Abstract
Reactive oxygen species (ROS)-inducing anticancer agents such as phenethylisothiocyanate (PEITC) activate stress pathways for killing cancer cells. Here we demonstrate that PEITC-induced ROS decreased expression of microRNA 27a (miR-27a)/miR-20a:miR-17-5p and induced miR-regulated ZBTB10/ZBTB4 and ZBTB34 transcriptional repressors, which, in turn, downregulate specificity protein (Sp) transcription factors (TFs) Sp1, Sp3, and Sp4 in pancreatic cancer cells. Decreased expression of miR-27a/miR-20a:miR-17-5p by PEITC-induced ROS is a key step in triggering the miR-ZBTB Sp cascade leading to downregulation of Sp TFs, and this is due to ROS-dependent epigenetic effects associated with genome-wide shifts in repressor complexes, resulting in decreased expression of Myc and the Myc-regulated miRs. Knockdown of Sp1 alone by RNA interference also induced apoptosis and decreased pancreatic cancer cell growth and invasion, indicating that downregulation of Sp transcription factors is an important common mechanism of action for PEITC and other ROS-inducing anticancer agents.
INTRODUCTION
Reactive oxygen species (ROS), including radicals such as superoxide, nitric oxide, and hydroxyl radicals and nonradical species such as hydrogen peroxide, ozone, and peroxynitrates, function in normal cells to maintain homeostasis via redox pathways (1–3). In some cancer cell lines, a modest increase in forms of ROS can enhance cell proliferation, survival, and drug resistance; however, further increases in ROS that cannot be attenuated by intracellular redox systems result in cell death (3). ROS levels are higher in cancer than in noncancer cells, and drug-induced elevation of ROS is a “way to selectively kill cancer cells without causing toxicity to normal cells” (3). Drug-induced ROS in cancer cells may be due to inhibition or inactivation of redox pathway enzymes or due to direct effects on mitochondria, which include opening of the permeability transition pore complex, resulting in decreased mitochondrial membrane potential (MMP) and activation of proapoptotic cascades (3–5).
Several anticancer drugs that induce ROS, including arsenic trioxide, the methyl ester of 2-cyano-3,12-dioxo-oleana-1,9-dien-28-oic acid (CDDO-Me), curcumin, betulinic acid, a synthetic nonsteroidal anti-inflammatory drug (NSAID) (GT-094), and celastrol also downregulate specificity protein (Sp) transcription factors Sp1, Sp3, and Sp4 and prooncogenic Sp-regulated genes (6–11). Similar effects have been reported for H2O2, t-butylhydroperoxide, and pharmacologic doses of ascorbate that induce H2O2 (12). Moreover, the effects of ROS inducers and prooxidants on growth inhibition, induction of apoptosis, and downregulation of Sp proteins and Sp-regulated genes are attenuated after cotreatment with antioxidants (6–12). Induction of ROS by CDDO-Me, GT-094, betulinic acid, and celastrol decreases expression of Sp transcription factors through downregulation of microRNA 27a (miR-27a) and/or miR-17/miR-20a and induction of the miR-regulated Sp repressors ZBTB10 and ZBTB4, respectively (6–9). The relationship between drug-induced ROS and disruption of miR-dependent suppression of ZBTB transcriptional repressors suggests that this may be an important underlying mechanism of action for other ROS-inducing anticancer agents.
Phenethylisothiocyanate (PEITC) and other isothiocyanates (ITCs) have been linked to the chemoprevention and anticancer activity associated with consumption of cruciferous vegetables (13–17). PEITC and other ITCs inhibit cancer cell growth, survival, and angiogenesis, and these activities are accompanied by downregulation of total and activated STAT3 protein (18), suppression of NF-κB (and p65 downregulation) (16, 17), and decreased expression of epidermal growth factor receptor (EGFR) (18), genes involved in migration/invasion (19, 20), and bcl-2 and cyclins (21–23). Silencing of Sp1 and Sp1/Sp3/Sp4 (combined) by RNA interference (RNAi) shows that many of these same genes are regulated by Sp transcription factors in cancer cell lines (8–11, 24). PEITC and the related benzyl analog (BITC) also induce ROS in many cancer cell lines (22, 25–29). In Ras-transformed ovarian cancer cells, PEITC decreases intracellular glutathione (GSH) and also inhibits glutathione peroxidase activity, resulting in the induction of ROS and decreased MMP (25). In this study, we have used PEITC as a model ROS inducer to test the hypothesis that an important underlying mechanism of action of anticancer agents that induce ROS is due to downregulation of Sp1, Sp3, Sp4, and Sp-regulated genes through disruption of miR-ZBTB interactions and that this pathway is dependent on ROS-mediated epigenetic modulation of Myc expression.
MATERIALS AND METHODS
Cell lines, reagents, and antibodies.
The Panc28 cell line was a generous gift from Paul Chiao (University of Texas M. D. Anderson Cancer Center, Houston, TX), and the L3.6pL cell line was kindly provided by I. J. Fidler (University of Texas M. D. Anderson Cancer Center). Panc1, SW480, and RKO cells were obtained from the American Type Culture Collection (Manassas, VA). The cells were maintained in Dulbecco's modified Eagle medium (DMEM)–Ham's F-12 nutrient mixture (Sigma-Aldrich, St. Louis, MO) with phenol red supplemented with 0.22% sodium bicarbonate, 5% fetal bovine serum (FBS), and 10 ml/liter 100× antibiotic/antimycotic solution (Sigma-Aldrich). Cells were grown in 150-cm2 culture plates in an air-CO2 (95:5) atmosphere at 37°C and passaged approximately every 3 to 5 days. Antibodies to survivin and CD1 were from Epitomics (Burlingame, CA); vascular endothelial growth factor (VEGF) was from Rockland (Gilbertsville, PA); cleaved poly(ADP-ribose) polymerase (cPARP), cMet, and cMyc were from Cell Signaling (Boston, MA); VEGF receptor 1 (VEGFR1) and ZBTB34 were from Abcam (Cambridge, MA); ZBTB10 was from Bethyl Laboratories Inc. (Montgomery, TX). All other antibodies were purchased from Santa Cruz (Santa Cruz, CA). Glutathione (GSH; 98% pure) and PEITC (99% pure) were purchased from Sigma-Aldrich. Sp1 and chemiluminescence reagents (Immobilon Western) for Western blot imaging were purchased from Millipore (Billerica, MA). Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA). HPDE cells were provided by M. S. Tsao, University of Toronto (Toronto, ON, Canada), and S. Srivastava, Texas Tech University (Amarillo, TX).
Cell proliferation assay.
Pancreatic cancer cells (4 × 104 per well) were plated in 12-well plates and allowed to attach for 24 h. The medium was then changed to DMEM–Ham's F-12 mixture without phenol red containing 2.5% charcoal-stripped FBS, and either vehicle (dimethyl sulfoxide [DMSO]) or different concentrations of compounds in DMSO were added. Cells were then trypsinized and counted after 24, 48, and 72 h using a Coulter Z1 cell counter. Pancreatic cancer cells (1.5 × 105 per well) were plated in 12-well plates and allowed to attach for 24 h. Cells were pretreated with GSH for 3 h and were either dosed with PEITC alone or cotreated with PEITC and GSH. Cells were trypsinized and counted after 24 h using the Coulter Z1 cell counter. Each experiment was done in triplicate, and results are expressed as means ± standard errors (SE) for each set of experiments.
Western blot analysis.
Pancreatic cancer cells were seeded in 6-well plates using 2.5% DMEM–Ham's F-12 medium, and after 24 h, cells were pretreated with GSH for 3 h. Cells were then treated with vehicle (DMSO) and PEITC alone or cotreated with PEITC and GSH for the indicated time, and Western blot analysis of whole-cell lysates was performed essentially as described previously (8, 10, 11).
Measurement of mitochondrial membrane potential.
MMP was measured with a mitochondrial membrane potential detection kit (Stratagene, Cedar Creek, TX) according to the manufacturer's protocol using JC-1 dye. Pancreatic cancer cells were plated on 2-well Lab-Tex Coverglass slides (Nunc, NY), and after 24 h, cells were treated with DMSO or PEITC alone or PEITC in combination with GSH for 24 h. Cells were then incubated with 1× JC-1 dye at 37°C for 15 min, washed twice with assay buffer, and analyzed microscopically for changes in MMP essentially as described previously (10).
ROS determination.
Cellular ROS levels were evaluated with the cell-permeable probe CM-H2DCFDA [5-(and 6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester]. Cells were seeded at a cellular density of 1.5 × 105 cells/ml in 6-well plates, allowed to attach for 24 h, pretreated with GSH for 3 h, and treated with vehicle (DMSO), PEITC alone, or PEITC plus GSH for the indicated times. ROS was determined by flow cytometry as described previously (6).
TEM.
Cells were seeded at a cellular density of 1 × 105 cells/ml using 2.5% DMEM–Ham's F-12 in Permanox 2-well chambered slides. Cells were pretreated with GSH and then dosed with vehicle (DMSO) and PEITC alone or cotreated with PEITC and GSH for the indicated time. After dosing, cell cultures were fixed in 2% glutaraldehyde, 2.5% paraformaldehyde, and 0.1 M sodium cacodylate buffer for 1 h at room temperature. After washing in buffer, the cell monolayers were poststained with 1% osmium tetroxide reduced by 0.2% potassium ferrocyanide for 1 h and then dehydrated in an ascending alcohol series and embedded in epoxy resin. Thin sections were cut en face, stained with 2% uranyl acetate and Reynold's lead citrate, and then examined with an FEI Morgagni 268 transmission electron microscope (TEM) at an accelerating voltage of 80 kV. Digital images were acquired with a MegaViewIII camera operated with iTEM software (Olympus Soft Imaging Systems, Germany) and then postprocessed with Adobe Photoshop.
Annexin V staining.
Panc1, Panc28, and L3.6pL cells (1.5 × 105/ml) were pretreated and dosed with either vehicle or PEITC alone or in combination with GSH for 24 h. Cells were then stained using the Vybrant apoptosis assay kit according to the manufacturer's protocol (Biotium, CA).
RT-PCR.
Total RNA was isolated using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's protocol. RNA was eluted with 100 μl of RNase-free water and stored at −80°C. RNA was reverse transcribed using the Verso cDNA synthesis kit (Thermo scientific, PA) according to the manufacturer's protocol. Real-time (RT)-PCR was carried out using Sybr green (Life Technologies, NY). The following primers were used: TBP (F), 5′-TGCACAGGAGCCAAGAGTGAA-3′; BP (R), 5′-CACATCACAGCTCCCCACCA-3′; ZBTB10 (F), 5′-GCTGGATAGTAGTTATGTTGC-3′; ZBTB10 (R), 5′-CTGAGTGGTTTGATGGACAGA-3′; ZBTB4 (F), 5′-ACCTGTGCAGGAATTTCCAC-3′; ZBTB4 (R), 5′-GAGCGGCCAAGTTACTGAAG-3′; ZBTB34 (F), 5′-GCCAGCTTTCTTCAGATGCAGTG-3′; and ZBTB34 (R), 5′-CTCTTCAGCACCGACGGTAACA-3′.
Quantification of miRNA (RNU6B and miR-17, -20a, and -27a) was done using the TaqMan miRNA assay kit (Life Technologies) according to the manufacturer's protocol with real-time PCR. U6 small nuclear RNA was used as a control to determine relative miRNA expression.
RNA interference and miR mimic assays.
Small interfering RNAs (siRNAs) for Myc and GL2 were purchased from Sigma-Aldrich. The siRNA complexes used in this study are as follows: siGL2-5′, CGU ACG CGG AAU ACU UCG A; siMyc, SASI_Hs01_00222676.
The Panc1, Panc28, and L3.6pL pancreatic cancer cell lines were seeded (6 × 104 per well) in 6-well plates in DMEM–Ham's F-12 medium supplemented with 2.5% charcoal-stripped FBS without antibiotic and left to attach for 1 day. The Myc siRNA knockdown with siGL2 as a control was performed using Lipofectamine 2000 transfection reagent as per the manufacturer's instructions. The effects of miR mimics and antagomirs were determined as previously described (6–9, 30, 31).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT Express magnetic chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Panc1 cells (5 × 106 cells) were treated with 20 μM PEITC for 3 h. Cells were then fixed with 1% formaldehyde, and the cross-linking reaction was stopped by addition of 0.125 M glycine. After washing twice with phosphate-buffered saline, cells were scraped and pelleted. Collected cells were hypotonically lysed, and nuclei were collected. Nuclei were then sonicated to the desired chromatin length (∼200 to 1,500 bp). The sonicated chromatin was immunoprecipitated with normal IgG (Santa Cruz), c-Myc (Abcam), H3K27me3 (Abcam), H3K4me3 (Abcam), H4K16Ac (Active Motif), or RNA polymerase II (pol II; GeneTex) antibodies and protein A-conjugated magnetic beads at 4°C for overnight. After the magnetic beads were extensively washed, protein-DNA cross-links were reversed and eluted. DNA was prepared by proteinase K digestion followed by PCR amplification. The primers for detection of the c-Myc promoter region were 5′-GCC CTT TCC CCA GCC TTA GC-3′ (sense) and 5′-AAC CGC ATC CTT GTC CTG TGA GTA-3′ (antisense), the primers for detection of the beta-actin (ACTB) promoter region were 5′-CTC CCT CCT CCT CTT CCT CA-3′ (sense) and 5′-TCG AGC CAT AAA AGG CAA CTT-3′ (antisense), the primers for detection of the Sp1 promoter region were 5′-CTA ACT CCA ATC ATA ACG TTC C-3′ (sense) and 5′-GAG CTG GAG ATG ATT GGC TTG-3′ (antisense), the primers for detection of the miR-23a/27a cluster promoter region were 5′-TAG AGG ACC ACG TGT TCA TTT TGC-3′ (sense) and 5′-TGT AAA ATG GAG CCA AGA GCC TC-3′ (antisense), and the primers for detection of the miR-17/92 cluster promoter region were 5′-ACT TTG CAG TCT CGG GTG TTC-3′ (sense) and 5′-CGG GAT AAA GAG TTG TTT CTC CAA-3′ (antisense). PCR products were resolved on a 2% agarose gel in the presence of ethidium bromide (EtBr).
Effects of Sp1 knockdown on cell proliferation, invasion, death, and cycle analysis.
Panc1 and L3.6pL cells were seeded in 12-well plates and permitted to attach for 24 h, and then cells were transfected with 100 nM siRNA control or siRNAs for Sp1 using Lipofectamine 2000 (Invitrogen, Grand Island, NY). Cells were trypsinized and counted at the indicated times using a Coulter Z1 cell counter (Beckman Coulter, Fullerton, CA). For cell cycle analysis, cells were stained with propidium iodide solution (50 μg/ml) and were analyzed by a FACSCalibur flow cytometer 24 h after transfection. Apoptosis was detected using a fluorescein isothiocyanate (FITC) annexin V staining kit (Life Technologies, Grand island, NY) followed by fluorescence-activated cell sorter (FACS) analysis according to the manufacturer's protocol. Panc1 and L3.6pL cells were transfected with Sp1 siRNAs or a nonspecific control siRNA, and after 32 h, a cell invasion assay was performed as previously described (32).
Xenograft study.
Female athymic nude mice, 4 to 6 weeks old, were purchased from Harlan Laboratories (Houston, TX). L3.6pL cells (3 × 105) in a 1:1 ratio of Matrigel (BD Biosciences) were injected into either side of the flank area of nude mice. Seven days after the tumor cell inoculation, mice were divided into two groups of 10 animals each. The first group received 100 μl vehicle (corn oil), and the second group of animals received an injection of 60 mg/kg of body weight/day of PEITC in corn oil every 2nd day for 18 days (9 doses) by intraperitoneal (i.p.) injection. The mice were weighed, and the tumor areas were measured throughout the study. After 20 days, the animals were sacrificed; final body and tumor weights were determined and plotted. Research involving animal experimentation was reviewed and approved by the Texas A&M University Institutional Animal Care and Use Committee.
Statistical analysis.
Statistical significance of differences between the treatment groups was determined by an analysis of variance and/or Student's t test, and levels of probability were noted. Fifty percent inhibitory concentrations (IC50s) were calculated using linear regression analysis and expressed in μM, at 95% confidence intervals.
RESULTS
Inhibition of cell and tumor growth and induction of ROS.
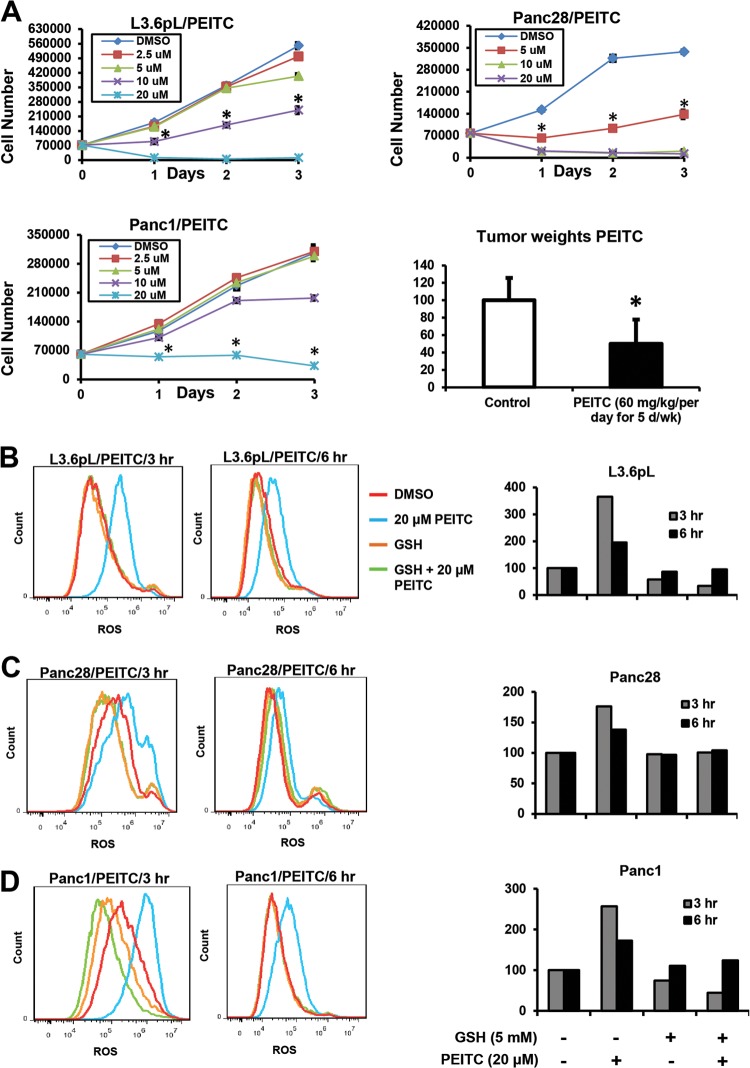
Initial studies showed that PEITC inhibited proliferation of Panc1, L3.6pL, and Panc28 pancreatic cancer cells after treatment for 1, 2, or 3 days. Growth inhibition after treatment for 24 h was observed for 20 μM PEITC in all cell lines, and 10 μM PEITC also significantly inhibited growth in L3.6pL and Panc28 cells (Fig. 1A). In contrast, only minimal inhibition of nontransformed HPDE pancreatic cells was observed after treatment with 10 or 20 μM PEITC (see Fig. S1A in the supplemental material). PEITC (60 mg/kg/day) also inhibited tumor growth in athymic nude mice bearing L3.6pL cells as xenografts (Fig. 1A). The concentrations of PEITC required for inhibition of pancreatic cell growth were slightly higher than previously reported in prostate and bladder cancer cells, and this was also confirmed in this study (see Fig. S1B and C in the supplemental material). Subsequent in vitro cell culture experiments primarily used 20 μM PEITC, since the major focus of this study was to investigate the mechanism of action of PEITC and key early events that occur within 24 h after treatment. Using the cell-permeant ROS-sensitive probe carboxy H2DCFDA, we observed by FACS analysis that ROS was induced by PEITC in L3.6pL, Panc1, and Panc28 cells after treatment for 3 or 6 h; in cells cotreated with PEITC plus the antioxidant glutathione (GSH), there was significant inhibition of ROS induction (Fig. 1B to D). These data are consistent with the results observed in transformed ovarian cancer cells, where PEITC rapidly depleted cellular GSH due, in part, to direct inhibition of glutathione peroxidase activity (25).
FIG 1.
PEITC inhibits pancreatic cancer cell growth and induces ROS. (A) L3.6pL, Panc28, and Panc1 cells were treated with different concentrations of PEITC for up to 72 h, and cells were counted as outlined in Materials and Methods. Relative tumor weights after treatment with PEITC or corn oil (control) were determined as outlined in Materials and Methods, and a significant (P < 0.05) decrease in weight is indicated (*) (10 animals per treatment group). L3.6pL (B), Panc28 (C), and Panc1 (D) cells were treated with 20 μM PEITC, GSH, or their combination for 3 and 6 h, and ROS was determined by FACS analysis using the cell-permeant CM-H2DCFDA dye as described in Materials and Methods; effects observed after 3 and 6 h were quantitated (similar results were obtained in duplicate experiments).
Effects of PEITC on mitochondrial structure, MMP, and apoptosis: role of ROS.
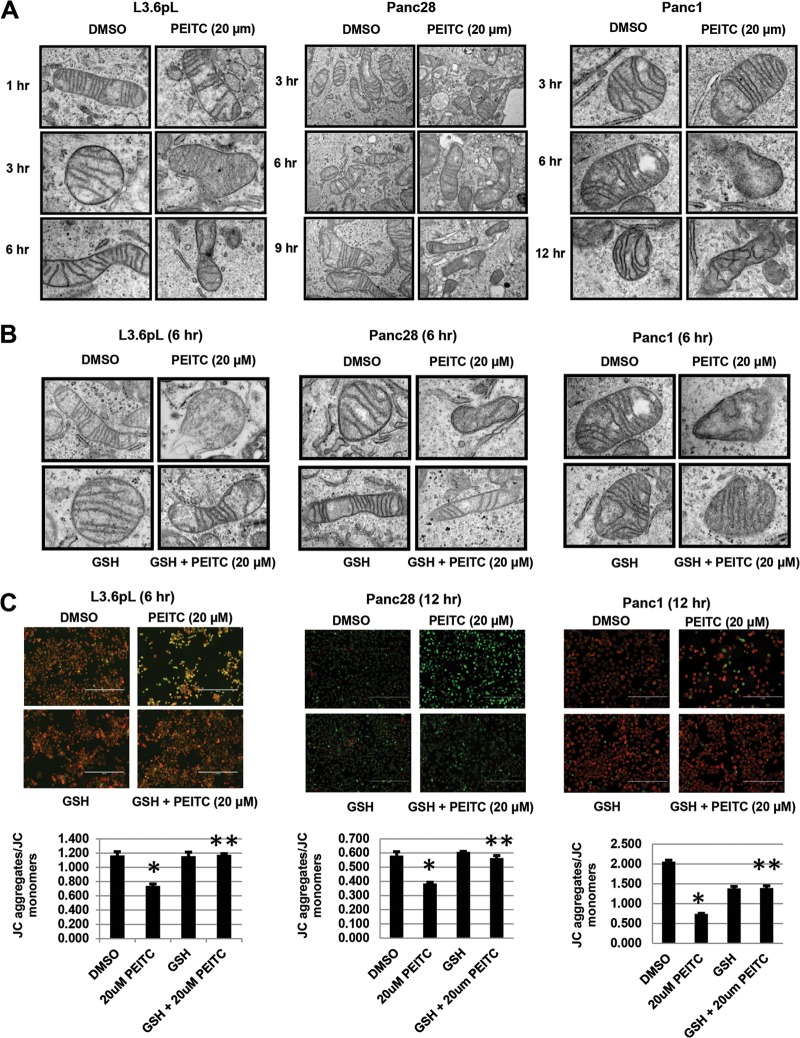
The effects of PEITC on mitochondrial structure and integrity were examined by TEM. Initial studies in L3.6pL cells showed that treatment with 20 μM PEITC for 1, 3, or 6 h resulted in a significant loss of mitochondrial architecture and crista structure only after 6 h, and this response was attenuated after cotreatment with GSH (Fig. 2A). A similar approach was used for Panc28 and Panc1 cells (Fig. 2A), and the PEITC-induced mitochondrial damage observed after treatment for 6 h was attenuated in cells cotreated with PEITC plus GSH (Fig. 2B). The effects of PEITC on mitochondria were further investigated by determining the effects of this compound on loss of mitichondrial membrane potential (MMP) using the JC-1 dye and determining JC aggregates (red mitochondrial fluorescence indicating high MMP)/JC monomer (green cytoplasmic fluorescence caused by collapse of mitochondrial potential) ratios (Fig. 2C). MMP was not decreased in any of the cell lines after treatment with PEITC for 3 h (data not shown); however, Fig. 2C illustrates the earliest time points at which PEITC decreased MMP in L3.6pL (6 h), Panc28 (12 h), and Panc1 (12 h) cells, and these effects were attenuated after cotreatment with GSH, confirming the importance of PEITC-induced ROS in mediating mitochondrial damage in pancreatic cancer cells. These results show that PEITC induces ROS prior to loss of MMP and damage to mitochondria, and these results are consistent with a previous report showing that PEITC rapidly induces extramitochondrial ROS in transformed cell lines (25).
FIG 2.
PEITC disrupts mitochondrial structure and decreases MMP. L3.6pL, Panc28, and Panc1 cells were treated with 20 μM PEITC alone (A) or in combination with GSH (B), and mitochondrial ultrastructure was determined by TEM as outlined in Materials and Methods. The widths of the fields are 2 μm (A) and 5.3 μm (B). (C) Cells were treated with DMSO (solvent), 20 μM PEITC, or 5 mM GSH or GSH plus PEITC for the indicated times, and changes in MMP were determined by JC-1 staining as outlined in Materials and Methods. Results are means ± SE (3 replicates per data point), and significant (P < 0.05) inhibition (*) and reversal of the effect by GSH (**) are indicated. Red, JC-1 aggregates; green, JC-1 monomers.
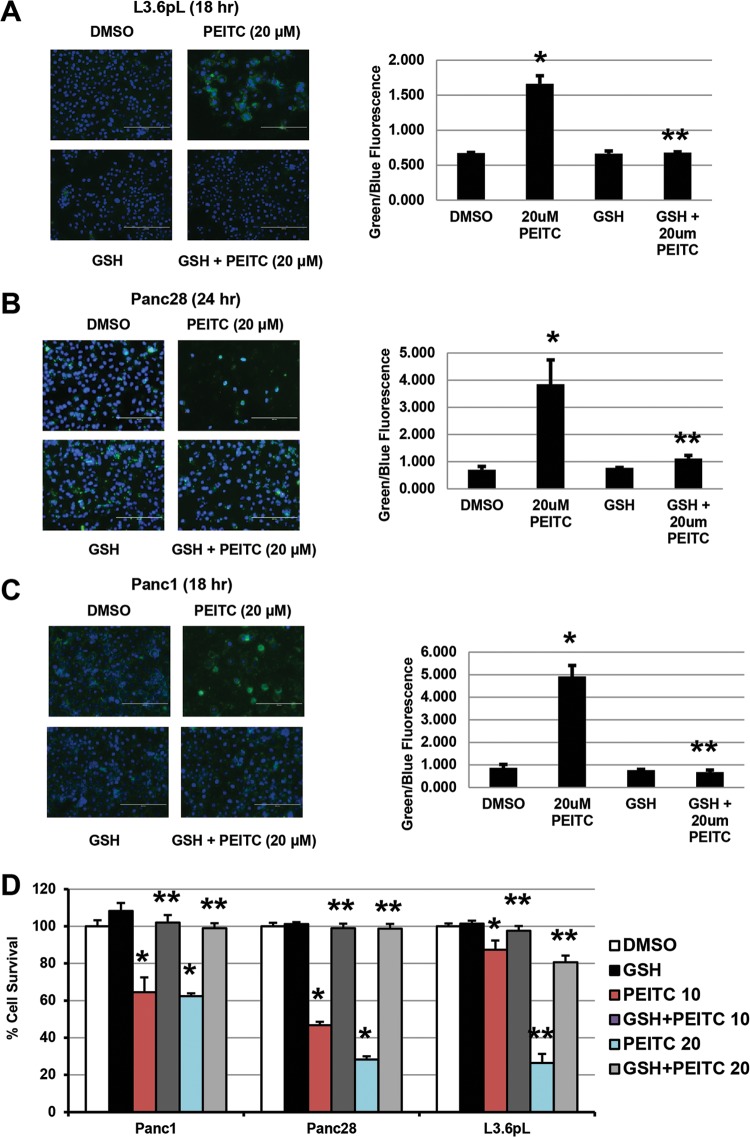
PEITC also induced annexin V staining in Panc1, L3.6pL, and Panc28 cells (Fig. 3A to C), and this apoptotic response was significantly blocked in cells cotreated with GSH; moreover, GSH also attenuated PEITC-induced growth inhibition (Fig. 3D), confirming that PEITC-induced ROS plays an essential role in the antineoplastic activity of this compound in pancreatic cancer cells, and similar results have previously been reported in other cancer cell lines treated with PEITC or BITC (22, 25–29). While it is clear that induction of ROS can induce mitochondrial damage, cellular stress, and apoptosis, other pathways and genes responsible for the cytotoxicities of PEITC and other ROS inducers are not well defined.
FIG 3.
Glutathione inhibits PEITC-induced apoptosis and growth inhibition. (A to C) L3.6pL (A), Panc28 (B), and Panc1 (C) cells were treated with 20 μM PEITC or GSH alone or in combination, and annexin V staining was determined as outlined in Materials and Methods. (D) Cell proliferation. Cells were treated as described for panels A to C for 24 h; cells were then counted as outlined in Materials and Methods. Results are means ± SE (3 replicates for each data point), and significant (P < 0.05) effects by PEITC (*) and attenuation by cotreatment with GSH (**) are indicated.
PEITC downregulates Sp1, Sp3, Sp4, and Sp-regulated gene products.
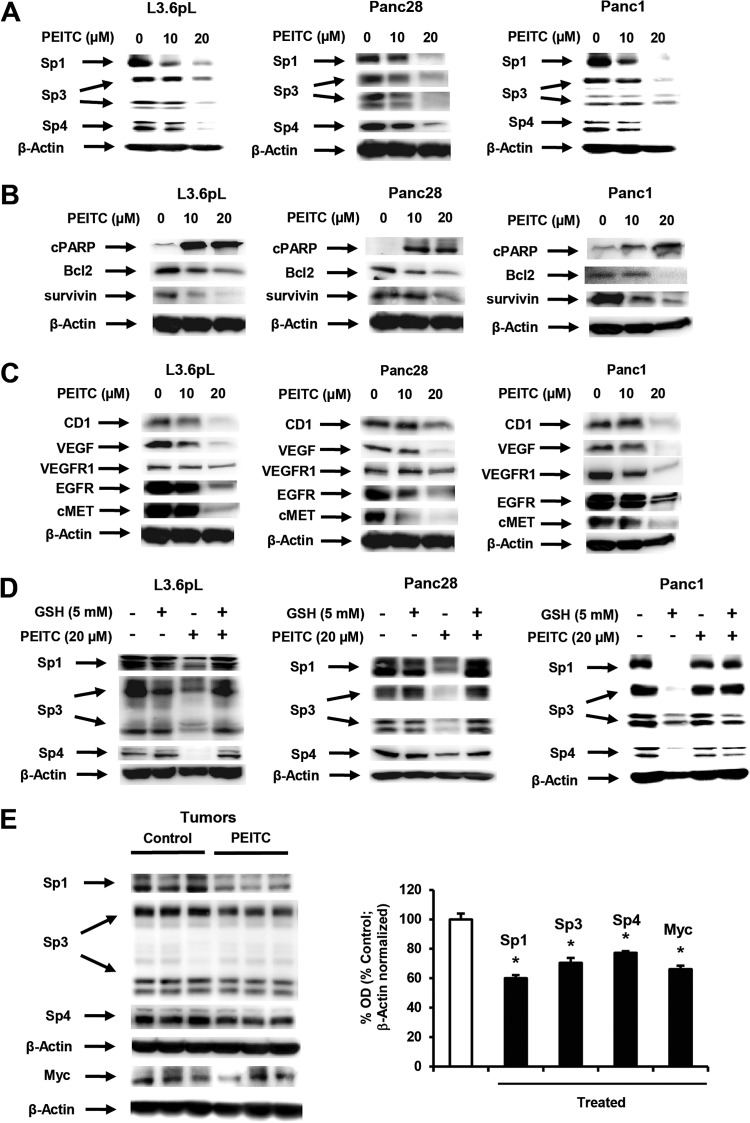
Previous studies in this laboratory show that ROS-inducing anticancer agents betulinic acid, celastrol, and CDDO-Me (all triterpenoids) and the nitro-NSAID GT-094 decrease expression of Sp transcription factors (6–12), and therefore, the effects of PEITC on expression of Sp1, Sp3, Sp4, and Sp-regulated gene products were also investigated. Treatment of L3.6pL, Panc1, and Panc28 cells with 10 or 20 μM PEITC for 24 h decreased levels of Sp1, Sp3 (low- and high-molecular-weight forms), and Sp4 proteins (Fig. 4A). PEITC also decreased expression of the survival gene products bcl-2 and survivin, poly(ADP-ribose) polymerase (PARP) cleavage (Fig. 4B), and downregulated growth-promoting (cyclin D1 and epidermal growth factor receptor [EGFR]) and angiogenic (vascular endothelial growth factor [VEGF] and VEGFR1) gene products (Fig. 4C). We also observed that PEITC-induced downregulation of Sp1, Sp3, and Sp4 proteins was reversed in L3.6pL, Panc1, and Panc28 cells cotreated with PEITC plus GSH (Fig. 4D), confirming a role for ROS in mediating repression of Sp transcription factors, and this correlated with the role of ROS in PEITC-induced apoptosis and growth inhibition in pancreatic cancer cells (Fig. 3). PEITC also decreased expression of Sp1, Sp3, and Sp4 proteins in tumors from athymic nude mice bearing L3.6pL cells as xenografts (Fig. 4E).
FIG 4.
PEITC downregulates Sp1, Sp3, Sp4, and Sp-regulated genes. (A to C) Pancreatic cancer cells were treated with different concentrations of PEITC for 24 h, and whole-cell lysates were analyzed for Sp1, Sp3, and Sp proteins (A), prosurvival proteins (B), and growth-promoting and angiogenic proteins (C) by Western blotting as outlined in Materials and Methods. (D) Cells were treated with PEITC or GSH alone or in combination, and cell lysates were analyzed by Western blotting as indicated for panels A to C. Results are typical of duplicate analyses, which gave comparable results. (E) Western blots of tumor lysates from control and PEITC-treated animals bearing L3.6pL cells as xenografts and protein quantitation relative to controls. Data shown in panels A to C were from the same experiment.
The effects of PEITC and other ROS inducers, including benzylisothiocyanate (BITC) and piperlongumine (33), on decreased expression of Sp1, Sp3, and Sp4 and the reversal of this response by GSH were confirmed in other cancer cell lines. PEITC and BITC decreased expression of Sp1, Sp3, and Sp4 in RKO and SW480 colon cancer cells (see Fig. S2 in the supplemental material), and similar effects were observed for PEITC and BITC in 253JB-V and KU7 bladder (see Fig. S3 in the supplemental material) and L3.6pL pancreatic and KU7 bladder (see Fig. S4 in the supplemental material) cancer cells, respectively. Piperlongumine also decreased Sp1, Sp3, and Sp4 expression in Panc1 and L3.6pL cells (see Fig. S5 in the supplemental material). The downregulation of Sp proteins by PEITC, BITC, and piperlongumine was attenuated after cotreatment with antioxidants, and we also observed induction of ROS by these compounds (data not shown).
Prooncogenic activity of Sp1, Sp3, and Sp4: RNAi studies.
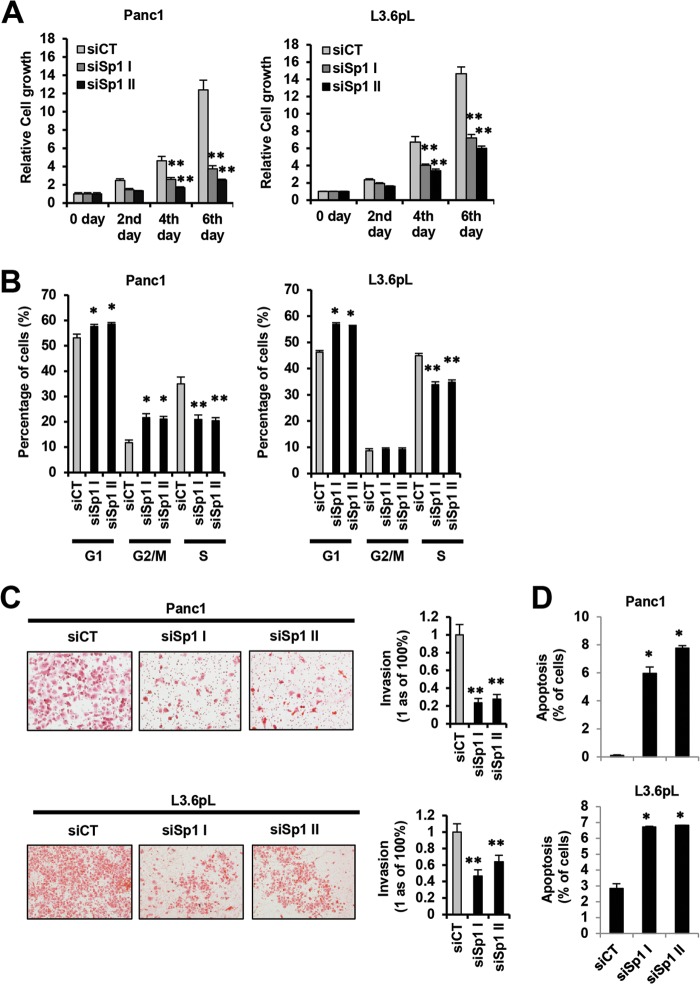
The functional importance of downregulation of Sp1, Sp3, Sp4, and prooncogenic Sp-regulated genes by ROS-inducing anticancer agents was further investigated by RNA interference (RNAi) in pancreatic cancer cells using Sp1 as a model. Silencing of Sp1 by RNAi in Panc1 and L3.6pL cells significantly decreased cell proliferation (Fig. 5A), cell cycle progression (Fig. 5B), and invasion in a Boyden chamber assay (Fig. 5C) and induced apoptosis (Fig. 5D). These results demonstrate that Sp1 is a prooncogenic factor, indicating that downregulation of Sp1, other Sp transcription factors, and Sp-regulated genes by ROS-inducing anticancer agents is an important underlying mechanism of action for these compounds.
FIG 5.
(A and B) Prooncogenic activity of Sp1 in pancreatic cancer cells. Cell growth (A) and cell cycle progression (B). Panc1 and L3.6pL cells were transfected with two different oligonucleotides that silence Sp1 (siSp1I and siSp1II), and cell proliferation and cell cycle progression were determined by cell counting or FACS analysis as outlined in Materials and Methods. (C and D) Pancreatic cancer cell invasion and quantitation (C) and induction of apoptosis (D). Panc1 and L3.6pL cells were transfected with siSpI or siSp1II, and inhibition of cell migration in a Boyden chamber assay and induction of apoptosis were determined and quantitated relative to the control (siCT) as outlined in Materials and Methods. Results in panels A to C are expressed as means ± SE for at least 3 replicate determinations for each treatment group, and significant (P < 0.05) increases (*) or decreases (**) compared to a control group (siCT) are indicated.
ROS-dependent disruption of miR-ZBTB interactions.
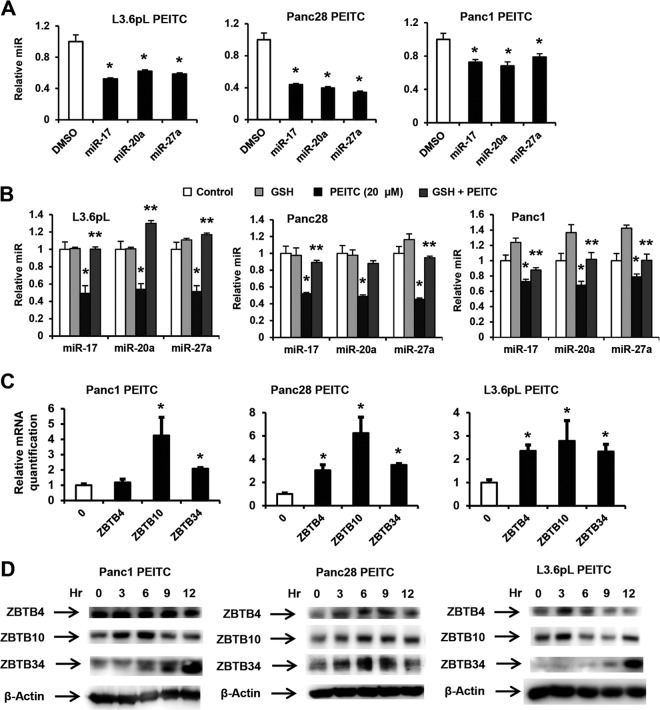
Previous studies show that ROS-dependent downregulation of Sp1, Sp3, and Sp4 in pancreatic and other cancer cells was due to downregulation of miR-27a and/or miR-20a/miR-17, resulting in the induction of miR-regulated “Sp repressors” ZBTB10 and ZBTB4, respectively (6–12). These transcriptional repressors competitively bind GC-rich Sp binding sites in promoters of Sp1, Sp3, Sp4, and Sp-regulated genes, resulting in decreased transactivation (30, 31). Figure 6A shows that PEITC also downregulated miR-27a, miR-20a, and miR-17 in L3.6pL, Panc28, and Panc1 cells, and PEITC-dependent downregulation of these miRs in L3.6pL, Panc28, and Panc1 cells was attenuated after cotreatment with GSH (Fig. 6B), confirming that downregulation of the miRs by PEITC was ROS dependent. PEITC induced ZBTB10 mRNA in all three cell lines and ZBTB4 mRNA in Panc28 and L3.6pL cells. We also examined other ZBTB family members that contained >6-bp sequences in their 3′ untranslated regions (UTRs) that complemented miR-27a and miR-20a/miR-17 lead sequence motifs and identified ZBTB34 as a potential miR-27a-regulated gene, and ZBTB34 mRNA was also induced by PEITC in Panc1, Panc28, and L3.6pL cells (Fig. 6C). Induction of ZBTB proteins by PEITC was time and cell context dependent (Fig. 6D); maximal levels of ZBTB10 and ZBTB34 were observed after 3 to 6 and 9 to 12 h, respectively, in Panc1 cells, whereas levels of ZBTB4 were relatively unchanged (0 to 12 h), and time-dependent induction of ZBTB proteins also varied in Panc28 and L3.6pL cells. Overexpression of ZBTB10 and ZBTB4 decreases expression of Sp1, Sp3, and Sp4 in cancer cell lines (30, 31), and the results shown in Fig. S6 in the supplemental material show that overexpression of ZBTB10, ZBTB4, and ZBTB34 also decreased expression of the three Sp proteins in Panc1, Panc28, and L3.6pL cells. Regulation of the ZBTB repressors was confirmed in studies using miR mimics that decrease ZBTB4, ZBTB10, and ZBTB34 mRNA expression (see Fig. S7A in the supplemental material) and antisense miRs that increase ZBTB4, ZBTB10, and ZBTB34 gene expression (see Fig. S7B in the supplemental material).
FIG 6.
PEITC disrupts miR-ZBTB interactions. (A) PEITC decreases miR expression. Pancreatic cancer cells were treated with 20 μM PEITC for 24 h, and miR expression was determined by real-time PCR as outlined in Materials and Methods. (B) GSH attenuates PEITC-mediated downregulation of miRs. Cells were treated with 20 μM PEITC, 5 mM GSH alone, or PEITC and GSH in combination for 24 h and analyzed for miR expression as outlined for panel A. (C) Induction of ZBTB expression. Cells were treated as described for panel B, and ZBTB4, ZBTB10, and ZBTB34 mRNA levels were determined by real-time PCR as outlined in Materials and Methods, and significant induction is indicated (*) (means ± SE for 3 replicate determinations). (D) ZBTB protein expression. Cells were treated with 20 μM PEITC for different times, and ZBTB protein expression was determined by Western blotting as outlined in Materials and Methods. Results for panels A to C are expressed as means ± SE for at least 3 replicate experiments for each treatment group; significant (P < 0.05) PEITC-induced responses (*) and attenuation by GSH (**) are indicated.
ROS-dependent downregulation of Myc.
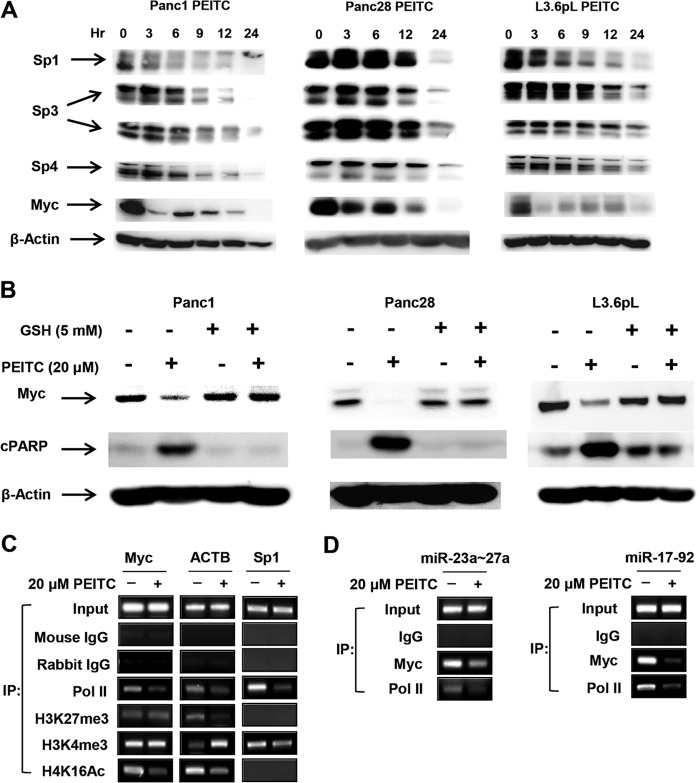
A recent study showed that H2O2 induced genome-wide shifts of repressor complexes from non-GC-rich to GC-rich sites in colon cancer cells, and this resulted in decreased expression of several genes, including Myc (34); it has previously been reported that Myc regulates expression of multiple miRs, including miR-27a and miR-20a/miR-17-5p, which are members of the miR-23a/27a/24-2 and miR-17-92 clusters (35–37). PEITC rapidly decreased Myc protein levels within 3 h in all three cell lines; however, the rate of decrease was slower in Panc28 cells. PEITC also decreased Myc RNA levels in Panc1, Panc28, and L3.6pL cells (see Fig. S7C). In addition, Sp1 (but not Sp3 or Sp4) protein was also decreased in L3.6pL and Panc1 cells within 3 h after treatment with PEITC (Fig. 7A). The rapid downregulation of Sp1 is consistent with a report showing that ROS induced chromatin-wide shifts of repressor complexes to some GC-rich sites (34) since the Sp1 gene promoter contains multiple GC boxes (38). We also observed that PEITC-mediated downregulation of Myc (and induction of PARP cleavage) was attenuated in cells cotreated with PEITC plus GSH (Fig. 7B).
FIG 7.
PEITC-induced responses are due to downregulation of Myc. (A) Time course effects on Sp transcription factors and Myc. Cells were treated with 20 μM PEITC for various times and analyzed by Western blotting as outlined in Materials and Methods. (B) GSH attenuates Myc downregulation by PEITC. Cells were treated with 20 μM PEITC, or 5 mM GSH alone, or PEITC and GSH in combination for 24 h, and whole-cell lysates were analyzed by Western blotting as outlined in Materials and Methods. (C and D) ChIP analysis of cMyc, Sp1, and ACTB (C) and the miR promoters (D). Cells were treated with 20 μM PEITC for 3 h, and histone methylation and acetylation marks on the Myc, ACTB, and Sp1 promoters (C) and Myc binding to the miR-23a/27a and miR-17-92 promoters (D) were determined in ChIP assays as outlined in Materials and Methods. Results are representative of replicate assays, and the various primers used (C and D) are listed in Materials and Methods.
Since ROS-mediated induction of Myc in SW480 colon cancer cells was due to genome-wide shifts of repressor complexes (34), we used a ChIP assay to determine changes in histone methylation/acetylation marks and pol II on the Myc gene in Panc1 cells treated with DMSO (control) or PEITC for 3 h (Fig. 7C). Methylation marks (H3K27me3 and H3K4me) were unchanged; however, the activation mark H4K16Ac and pol II were decreased after treatment with PEITC, and this is consistent with decreased Myc expression. In contrast, pol II and H3K4me3 were decreased on the Sp promoter, whereas pol II and the activation mark H4K16Ac were decreased on the ACTB promoter and ACTB mRNA levels were also decreased in SW480 cells treated with H2O2 (34). These results suggest that changes in histone acetylation/methylation marks in response to ROS are both gene and cell context specific. We also observed that treatment of Panc1 cells with PEITC resulted in loss of Myc (and pol II) from the miR-23a/27a/24-2 and miR-17-92 promoters (Fig. 7D), which is consistent with PEITC-mediated downregulation of Myc (Fig. 7A) and the corresponding miRs (Fig. 6A).
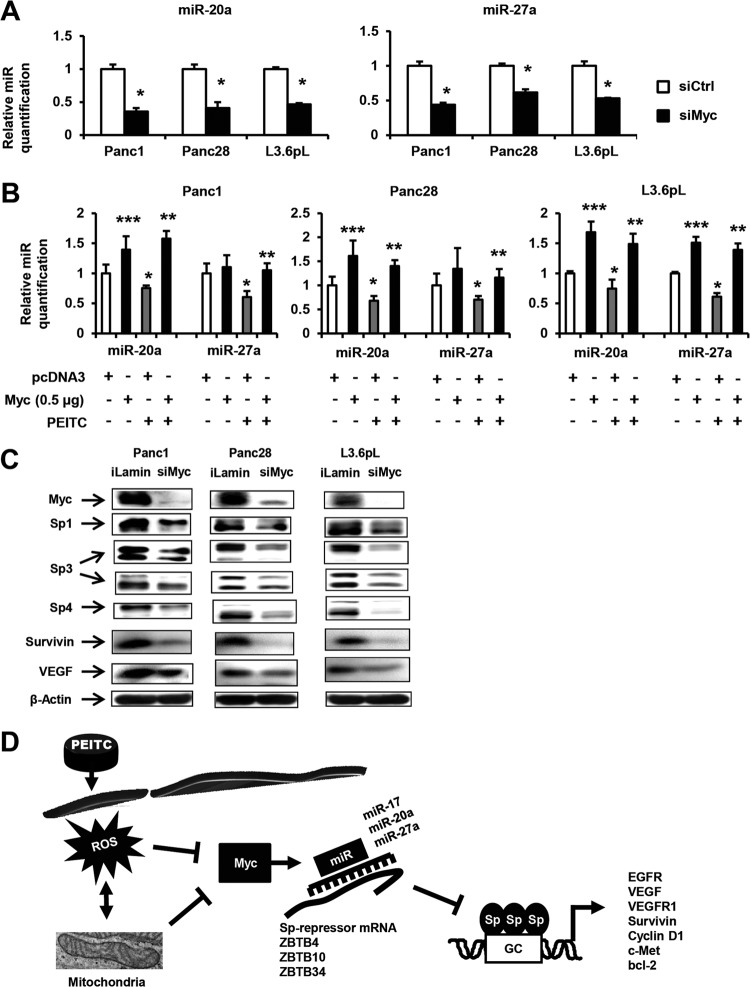
These results indicate that Myc plays a key role in PEITC/ROS-induced responses, and this was further investigated by RNAi. Transfection of pancreatic cancer cells with siMyc decreased expression of miR-20a and miR-27a in Panc1, Panc28, and L3.6pL cells (Fig. 8A). siMyc also induced expression of ZBTB10, ZBTB4, and ZBTB34 mRNA and protein in these cells; however, the magnitudes of these responses were variable (data not shown) due to the time-dependent induction of these repressors (Fig. 6D). Since PEITC-/ROS-mediated downregulation of miRs triggers the downstream effects on ZBTB and Sp proteins, we further confirmed the role of Myc in this response by overexpression studies. PEITC-induced downregulation of miR-27a and miR-20a in Panc1, Panc28, and L3.6pL cells was significantly reversed by overexpression of Myc (Fig. 8B); moreover, Myc overexpression also increased basal miR expression in these cells. The results shown in Fig. 8C show that knockdown of Myc decreased Sp1, Sp3, and Sp4 expression in Panc1, Panc28, and L3.6pL cells, and this was accompanied by decreased expression of survivin and VEGF in the three cell lines and of bcl-2 in L3.6pL and Panc28 cells. Like Myc, Sp1 expression is also rapidly decreased in L3.6pL cells (Fig. 7A) due to ROS-induced changes in histone marks (Fig. 7C), and knockdown of Sp1 also decreases miR-27a/miR-20a expression in these cell lines. Figure 8D summarizes the pathway resulting from induction of ROS by PEITC, and our results suggest that this is a pathway common to ROS-inducing anticancer drugs.
FIG 8.
Myc knockdown disrupts miR-ZBTB interactions and downregulates Sp proteins. (A) Knockdown of Myc by RNA interference (siMyc) in pancreatic cancer cells decreases expression of miRs. Panc1, Panc28, and L3.6pL cells were transfected with siMyc or a nonspecific oligonucleotide (siCtrl), and expression levels of miR-20a and miR-27a were determined by real-time PCR as outlined in Materials and Methods. (B) Myc overexpression inhibits PEITC-mediated suppression of miR-20a and miR-27a. Panc1, Panc28, and L3.6pL cells were treated with 20 μM PEITC and transfected with a Myc expression plasmid (pMyc) or empty vector (pcDNA3), and miR-20a and miR-27a expression was determined by real-time PCR as outlined in Materials and Methods. (C) Myc knockdown decreases Sp proteins in pancreatic cancer cells. Panc1, Panc28, and L3.6pL cells were transfected with siMyc or siLamin (control), and whole-cell lysates were isolated and analyzed by Western blotting as outlined in Materials and Methods. (D) Proposed mechanism of ROS-induced downregulation of Sp transcription factors. Results shown in panels A and B are means ± SE for replicate determinations, and significantly (P > 0.05) decreased expression (*) and rescue (**) are indicated. Significant upregulation by Myc overexpression is indicated (***).
DISCUSSION
Sp1, Sp3, and Sp4 transcription factors are highly expressed in cancer cells and tumors (6–12), and studies focused on Sp1 show that this protein is a negative-prognostic factor for pancreatic and gastric cancer patient survival (39–41) and that Sp1 expression is critical for malignant transformation of human fibroblast cells (42). Expression of Sp1 in rodents and humans decreases with age (43–45), and the high tumor/nontumor ratio of Sp1 suggests that Sp transcription factors are important drug targets for cancer chemotherapy. Knockdown of Sp1, Sp3, and Sp4 alone or in combination with RNAi also decreases expression of several gene products involved in cancer cell growth (cyclin D1, C-MET, and EGFR), survival (bcl-2 and survivin), angiogenesis (VEGF and VEGF receptors), and inflammation (p65 and NF-κB) (6–12, 24); many of these genes are themselves individual targets for anticancer drugs. Moreover, knockdown of Sp1 alone in pancreatic cancer cells also inhibits growth, cell cycle progression, and migration and induces apoptosis (Fig. 5). Carcinogen-induced transformed fibrosarcoma cells that form tumors in nude mice lose their tumorigenicity after knockdown of Sp1 (42). Several clinically used and experimental anticancer drugs downregulate Sp1, Sp3, and Sp4 proteins and prooncogenic Sp-regulated gene products in cancer cell lines through multiple pathways that are dependent on the drug and cell context. For example, curcumin induces proteasome-dependent downregulation of Sp proteins in bladder cancer cells (46), whereas in pancreatic cancer cells, the effects of curcumin on decreased expression of Sp1, Sp3, and Sp4 are ROS dependent (11), and these effects are attenuated after cotreatment with antioxidants.
Previous studies with PEITC and related ITCs show that these compounds inhibit cancer cell growth and angiogenesis/invasion, induce apoptosis, and decrease expression of several gene products, such as p65 (NF-κB), bcl-2, cyclin, and EGFR, that are also decreased in cancer cells after silencing Sp1 or Sp1/Sp3/Sp4 (combined) (8–11, 24). The mechanisms of action of PEITC and related ITCs are either ROS dependent or independent in different cancer cell lines, and the ROS-mediated effects have been extensively investigated (25–29). For example, PEITC rapidly depleted intracellular GSH and inhibited glutathione peroxidase activity in Ras-transformed ovarian cells, and the subsequent induction of ROS decreased MMP and activated the intrinsic apoptosis pathway (25). PEITC also induced ROS and decreased MMP in bladder and prostate cancer cells (26–28), and this was accompanied by cytochrome c release from mitochondria and changes in mitochondrial proteins. PEITC also affected mitochondrial oxidative phosphorylation in prostate cancer cells. In pancreatic cancer cells, PEITC rapidly induced ROS (within 3 h) (Fig. 1B to D), and using TEM, we also showed that PEITC did not significantly affect mitochondrial ultrastructure until 6 h after treatment (Fig. 2). Similar results were observed for MMP, which was decreased only after ≥6 h (Fig. 2). These results, coupled with the protective effects of the antioxidant glutathione, are comparable to the induction of extramitochondrial ROS by PEITC in Ras-transformed ovarian epithelial cells, which was then followed by ROS-mediated mitochondrial damage (25).
Prooxidants such as H2O2 and t-butylhydroperoxide and pharmacological doses of ascorbate decrease Sp1, Sp3, Sp4, and Sp-regulated genes in cancer cell lines (10, 12), and ROS-inducing anticancer agents, including betulinic acid, curcumin, CDODA-Me (methyl 2-cyano-3,11-dioxo-18-olean-1,12-dien-30-oate), arsenic trioxide, and celastrol, downregulate Sp transcription factors (6–12). PEITC also induced ROS and ROS-dependent downregulation of Sp1, Sp3, and Sp4 in pancreatic cancer cells (Fig. 4). Similar results were observed for PEITC, BITC, and piperlongumine in multiple cancer cell lines (see Fig. S2 to S5 in the supplemental material). These data, coupled with the functional effects of Sp1 knockdown in Panc1 cells (Fig. 5), support the hypothesis that an important underlying mechanism of action of ROS-inducing anticancer agents is due to downregulation of Sp transcription factors. Thus, the effectiveness of ROS-inducing antineoplastic drugs is due not only to ROS-dependent activation of proapoptotic pathways, including mitochondrial damage and activation of other stress response pathways, but also to downregulation of Sp transcription factors and Sp-regulated genes.
High expression of Sp1, Sp3, and Sp4 in cancer cell lines is due, in part, to microRNA-dependent regulation of the transcriptional repressors ZBTB10 and ZBTB4, which do not contain transactivation domains and competitively bind GC-rich promoter sites to displace Sp proteins (30, 31). ZBTB10 expression is repressed by miR-27a (30), and ZBTB4 is repressed by miR-20a and miR-17-5-p (part of the miR-17-92 cluster) and other paralogs (31). PEITC-/ROS-mediated downregulation of miR-27a and miR-20a/miR-17-5p results in induction of ZBTB10 and ZBTB4, respectively, and subsequent downregulation of Sp proteins, and these responses are attenuated in cells cotreated with GSH. miR-27a/20a antagomirs or overexpression of ZBTB10 or ZBTB4 also decreases expression of Sp1, Sp3, and Sp4 proteins (30, 31), and in this study, we also show that ZBTB34 is another miR-27a-regulated Sp repressor that decreases expression of Sp1, Sp3, and Sp4 (see Fig. S6 in the supplemental material). Thus, miR-27a plays a particularly important role in maintaining high levels of Sp1, Sp3, and Sp4 expression in all three cell lines by regulating ZBTB10 and ZBTB34 expression.
The key unknown link between induction of ROS and the ZBTB repressors that downregulate expression of Sp1, Sp3, and Sp4 is the mechanism associated with miR downregulation. A recent study reported that treatment of colon cancer cells with H2O2 results in relocation of large chromatin-associated repressor complexes from non-GC-rich to high-GC-rich sites; genes with GC-rich promoters such as Myc and ACTB are downregulated (34). Treatment of L3.6pL, Panc1, and Panc28 cells with PEITC resulted in rapid downregulation of Myc in all three cell lines, and this was attenuated by GSH (Fig. 7B). Interestingly, Sp1 was also decreased in Panc1 and L3.6pL cells within 3 h after PEITC treatment (Fig. 7A), and these data are consistent with results obtained for H2O2 in colon cancer cells (34) since Sp1 also has several GC-rich cis-elements in its 5′ region (38). The PEITC-/ROS-mediated shift in histone marks associated with nuclear relocalization of repressor complexes was also confirmed by ChIP assays on the Myc and Sp1 promoters (Fig. 7C). These data, coupled with the effects of Myc knockdown on expression of Sp1, Sp3, Sp4, and Sp-regulated gene products (Fig. 8C), suggest that in pancreatic cancer cells, ROS also induces shifts in repressor complexes as observed in colon cancer cells (34). Moreover, ROS-dependent repression of Myc and decreased Myc binding to the miR-23a/27a/24-2 and miR-17-92 promoters (Fig. 7D) are consistent with downregulation of miR-27a and miR-20a/miR-17-5p (Fig. 6A), which are members of two Myc-regulated miR clusters (35–37).
In summary, results of this study demonstrate that PEITC and other ROS-inducing anticancer agents decrease expression of Sp1, Sp3, Sp4, and prooncogenic Sp-regulated genes in pancreatic and other cancer cell lines. Since knockdown of Sp1 alone by RNAi decreases cancer cell growth and invasion and induces apoptosis (Fig. 5), this indicates that downregulation of Sp transcription factors by ROS-inducing anticancer agents contributes to their anticancer activity. A key element in triggering this response is due to ROS-mediated repression of Myc (and Sp1 in some cells) due to epigenetic effects associated with changes in histone marks; this is consistent with results of a recent study showing comparable effects with H2O2 in colon cancer cells (34). The ROS-dependent loss of Myc decreases miR expression and disrupts miR-ZBTB interactions, resulting in the induction of ZBTB repressors that decrease expression of Sp1, Sp3, and Sp4 (Fig. 8D). These results indicate that ROS-inducing anticancer agents may be particularly efficacious for treating cancer patients that overexpress Sp1 (41) and other Sp proteins, and the identification of this ROS-induced pathway will be important for the design of combination therapies, particularly since Sp proteins decrease expression of drug-resistant genes such as those encoding survivin and multidrug-resistant proteins (10, 12, 47).
Supplementary Material
ACKNOWLEDGMENTS
Financial support of this work by Public Health Service grant P30 ES023512 (from the National Institute of Environmental Health, National Institutes of Health) and Texas AgriLife Research is gratefully acknowledged.
Footnotes
Published ahead of print 14 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01602-13.
REFERENCES
- 1.Lydon N. 2009. Attacking cancer at its foundation. Nat. Med. 15:1153–1157. 10.1038/nm1009-1153 [DOI] [PubMed] [Google Scholar]
- 2.Fruehauf JP, Meyskens FL., Jr 2007. Reactive oxygen species: a breath of life or death? Clin. Cancer Res. 13:789–794. 10.1158/1078-0432.CCR-06-2082 [DOI] [PubMed] [Google Scholar]
- 3.Trachootham D, Alexandre J, Huang P. 2009. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 8:579–591. 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- 4.Engel RH, Evens AM. 2006. Oxidative stress and apoptosis: a new treatment paradigm in cancer. Front. Biosci. 11:300–312. 10.2741/1798 [DOI] [PubMed] [Google Scholar]
- 5.Schumacker PT. 2006. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell 10:175–176. 10.1016/j.ccr.2006.08.015 [DOI] [PubMed] [Google Scholar]
- 6.Chintharlapalli S, Papineni S, Lei P, Pathi S, Safe S. 2011. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 11:371. 10.1186/1471-2407-11-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pathi SS, Jutooru I, Chadalapaka G, Sreevalsan S, Anand S, Thatcher GR, Safe S. 2011. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol. Cancer Res. 9:195–202. 10.1158/1541-7786.MCR-10-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jutooru I, Chadalapaka G, Abdelrahim M, Basha MR, Samudio I, Konopleva M, Andreeff M, Safe S. 2010. Methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate decreases specificity protein transcription factors and inhibits pancreatic tumor growth: role of microRNA-27a. Mol. Pharmacol. 78:226–236. 10.1124/mol.110.064451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadalapaka G, Jutooru I, Safe S. 2012. Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis 33:886–894. 10.1093/carcin/bgs102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jutooru I, Chadalapaka G, Sreevalsan S, Lei P, Barhoumi R, Burghardt R, Safe S. 2010. Arsenic trioxide downregulates specificity protein (Sp) transcription factors and inhibits bladder cancer cell and tumor growth. Exp. Cell Res. 316:2174–2188. 10.1016/j.yexcr.2010.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jutooru I, Chadalapaka G, Lei P, Safe S. 2010. Inhibition of NFkappaB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J. Biol. Chem. 285:25332–25344. 10.1074/jbc.M109.095240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathi SS, Lei P, Sreevalsan S, Chadalapaka G, Jutooru I, Safe S. 2011. Pharmacologic doses of ascorbic acid repress specificity protein (Sp) transcription factors and Sp-regulated genes in colon cancer cells. Nutr. Cancer 63:1133–1142. 10.1080/01635581.2011.605984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murillo G, Mehta RG. 2001. Cruciferous vegetables and cancer prevention. Nutr. Cancer 41:17–28. 10.1080/01635581.2001.9680607 [DOI] [PubMed] [Google Scholar]
- 14.Cohen JH, Kristal AR, Stanford JL. 2000. Fruit and vegetable intakes and prostate cancer risk. J. Natl. Cancer Inst. 92:61–68. 10.1093/jnci/92.1.61 [DOI] [PubMed] [Google Scholar]
- 15.Sahu RP, Srivastava SK. 2009. The role of STAT-3 in the induction of apoptosis in pancreatic cancer cells by benzyl isothiocyanate. J. Natl. Cancer Inst. 101:176–193. 10.1093/jnci/djn470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Shen G, Chen C, Gelinas C, Kong AN. 2005. Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene 24:4486–4495. 10.1038/sj.onc.1208656 [DOI] [PubMed] [Google Scholar]
- 17.Kallifatidis G, Rausch V, Baumann B, Apel A, Beckermann BM, Groth A, Mattern J, Li Z, Kolb A, Moldenhauer G, Altevogt P, Wirth T, Werner J, Schemmer P, Buchler MW, Salnikov AV, Herr I. 2009. Sulforaphane targets pancreatic tumour-initiating cells by NF-kappaB-induced antiapoptotic signalling. Gut 58:949–963. 10.1136/gut.2008.149039 [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Xu C, Keum YS, Reddy B, Conney A, Kong AN. 2006. Inhibition of EGFR signaling in human prostate cancer PC-3 cells by combination treatment with beta-phenylethyl isothiocyanate and curcumin. Carcinogenesis 27:475–482. 10.1093/carcin/bgi272 [DOI] [PubMed] [Google Scholar]
- 19.Yang MD, Lai KC, Lai TY, Hsu SC, Kuo CL, Yu CS, Lin ML, Yang JS, Kuo HM, Wu SH, Chung JG. 2010. Phenethyl isothiocyanate inhibits migration and invasion of human gastric cancer AGS cells through suppressing MAPK and NF-kappaB signal pathways. Anticancer Res. 30:2135–2143 [PubMed] [Google Scholar]
- 20.Lai KC, Huang AC, Hsu SC, Kuo CL, Yang JS, Wu SH, Chung JG. 2010. Benzyl isothiocyanate (BITC) inhibits migration and invasion of human colon cancer HT29 cells by inhibiting matrix metalloproteinase-2/-9 and urokinase plasminogen (uPA) through PKC and MAPK signaling pathway. J. Agric. Food Chem. 58:2935–2942. 10.1021/jf9036694 [DOI] [PubMed] [Google Scholar]
- 21.Srivastava SK, Xiao D, Lew KL, Hershberger P, Kokkinakis DM, Johnson CS, Trump DL, Singh SV. 2003. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits growth of PC-3 human prostate cancer xenografts in vivo. Carcinogenesis 24:1665–1670. 10.1093/carcin/bgg123 [DOI] [PubMed] [Google Scholar]
- 22.Xiao D, Vogel V, Singh SV. 2006. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol. Cancer Ther. 5:2931–2945. 10.1158/1535-7163.MCT-06-0396 [DOI] [PubMed] [Google Scholar]
- 23.Kim SH, Nagalingam A, Saxena NK, Singh SV, Sharma D. 2011. Benzyl isothiocyanate inhibits oncogenic actions of leptin in human breast cancer cells by suppressing activation of signal transducer and activator of transcription 3. Carcinogenesis 32:359–367. 10.1093/carcin/bgq267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chadalapaka G, Jutooru I, Burghardt R, Safe S. 2010. Drugs that target specificity proteins downregulate epidermal growth factor receptor in bladder cancer cells. Mol. Cancer Res. 8:739–750. 10.1158/1541-7786.MCR-09-0493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. 2006. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell 10:241–252. 10.1016/j.ccr.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 26.Xiao D, Powolny AA, Moura MB, Kelley EE, Bommareddy A, Kim SH, Hahm ER, Normolle D, Van Houten B, Singh SV. 2010. Phenethyl isothiocyanate inhibits oxidative phosphorylation to trigger reactive oxygen species-mediated death of human prostate cancer cells. J. Biol. Chem. 285:26558–26569. 10.1074/jbc.M109.063255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powolny AA, Singh SV. 2010. Differential response of normal (PrEC) and cancerous human prostate cells (PC-3) to phenethyl isothiocyanate-mediated changes in expression of antioxidant defense genes. Pharm. Res. 27:2766–2775. 10.1007/s11095-010-0278-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahu RP, Zhang R, Batra S, Shi Y, Srivastava SK. 2009. Benzyl isothiocyanate-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of MAPK in human pancreatic cancer cells. Carcinogenesis 30:1744–1753. 10.1093/carcin/bgp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang L, Zhang Y. 2005. Mitochondria are the primary target in isothiocyanate-induced apoptosis in human bladder cancer cells. Mol. Cancer Ther. 4:1250–1259. 10.1158/1535-7163.MCT-05-0041 [DOI] [PubMed] [Google Scholar]
- 30.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. 2007. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 67:11001–11011. 10.1158/0008-5472.CAN-07-2416 [DOI] [PubMed] [Google Scholar]
- 31.Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, Park YY, Lee JS, Safe S. 2012. Identification of oncogenic microRNA-17–92/ZBTB4/specificity protein axis in breast cancer. Oncogene 31:1034–1044. 10.1038/onc.2011.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. 2013. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32:1616–1625. 10.1038/onc.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, Stern AM, Mandinova A, Schreiber SL, Lee SW. 2011. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 475:231–234. 10.1038/nature10167 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. 2011. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20:606–619. 10.1016/j.ccr.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051–4060. 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods K, Thomson JM, Hammond SM. 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282:2130–2134. 10.1074/jbc.C600252200 [DOI] [PubMed] [Google Scholar]
- 37.van Haaften G, Agami R. 2010. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 24:1–4. 10.1101/gad.1887110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolas M, Noe V, Jensen KB, Ciudad CJ. 2001. Cloning and characterization of the 5′-flanking region of the human transcription factor Sp1 gene. J. Biol. Chem. 276:22126–22132. 10.1074/jbc.M010740200 [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, Ajani J, Xie K. 2003. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 9:6371–6380 [PubMed] [Google Scholar]
- 40.Yao JC, Wang L, Wei D, Gong W, Hassan M, Wu TT, Mansfield P, Ajani J, Xie K. 2004. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin. Cancer Res. 10:4109–4117. 10.1158/1078-0432.CCR-03-0628 [DOI] [PubMed] [Google Scholar]
- 41.Jiang NY, Woda BA, Banner BF, Whalen GF, Dresser KA, Lu D. 2008. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 17:1648–1652. 10.1158/1055-9965.EPI-07-2791 [DOI] [PubMed] [Google Scholar]
- 42.Lou Z, O'Reilly S, Liang H, Maher VM, Sleight SD, McCormick JJ. 2005. Down-regulation of overexpressed sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 65:1007–1017 [PubMed] [Google Scholar]
- 43.Zannetti A, Del Vecchio S, Carriero MV, Fonti R, Franco P, Botti G, D'Aiuto G, Stoppelli MP, Salvatore M. 2000. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 60:1546–1551 [PubMed] [Google Scholar]
- 44.Chiefari E, Brunetti A, Arturi F, Bidart JM, Russo D, Schlumberger M, Filetti S. 2002. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer 2:35. 10.1186/1471-2407-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, Nagawa H, Suzuki N. 2004. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 25:461–468. 10.3892/ijo.25.2.461 [DOI] [PubMed] [Google Scholar]
- 46.Chadalapaka G, Jutooru I, Chintharlapalli S, Papineni S, Smith R, III, Li X, Safe S. 2008. Curcumin decreases specificity protein expression in bladder cancer cells. Cancer Res. 68:5345–5354. 10.1158/0008-5472.CAN-07-6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noratto GD, Jutooru I, Safe S, Angel-Morales G, Mertens-Talcott SU. 2013. The drug resistance suppression induced by curcuminoids in colon cancer SW-480 cells is mediated by reactive oxygen species-induced disruption of the microRNA-27a-ZBTB10-Sp axis. Mol. Nutr. Food Res. 57:1638–1648. 10.1002/mnfr.201200609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.