Abstract
Mutations in the leucine-rich repeat kinase 2 gene (LRRK2) are associated with familial and sporadic Parkinson's disease (PD). LRRK2 is a complex protein that consists of multiple domains, including predicted C-terminal WD40 repeats. In this study, we analyzed functional and molecular features conferred by the WD40 domain. Electron microscopic analysis of the purified LRRK2 C-terminal domain revealed doughnut-shaped particles, providing experimental evidence for its WD40 fold. We demonstrate that LRRK2 WD40 binds and sequesters synaptic vesicles via interaction with vesicle-associated proteins. In fact, a domain-based pulldown approach combined with mass spectrometric analysis identified LRRK2 as being part of a highly specific protein network involved in synaptic vesicle trafficking. In addition, we found that a C-terminal sequence variant associated with an increased risk of developing PD, G2385R, correlates with a reduced binding affinity of LRRK2 WD40 to synaptic vesicles. Our data demonstrate a critical role of the WD40 domain within LRRK2 function.
INTRODUCTION
Parkinson's disease (PD) is the second most common age-related neurodegenerative disease and is clinically characterized by movement impairments, bradykinesia, rigidity, and resting tremor and pathologically by the progressive loss of dopaminergic neurons in the substantia nigra and the formation of Lewy bodies (1, 2). Although the majority of cases are sporadic, mutations in the leucine-rich repeat kinase 2 (LRRK2) gene (PARK8; Online Mendelian Inheritance in Man [OMIM] accession number 609007) had been unequivocally linked to late-onset autosomal dominant PD. LRRK2 mutations account for up to 13% of familial PD cases compatible with dominant inheritance and are also found in 1 to 2% of sporadic PD patients (62–64). LRRK2 is a complex 286-kDa protein that consists of multiple domains, including (in order, from the amino to carboxyl terminus) armadillo, ankyrin, and the namesake leucine-rich repeats (LRRs), followed by an ROC (Ras of complex proteins) GTPase domain, a COR (C-terminal of ROC) dimerization domain, a kinase domain, and a predicted C-terminal WD40 repeat domain (4–6). Several single-nucleotide alterations have been identified in LRRK2, but only five missense mutations within the ROC, COR, and kinase domains clearly segregate with PD in large family studies (7, 8). It has recently been shown that the WD40 domain is required to stabilize the LRRK2 dimer and to execute LRRK2-associated kinase activity as well as neurotoxicity (9, 10), but the role of this domain within LRRK2 physiological and pathological function has not yet been completely defined. The beta-propeller-forming WD40 domains are among the 10 most abundant domain types across eukaryotic proteomes (11) and constitute platforms where multiprotein complexes assemble reversibly (12). Here, we systematically analyzed the protein-protein interactions conferred by the LRRK2 WD40 domain. The nature of the LRRK2 WD40 interactors and the finding that the LRRK2 WD40 domain is able to bind to synaptic vesicles (SV) contribute to accumulating evidence suggesting that LRRK2 serves as a scaffold protein connecting vesicle trafficking and cytoskeleton (13). Strong genetic association indicates that the substitution of arginine for glycine 2385 (G2385R) within the LRRK2 WD40 domain is a pathologically relevant variant (14). This variant is considered a common risk factor for sporadic PD in Chinese Han and Korean ethnicity, but its functional impact is largely unknown (15, 16). We demonstrate that the G2385R variant alters LRRK2 WD40 binding to synaptic vesicles. Altogether, these data suggest that the LRRK2 WD40 domain is a determinant for LRRK2 physiological and pathological activities.
MATERIALS AND METHODS
Cell cultures.
Cortical neuron cultures were prepared from embryonic day 15.5 to 16.5 mouse cortexes (C57BL/6). Medium-density (150 to 200 cells/mm2) neuron cultures were plated and grown in Neurobasal culture medium supplemented with 2% B27 and 0.1% gentamicin on 24-well plastic tissue culture plates (Iwaki; Bibby Sterilin, Staffordshire, United Kingdom). In such cultures glial growth is reduced to less than 0.5% of the nearly pure neuronal population (17). HEK293T (ATCC CRL-11268) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin mix according to standard protocols. All media were purchased from Life Technologies.
Plasmids and transfection.
Human LRRK2 LRR (amino acids [aa] 921 to 1356), LRRK2 WD40 (aa 2124 to 2527), full-length RACK1, and full-length human LRRK2 were subcloned into pDEST-15 (N-terminal glutathione S-transferase [GST] tag) and/or pDEST-733 (N-terminal red fluorescent protein [RFP] tag) using the Gateway system (Life Technologies) as described previously (18). The LRRK2 WD40 G2385R variant as well as FLAG- and RFP-tagged LRRK2 consisting of residues 1 to 2141 (hereinafter termed LRRK2 1–2141) were generated by site-directed mutagenesis using a QuikChange mutagenesis kit (Stratagene). Cloning of pGEX-RACK1 and full-length FLAG-LRRK2 was described previously (18–20). LRRK2 WD40 consisting of aa 2148 to 2527 with six copies of a His tag (6×His) was cloned into pETM11. pGFP (where GFP is green fluorescent protein) and pDsRed2-1 were purchased from Clontech laboratories. Neurons were transfected at 10 days of in vitro culture (DIV10) with GFP and RFP-tagged constructs or DsRed in a 1:3 ratio by calcium phosphate precipitation as previously described (21) and processed when indicated in the text.
Purification of GST- and His-tagged proteins.
GST fusion proteins containing single LRRK2 domains and GST-RACK1 were expressed in the Escherichia coli BL21 strain (Life Technologies), purified as described earlier (22). The purification procedure for the N-terminal 6×His-tagged LRRK2 WD40 construct (LRRK2 2148-end), used for electron microscopy (EM) analysis, has been adapted to that used for the GST fusion proteins. Briefly, for expression, E. coli cells were grown in terrific broth (TB) medium with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction overnight at 18°C. Cells were lysed in 20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM beta-mercaptoethanol (BME), 0.7% Sarkosyl, and 2% Triton X-100 by sonication. The lysate was loaded onto a nickel column, and the column was then washed with 20 column volumes each of lysis buffer and high-salt buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 350 mM KCl, 5 mM MgCl2, 1 mM ATP). The His-tagged protein was finally eluted with lysis buffer containing 150 mM imidazole. The protein was then dialyzed overnight against 20 mM Tris, pH 7.5, 150 mM NaCl, and 5 mM dithiothreitol (DTT).
Subcellular fractionation and synaptic vesicle binding assay.
Subcellular fractionation of rat forebrain tissue was carried out as previously described in the presence of protease inhibitors (23). Briefly, the freshly dissected cerebral cortex was homogenized with a glass-Teflon homogenizer in ice-cold buffered sucrose (0.32 M sucrose, 5 mM HEPES, pH 7.4) (homogenate) and centrifuged at 800 × g for 10 min. The nuclear pellet was discarded, and the postnuclear supernatant (containing cell membrane, cytosol, and organelles; S1 fraction) was centrifuged at 9,200 × g for 15 min to give a supernatant fraction (containing cytosol and microsomes; S2 fraction) and a crude mitochondrial pellet (containing mitochondria and synaptosomes; P2 fraction). The P2 fraction was subjected to osmotic lysis by homogenization in 10 volumes of ice-cold water and centrifuged at 25,000 × g for 20 min to yield a lysate pellet (LP1) enriched in presynaptic plasma membranes and a lysate supernatant (LS1). The LS1 fraction was further centrifuged at 16,500 × g for 2 h to yield a synaptosolic fraction (LS2) and a crude SV pellet (LP2) containing synaptic vesicles and small presynaptic plasma membranes. The LP2 fraction was further fractionated by centrifugation through a continuous sucrose gradient and chromatography through a controlled-pore glass column to yield highly purified SV (untreated SV [US]) and a column flowthrough (FT). When required, purified SV were partially depleted of endogenous proteins by dilution in 0.2 M NaCl (salt-treated SV, SSV). SV were centrifuged at 200,000 × g for 2 h after 2 h of incubation at 0°C. After centrifugation, SV were resuspended in 0.3 M glycine, 5 mM HEPES-NaOH, pH 7.4, at a protein concentration of 1.5 to 2 mg/ml. The binding of GST fusion proteins to SV was carried out using a high-speed sedimentation assay (24). Briefly, SV (5 to 10 μg of total protein) were incubated for 1 h at 0°C with increasing amounts of a GST fusion protein in a buffer containing 220 mM glycine, 30 mM NaCl, 5 mM Tris-HCl, 4 mM HEPES (pH 7.4), 0.22 mM NaN3, and 100 μg/ml of bovine serum albumin (BSA). After the incubation, GST fusion protein which bound to SV was separated by high-speed centrifugation (400,000 × g for 30 min). Aliquots of the resuspended pellets were subjected to SDS-PAGE and subsequent Western blotting with GST-specific antibodies. The amount of GST protein was determined as a function of optical density in comparison to known amounts of fusion proteins. The recovery of SV, used to correct the amounts of fusion protein bound to SV, was determined by Western blotting with antisynaptophysin antibodies. FLAG-LRRK2 was purified via affinity chromatography using FLAG-M2 agarose beads (Sigma-Aldrich) from HEK293T cells transfected by lipofection using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. The binding of FLAG-LRRK2 to SV was performed as described above with minor modifications: only one concentration of fusion protein (50 nM) was assayed, and FLAG-LRRK2 yield was evaluated via Western blotting with FLAG-specific antibodies.
Pulldown, immunoprecipitation, and antibodies.
For pulldowns, 5 μg of each GST fusion protein was loaded onto glutathione-Sepharose resin (GE Healthcare) and coincubated with adult mouse brain lysate or the LS1 fraction (1 mg of total protein). In immunoprecipitation assays, 10 μg of 1E11 anti-LRRK2 antibody was incubated with 1 mg of protein lysate and loaded onto protein G-Sepharose resin (GE Healthcare). In both procedures, resins were extensively washed in Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 0.2% Triton X-100), followed by final elution of the samples with Laemmli buffer. For protein identification by Western blotting, samples were loaded onto 4 to 12% NuPAGE gels (Invitrogen); the proteins were transferred onto a nitrocellulose membrane (Sigma-Aldrich) at 80 V for 120 min at 4°C. The primary antibodies were applied overnight in blocking buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20, and 5% nonfat dry milk); primary antibodies (source in parentheses) included rat monoclonal anti-LRRK2 1E11 at 1:1,000 (that recognizes an epitope within the LRRK2 kinase domain [25]), mouse anti-synapsin I at 1:1,000 (Synaptic System), mouse anti-SNAP-25 at 1:1,000 (Chemicon), rabbit anti-MAP2 at 1:1,000, mouse anti-MAP6 at 1:1,000, rabbit anti-N-ethylmaleimide-sensitive factor (anti-NSF; 1:1,000 [Cell Signaling]), mouse anti-FLAG at 1:2,000, mouse anti-Rac1 at 1:1,000, mouse anti-β-actin at 1:1,000 mouse anti-syntaxin 1A at 1:1,000, mouse anti-Rab3A at 1:1,000, and mouse anti-α-tubulin at 1:1,000 (Sigma-Aldrich). Secondary antibodies (horseradish peroxidase [HRP]-conjugated anti-mouse, anti-rabbit, or anti-rat antibodies; Jackson ImmunoResearch) were used in a ratio of 1:8,000. The signal was detected using an enhanced chemiluminescence (ECL) detection system (GE Healthcare). Films (Hyperfilm ECL; GE Healthcare) were digitalized using a GS-800 densitometer (Bio-Rad) calibrated according to the manufacturer's instructions, and protein abundance was estimated as a function of the optical density of a specific band quantified by ImageJ software (NIH).
MS/MS identification.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed on an Ultimate 3000 Nano high-performance liquid chromatography (HPLC) system (Dionex) coupled to a linear trap quadrupole (LTQ) OrbitrapXL mass spectrometer (Thermo Fisher Scientific) by a nanospray ion source. Tryptic peptide mixtures were automatically injected and loaded at a flow rate of 30 μl/min in 95% buffer C (0.5% trifluoroacetic acid in HPLC-grade water) and 5% buffer B (98% acetonitrile and 0.1% formic acid in HPLC-grade water) onto a nanotrap column (100-μm interior diameter [i.d.] by 2 cm; Acclaim PepMap100 C18 column, 5-μm particle diameter, 100-Å pore size [Dionex]). After 5 min, peptides were eluted and separated on the analytical column (75-μm i.d. by 15 cm; Acclaim PepMap100 C18 column, 3-μm particle diameter, 100-Å pore size [Dionex]) by a linear gradient from 5% to 40% of buffer B in buffer A (2% acetonitrile and 0.1% formic acid in HPLC grade water) at a flow rate of 300 nl/min over 90 min. Remaining peptides were eluted by a short gradient from 40% to 100% buffer B in 5 min. The eluted peptides were analyzed by the LTQ OrbitrapXL mass spectrometer. From the high-resolution mass spectrometry prescan with a mass range of 300 to 1,500, the 10 most intense peptide ions were selected for fragment analysis in the linear ion trap if they exceeded an intensity of at least 200 counts and if they were at least doubly charged. The normalized collision energy for collision-induced dissociation was set to a value of 35, and the resulting fragments were detected with normal resolution in the linear ion trap. The lock mass option was activated; the background signal with a mass of 445.12002 was used as lock mass (26). Every ion selected for fragmentation was excluded for 30 s by dynamic exclusion. The acquired spectra were processed and analyzed by using Mascot Daemon (version 2.4.0) in combination with ExtractMSN (Thermo-Fisher) with the following settings: cysteine carbamidomethylation as a fixed modification and methionine oxidation and asparagine/glutamine deamidation as variables with a maximum of three modifications per peptide allowed. Mass tolerances for parent and fragment peptides were set to 10 ppm and 1.00 Da, respectively. The database used consisted of a combined set of mouse and E. coli subsets of the Swiss-Prot database (UniProt release 2012_7, published on 11 July 2012; 20,847 entries plus decoy) with spiked-in bait proteins (human LRRK2 and GST of Schistosoma japonicum). Reversed sequences generated by Scaffold were used as a decoy. Mascot result files were analyzed by the Scaffold software (version 4.1.1) (Proteome Software, Inc., Portland, OR) to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 90% probability as specified by the Peptide Prophet algorithm (27). Protein identifications were accepted if they could be established at greater than 95% probability and contained at least two identified unique peptides. Protein probabilities were assigned by the Protein Prophet algorithm (28). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The final data sets describing the LRRK2 interactome were generated based on at least four independent experiments for each bait (LRRK2 WD40, LRRK2 LRR, and GST control). Proteins were considered specific LRRK2 WD40 interactors if they were found in at least two independent pulldown experiments and if they were, based on spectral counting, at least 2-fold enriched in the LRRK2 WD40 pulldown set compared to the GST control set. Typical contaminants, including E. coli proteins, ribosomal proteins, and keratins, were excluded.
Exoendocytotic assay, immunocytochemistry, and quantification.
The endocytosis assay to monitor SV recycling was performed as described previously with minor modifications (13, 29) in neurons transfected at DIV12 with RFP-WD40 or RFP-RACK1 or DsRed plus GFP to visualize cellular processes. Briefly, rabbit polyclonal antibodies directed against the intravesicular domain of synaptotagmin 1 (Synaptic System) were diluted 1:400 in Tyrode solution (124 mM NaCl, 5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 30 mM glucose, 25 mM HEPES, pH 7.4) and applied for 5 min at room temperature (RT) on living cultures. After extensive washing with Tyrode solution, neurons were fixed in 4% paraformaldehyde and 4% sucrose at room temperature. Where indicated in the figure legends, rat anti-FLAG (1:100; kindly provided by E. Kremmer, Institute of Molecular Immunology, Helmholtz Zentrum München), rabbit anti-SV2A (1:400; Synaptic System), or mouse anti-SNAP-25 (1:1,000; Sigma-Aldrich) antibody was applied in GDB buffer (30 mM phosphate buffer, pH 7.4, containing 0.2% gelatin, 0.5% Triton X-100, and 0.8 M NaCl) overnight at 4°C. Cy5-coupled secondary antibodies and 4′,6′-diamidino-2-phenylindole (DAPI; Life Technologies) were diluted 1:1,000 in GDB buffer and applied for 1 h. Transfected neurons were randomly chosen in at least four independent experiments for each condition. The fluorescence images were acquired using an LSM Zeiss 510 confocal microscope with a Zeiss 63× objective (Karl Zeiss, Jena, Germany) at a resolution of 1 pixel equal to 0.098 μm. Image analysis was performed using ImageJ (NIH). To quantify synaptotagmin- or SV2A-positive clusters, GFP-positive processes were manually tracked, and the number of positive clusters in the region of interest was automatically counted. To evaluate neuron morphology, neurites were randomly selected and manually traced, and length and number of processes were automatically determined and logged into Microsoft Excel as described previously (30).
Electron microscopy.
Purified GST- or 6×His-tagged fusion proteins were adsorbed onto carbon-coated grids that were glow discharged in air before the application of 5 μl of protein solution. Excess protein solution was blotted off after 2 min. The adsorbed molecules were negatively stained for 30 s using 5 μl of stain solution as indicated in the figure legends. Micrographs were recorded on Kodak SO-163 image film (Sigma-Aldrich) using JEOL JEM 2010 and JEOL JEM 100CX electron microscopes operated at 120 and 100 kV, respectively, at defocus values of 600 to 900 nm. Suitable micrographs were selected by optical diffraction and digitized using a Flextight Precision II (Hasselblad) scanner, resulting in pixel sizes of 1.59 or 1.28 Å at the specimen level. Particles were selected and extracted using EMAN1 (31) and classified using IMAGIC, version 5.0 (Image Science Software).
Homology modeling.
Modeling templates were identified in the Protein Data Bank (PDB) (32, 33) using the profile-profile alignment program Phyre2 (3). The initial homology model was built based on the WD40 domain structure of RACK1 from Arabidopsis thaliana (PDB code 3DM0) (34). The model structure was further refined using the YASARA energy minimization server to increase the model accuracy (35). The quality of the final structure model was validated using PROCHECK (36). PyMOL was used for visualization of the final model.
Statistical analysis.
All data are expressed as means ± standard error of the means (SE). Data were analyzed with an unpaired Student's t test (two classes) or by analysis of variance (ANOVA) followed by Tukey's post hoc test (more than two classes). The indication of the number of experiments (n) and level of significance (P) are given throughout the text.
Database accession number.
The interactome data set has been submitted to the IntAct database under accession number IM-20537 (http://www.ebi.ac.uk/intact/search/do/search?searchString=imex:IM-20537).
RESULTS
WD40 links LRRK2 to presynaptic proteins.
Previous findings, including our own work, suggest that LRRK2 regulates trafficking of synaptic vesicles (SV) (13, 37, 38). Here, we investigated the interaction between LRRK2 and SV at the molecular level. For this purpose, we first tested the subcellular distribution of LRRK2. Subcellular fractionation of mouse brain lysate revealed cosedimentation of LRRK2 with synapsin I and synaptophysin in a biochemically defined fraction enriched in SV (Fig. 1A, lane US). Interestingly, mild salt treatment, reported to partially remove associated proteins such as synapsin I from SV (23), did not reduce the amount of LRRK2 or synaptophysin found in the SV fraction (Fig. 1A, lane SSV). Next, we incubated full-length FLAG-LRRK2 at a nanomolar concentration with purified SV and tested the extent of binding in a high-speed sedimentation assay (24). Western blot analysis using a FLAG-specific antibody revealed that recombinant LRRK2 associates to native SV. Interestingly, LRRK2 showed a binding affinity similar to that of SSV (Fig. 1B and C). In the same SV population we described a strong reduction in the amount of synapsin I upon salt treatment (Fig. 1B and D). These data suggest that LRRK2 forms a complex with SV. Next, we investigated if LRRK2 associates to SV through specific protein-protein interactions. The immunoprecipitation of LRRK2 showed that well-described SV proteins such as NSF, syntaxin 1A, synapsin I, dynamin-1, MUNC18-1, and VAMP2 as well as actin and tubulin are LRRK2-interacting proteins (Fig. 1E) (13, 39). LRRK2 contains two domains that typically mediate protein-protein interactions, an N-terminal LRR and a C-terminal WD40 domain (5, 10, 37). Thus, we proceeded with a systematic analysis of protein-protein interactions conferred by LRRK2 LRR and WD40 domains. To this aim, we expressed the LRRK2 LRR and WD40 domains as GST fusion proteins (herein termed LRRK2 LRR and LRRK2 WD40) (see Fig. S1A in the supplemental material). GST-only served as a control in order to detect false positives caused by unspecific binding to the affinity tag or matrix. Equal amounts of each protein (baits and control) were loaded onto glutathione-Sepharose resin and incubated with adult mouse brain lysates. Interacting proteins were identified by silver staining (see Fig. S1B), Western blot assays (see Fig. S1C), and MS/MS spectrometry (see Table S1 in the supplemental material). This combined approach revealed a panel of 17 and 42 putative interactors for LRRK2 LRR and LRRK2 WD40, respectively, and suggested the C-terminal WD40 domain as an important hub for protein-protein interaction within LRRK2. Thus, we performed a more detailed investigation of the interactions conferred by LRRK2 WD40 combining Western blot assays and MS/MS spectrometry. As a protein source, we used mouse brain lysate or the LS1 fraction, partially enriched in SV. Our analysis identified 84 putative LRRK2 WD40 interactors (Table 1); in particular, we demonstrated that LRRK2 WD40 has affinity for NSF, syntaxin 1A, synapsin I, VAMP2, Rab3A, MAP2, Rac1, actin, tubulin, and Hsp90 but not for synaptophysin (Fig. 2). Surprisingly, we found that LRRK2 WD40 binds endogenous LRRK2, implying a role for this domain in LRRK2 dimerization (10). WD40 repeats commonly constitute a molecular platform for protein-protein interactions (12). In order to determine the specificity/exclusiveness of the protein interactions determined by LRRK2 WD40, we investigated the interactome of RACK1, an unrelated protein encompassing a well-defined WD40 repeat domain (40). We performed pulldown experiments using full-length RACK1 fused to GST as a bait protein and analyzed the coprecipitated proteins by Western blotting (see Fig. S2A and B in the supplemental material) and mass spectrometry (see Table S2). As a result of this analysis, we found that only seven putative LRRK2 WD40 binding partners demonstrated affinity also for RACK1. Notably, among them, we did not identify LRRK2 itself, MAP2, actin, syntaxin 1A, and NSF, indicating that these proteins are bona fide specific binders of LRRK2 WD40.
FIG 1.
LRRK2 binds presynaptic proteins. (A) Distribution of LRRK2, synapsin I, and synaptophysin immunoreactivities in subcellular fractions of rat forebrain. LRRK2 is present in highly purified vesicle fractions (US), and its association with the vesicle membrane is not affected by salt treatment (SSV). H, homogenate. (B) LRRK2 binds to SV. Full-length FLAG-LRRK2, purified from transfected HEK293T cells, was incubated with unstripped SV (US) or salt-stripped SV (SSV) before high-speed sedimentation. Representative Western blots stained with anti-FLAG antibody show the initial amount of FLAG-LRRK2 protein (total), the yield of protein precipitated by US or SSV (bound), and the amount of endogenous synapsin I associated to SV. Fusion proteins were incubated with equal amounts of SV (monitored by antisynaptophysin staining). (C) The graph reports the yield of FLAG-LRRK2 precipitated by US and SSV, expressed as a fraction of FLAG-LRRK2 total protein and normalized against the SV total protein amount. (D) The graph reports the amount of endogenous synapsin I associated to US or SSV. Data are expressed as optical density (OD) in arbitrary units. *, P < 0.05, n = 4 (Student's t test). (E) Immunoprecipitation of endogenous LRRK2 from adult brain lysate confirms the interaction between LRRK2 and selected presynaptic and cytoskeletal proteins.
TABLE 1.
Identification of LRRK2 WD40 interactors
| Accession no.a | Protein name | Gene | Experiment(s)b | Mass (kDa) | Coverage (%)c | No. of WD40 repeatsd | Sourcee |
|---|---|---|---|---|---|---|---|
| P60711 (P60710) | Actin, beta | Actb | MS, WB, IP | 42 | 46.0 | 17 | LS1, total brain |
| P85515 | Alpha-centractin | Actr1a | MS, WB | 43 | 16.0 | 4 | LS1 fraction |
| P39069 | Adenylate kinase 1 | Ak1 | MS | 22 | 14.0 | 2 | LS1 fraction |
| Q9DBG3 | Adaptor-related protein complex 2, beta 1 subunit | Ap2b1 | MS | 105 | 4.0 | 4 | Total brain |
| Q08163 | Adenylyl cyclase-associated protein 1 | Cap1 | MS | 52 | 6.0 | 2 | LS1 fraction |
| P84079 | ADP-ribosylation factor 1 | Arf1 | MS | 21 | 15.0 | 2 | LS1 fraction |
| Q5XI73 | Rho GDP dissociation inhibitor (GDI) alpha | Arhgdia | MS | 23 | 33.0 | 4 | LS1 fraction |
| Q8VDN2 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | Atp1a1 | MS | 113 | 17.0 | 4 | Total brain |
| Q6PIE5 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | Atp1a2 | MS | 112 | 17.0 | 5 | Total brain |
| P06687 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | Atp1a3 | MS | 112 | 12.0 | 10 | LS1 fraction |
| Q9R0K7 | ATPase, Ca2+ transporting, plasma membrane 2 | Atp2b2 | MS | 133 | 4.0 | 3 | Total brain |
| Q03265 | ATP synthase alpha subunit, isoform 1 | Atp5a1 | MS | 60 | 21.0 | 12 | LS1 fraction |
| P56480 (P10719) | ATP synthase beta subunit | Atp5b | MS | 56 | 13.0 | 5 | LS1 fraction, total brain |
| P62815 | V-type proton ATPase subunit B, brain isoform | Atp6v1b2 | MS | 57 | 36.0 | 14 | LS1 fraction |
| Q6PCU2 | ATPase, H+ transporting, lysosomal V1 subunit E1 | Atp6v1e1 | MS | 26 | 15.0 | 4 | LS1 fraction |
| P47728 | Calbindin 2 | Calb2 | MS | 31 | 14.0 | 3 | LS1 fraction |
| P28480 | T-complex protein 1 subunit alpha | Tcp1 | MS | 60 | 11.0 | 5 | LS1 fraction |
| Q5XIM9 | T-complex protein 1 subunit beta | Cct2 | MS | 57 | 18.0 | 6 | LS1 fraction |
| Q68FD5 | Clathrin, heavy polypeptide (Hc) | Cltc | MS | 192 | 24.0 | 28 | Total brain |
| P63041 | Complexin 1 | Cplx1 | MS | 15 | 17.0 | 2 | LS1 fraction |
| P39053 | Dynamin 1 | Dnm1 | MS, IP | 98 | 8.0 | 7 | Total brain |
| Q62952 | Dihydropyrimidinase-like 3 | Dpysl3 | MS | 62 | 31.0 | 7 | LS1 fraction |
| P38650 | Dynein cytoplasmic 1 heavy chain 1 | Dync1h1 | MS | 532 | 18.0 | 4 | LS1 fraction |
| P85845 | Fascin homolog | Fscn1 | MS | 54 | 9.0 | 3 | LS1 fraction |
| P50398 | Rab GDP dissociation inhibitor alpha | Gdi1 | MS | 51 | 49.0 | 16 | LS1 fraction |
| P50399 | Rab GDP dissociation inhibitor beta | Gdi2 | MS | 51 | 25.0 | 5 | LS1 fraction |
| P59215 (P18872) | Guanine nucleotide-binding protein G(o) subunit alpha | Gnao1 | MS | 40 | 12.0 | 3 | LS1 fraction, total brain |
| P07901 | Heat shock protein 90, alpha | Hsp90aa1 | MS, WB | 85 | 5.0 | 2 | Total brain |
| P11499 | Heat shock protein 90, beta | Hsp90ab1 | MS, WB | 83 | 7.0 | 2 | Total brain |
| O35814 | Stress-induced phosphoprotein 1 | Stip1 | MS | 63 | 10.0 | 4 | LS1 fraction |
| Q2PQA9 | Kinesin-1 heavy chain | Kif5b | MS | 110 | 4.0 | 3 | LS1 fraction |
| Q9WV63 | Kinesin-like protein KIF2A | Kif2a | MS | 80 | 5.0 | 3 | LS1 fraction |
| P63086 | Mitogen-activated protein kinase 1 | Mapk1 | MS | 41 | 20.0 | 6 | LS1 fraction |
| Q3B8Q0 | Microtubule-associated protein RP/EB family member 2 | Mapre2 | MS | 37 | 11.0 | 2 | LS1 fraction |
| P19332 | Microtubule-associated protein tau | Mapt | MS | 76 | 7.0 | 5 | LS1 fraction |
| P34926 | Microtubule-associated protein 1A | Map1a | MS | 300 | 2.0 | 3 | LS1 fraction |
| P15205 | Microtubule-associated protein 1B | Map1b | MS | 270 | 2.0 | 5 | LS1 fraction |
| P15146 (P20357) | Microtubule-associated protein 2 | Map2 | MS | 199 | 6.0 | 6 | LS1 fraction, total brain |
| Q9QYF3 | Myosin VA | Myo5a | MS | 212 | 5.0 | 8 | LS1 fraction |
| Q63610 | Tropomyosin alpha-3 | Tpm3 | MS | 29 | 19.0 | 4 | LS1 fraction |
| P85969 | Beta-soluble NSF attachment protein | Napb | MS | 34 | 11.0 | 3 | LS1 fraction |
| Q9Z0E0 | Neurochondrin | Ncdn | MS | 79 | 4.0 | 2 | LS1 fraction |
| P46460 | N-Ethylmaleimide sensitive fusion protein | Nsf | MS, WB, IP | 83 | 10.0 | 6 | Total brain |
| Q9QXU9 | ProSAAS | Pcsk1n | MS | 27 | 18.0 | 5 | LS1 fraction |
| Q9EPC6 | Profilin 2 | Pfn2 | MS | 15 | 18.0 | 3 | LS1 fraction |
| P60203 | Proteolipid protein (myelin) 1 | Plp1 | MS | 30 | 9.0 | 2 | LS1 fraction |
| P63329 | PP2B catalytic subunit alpha isoform | Ppp3ca | MS | 59 | 28.0 | 11 | LS1 fraction |
| Q63716 | Peroxiredoxin 1 | Prdx1 | MS | 22 | 20.0 | 3 | LS1 fraction |
| P35704 | Peroxiredoxin 2 | Prdx2 | MS | 22 | 31.0 | 8 | LS1 fraction |
| O35244 | Similar to peroxiredoxin 6 | Prdx6 | MS | 25 | 32.0 | 5 | LS1 fraction |
| P63011 | Ras-related protein Rab-3A | Rab3a | WB | 25 | 31.0 | 7 | Total brain |
| Q9WU34 | Neuron-specific septin-3 | Sept3 | MS | 41 | 11.0 | 4 | LS1 fraction |
| Q9JJM9 | Septin 5 | Sept5 | MS | 43 | 10.0 | 3 | LS1 fraction |
| Q9WVC0 | Septin 7 | Sept7 | MS | 51 | 15.0 | 5 | LS1 fraction |
| O35179 | Endophilin-A1 | Sh3gl2 | MS | 40 | 29.0 | 9 | LS1 fraction |
| Q5PPJ9 | Endophilin-B2 | Sh3glb2 | MS | 45 | 8.0 | 3 | LS1 fraction |
| P60881 | Synaptosome-associated protein 25 | Snap25 | MS, WB | 23 | 14.0 | 2 | LS1 fraction |
| Q61548 | Synaptosome-associated protein 91 | Snap91 | MS | 92 | 4.0 | 3 | Total brain |
| P37377 | Synuclein, alpha | Snca | MS | 14 | 39.0 | 3 | LS1 fraction |
| Q91ZZ3 | Synuclein, beta | Sncb | MS | 14 | 50.0 | 5 | LS1 fraction |
| P16546 | Spectrin alpha 2 | Spna2 | MS | 285 | 4.0 | 9 | Total brain |
| Q62261 | Spectrin beta 2 | Spnb2 | MS | 274 | 1.0 | 2 | Total brain |
| P63039 | 60-kDa heat shock protein | Hspd1 | MS | 61 | 46.0 | 25 | LS1 fraction |
| O35814 | Stress-induced phosphoprotein 1 | Stip1 | MS | 63 | 10.0 | 4 | LS1 fraction |
| P13668 | Stathmin | Stmn1 | MS | 17 | 36.0 | 5 | LS1 fraction |
| P61264 | Syntaxin 1B | Stx1b | MS | 33 | 13.0 | 3 | Total brain |
| P61765 (O08599) | Syntaxin-binding protein 1 | Stxbp1 | MS, IP | 68 | 19.0 | 10 | LS1 fraction, total brain |
| Q9JIS5 | Synaptic vesicle glycoprotein 2A | Sv2a | MS | 83 | 5.0 | 2 | Total brain |
| O88935 | Synapsin-1 | Syn1 | WB, IP | 75 | 16.0 | 5 | Total brain |
| Q63537 (Q64332) | Synapsin-2 | Syn2 | WB, IP | 63 | 17.0 | 7 | LS1 fraction, total brain |
| Q62910 | Synaptojanin 1 | Synj1 | MS | 173 | 4.0 | 5 | LS1 fraction |
| Q6P9V9 (Q99KA2) | Tubulin alpha-1B chain | Tuba1b | MS, WB, IP | 50 | 45.0 | 21 | LS1 fraction, total brain |
| Q5XIF6 | Tubulin alpha-4A chain | Tuba4a | MS | 50 | 36.0 | 5 | LS1 fraction |
| P85108 | Tubulin beta-2A chain | Tubb2a | MS | 50 | 64.0 | 2 | LS1 fraction |
| Q3KRE8 | Tubulin beta-2B chain | Tubb2b | MS | 50 | 64.0 | 10 | LS1 fraction |
| Q4QRB4 | Tubulin beta-3 chain | Tubb3b | MS | 50 | 66.0 | 12 | LS1 fraction |
| Q6P9T8 | Tubulin beta-4B chain | Tubb4b | MS | 50 | 63.0 | 28 | LS1 fraction |
| P69897 | Tubulin beta-5 chain | Tubb5b | MS | 50 | 63.0 | 6 | LS1 fraction |
| Q00981 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | Uchl1 | MS | 25 | 37.0 | 6 | LS1 fraction |
| P63045 | Vesicle-associated membrane protein 2 | Vamp2 | MS, WB, IP | 13 | 46.0 | 6 | LS1 fraction |
| P35213 | 14-3-3 protein beta polypeptide | Ywhab | MS | 28 | 30.0 | 2 | LS1 fraction |
| P62259 | 14-3-3 protein epsilon polypeptide | Ywhae | MS | 29 | 16.0 | 2 | Total brain |
| P68511 | 14-3-3 protein eta polypeptide | Ywhah | MS | 28 | 52.0 | 7 | LS1 fraction |
| P68255 | 14-3-3 protein theta | Ywhaq | MS | 28 | 30.0 | 4 | LS1 fraction |
UniProtKB/Swiss-Prot accession numbers.
Interactors were found by immunoprecipitation of endogenous LRRK2 (IP) or LRRK2 WD40 pulldown assays analyzed by Western blotting (WB) and mass spectrometry (MS).
Sequence coverage for proteins identified by LC-MS/MS.
Number of identified unique peptides.
Lysate from adult mouse brain (total brain) or the LS1 fraction.
FIG 2.
The C-terminal WD40 domain of LRRK2 represents a hub for protein-protein interactions. Western blots are shown, confirming the specific interaction of proteins following a domain-based pulldown assay for the LRRK2 WD40 domain. s-physin, synaptophysin.
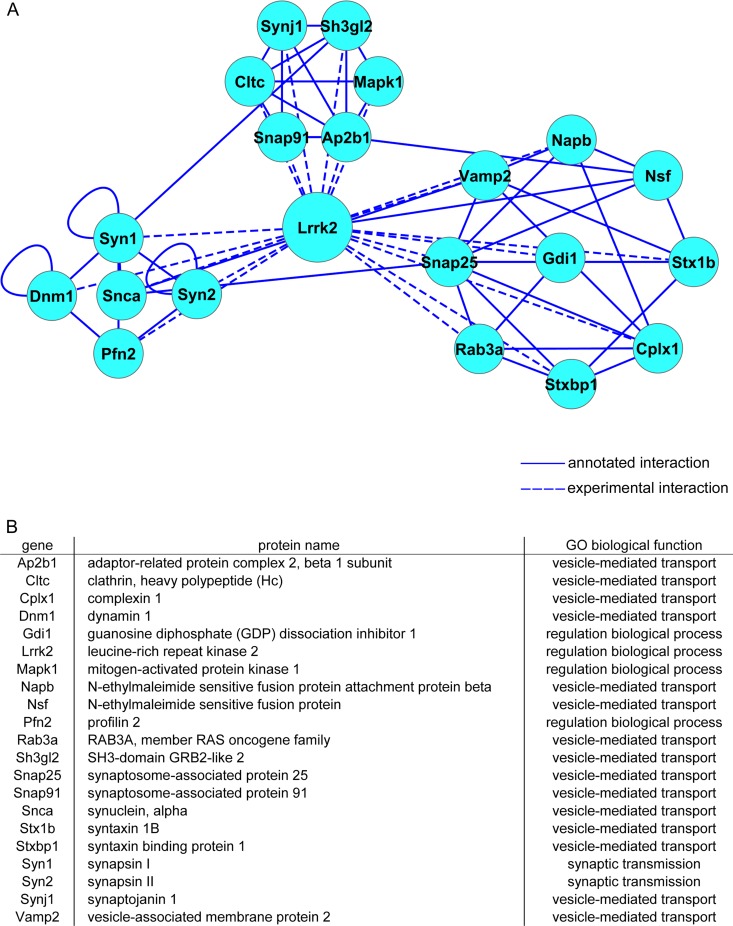
For further analysis, we elaborated the interactome conferred to LRRK2 by its WD40 domain, in silico, using the STRING protein database tools (accessible online at http://string-db.org) (41). The resulting scale-free network was visualized by Cytoscape software (42). The network included 85 nodes (84 interactors plus LRRK2) connected by 160 edges (see Fig. S3 and Table S3 in the supplemental material). In order to identify major hubs within the network, the complete data set was filtered for those nodes qualified by a degree of connectivity higher than four. The filtered data set formed a subnetwork of 21 nodes associated through 62 edges (Fig. 3A). Interestingly, proteins represented in this subnetwork are key determinants of SV trafficking (Fig. 3B). Taken together, our data suggest that the C-terminal WD40 domain of LRRK2 serves as a major hub for its interaction with other proteins and that LRRK2 is part of a highly interconnected protein network involved in synaptic vesicle trafficking.
FIG 3.
LRRK2 interacts with presynaptic proteins. (A) The network of LRRK2 interactors was modeled from STRING annotation on the Cytoscape representation. annotated, interactions annotated on STRING; experimental, interactions described in the manuscript. (B) Gene symbols, names, and gene ontology (GO) terms for the proteins included in the network.
The LRRK2 WD40 domain induces neurotoxicity.
It has been reported that the C-terminal LRRK2 WD40 domain is required for LRRK2-associated neurotoxicity (10). To gain a better understanding of this phenomenon, we cotransfected cortical neurons at DIV3 with GFP and vectors expressing either DsRed, the isolated LRRK2 WD40 domain (RFP-LRRK2 WD40), truncated LRRK2 lacking the C-terminal domain (RFP LRRK2 1–2141), full-length LRRK2 (RFP-LRRK2), or full-length RACK1 (RFP-RACK1) and analyzed neuron morphology at DIV16 (Fig. 4). The overexpression of the full-length RFP LRRK2 severely reduced the number of processes compared to control neurons expressing DsRed. This outcome might be related to the high expression of recombinant full-length LRRK2 achieved in our model (43). Interestingly, while ectopic LRRK2 1–2141 expression did not significantly alter neuron morphology, overexpression of the single LRRK2 WD40 domain significantly impaired process number. Given the severe neurotoxicity observed upon expression of RFP WD40 and RFP LRRK2 from DIV3 to DIV16, we modified our experimental setup and investigated the effect of ectopic LRRK2 construct expression in neurons transfected at DIV10 and imaged at DIV16 (Fig. 5A). Given that we experienced an extremely low efficiency when transfecting full-length LRRK2 in primary cultures at DIV10 (data not shown), we excluded this condition in subsequent experiments. While ectopic DsRed, RACK1, and LRRK2 1–2141 were diffusely distributed in neuronal soma and along neuronal processes, partially colocalizing with the synaptic marker SNAP-25, we found that the ectopically expressed LRRK2 WD40 domain was mainly localized in a perinuclear somatic region, with low colocalization with SNAP-25 (see Fig. S4 in the supplemental material). Next, we considered neuron morphology. The overexpression of RFP-LRRK2 WD40 significantly reduced the number of processes and increased the amount of swollen or fragmented neurites compared to control neurons. In contrast, ectopic LRRK2 1–2141 expression did not influence total neurite number and was associated to a milder increase of swollen processes than LRRK2 WD40. Finally, we found an increased number of processes in RACK1-overexpressing neurons (Fig. 5B and C). Given that neuronal fragmentation represents the first sign of neuronal sufferance (44), our data indicate that the prolonged expression of the isolated LRRK2 WD40 domain induces toxicity in neurons.
FIG 4.
Expression of an LRRK2 WD40 domain construct is sufficient to induce neurotoxicity in primary neurons. Neurons were transfected at DIV3 and imaged at DIV16. Long-lasting overexpression of RFP-LRRK2 WD40 and full-length RFP-LRRK2 significantly reduced the number of processes and increased the amount of swollen or fragmented neurites compared to levels in DsRed-, RFP LRRK2 1–2141-, or RFP-RACK1-transfected neurons. Images show signals acquired for GFP (channel 1, CH1), DsRed or RFP (channel 2, CH2), superimposed channel signals (merge), and tracings. Arrow heads indicate an RFP-LRRK2 WD40-positive cell. Numbers in the tracing panel indicate total process numbers and are expressed as means ± SE. **, P < 0.01 versus DsRed; #, P < 0.01 versus full-length RFP-LRRK2 (ANOVA; Tukey's post hoc test, n = 3; 8 neurons were analyzed for each experimental case). Panel size shown is 200 by 200 μm.
FIG 5.
Expression of an LRRK2 WD40 domain construct induces neurotoxicity in primary neurons. (A) Neurons were transfected at DIV10 and imaged at DIV16. Long-lasting overexpression of RFP-LRRK2 WD40 significantly reduced the number of processes and increased the amount of swollen or fragmented neurites compared to levels in DsRed-, LRRK2 1–2141-, or RFP-RACK1-transfected neurons. Images show signals acquired for GFP (channel 1, CH1), DsRed or RFP (channel 2, CH2), superimposed channel signals (merge), and tracings. Graphs report the number of total processes (B) and number of fragmented processes (C) (means ± SE). *, P < 0.05 versus DsRed; **, P < 0.01 versus DsRed; #, P < 0.05 versus LRRK2 WD40 (ANOVA; Tukey's post hoc test, n = 4; 7 neurons were analyzed for each experimental case). Panel size shown is 200 by 200 μm.
LRRK2 WD40 domain and RACK1 alter SV trafficking.
Given the interactions described between the LRRK2 WD40 domain and presynaptic proteins, we wondered whether the LRRK2 WD40 domain affects proper SV trafficking and distribution. To test this hypothesis, we cotransfected cortical neurons at DIV10 with GFP and either DsRed, RFP-LRRK2 WD40, RFP-RACK1, or LRRK2 1–2141 expression vectors. At DIV12 we determined the SV exoendocytic rate by exposing living cortical neurons to an antisynaptotagmin antibody as previously described (13). Transfected neurons were then tracked via laser confocal microscopy. Recycling vesicles appeared as synaptotagmin-positive clusters along GFP-positive neuronal processes (Fig. 6A). The total vesicle pool was estimated by staining with antibodies directed against an integral SV protein (SV2A) after fixation and permeabilization of the cells (Fig. 6B). While ectopic LRRK2 1–2141 did not influence synaptotagmin uptake, the overexpression of RFP-LRRK2 WD40 as well as RFP-RACK1 induced a significant decrease in the number of synaptotagmin-positive clusters (Fig. 6C). At the same time, neither LRRK2 WD40 nor RACK1 nor LRRK2 1–2141 overexpression affected the amount of SV2A-positive clusters compared to DsRed transfected neurons (Fig. 6D). Next, we examined if the expression of LRRK2 WD40, RACK1, or LRRK2 1–2141 had an impact on the distribution of cycling or total SV pools. To this end, we tracked the distribution of synaptotagmin (cycling SV) and SV2A-positive (total SV) clusters along GFP-positive processes. Interestingly, in LRRK2 WD40 transfected neurons, synaptotagmin-positive clusters were mainly distributed proximally to the cellular soma while SV2A-positive clusters were homogeneously diffused along the entire length of the neurites (Fig. 6E and F). Taken together, these data strongly indicate that ectopic expression of the LRRK2 WD40 domain influences trafficking, distribution, and topology of the SV cycling pool.
FIG 6.
The LRRK2 WD40 domain sequesters cycling synaptic vesicles. The exoendocytotic assay was performed at DIV12 on cortical neurons transfected at DIV10. (A) Cycling SV appear as synaptotagmin (s-tagmin)-positive clusters along neuron processes. Images show signals acquired for synaptotagmin (channel 1, CH1) and DsRed or RFP or FLAG (channel 2, CH2) and their superimposition plus GFP (merge). (B) The total SV pool was revealed by staining with anti-SV2A antibodies upon permeabilization. Images show signals acquired for SV2A (channel 1, CH1) and DsRed or RFP (channel 2, CH2) and their superimposition plus GFP (merge). (C) SV cycling is strongly reduced upon either LRRK2 WD40 or RACK1 overexpression. The graph reports the number of synaptotagmin-positive clusters per 10 μm of GFP-positive process. (D) The total number of SV pools was not altered by LRRK2 WD40, RACK1, or LRRK2 1–2141 overexpression. (E and F) Ectopically expressed LRRK2 WD40 confines cycling SV within a perisomatic region, while total SV pool distribution is not affected. The graph reports the percentage of synaptotagmin- or SV2A-positive clusters distributed within the proximal half of the process. Data are expressed as means ± SE. *, P < 0.01 versus DsRed (ANOVA; Tukey's post hoc test, n = 4; 7 neurons were analyzed for each experimental case). Panel length is 10 μm.
The G2385R PD risk variant alters LRRK2 WD40 binding properties to synaptic vesicles.
The critical role conferred by the LRRK2 WD40 domain is suggested by the existence of the G2385R polymorphism, described as a risk factor for the development of PD (14–16).
Sequence alignments and secondary structure predictions suggest that the LRRK2 C terminus contains a WD40 propeller domain composed of seven β-blades. We assessed the structure of the region comprising LRRK2 residues 2124 to 2499 using homology modeling in order to gain information about the structural features of the LRRK2 C terminus (Fig. 7A; see also Fig. S5A in the supplemental material). Our model, based on the structure of the WD40 protein RACK1 from Arabidopsis thaliana (34), shows that the LRRK2 C-terminal domain is compatible with the characteristic structure of WD40 domains, i.e., seven β-propeller repeats combined to a cleft of basic residues (5, 15). A more detailed analysis of the region surrounding the residue G2385 revealed that G2385 lies in close vicinity of two hydrophobic residues, V2375 and L2439, on one side and with R2442 and R2443 on the other side. Even with its limitations, the structural model suggests that the substitution of G2385 with a bulky and positively charged arginine residue would lead to unfavorable charge repulsions and potentially conformational changes of the protein near the interface between β-blades 5 and 6. This might not only alter the local conformation but also affect the binding surface of WD40 for interaction with interacting proteins and thus impair LRRK2 activity. In order to experimentally assess the structural properties of the LRRK2 C terminus and the alterations caused by the G2385R variant, purified GST fusion proteins LRRK2 WD40 and LRRK2 WD40 G2385R were analyzed by transmission electron microscopy (EM). To meet the demand of EM for highly pure material, purity and concentration of the GST fusion proteins were confirmed by SDS-PAGE (see Fig. S2A in the supplemental material). While particles with distinct shapes were discernible on electron micrographs of negatively stained LRRK2 WD40 fusion proteins, similar structures were not visible for the GST tag alone (see Fig. S5B). Single-particle classification and averaging indicated the existence of particle populations with an annular, doughnut-like appearance and diameters ranging from 5 to 8 nm for LRRK2 WD40 (Fig. 7B). Structures similar in size and shape were also detected for the GST-RACK1 fusion protein (Fig. 7B), indicating that the fold of the LRRK2 WD40 domain is comparable despite limited sequence homology. To ensure that the fusion of the WD40 domains to the GST tag does not induce ring-like structures, N-terminal 6×His-tagged LRRK2 WD40 was subjected to EM (see Fig. S5C in the supplemental material). Although the protein lacking the GST tag turned out to be more aggregation prone, doughnut-like structures similar in size and appearance to those observed in GST fusion proteins were still present. The analysis of the LRRK2 WD40 G2385R variant by EM revealed strong similarity to the other WD40 domain constructs, however, with a wider size distribution ranging from 5 to 13 nm. This larger size heterogeneity may be due to structural alterations in WD40 induced by electrostatic repulsion in the β-propeller fold, as suggested by the structural model.
FIG 7.
The G2385R substitution impacts the local structure of LRRK2 WD40. (A) Combined stick-and-ribbon representation showing a structural homology model (based on RACK1 from Arabidopsis thaliana, PDB 3DM0) for the LRRK2 WD40 domain. The N terminus, C terminus, and the position of G2385 (red sphere) are indicated. (B) Transmission EM images of negatively stained (1% uranyl acetate) LRRK2 WD40, LRRK2 WD40 G2385R, and RACK1 proteins show doughnut-shaped particles consistent with the characteristic structure of WD40 folds. Four representative averaged single-particle two-dimensional projections are shown for each protein (scale bar, 5 nm). (C) LRRK2 WD40 binds SV. Increasing nanomolar amounts of LRRK2 WD40, LRRK2 WD40 G2385R domains, and RACK1 protein were incubated with unstripped SV (US) or salt-stripped SV (SSV) before high-speed sedimentation. Representative Western blots show initial amount of fusion protein (total) and the yield of GST fusion proteins precipitated by US or SSV (bound). Fusion proteins were incubated with equal amounts of SV (monitored by antisynaptophysin staining). (D) The table reports the dissociation constant (KD) describing the binding between the indicated fusion protein and US or SSV. Data are expressed as means ± SE. *, P < 0.05 versus LRRK2 WD40 G2385R binding to US; °, P < 0.01 versus LRRK2 WD40 binding to SSV (ANOVA; Tukey's post hoc test, n = 4). (E) The graph reports the yield of precipitated GST fusion protein normalized versus the SV total protein amount (average data plus fitting) on the y axis and the initial amount of GST fusion protein on the x axis.
In order to further characterize the G2385R variant, we investigated the impact of LRRK2 WD40 G2385R overexpression on neurons. Ectopic LRRK2 WD40 G2385R induced neuronal toxicity and sequestered cycling SV to a similar extent as LRRK2 WD40 (see Fig. S6 in the supplemental material).
The data presented above suggest that the WD40 domain plays a major role in tethering LRRK2 to SV via protein interactions; thus, we asked if the G2385R variant can influence LRRK2 binding to SV. To explore this hypothesis, we incubated LRRK2 WD40 and LRRK2 WD40 G2385R as well as RACK1 GST fusion proteins at nanomolar concentrations with purified SV and tested the extent of binding by the means of a high-speed sedimentation assay (24). Western blot analysis using a GST-specific antibody revealed that LRRK2 WD40 as well as LRRK2 WD40 G2385R and RACK1 binds to purified SV (Fig. 7C to E) while GST alone failed to bind SV at significant rates (see Fig. S5D in the supplemental material). As mild salt treatment partially washes away peripheral SV proteins, we exploited this feature to further investigate the binding properties of the three fusion proteins. Interestingly, the salt treatment did not alter the SV association of LRRK2 WD40 and RACK1, whereas the affinity/binding strength of the LRRK2 WD40 G2385R domain to salt-treated SV was significantly reduced (Fig. 7C to E).
Taken together, these results suggest that the structural alteration induced by the G2385R substitution functionally disturbs LRRK2 WD40 binding properties to SV.
DISCUSSION
Since the first description of LRRK2 as a PD-causative gene, major attention has been devoted to its GTPase and kinase activity, linking disease-associated mutations to altered functional and (patho)physiological enzymatic properties of the protein (45–50). However, LRRK2 C-terminal deletion mutants fail to induce apoptosis and toxicity and demonstrate a reduced kinase activity (Fig. 4 and 5) (10, 51). These reports, however, do not address the question of whether the kinase domain or WD40 domain or both are causative for PD pathology. We, on the other hand, observed that the severe toxicity induced by overexpression of full-length LRRK2 in primary cultures is mimicked by ectopic expression of a construct containing only the C-terminal WD40 domain of LRRK2. Noticeably, in contrast to LRRK2 WD40, RACK1, another WD40 protein, has a positive effect on neurite complexity. These data suggest that the C-terminal WD40 domain has a major role in LRRK2-associated toxicity (43).
From structural prediction it appears likely that the LRRK2 WD40 domain folds as a seven-bladed propeller (15, 52). However, sequence homology is considerably low, and no experimental evidence for this assumption has existed so far. According to our EM analysis, the purified LRRK2 WD40 fusion protein forms doughnut-like structures with an average diameter of 5 to 8 nm, resembling EM structures reported for known WD40-containing proteins such as the splicing factor Prp19p (53) or RACK1 (this paper). These findings suggest that the LRRK2 C terminus may indeed form a propeller-like structure, in agreement with a WD40 fold. WD repeat-containing proteins execute a broad spectrum of critical functions. They participate in organizing cytoskeleton assembly, mitotic spindle formation, and vesicular trafficking (11, 12, 54, 55). Furthermore, increasing evidence describes WD40 repeat domains as molecular hubs orchestrating complex protein-protein interactions. Structurally, this feature is based on the evolutionary principle to generate binding epitopes with different specificities by concatenation of stable folded repeats and loops with variable sequences (11). Following the “guilt by association” principle (56), we systematically analyzed the domain-specific interactome of the LRRK2 C terminus in order to assign specific physiological functions to this domain. Through its WD40 domain, LRRK2 interacts with critical players of the SV cycle such as NSF, syntaxin 1A, synapsin I, dynamin-1, MUNC18-1, VAMP2, synaptojanin, and synuclein. Thus, the ability of LRRK2 to influence vesicle trafficking (13, 37, 38) is likely to involve its WD40 domain. Accordingly, we observed a severe reduction in the transport of synaptotagmin-labeled (cycling) SV to distal parts of neuronal processes upon overexpression of LRRK2 WD40 domain constructs; such output might be read as a dominant negative effect of ectopic LRRK2 WD40 executed on endogenous LRRK2 function. RACK1 is a seven-bladed WD40 propeller protein (19, 40), reported to bind and anchor recycling endosomal vesicles to centrosomes (57). Accordingly, we reported that RACK1 significantly reduces SV recycling (but not spatial segregation) once it is overexpressed in neurons and that it cosediments with pure SV. Thus, the ability to bind and sequester SV might be a biochemical property common to several WD40-containing proteins or, alternatively, might arise as an in vitro effect due to high local concentrations of WD40 domains. Even if we cannot completely rule out the latter explanation, our data suggest a peculiar physiological role for LRRK2 at the synaptic site. In fact, we demonstrated here that endogenous LRRK2 can be detected in a pure SV fraction, that full-length LRRK2 binds SV, and that LRRK2 WD40, but not LRRK2 LRR or full-length RACK1, interacts with several proteins involved in SV trafficking. Thus, we propose that LRRK2 association with SV is mediated by the interaction of its WD40 domain with SV-integral and -associated proteins.
The description of the G2385R point mutation within the WD40 domain as the main PD risk variant in the Chinese Han and Korean population further underlines the functional and pathological role of this domain (14). G2385R carrier patients demonstrate clinical features similar to noncarrier patients; however, the G2385R variant does correlate to a small but significant effect in lowering the age of PD onset (16). It has recently been reported that G2385R has a mild impact on LRRK2 biochemical properties, such as reduced LRRK2 kinase activity and interactions with other proteins (9), while it neither influences LRRK2 toxicity in neuron cultures nor affects overall autophosphorylation (46). Accordingly, we reported here that LRRK2 WD40 G2385R behaved similarly to LRRK2 WD40 once it was overexpressed in primary neurons. Comparative protein models predict that the glycine 2385 residue stays at the surface of the WD40 domain (Fig. 7A) (15). The G2385R variant replaces the glycine with a long, positively charged arginine residue, thus increasing the net positive charge of the domain and likely inducing an electrostatic repulsion between WD40 repeats 5 and 6. This may explain the increased mean diameter observed for LRRK2 WD40 G2385R in our EM analysis. The changes in surface charge and local structural features in the WD40 fold are expected to result in altered biochemical properties which affect protein-protein interaction strength and quality. Indeed, we reported that the association with SV is partially impaired by the glycine-to-arginine change. In particular, while LRRK2 WD40 showed a high affinity toward both native and salt-treated SV, LRRK2 WD40 G2385R binding to SV was significantly sensitive to salt treatment. In line with our proposal of LRRK2 as a synaptic scaffold protein involved in vesicular trafficking and vesicle storage, our data strongly support the idea that in addition to increased kinase activity, other molecular mechanisms, such as altered protein binding, may underlie LRRK2-associated forms of PD. In particular, given the recent independent evidence linking LRRK2 dysfunction to neurotransmission defects in PD models (58–60) and in patients carrying LRRK2 mutations (61), an altered presynaptic vesicle transport, storage, and release kinetics may arise as a common pathway disturbed by the different LRRK2 pathological mutations described so far and become a future target for pharmacological treatment.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Nathalie Théret (INSERM, Rennes, France) for reagents, to Sandra Helm for technical assistance, and to Peijian Zou and Arie Geerlof for initial bacterial expression clones of the WD40 domain. In addition we thank Pablo Porras and Henning Hermjakob (EMBL-EBI) for data integration into the IntAct database.
This work was supported by the National Genome Research Framework program NGFN-Plus (grant 01GS08140, subproject 12), the Helmholtz Alliance for Mental Health in an Aging Society (grant HA-215, topic 3 WP11), the European Community's Seventh Framework Program FP7/2009 under grant agreement number 241955, number 278568, PRIMES, and number 241481, AFFINOMICS (to M.U.), and the LRRK2 Biology LEAPS 2012 award of the Michael J. Fox Foundation to G.P., C.J.G., M.S., and M.U. G.P. and F.O. are supported by Fondazione Cariplo (grant 2011-0540), MJFF, and Fondazione Telethon (grant GGP12237). G.P. is supported also by the FIRB program (grant RBFR08F82X_002) and Fondazione Grigioni per il morbo di Parkinson. F.O. is grateful to PRIN 2010-11. M.M. is supported by Fondazione Cariplo (grant 2008-3184).
Author contributions are as follows. G.P., C.J.G., F.O. and M.U. designed the study. G.P., C.J.G., and M.U. wrote the paper. G.P., M.D.C., A.M., F.P., F.A., F.G., P.J., C.J.K., F.V.Z., and A.K. performed experiments. G.P., C.J.G., C.S., C.J.O.K., M.M., A.V., C.J.O.K., L.P., M.Z, S.W., M.S., and M.U. analyzed the data and/or provided data analysis expertise.
Footnotes
Published ahead of print 31 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00914-13.
REFERENCES
- 1.Clarke C, Moore AP. 2005. Parkinson's disease. Clin. Evid. 13:1658–1677 [PubMed] [Google Scholar]
- 2.Fahn S. 2003. Description of Parkinson's disease as a clinical syndrome. Ann. N. Y. Acad. Sci. 991:1–14. 10.1111/j.1749-6632.2003.tb07458.x [DOI] [PubMed] [Google Scholar]
- 3.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 4.Bosgraaf L, Van Haastert PJ. 2003. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta 1643:5–10. 10.1016/j.bbamcr.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 5.Mills RD, Mulhern TD, Cheng H-C, Culvenor JG. 2012. Analysis of LRRK2 accessory repeat domains: prediction of repeat length, number and sites of Parkinson's disease mutations. Biochem. Soc. Trans. 40:1086–1089. 10.1042/BST20120088 [DOI] [PubMed] [Google Scholar]
- 6.Guo L, Wang W, Chen SG. 2006. Leucine-rich repeat kinase 2: relevance to Parkinson's disease. Int. J. Biochem. Cell Biol. 38:1469–1475. 10.1016/j.biocel.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 7.Goldwurm S, Zini M, Di Fonzo A, De Gaspari D, Siri C, Simons EJ, van Doeselaar M, Tesei S, Antonini A, Canesi M, Zecchinelli A, Mariani C, Meucci N, Sacilotto G, Cilia R, Isaias IU, Bonetti A, Sironi F, Ricca S, Oostra BA, Bonifati V, Pezzoli G. 2006. LRRK2 G2019S mutation and Parkinson's disease: a clinical, neuropsychological and neuropsychiatric study in a large Italian sample. Parkinsonism Relat. Disord. 12:410–419. 10.1016/j.parkreldis.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 8.Bonifati V. 2006. Parkinson's disease: the LRRK2–G2019S mutation: opening a novel era in Parkinson's disease genetics. Eur. J. Hum. Genet. 14:1061–1062. 10.1038/sj.ejhg.5201695 [DOI] [PubMed] [Google Scholar]
- 9.Rudenko IN, Kaganovich A, Hauser DN, Beylina A, Chia R, Ding J, Maric D, Jaffe H, Cookson MR. 2012. The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson's disease is a partial loss-of-function mutation. Biochem. J. 446:99–111. 10.1042/BJ20120637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen ND, Peng Y, Ho CC-Y, Rideout HJ, Petrey D, Liu P, Dauer WT. 2009. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One 4:e8463. 10.1371/journal.pone.0008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem. Sci. 35:565–574. 10.1016/j.tibs.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 12.Smith TF. 2008. Diversity of WD-repeat proteins. Subcell. Biochem. 48:20–30. 10.1007/978-0-387-09595-0_3 [DOI] [PubMed] [Google Scholar]
- 13.Piccoli G, Condliffe SB, Bauer M, Giesert F, Boldt K, De Astis S, Meixner A, Sarioglu H, Vogt-Weisenhorn DM, Wurst W, Gloeckner CJ, Matteoli M, Sala C, Ueffing M. 2011. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J. Neurosci. 31:2225–2237. 10.1523/JNEUROSCI.3730-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan E-K. 2006. Identification of a common genetic risk variant (LRRK2 Gly2385Arg) in Parkinson's disease. Ann. Acad. Med. Singapore 35:840–842 [PubMed] [Google Scholar]
- 15.Tan EK, Zhao Y, Skipper L, Tan MG, Di Fonzo A, Sun L, Fook-Chong S, Tang S, Chua E, Yuen Y, Tan L, Pavanni R, Wong MC, Kolatkar P, Lu CS, Bonifati V, Liu JJ. 2007. The LRRK2 Gly2385Arg variant is associated with Parkinson's disease: genetic and functional evidence. Hum. Genet. 120:857–863. 10.1007/s00439-006-0268-0 [DOI] [PubMed] [Google Scholar]
- 16.Tan EK, Peng R, Wu YR, Wu RM, Wu-Chou YH, Tan LC, An XK, Chen CM, Fook-Chong S, Lu CS. 2009. LRRK2 G2385R modulates age at onset in Parkinson's disease: A multi-center pooled analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B:1022–1023. 10.1002/ajmg.b.30923 [DOI] [PubMed] [Google Scholar]
- 17.Brewer GJ, Torricelli JR, Evege EK, Price PJ. 1993. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35:567–576. 10.1002/jnr.490350513 [DOI] [PubMed] [Google Scholar]
- 18.Gloeckner CJ, Boldt K, Schumacher A, Ueffing M. 2009. Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Proteomics 564:359–372. 10.1007/978-1-60761-157-8_21 [DOI] [PubMed] [Google Scholar]
- 19.Bourd-Boittin K, Le Pabic H, Bonnier D, L'Helgoualc'h A, Théret N. 2008. RACK1, a new ADAM12 interacting protein. Contribution to liver fibrogenesis. J. Biol. Chem. 283:26000–26009. 10.1074/jbc.M709829200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. 2006. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 15:223–232. 10.1093/hmg/ddi439 [DOI] [PubMed] [Google Scholar]
- 21.Xia Z, Dudek H, Miranti CK, Greenberg ME. 1996. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J. Neurosci. 16:5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frangioni JV, Neel BG. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179–187. 10.1006/abio.1993.1170 [DOI] [PubMed] [Google Scholar]
- 23.Huttner WB, Schiebler W, Greengard P, De Camilli P. 1983. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 96:1374–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messa M, Congia S, Defranchi E, Valtorta F, Fassio A, Onofri F, Benfenati F. 2010. Tyrosine phosphorylation of synapsin I by Src regulates synaptic-vesicle trafficking. J. Cell Sci. 123:2256–2265. 10.1242/jcs.068445 [DOI] [PubMed] [Google Scholar]
- 25.Gillardon F, Kremmer E, Froehlich T, Ueffing M, Hengerer B, Gloeckner CJ. 2013. ATP-competitive LRRK2 inhibitors interfere with monoclonal antibody binding to the kinase domain of LRRK2 under native conditions. A method to directly monitor the active conformation of LRRK2? J. Neurosci. Methods 214:62–68. 10.1016/j.jneumeth.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 26.Olsen JV. 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4:2010–2021. 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- 27.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 29.Matteoli M, Takei K, Perin MS, Sudhof TC, De Camilli P. 1992. Exo-endocytotic recycling of synaptic vesicles in developing processes of cultured hippocampal neurons. J. Cell Biol. 117:849–861. 10.1083/jcb.117.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccoli G, Verpelli C, Tonna N, Romorini S, Alessio M, Nairn AC, Bachi A, Sala C. 2007. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J. Prot. Res. 6:3203–3215. 10.1021/pr0701308 [DOI] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 32.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. 2000. The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 7(Suppl):957–959. 10.1038/80734 [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. 2002. The Protein Data Bank. Acta Crystallogr. D. Biol. Crystallogr. 58:899–907. 10.1107/S0907444902003451 [DOI] [PubMed] [Google Scholar]
- 34.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. 2008. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 17:1771–1780. 10.1110/ps.035121.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieger E, Koraimann G, Vriend G. 2002. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 47:393–402. 10.1002/prot.10104 [DOI] [PubMed] [Google Scholar]
- 36.Laskowski R, Rullmann JA, MacArthur M, Kaptein R, Thornton J. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477–486 [DOI] [PubMed] [Google Scholar]
- 37.Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim C-H, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. 2008. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 314:2055–2065. 10.1016/j.yexcr.2008.02.015 [DOI] [PubMed] [Google Scholar]
- 38.Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. 2010. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 6:e1000902. 10.1371/journal.pgen.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meixner A, Boldt K, Van Troys M, Askenazi M, Gloeckner CJ, Bauer M, Marto JA, Ampe C, Kinkl N, Ueffing M. 2011. A QUICK screen for Lrrk2 interaction partners-leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol. Cell Proteomics 10:M110.001172. 10.1074/mcp.M110.001172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sengupta J, Nilsson J, Gursky R, Spahn CMT, Nissen P, Frank J. 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 10:957–962. 10.1038/nsmb822 [DOI] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:D561–D568. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. 2014. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J. Neurosci. 34:418–433. 10.1523/JNEUROSCI.2712-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chernova T, Steinert JR, Guerin CJ, Nicotera P, Forsythe ID, Smith AG. 2007. Neurite degeneration induced by heme deficiency mediated via inhibition of NMDA receptor-dependent extracellular signal-regulated kinase 1/2 activation. J. Neurosci. 27:8475–8485. 10.1523/JNEUROSCI.0792-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. 2009. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J. Neurochem. 109:959–968. 10.1111/j.1471-4159.2009.06024.x [DOI] [PubMed] [Google Scholar]
- 46.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. 2007. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 16:223–232. 10.1093/hmg/ddl471 [DOI] [PubMed] [Google Scholar]
- 47.Lee BD, Shin J-H, VanKampen J, Petrucelli L, West AB, Ko HS, Lee Y-I, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, Dawson VL, Dawson TM. 2010. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat. Med. 16:998–1000. 10.1038/nm.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kett LR, Boassa D, Ho CC-Y, Rideout HJ, Hu J, Terada M, Ellisman M, Dauer WT. 2012. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 21:890–899. 10.1093/hmg/ddr526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. 2006. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 23:329–341. 10.1016/j.nbd.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 50.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. 2008. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 27:2432–2443. 10.1038/emboj.2008.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iaccarino C, Crosio C, Vitale C, Sanna G, Carri MT, Barone P. 2007. Apoptotic mechanisms in mutant LRRK2-mediated cell death. Hum. Mol. Genet. 16:1319–1326. 10.1093/hmg/ddm080 [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Jiang F, Zhuo Z, Wu X-H, Wu Y-D. 2013. A method for WD40 repeat detection and secondary structure prediction. PLoS One 8:e65705. 10.1371/journal.pone.0065705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohi MD, Kooi CWV, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, Walz T, Gould KL. 2005. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 25:451–460. 10.1128/MCB.25.1.451-460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pryer NK, Salama NR, Schekman R, Kaiser CA. 1993. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J. Cell Biol. 120:865–875. 10.1083/jcb.120.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaisman N, Tsouladze A, Robzyk K, Ben-Yehuda S, Kupiec M, Kassir Y. 1995. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol. Gen. Genet. 247:123–136. 10.1007/BF00705642 [DOI] [PubMed] [Google Scholar]
- 56.Wang PI, Marcotte EM. 2010. It's the machine that matters: predicting gene function and phenotype from protein networks. J. Proteomics 73:2277–2289. 10.1016/j.jprot.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ai E, Skop AR. 2009. Endosomal recycling regulation during cytokinesis. Commun. Integr. Biol. 2:444–447. 10.4161/cib.2.5.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Patel JC, Wang J, Avshalumov MV, Nicholson C, Buxbaum JD, Elder GA, Rice ME, Yue Z. 2010. Enhanced striatal dopamine transmission and motor performance with LRRK2 overexpression in mice is eliminated by familial Parkinson's disease mutation G2019S. J. Neurosci. 30:1788–1797. 10.1523/JNEUROSCI.5604-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. 2009. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc. Natl. Acad. Sci. 106:14622–14627. 10.1073/pnas.0906334106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, Beal MF, Burke RE, Li C. 2009. Mutant LRRK2R1441G BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat. Neurosci. 12:826–828. 10.1038/nn.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sossi V, de la Fuente-Fernández R, Nandhagopal R, Schulzer M, McKenzie J, Ruth TJ, Aasly JO, Farrer MJ, Wszolek ZK, Stoessl JA. 2010. Dopamine turnover increases in asymptomatic LRRK2 mutations carriers. Mov. Disord. 25:2717–2723. 10.1002/mds.23356 [DOI] [PubMed] [Google Scholar]
- 62.Berg D, Schweitzer KJ, Leitner P, Zimprich A, Lichtner P, Belcredi P, Brüssel T, Schulte C, Maass S, Nägele T, Wszolek ZK, Gasser T. 2005. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease*. Brain 128:3000–3011. 10.1093/brain/awh666 [DOI] [PubMed] [Google Scholar]
- 63.Brice A. 2005. Genetics of Parkinson's disease: LRRK2 on the rise. Brain 128:2760–2762. 10.1093/brain/awh676 [DOI] [PubMed] [Google Scholar]
- 64.Hardy J, Lewis P, Revesz T, Lees A, Paisan-Ruiz C. 2009. The genetics of Parkinson's syndromes: a critical review. Curr. Opin. Genet. Dev. 19:254–265. 10.1016/j.gde.2009.03.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.