron 7, respectively. ΔE6, ΔE7, and ΔE8 contain short deletions (46 to 110 nt) made in exon 6, exon 7, or exon 8, respectively. We reasoned that deletion of either SRSF10-dependent exonic or intronic sequences would likely minimize the difference of lipin1 minigene splicing between WT and KO cells.
While ΔE7 has a negligible effect on lipin1 splicing compared to the WT construct (compare Fig. 4E, lane 5, with C, lane 1), the other four deletions either significantly increased or decreased the activation of exon 7 in WT cells (compare Fig. 4E, lanes 1, 3, 7, and 9, with C, lane 1), indicating critical roles for each of the deleted regions in exon 7 splicing. However, four of the five deletion mutants still remained responsiveness to SRSF10 deficiency: there was a 2- to 3-fold increase in the E7+/E7 ratio upon loss of SRSF10 compared with the ratio obtained in the presence of SRSF10 (compare lane 1 with lane 2, lane 3 with lane 4, lane 5 with lane 6, and lane 7 with lane 8). These findings indicate that the genomic region of lipin1 exons 6 and 7 plus intron 7 sequences contains various regulatory elements that are involved in exon 7 splicing, but they function in an SRSF10-independent way. Interestingly, the ΔE8 minigene that comprises only the first 15 bases of exon 8 (i.e., lacking the last 110-bp sequence) was spliced with similar ratios of exon inclusion to skipping in WT and KO cells. This suggests that the 110-bp sequences within exon 8 likely function as an SRSF10-dependent cis element in inhibiting exon 7 inclusion.
SRSF10 binds to exon 8 sequences and represses exon 7 inclusion.
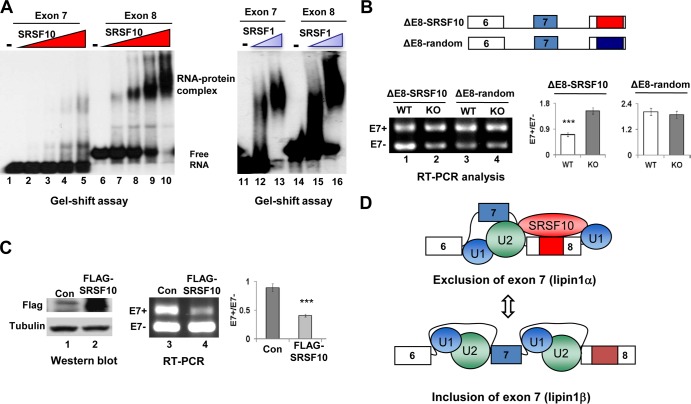
SRSF10 is a sequence-specific splicing regulator, so we next investigated whether SRSF10 binds to the exon 8 sequences and thus represses exon 7 inclusion. To this end, we prepared 32P-labeled full-length exon 8 RNA and compared its binding activity in gel shift assays with exon 7 RNA prepared in a similar fashion. The results indicate that SRSF10 strongly binds to exon 8 RNA in a concentration-dependent manner. In contrast, it displays only weak binding to exon 7 RNA (Fig. 5A, compare lanes 1 to 5 with 6 to 10). SRSF10 bound to exon 8 RNA with an affinity comparable to its affinity for the GluR-B flop exon (10) (data not shown). As a control, another SR protein (SRSF1) did not show significant differences in binding to either RNA (Fig. 5A, compare lanes 11 to 13 with 14 to 16). We therefore concluded that SRSF10 binds to exon 8 RNA in a highly specific manner.
FIG 5.
Characteristics of SRSF10-regulated lipin1α/β splicing. (A) The indicated 32P-labeled RNAs were incubated with increasing amounts of recombinant His-SRSF10 (75 ng, 150 ng, 300 ng, and 600 ng), and complexes were resolved by nondenaturing PAGE (left). Each of the indicated RNA probes was incubated with 150 ng or 300 ng of recombinant SRSF1 and analyzed (right). (B) Three copies of SRSF10 consensus sequences or random control sequences were inserted downstream of the ΔE8 minigene to construct ΔE8-SRSF10 and ΔE8-random derivatives (top). F2/R2 primer pairs were used for in vivo splicing analysis, and quantification was performed as described for Fig. 4C (bottom). (C) Cotransfection of KO immortalized MEF cells with lipin1 minigene in the presence of either vector or FLAG-SRSF10 plasmid. Whole-cell lysates were prepared and analyzed by Western blotting using anti-FLAG and antitubulin antibodies (left). RNA was isolated and analyzed by RT-PCR, and quantification was performed as described for Fig. 4C (right). (D) Model for inhibition of lipin1 exon 7 inclusion by SRSF10. (Top) SRSF10 binding to lipin1 exon 8 containing the SRSF10 binding region (red box) stimulates or stabilizes interactions of general splicing factor U1 or U2 snRNPs at the nearby constitutive splice sites, thereby decreasing the competitiveness of the competing alternative sites and causing exon 7 skipping. (Bottom) In contrast, when binding of SRSF10 to exon 8 is disrupted, stabilization of U1 or U2 snRNPs at alternative splice sites for exon 7 is relatively stimulated, hence leading to exon 7 inclusion.
To provide direct evidence that binding of SRSF10 to the downstream constitutive exon is responsible for exon 7 exclusion, we reintroduced SRSF10 binding sequences identified by SELEX (24) in exon 8 and examined whether they would be sufficient to repress exon 7 inclusion. Specifically, two derivatives (designated ΔE8-SRSF10 and ΔE8-random) were made based on the lipin1 ΔE8 construct to allow for the insertion of three copies of the SRSF10 consensus motif or random sequences just downstream of the 3′ splice site in exon 8 (Fig. 5B). When RNA from the transfected cells was analyzed by RT-PCR, approximately 60% of exon 7 was excluded when SRSF10 binding motifs were inserted in exon 8, compared to at most 30% exon exclusion observed with the control sequence (Fig. 5B, compare lane 1 and lane 3). However, the differential effects on exon 7 splicing between the two inserted sequences were completely diminished upon SRSF10 depletion (Fig. 5B, compare lane 2 and lane 4), confirming the direct involvement of SRSF10 bound to exon 8 sequences in repressing exon 7 splicing. Furthermore, exogenous expression of FLAG-tagged SRSF10 in KO MEF cells also inhibits exon 7 inclusion (Fig. 5C). Taken together, these results strongly indicate that binding of SRSF10 to the downstream constitutive sequences plays a critical role in repressing exon 7 inclusion (Fig. 5D).
Overexpression of lipin1α could partially rescue adipogenic defects caused by SRSF10 depletion.
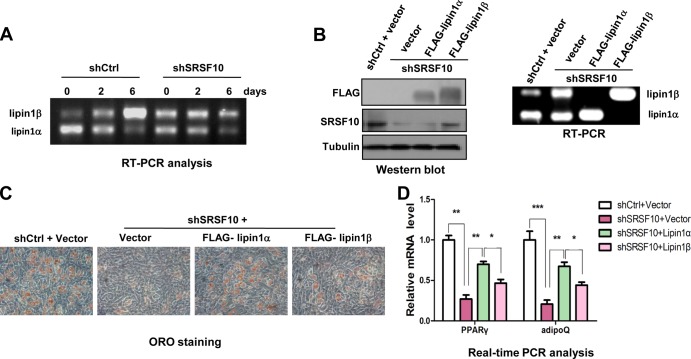
We next examined whether the lipin1α/β ratio was altered in shSRSF10-transdued C3H10T1/2 cells. As shown in Fig. 6A, lipin1α is still the major isoform expressed in C3H10T1/2 cells, and expression patterns of the two lipin1 isoforms during differentiation were similar to that observed in WT MEF cells (lanes 1 to 3). Knockdown of SRSF10 could also significantly decrease lipin1α while increasing lipin1β, and the latter remained virtually the same in the differentiation process (lanes 4 to 6). In addition, other splicing changes were also observed in C3H10T1/2 cells upon SRSF10 knockdown as in the KO MEF cells (see Fig. S5 in the supplemental material).
FIG 6.
Exogenous expression of the lipin1α isoform partially rescues the adipogenic defect caused by SRSF10 depletion. (A) RNA was extracted from shSRSF10 or shCtrl C3H10T1/2 stable cells at different time points after induction. Expression patterns of endogenous lipin1α/β were determined by RT-PCR. (B) shSRSF10 C3H10T1/2 cells were transfected with control vector, FLAG-lipin1α, or FLAG-lipin1β plasmids. shCtrl C3H10T1/2 cells were also transfected with control vector. Exogenous expression of lipin1α/β was confirmed either at the protein level by Western blotting using anti-FLAG antibodies (left) or at the mRNA level by RT-PCR (right). (C) Hormone-induced adipogenesis of each of the transfected cells in panel B. Microscopic views of ORO-stained cells after 6 days of induction are shown. (D) RNAs were extracted from cells described in panel C. Relative PPARγ (left) and AdipoQ (right) mRNA levels were measured by real-time PCR. All data are normalized to 36b4 mRNA. Values shown are the mean ± SD (n = 3). The significance of each detected change was evaluated by Student's t test.
We then examined the ability of the two lipin1 isoforms to complement the adipogenic defect in SRSF10-knockdown C3H10T1/2 cells, along with differentiation of cells expressing a control shRNA for comparison. Western blotting and RT-PCR analysis both confirmed respective overexpression of the two lipin1 isoforms (Fig. 6B). Compared to the effects of lipin1β, lipin1α isoform expression in these cells could significantly restore differentiation as measured by ORO staining (Fig. 6C) and real-time PCR of two adipocyte markers (Fig. 6D). This finding is consistent with previous reports that the lipin1α isoform is more necessary for adipocyte differentiation than the lipin1β isoform (8). Most importantly, this observation provides direct evidence that SRSF10 promotes adipogenesis by repressing exon 7 inclusion, thus maintaining lipin1α as the predominant isoform in undifferentiated cells. However, we also observed that shSRSF10 C3H10T1/2 cells with expression of the lipin1α isoform do not differentiate into adipocytes as well as the controls cells (Fig. 6C and D). This probably indicates that SRF10 works in part through regulation of lipin1α isoform levels.
DISCUSSION
During adipocyte differentiation, cells undergo dramatic morphological changes and critical modification in gene expression. Here we have provided evidence that the splicing factor SRSF10 is required for adipocyte differentiation and maturation and that its loss leads to changes in splicing of lipin1 pre-mRNA and severe adipogenic defects. We propose a model in which SRSF10 represses exon 7 inclusion and favors generation of the lipin1α isoform, which, in turn, facilitates adipocyte differentiation (Fig. 6D). Below we discuss how SRSF10 functions as an important regulator of alternative splicing during adipogenesis and how loss of SRSF10 leads to deregulated lipin1 splicing in this process.
We previously reported that SRSF10 functions as a sequence-specific splicing activator by directly binding to exonic splicing enhancers (10). However, this activation model seems unlikely to explain why SRSF10 represses inclusion of exon 7 in lipin1 pre-mRNA splicing. By using a lipin1 minigene system combined with a series of mutagenesis and gel shift assays, we demonstrated that skipping of exon 7 is controlled by SRSF10-regulated cis element located in the constitutive exon 8. The activity of this element depends on the binding of SRSF10 and correlates with the relative abundance of lipin1α mRNA. Therefore, it is reasonable to propose that when binding occurs in exon 8, SRSF10 could activate the nearby splice sites and reduce the relative competitiveness of alternative sites, thereby preventing the exon 7 from inclusion (Fig. 6D). A similar principle has also been employed by the typical SR protein SRSF1, as it was shown to activate exon exclusion of the Ron proto-oncogene by directly binding to an enhancer element located within the downstream constitutive exon (25).
Lipin1 plays a critical role in adipocyte development and maturation by functioning as a critical enzyme in glycerolipid biosynthesis and transcriptional coactivator (15). Our findings demonstrated that loss of SRSF10 changes the relative ratio of lipin1α to lipin1β, and that expression of lipin1α results in visually significant adipogenic “rescue” compared to lipin1β expression. This likely suggests that the balance between lipin1α and lipin1β expression levels in undifferentiated cells plays a critical role in driving preadipocyte differentiation. In addition, lipin1β expression was found to increase dramatically over the course of adipocyte differentiation; however, this increase was much repressed in differentiation of SRSF10-depleted cells (Fig. 4A and 6A). Therefore, we reasoned that attenuated adipogenesis of SRSF10-depleted cells could also be attributed to the modest expression of lipin1β during differentiation, as well as changed levels of the two lipin1 isoforms at the undifferentiated stage. The possible regulation of lipin1β expression during adipogenesis by SRSF10 is unclear and will be a focus of future study.
Beyond the involvement of lipin1 in mediating effects of SRSF10 on adipogenesis, other dysregulated splicing targets could also contribute to this attenuated adipogenesis. Ectopic expression of axin1 in 3T3-L1 preadipocytes promoted their differentiation into adipocytes (11). ATP-citrate lyase (ACLY), an enzyme that catalyzes the formation of acetyl coenzyme A (acetyl-CoA), played a central role in adipogenic differentiation (12) (13). Upf1 facilitated adipocyte differentiation by being involved in staufen1-mediated mRNA decay in adipogenesis (14). It is likely that SRSF10 facilitates adipogenesis by controlling normal levels of these WT splice isoforms during adipogenesis. In contrast, SRSF10 deletion results in isoform switching in these events, which in turn could reduce the cells' capacity for differentiation. The PKM2 isoform appears to be critical for cancer metabolism, tumor growth, and C2C12 cell differentiation (26, 27); however, specific knockdown of PKM2 expression using a retrovirus-mediated strategy did not change the adipogenic capability of C3H10T1/2 cells (see Fig. S6 in the supplemental material). Further evaluation of these splice isoforms resulting from SRSF10 deficiency in adipogenesis will be informative.
In summary, the loss of splicing factor SRSF10 leads to greatly impaired development of WAT in vivo and severe adipogenic defects in cultured cells. By regulating alternative splicing, SRSF10 was demonstrated to be a key player during adipocyte differentiation and maturation.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. L. Manley (Columbia University) for his generous gifts of SRSF10 mice and anti-SRSF10 antibodies.
This work was supported by grants from the Ministry of Science and Technology of China (2012CB524900), the National Natural Science Foundation (31370786 and 31170753), and the One Hundred Talents Program of the Chinese Academy of Sciences.
We declare no conflict of interest.
Footnotes
Published ahead of print 7 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01674-13.
REFERENCES
- 1.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21:1636–1652. 10.1101/gad.1558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. 2009. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev. Cell 16:528–538. 10.1016/j.devcel.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. 2001. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7:331–342. 10.1016/S1097-2765(01)00181-2 [DOI] [PubMed] [Google Scholar]
- 4.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, Rosenfeld MG, Manley JL, Ross J, Jr, Chen J, Xiao RP, Cheng H, Fu XD. 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59–72. 10.1016/j.cell.2004.11.036 [DOI] [PubMed] [Google Scholar]
- 5.Scherer PE. 2006. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 55:1537–1545. 10.2337/db06-0263 [DOI] [PubMed] [Google Scholar]
- 6.Kershaw EE, Flier JS. 2004. Adipose tissue as an endocrine organ. J. Clin. Endocr. Metab. 89:2548–2556. 10.1210/jc.2004-0395 [DOI] [PubMed] [Google Scholar]
- 7.Goodson ML, Mengeling BJ, Jonas BA, Privalsky ML. 2011. Alternative mRNA splicing of corepressors generates variants that play opposing roles in adipocyte differentiation. J. Biol. Chem. 286:44988–44999. 10.1074/jbc.M111.291625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterfy M, Phan J, Reue K. 2005. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 280:32883–32889. 10.1074/jbc.M503885200 [DOI] [PubMed] [Google Scholar]
- 9.Huot ME, Vogel G, Zabarauskas A, Ngo CT, Coulombe-Huntington J, Majewski J, Richard S. 2012. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol. Cell 46:187–199. 10.1016/j.molcel.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Chen M, Manley JL. 2008. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 15:1040–1048. 10.1038/nsmb.1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950–953. 10.1126/science.289.5481.950 [DOI] [PubMed] [Google Scholar]
- 12.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076–1080. 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Solana B, Li DQ, Kumar R. 2014. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochim. Biophys. Acta 1843:316–326. 10.1016/j.bbamcr.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK. 2012. Staufen1-mediated mRNA decay functions in adipogenesis. Mol. Cell 46:495–506. 10.1016/j.molcel.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 15.Reue K, Brindley DN. 2008. Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49:2493–2503. 10.1194/jlr.R800019-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuelson LC, Metzger JM. 2006. Isolation and freezing of primary mouse embryonic fibroblasts (MEF) for feeder plates. Cold Spring Harb. Protoc. 10.1101/pdb.prot4482 [DOI] [PubMed] [Google Scholar]
- 17.Ory DS, Neugeboren BA, Mulligan RC. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U. S. A. 93:11400–11406. 10.1073/pnas.93.21.11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. 1999. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4:611–617. 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- 19.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Wang Z, Zhou X, Cheng Y, Xie Z, Manley JL, Feng Y. 2013. Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. Proc. Natl. Acad. Sci. U. S. A. 110:E2687–2695. 10.1073/pnas.1310607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asaki T, Konishi M, Miyake A, Kato S, Tomizawa M, Itoh N. 2004. Roles of fibroblast growth factor 10 (Fgf10) in adipogenesis in vivo. Mol. Cell. Endocrinol. 218:119–128. 10.1016/j.mce.2003.12.017 [DOI] [PubMed] [Google Scholar]
- 22.Tang QQ, Lane MD. 2012. Adipogenesis: from stem cell to adipocyte. Annu. Rev. Biochem. 81:715–736. 10.1146/annurev-biochem-052110-115718 [DOI] [PubMed] [Google Scholar]
- 23.Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7:1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin C, Manley JL. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407–417. 10.1016/S0092-8674(02)01038-3 [DOI] [PubMed] [Google Scholar]
- 25.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20:881–890. 10.1016/j.molcel.2005.10.026 [DOI] [PubMed] [Google Scholar]
- 26.David CJ, Chen M, Assanah M, Canoll P, Manley JL. 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463:364–368. 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452:230–U274. 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.