Abstract
MET, the receptor for hepatocyte growth factor (HGF), plays an important role in signaling normal and tumor cell migration and invasion. Here, we describe a previously unrecognized mechanism that promotes MET expression in multiple tumor cell types. The levels of the Pim-1 protein kinase show a positive correlation with the levels of MET protein in human tumor cell lines and patient-derived tumor materials. Using small interfering RNA (siRNA), Pim knockout mice, small-molecule inhibitors, and overexpression of Pim-1, we confirmed this correlation and found that Pim-1 kinase activity regulates HGF-induced tumor cell migration, invasion, and cell scattering. The novel biochemical mechanism for these effects involves the ability of Pim-1 to control the translation of MET by regulating the phosphorylation of eukaryotic initiation factor 4B (eIF4B) on S406. This targeted phosphorylation is required for the binding of eIF4B to the eIF3 translation initiation complex. Importantly, Pim-1 action was validated by the evaluation of patient blood and bone marrow from a phase I clinical trial of a Pim kinase inhibitor, AZD1208. These results suggest that Pim inhibitors may have an important role in the treatment of patients where MET is driving tumor biology.
INTRODUCTION
MET is a cell surface receptor tyrosine kinase that is expressed primarily on epithelial and endothelial cells. The ligand for MET, hepatocyte growth factor/scatter factor (HGF/SF), was first described as a growth factor for hepatocytes and as a fibroblast-derived cell motility or scatter factor for epithelial cells (1). Binding of HGF to MET activates multiple signaling cascades that induce cell growth, survival, and motility (1–3). Hyperactivity of the HGF-MET signaling axis occurs in many different types of cancer and has been associated with the uncontrolled growth of tumor cells, the epithelial-to-mesenchymal transition, invasiveness, and metastasis (1–3). Because of the importance of MET in driving tumor growth and as a mechanism of resistance to chemotherapy, specific targeted agents are now in human clinical trials (4).
Several different mechanisms that can lead to the overactivation of the HGF-MET axis in tumor cells have been identified, including point mutations, copy number alterations, and increased transcription of the met gene (5). Patients with renal papillary, hepatocellular, or gastric cancer carry point mutations in MET (6, 7) that activate its signaling whereas in patients with gastric or esophageal cancer and in some patients with lung cancer an increased gene copy number leads to increased MET expression (4, 5). Transcriptional mechanisms are responsible for increased MET expression and have been found in many tumor types (5). However, translational mechanisms for the control of MET levels could be of importance and have not been well investigated.
Several factors can stimulate the MET signaling cascade. Autocrine secretion of HGF has been shown to activate the MET signaling cascade in acute myeloid leukemia (AML) patient samples (8). More recently, it has been noted that targeted inhibition of specific signaling pathways, e.g., inhibition of the epidermal growth factor (EGF) receptor in lung cancer, can lead to increased expression of MET, which then plays a critical role in driving tumor growth (9, 10). We demonstrated recently that AKT inhibitors induce upregulation of receptor tyrosine kinases, including MET, in prostate cancer in a Pim kinase-dependent cap-independent fashion (12). However, the role of Pim kinase-regulated translational control in tumorigenesis, the potential clinical relevance of this effect, and the mechanisms involved have not been fully elucidated.
The Pim family of serine/threonine kinases includes three isoforms, Pim-1, -2, and -3, which are known to modulate cell survival pathways and regulate the progression and growth of human cancers, including prostate cancer and hematologic malignancies (11). Both Pim-1 and -2 have been shown to cooperate with c-Myc in the induction of lymphomas (11). Known Pim substrates include BAD, Bcl-2, Bcl-xl, p27Kip1, and Cdc25A (11), suggesting a role for Pim kinase in regulating both apoptosis and the cell cycle transition, which is consistent with the observation that inhibitors of Pim kinases induce cell cycle arrest at the G1 phase (12). We found that the AKT inhibitor-induced upregulation of receptor tyrosine kinases in prostate cancer occurred in a Pim-1-dependent, cap-independent manner, suggesting that Pim-1 may regulate MET protein translation (13). However, the translational apparatus is complex and the exact biochemical mechanisms used by Pim-1 to control MET levels have not been elucidated.
Here, we report that Pim-1 levels correlate with MET levels in normal cells and a wide variety of tumor cells. Manipulation of Pim-1 levels and blockade of Pim activity demonstrate that Pim-1 kinase activity plays a central role in regulating the levels of MET protein. Moreover, this regulation is physiologically relevant, as we found that as a result of its ability to control MET expression, Pim-1 regulates the HGF-MET signaling pathway and associated effects on cell functions, including cell motility, invasion, and scattering. The Pim-mediated regulation of MET is controlled by Pim-1 phosphorylation of the eukaryotic translation initiation factor 4B (eIF4B) specifically on S406, enhancing the ability of this protein to bind to the translational apparatus. Blocking this phosphorylation inhibited the translation of MET. The results were validated using human cell lines and patient-derived tissues, including fresh human leukemic cells and bone marrow and blood cells acquired from a phase I trial of the Pim kinase inhibitor AZD1208 in humans.
MATERIALS AND METHODS
Antibodies and reagents.
The following antibodies were purchased from Cell Signaling Technology: anti-Pim-1 (catalog no. 3247), anti-MET (catalog no. 3127 and 8198), anti-phospho-MET (catalog no. 3077), anti-eIF4B (catalog no. 3592), anti-phospho-eIF4B (S406, catalog no. 8151), anti-phospho-eIF4B (S422, catalog no. 3591), anti-eIF3A (catalog no. 3411), anti-eIF4G (catalog no. 2498), anti-eIF4E (catalog no. 9742), anti-AKT (pan; catalog no. 4691), anti-phospho-AKT (S473, catalog no. 4058), anti-phospho-4E-BP1 (catalog no. 2855), anti-4E-BP1 (catalog no. 9452), anti-phospho-S6 (catalog no. 2215), anti-extracellular signal-regulated kinase (anti-ERK) (catalog no. 9102), anti-phospho-ERK (catalog no. 9101), anti-phospho-histone H3 (catalog no. 3377), and anti-Myc (catalog no. 9402). Anti-eIF3B (catalog no. sc-16378) and anti-phospho-p70S6K (catalog no. sc11759) antibodies were purchased from Santa Cruz Biotechnology. Biotinylated anti-MET antibody (catalog no. BAF358), for detecting MET in AML cell lines and patient samples, was from R&D Systems. Anti-Bcl-2 antibody was purchased from BD Transduction Laboratories (catalog no. 610539). Anti-β-actin (catalog no. A3854), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (catalog no. G9295), and anti-FLAG (catalog no. F1804) antibodies were purchased from Sigma. Horseradish peroxidase (HRP)-linked enhanced chemiluminescence (ECL) mouse antibody (catalog no. NA931V) and rabbit IgG (catalog no. NAV934V) were purchased from GE Healthcare Life Sciences.
The small-molecule inhibitors MK2206, PP242, AZD8055, BI-D1870, and BEZ235 were purchased from Selleck Biochemicals. U0126, rapamycin, and cycloheximide (CHX) were from Sigma. SMI-4a and SMI-16a were synthesized in the laboratory of Charles Smith at the Medical University of South Carolina. GNE-652 and AZD1208 were provided by Genentech and AstraZeneca, respectively.
Human recombinant insulin (catalog no. 12585-014) and EGF (catalog no. PHG0311) were purchased from Life Technologies. Human recombinant HGF was from Antigenix America (catalog no. HC66602B).
Cell culture.
AML cell lines Molm-14, CMK, Mono-mac, OCI-M1, NB4, and MV4-11 were obtained from the German Collection of Microorganisms and Cell Cultures; KG1 and HEL were from the American Type Culture Collection; Molm-16 and OCI-AML2 were provided by Marina Konopleva and Juliana Benito (M. D. Anderson Cancer Center). PC3-LN4 cells were described before (14). Wild-type (WT), triple-knockout (TKO), and Pim-1−/− murine embryonic fibroblasts (MEFs) and TKO MEFs expressing Pim-1 were established as described previously (15). Mouse prostate epithelial cells (MPECs) expressing Pim-1 (WFU8, -10, -11, and -12) were gifts from Scott D. Cramer (University of Colorado School of Medicine). BPH1 cells were provided by Simon W. Hayward (Vanderbilt University). All other cell lines were supplied by the American Type Culture Collection. Cells were grown in RPMI medium (PC3-LN4, DU145, BT474, H1993, MKN45, and all AML cell lines) or Dulbecco modified Eagle medium (HeLa, U2OS, 293T, and MEFs) supplemented with 2 mM Glutamax (Life Technologies) and 10% fetal bovine serum (BioAbChem) at 37°C under 5% CO2. In some experiments, cells were cultured in the presence of 0.2% fetal bovine serum for 24 h before insulin, HGF, EGF, or fetal bovine serum was added.
Plasmids and siRNAs.
The Pim-1-expressing construct as well as its ΔN81 and K67M kinase-dead mutants was described previously (16). The plasmids expressing eIF4B and its S406A, S422A, and S406/422A mutants were previously described (17). The S406D and S406E mutants were obtained using a site-directed mutagenesis service provided by Genewiz Inc. The dicistronic luciferase construct pR-MET-F was described elsewhere (13). Small interfering RNAs (siRNAs) targeting Pim-1 and eIF4B were from Dharmacon, and the siRNA aimed at AKT1/2 was obtained from Cell Signaling Technology. Cells were transfected with Lipofectamine 2000 reagent with both plasmids and siRNAs according to the manufacturer's instructions.
Immunoblotting and immunoprecipitation.
Cells were harvested in lysis buffer consisting of 50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 5 mM EDTA. Following 30 min of incubation in lysis buffer at 4°C, lysates were cleared by centrifugation at 13,000 rpm for 10 min at 4°C, and then protein concentrations were determined by the DC protein assay (Bio-Rad). FLAG, MET, ERK, or eIF3B was immunoprecipitated in a buffer containing 20 mM HEPES, pH 7.5, 120 mM NaCl, 1% Triton X-100, 1 mM EDTA, and 10% glycerol with anti-FLAG (Sigma; catalog no. F1804), anti-eIF3B (Santa Cruz; catalog no. sc16378), anti-ERK (Cell Signaling; catalog no. 9102), or anti-MET (Cell Signaling; catalog no. 8198) antibodies and protein A/G-agarose (Pierce). Quantification was performed by densitometry using ImageJ software.
Luciferase assays.
Firefly luciferase and Renilla luciferase activities were measured using a luminometer (Model TD 20/20; Turner Designs) and the reagents provided with the Dual Luciferase Reporter kit (Promega). When a dicistronic vector was used, transfection efficiency was corrected by normalizing the data to the β-galactosidase activity from a cotransfected plasmid carrying this enzyme.
m7-GTP cap binding assay.
After treatment, 5 × 105 cells were washed in phosphate-buffered saline and then resuspended in lysis buffer. After centrifugation (16,000 × g for 10 min at 4°C), 200 μg of protein was applied to 20 μl of 7-methyl (m7)-GTP-Sepharose 4B beads (GE Healthcare) and incubated for 3 h at 4°C. The beads were then washed with lysis buffer three times followed by boiling in Laemmli sample buffer.
In vitro kinase assay.
A 2.5-μg amount of glutathione S-transferase (GST)-tagged eIF4B (Abnova) was incubated with 100 ng of active Pim-1 or AKT1 (SignalChem) in kinase buffer (20 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 2 mM ATP) in the presence or absence of GNE-652 (0.1 μM) or GSK690693 (0.1 μM) at 37°C for 30 min. After incubation, 5× sample buffer was added to a final concentration of 1× (60 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 2% β-mercaptoethanol, and bromophenol blue) and boiled for 5 min. Samples were separated by SDS-PAGE followed by immunoblotting and mass spectrometry analyses.
Mass spectrometry.
After an in vitro kinase assay and SDS electrophoresis, Coomassie blue-stained eIF4B bands were excised, enzymatically digested, and analyzed by liquid chromatography (LC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) using an Orbitrap Elite mass spectrometer (Thermo) coupled to a Dionex 3000 nano-LC system in the Mass Spectrometry Facility at the Medical University of South Carolina (MUSC).
Methionine incorporation assay.
Cells were transfected with a plasmid expressing eIF4B or its mutants for 48 h prior to labeling with 20 μCi of [35S]methionine per ml (Easytag Express protein labeling mix; PerkinElmer) in RPMI 1640 medium for 1 h, after which cold methionine was added. Three hours prior to labeling, dimethyl sulfoxide (DMSO), AZD1208 (3 μM), BEZ235 (0.5 μM), or cycloheximide (CHX; 100 μg/ml) was added. After completion of the experiment, the cells were washed twice with phosphate-buffered saline (PBS) and lysed in lysis buffer A. Lysates were clarified by centrifugation for 10 min at 13,000 × g and then subjected to immunoprecipitation using an anti-MET antibody. The 35S-labeled proteins were visualized by autoradiography.
Flow cytometry analyses.
All flow cytometry service was accomplished by the Cell Evaluation and Therapy Shared Resource of the Hollings Cancer Center at MUSC.
For cell cycle analysis, HeLa cells were transfected with Pim-1 siRNA or a nontargeting control and synchronized with a double thymidine protocol. Cells were then released into fresh medium and harvested every 4 h. Cell cycle distribution was monitored by fluorescence-activated cell sorting (FACS) analysis of ethanol-fixed, propidium iodide-stained cells using a Becton, Dickinson FACSCalibur analytical flow cytometer.
For detecting MET expression on the plasma membrane, cells were first trypsinized and fixed with 75% ethanol. Cells (1 × 105) were washed twice with PBS containing 1% bovine serum albumin (BSA) and incubated with anti-MET antibody (Cell Signaling; catalog no. 8198) on ice for 1 h. After being washed twice with PBS containing 1% BSA, cells were stained with appropriate R-phycoerythrin (PE)-conjugated secondary antibody (Jackson ImmunoReserach; catalog no. 711-116-152) on ice for 30 min. Afterward, cells were washed twice with PBS containing 1% BSA and subjected to flow cytometric analysis.
In vitro cell motility assays.
We evaluated cell migration in transwell chambers (Corning) coated with fibronectin and invasion in chambers coated with Matrigel. Cells were starved for 24 h before seeding in transwell chambers. Cells in the upper chamber were incubated in the presence of 10 μg/ml mitomycin C (Santa Cruz) for 3 h prior to the beginning of the experiment. HGF (100 ng/ml) was added to the bottom well for 24 h. Cells that migrated were fixed with 1% paraformaldehyde, stained with crystal violet, and counted under a microscope. For the scratch assays, cells were seeded at 80% confluence in 6-well dishes and grown for an additional 24 h. Cells were starved for 24 h at this point. Cells were incubated in the presence of 10 μg/ml mitomycin for 3 h prior to the beginning of the experiment. A linear scratch was done using a pipette tip across the diameter of the plate and rinsed with phosphate-buffered saline (PBS). Cells were fed with HGF at a final concentration of 100 ng/ml. Cells were incubated for 24 h, rinsed with PBS, and fixed in 1% paraformaldehyde at room temperature. Pictures were taken on a microscope (Nikon) at a magnification of ×4. For scatter assays, DU145 cells were starved for 24 h and incubated with various concentrations of HGF for 16 h. Pictures were taken on a microscope (Nikon) at a magnification of ×4.
Immunohistochemistry and tissue microarray.
A high-density human tissue microarray was assembled in the Biorepository and Research Pathology Services of the Hollings Cancer Center at MUSC. Initial sections were stained for hematoxylin and eosin to verify the histology. Standard biotin-avidin complex immunohistochemistry was performed. Anti-Pim-1 antibody (19F7) was produced in this laboratory (16). The anti-MET antibody used in these experiments was purchased from Santa Cruz (sc-161). Immunostaining intensity was scored by a qualified pathologist as 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, and 5. Scoring was performed in a blind manner using a telepathology system without knowledge of overall Gleason score, tumor size, or clinical outcome. A total of 35 tissue samples from 27 patients were examined.
Polysome profiling and reverse transcription-PCR (RT-PCR) analysis.
MEF cell extracts used for polysome gradient centrifugation were prepared as described previously (18). In brief, Pim kinase wild-type and TKO cells cultured in 10-mm culture dishes were harvested after replacing the culture medium with fresh medium containing cycloheximide (Sigma; 100 μg/ml) for 15 min. Cells were washed with PBS and then directly lysed in TMK100 buffer (10 mM Tris-HCl [pH 7.4], 100 mM KCl, 5 mM MgCl2, 1% [vol/vol] Triton X-100, 0.5% [wt/vol] deoxycholate, 2 mM dithiothreitol) on ice for 10 min. The solution was centrifuged for 10 min at 10,000 × g at 4°C, and supernatants were layered on top of linear 10% to 50% (wt/vol) sucrose gradients. Centrifugation was carried out in a Beckman SW41Ti rotor at 35,000 rpm for 3 h at 4°C. Polysome profiles were monitored by A254.
Total RNA was isolated by TRIzol extraction. One-step RT-PCR was performed using the MyTaq one-step RT-PCR kit (Bioline) on an Eppendorf MasterCycler. All primers were designed and synthesized by Integrated DNA Technologies, Inc.
Human subjects.
For ex vivo treatment, bone marrow and peripheral blood mononuclear cells were obtained from patients with AML in accordance with the Declaration of Helsinki. All subjects gave written informed consent for their blood products to be used for research under an institutional review board (IRB)-approved protocol. Blood was collected at the Medical University of South Carolina Hospital. Mononuclear cells were isolated immediately as described previously (19), and Pim inhibitors were added for 24 h.
For AZD1208 phase I patient samples, bone marrow and peripheral blood samples were obtained under an IRB-approved clinical protocol with written consent. Following sample collection in a heparin-coated CPT tube (BD Biosciences), mononuclear cells were isolated and washed once and cell pellets were frozen. For lysate preparation, cell pellets were lysed in bicine-CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer supplemented with protease and phosphatase inhibitors (Protein Simple). Following a 30-min incubation on ice, lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4°C and then protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Pierce).
Statistical analysis.
The results of quantitative studies are reported as means ± standard deviations (SDs). Differences were analyzed by Student's t test. P values of <0.05 were regarded as significant.
RESULTS
The Pim-1 protein kinase regulates MET protein levels.
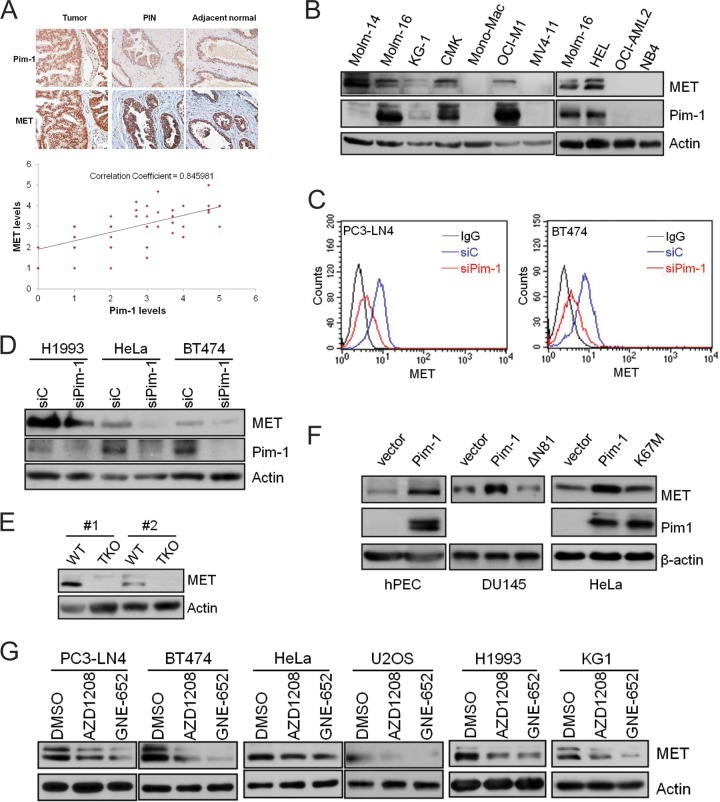
Because the MET tyrosine kinase drives the growth and metastasis of multiple human cancer types (5, 20), we first examined whether the levels of Pim-1 correlate with the level of MET in tumor cells. Immunohistochemical analysis of a high-density tissue microarray representing tissues from 27 patients with newly diagnosed prostate cancer indicated that the levels of Pim-1 and MET are highly correlated (correlation coefficient [R] = 0. 85 [Fig. 1A]). Western blot analysis of acute myeloid leukemia cell lines revealed that 5 of the 10 cell lines expressed Pim-1 and that MET protein was detectable in all of these cell lines. Moreover, MET was not detectable in four of the five cell lines that did not express Pim-1. MET was detectable in the absence of Pim-1 expression in only one of the cell lines examined (Molm-14) (Fig. 1B). The expression of MET in this cell line could be controlled by Pim-2 and/or Pim-3 or even another mechanism.
FIG 1.
The Pim-1 kinase regulates MET expression. (A) Representative images of a human prostate tissue microarray stained with anti-MET and anti-Pim-1 antibodies against normal, prostatic intraepithelial neoplasia (PIN), and tumor tissues. The relative strength of antibody staining was plotted as Pim-1 versus MET. The correlation coefficient (R) was derived by Microsoft Excel analysis. (B) Cell lysates from a panel of acute myeloid leukemia cell lines were analyzed by immunoblot assays using the indicated antibodies. (C) PC3-LN4 and BT474 cells were treated with siRNA targeting Pim-1 (siPim-1) or a nontargeting control (siC) for 72 h. The level of cell surface MET expression was visualized by flow cytometry using anti-MET antibody and R-phycoerythrin (PE)-conjugated secondary antibody. (D) Cell lysates from H1993, HeLa, and BT474 cells treated with siRNA targeting Pim-1 (siPim-1) or a nontargeting control (siC) for 72 h were analyzed by immunoblot assays using the indicated antibodies. (E) Cell lysates from wild-type (WT) and Pim TKO mouse embryonic fibroblasts were analyzed by immunoblot assays. Two different pairs of WT and TKO cells (#1 and #2) isolated separately from different embryos are examined. (F) Human prostate epithelial cells (hPEC), DU145 cells, or HeLa cells were transfected with plasmids expressing Pim-1 or its kinase-dead mutant ΔN81 or K67M. After 48 h, the levels of Pim-1, MET, and actin were examined by Western blotting. (G) Various tumor cell lines were treated with Pim inhibitor AZD1208 (3 μM) or GNE-652 (1 μM) for 24 h. Cell lysates were analyzed by immunoblot assays using the indicated antibodies.
To determine whether Pim-1 expression has a direct effect on MET protein levels, we used several different approaches. RNA interference-mediated silencing of Pim-1 expression markedly reduced both the total (13) and cell surface (Fig. 1C) expression of MET in the prostate tumor cell line PC3-LN4 as well as in tumor cell lines derived from other types of cancer, including H1993 (lung), HeLa (cervical), and BT474 (breast) (Fig. 1C and D). Murine embryonic fibroblasts (MEFs) obtained from mice with genetic knockout for all three Pim kinases expressed significantly lower levels of MET protein than did wild-type MEF cells (Fig. 1E). Conversely, overexpression of wild-type Pim-1 in DU145 and HeLa cells as well as normal human prostate epithelial cells (hPEC) resulted in increased MET expression (Fig. 1F). Pim kinase activity was required for the Pim-1-induced MET expression, as the addition of small-molecule Pim inhibitors to cultures of PC3-LN4, BT474, HeLa, U2OS, H1993, and KG1 cells reduced both the total cellular (Fig. 1G) and the cell surface (see Fig. S1 in the supplemental material) levels of MET. Moreover, although the overexpression of wild-type Pim-1 in DU145 cells, hPEC, and HeLa cells resulted in increased levels of MET protein, the overexpression of kinase-dead Pim-1 (ΔN81 and K67M) did not (Fig. 1F). Collectively, these results demonstrate that Pim-1 expression plays a key role in regulating the levels of MET protein and that this regulation is dependent on the kinase activity of Pim-1.
Pim-1 regulates the HGF-MET signaling pathway and cell motility.
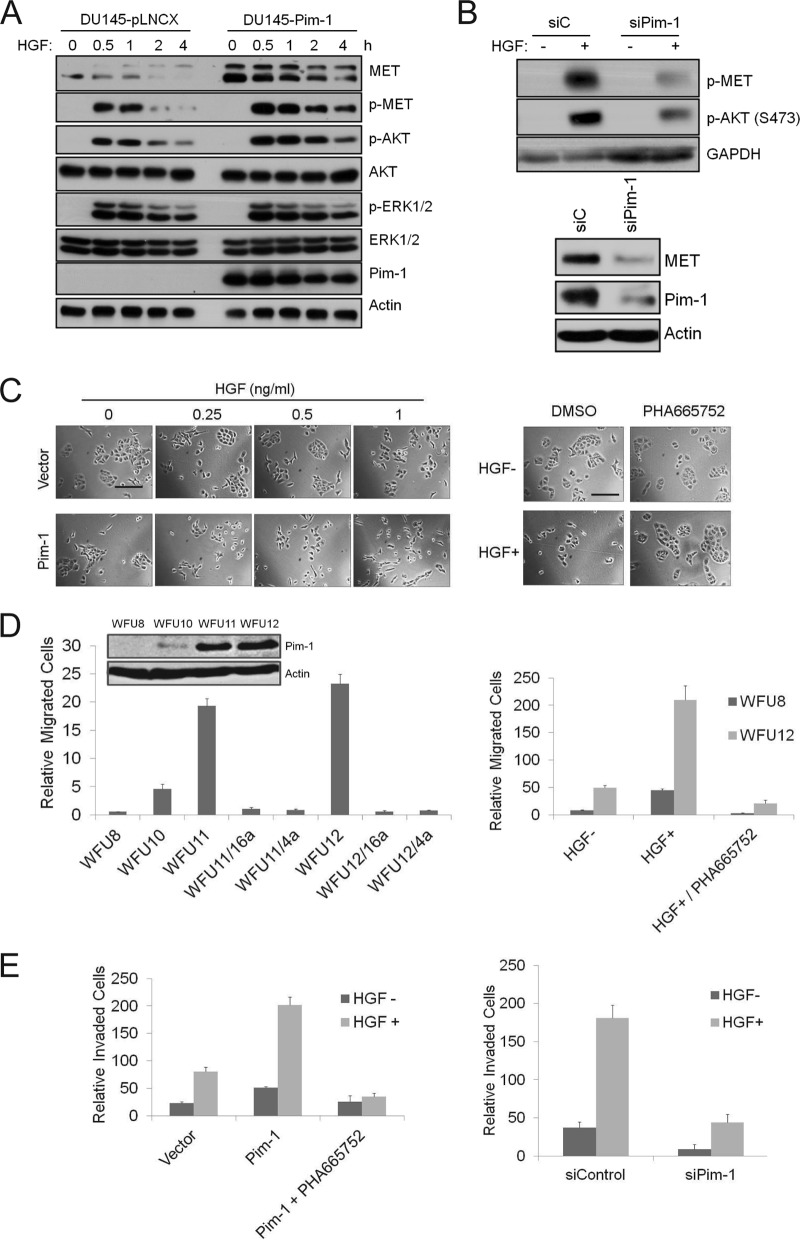
Because MET plays a critical role in HGF signaling, the ability of Pim-1 activity to regulate MET expression suggested the possibility that Pim-1 plays an important role in regulating the HGF-MET signaling pathways. Overexpression of Pim-1 in DU145 cells increased the levels of MET protein (Fig. 2A). The phosphorylation of MET and AKT in the presence of HGF was enhanced in the Pim-1-overexpressing cells (Fig. 2A). This effect was specific, as there was no difference in ERK phosphorylation between the overexpressor and wild-type cell lines (Fig. 2A). Conversely, in PC3-LN4 cells in which Pim-1 levels were reduced using siRNA, the HGF-induced phosphorylation of MET and AKT was lower than that in control cells (Fig. 2B). This cell line was chosen because it displays a higher metastatic potential (14) and increased expression of Pim-1 (unpublished data) than does the parental cell line PC3.
FIG 2.
Pim-1 regulates HGF-MET signaling and cell motility. (A) DU145 cells expressing an empty vector or Pim-1 were serum starved for 24 h before treatment with 100 ng/ml HGF for indicated times. Cell lysates were analyzed by immunoblot assays using the indicated antibodies. (B) (Top) PC3-LN4 cells were transfected with Pim-1 siRNA or a control siRNA for 48 h. Cells were serum starved for 24 h before treatment with 100 ng/ml HGF for 30 min. (Bottom) PC3-LN4 cells were transfected with Pim-1 siRNA or a control siRNA for 72 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (C) (Left) DU145 cells as described for panel A were treated with increasing concentrations of HGF for 24 h. (Right) DU145 cells expressing Pim-1 were treated with 0.25 ng/ml of HGF for 24 h in the presence of PHA665752 (1 μM) or DMSO. Microphotographs show cell scattering effect. Bars, 100 μm. (D) Immunoblots of mouse prostate epithelial cells transduced with a control vector (WFU8) or a Pim-1-expressing construct (WFU10, WFU11, and WFU12) are shown. The migration of these cells was examined for HGF-induced migration over 24 h. HGF (100 ng/ml) was added to the lower chamber. SMI-4a (10 μM), SMI-16a (10 μM), or PHA665752 (1 μM) was added to the upper chamber. Cells migrating through the membrane were counted, and the average ± SD is shown. (E) The invasion of PC3-LN4 cells was assayed using a chamber coated with Matrigel. HGF (100 ng/ml) was added to the lower chamber, and in specific experiments, PHA665752 (1 μM) was added to the upper chamber. Cells were transfected with a Pim-1-expressing plasmid (left) or Pim-1 siRNA (right) for 48 h prior to HGF addition. The average ± SD is shown.
Potentially, Pim-1 could regulate MET protein levels without having a significant physiologically relevant effect on the response to HGF. HGF induces scattering of DU145 cells which appears as loss of adherent cell “islands” or “clusters” (21). Culture of the Pim-1-overexpressing DU145 cells with HGF resulted in a pattern of scattering of the cells similar to that observed in HGF-treated wild-type cells; however, scattering was observed at a lower concentration of HGF (0.25 ng/ml versus 1 ng/ml) (Fig. 2C, left). Pretreatment with a MET inhibitor (PHA665752) blocked the HGF-induced cell scattering in the Pim-1-overexpressing cells (Fig. 2C, right), suggesting that this Pim-1 effect is mediated through the MET. Taken together, these data suggest that the Pim-1-induced increase in the MET levels enhances the ability of HGF to signal in these tumor cells.
The MET/HGF axis also plays an important role in mediating cell migration and invasion (5). To examine the ability of Pim-1 to modulate these HGF-induced activities, we first utilized mouse prostate epithelial cells (MPECs) that are immortalized but nontransformed and express different levels of Pim-1 protein (Fig. 2D, left). The MPECs that expressed higher levels of Pim-1 exhibited greater HGF-induced increases in migration than did the MPECs expressing lower levels of Pim-1 (Fig. 2D, left). The Pim-1 dependency of this biologic activity was also demonstrated by the addition of two Pim inhibitors, SMI-4a (4a) and SMI-16a (16a) (22), which blocked migration of the MPECs expressing higher levels of Pim-1 (WFU11 and WFU12, Fig. 2D, left). The MET inhibitor (PHA665752) inhibited this migration as well (Fig. 2D, right). Similar results were found in a scratch assay using the same MPECs. HGF-induced cellular migration into the scratch (also known as wound healing) was enhanced dramatically in the cells that expressed higher levels of Pim-1, and this effect was blocked by the addition of either a MET or a Pim inhibitor to the culture (see Fig. S2 in the supplemental material). Finally, to evaluate the ability of Pim-1 to modulate HGF-induced invasion of tumor cells, PC3-LN4 cells were placed in the upper chamber of a Boyden chamber in which the membrane had been coated with Matrigel, and HGF was added to the medium in the lower chamber. In this assay, the Pim-1-overexpressing PC3-LN4 cells demonstrated increased invasion, and this invasion was blocked by treatment with PHA665752 (Fig. 2E, left) or knockdown of Pim-1 expression by siRNA (Fig. 2E, right). Collectively, these results demonstrate that Pim-1 can control signaling through the HGF-MET axis and that this affects HGF-induced cell motility and invasion of both normal and tumor cells.
Pim-1 phosphorylates eIF4B at S406, a translation initiation factor that is required for MET translation.
Identification of the mechanisms by which Pim-1 controls the expression of MET requires a more detailed understanding of the mechanisms underlying translation of the MET protein. The 5′ untranslated region (UTR) of MET is relatively long (408 nucleotides [nt]) and guanine-cytosine rich (23), which is consistent with the possibility that this region functions as an internal ribosome entry site (13). Modeling of the secondary structure using the MFOLD program (24) indicated that the 5′ UTR of MET (23) is highly structured (see Fig. S3A in the supplemental material), with the overall folding energy of the most stable predicted structure being −293.3 kcal/mol. The translation initiation factor eIF4B preferentially promotes the translation of mRNAs containing such complex secondary structures in their 5′ UTRs (25). To determine whether eIF4B is required for MET expression, we transfected HeLa cells with siRNAs targeting eIF4B. Transfection with eIF4B siRNA led to reduced levels of MET protein as well as a marked fall in the levels of both Myc and Bcl-2 (see Fig. S3B), two proteins whose levels are, at least in part, translationally controlled by eIF4B (25). Moreover, eIF4B siRNA reduced the ability of the MET 5′ UTR (13) to drive firefly luciferase expression in both HeLa and U2OS cells (see Fig. S3C), suggesting that eIF4B plays an important role in regulating the translation of MET.
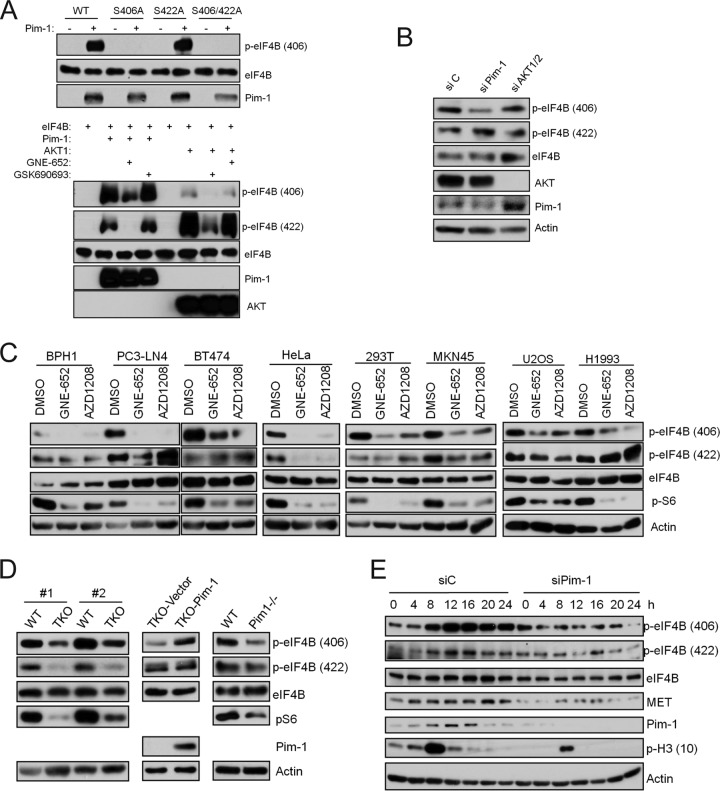
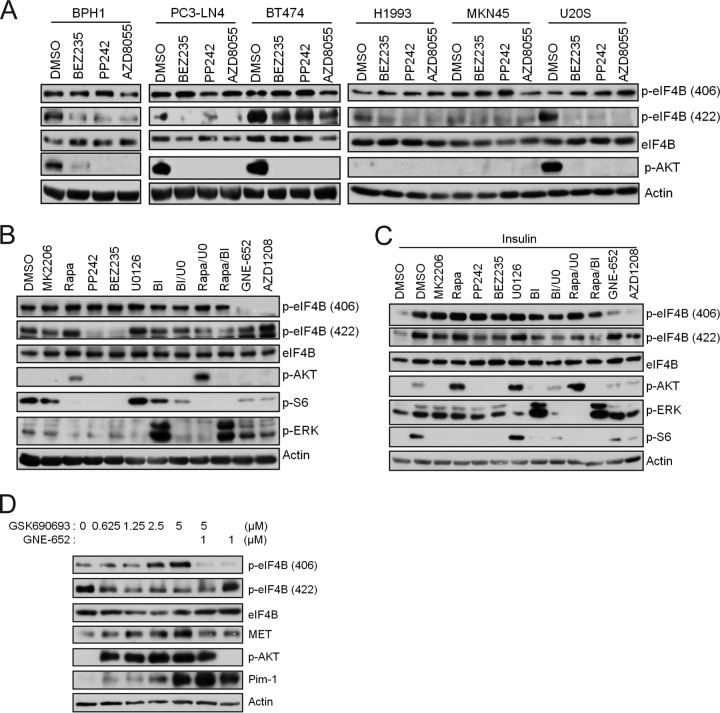
Using in vitro kinase assays, we found that Pim-1 phosphorylated wild-type eIF4B (Fig. 3A, top). To identify the phosphorylation site(s), we transfected the cells with eIF4B constructs with S406A and S422A mutations, which prevent phosphorylation at these sites. Pim-1 kinase was capable of phosphorylating eIF4B with an S422A mutation but not eIF4B with an S406A mutation (Fig. 3A, top). Similar results were obtained using commercially available recombinant eIF4B with Pim-1 phosphorylating eIF4B at S406 and to a lesser extent at S422 (Fig. 3A, bottom). Phosphorylation at both these sites was inhibited by the small-molecule pan-Pim kinase inhibitor GNE-652 (Fig. 3A, bottom). In marked contrast, and in agreement with a previous report (17), we found that AKT1 preferentially phosphorylated eIF4B at S422 and confirmed that this phosphorylation was inhibited by GSK690693, a small-molecule AKT inhibitor (Fig. 3A, bottom). The results were validated by mass spectrometric analysis of in vitro phosphorylation (see Fig. S4A in the supplemental material), which confirmed that Pim-1 directly phosphorylates S406 whereas AKT1 phosphorylates S422 in eIF4B.
FIG 3.
Pim-1 controls eIF4B phosphorylation at S406. (A) (Top) FLAG-tagged eIF4B as well as its mutants was expressed in U2OS cells and immunoprecipitated using anti-FLAG antibody with protein A/G beads. The beads were extensively washed and incubated with 100 ng of purified Pim-1 proteins at 37°C for 30 min. Samples were analyzed by immunoblot assays using indicated antibodies. (Bottom) Purified eIF4B and Pim-1 or AKT1 were incubated in the presence or absence of GNE-652 (0.1 μM) or GSK690693 (0.1 μM). Samples were subjected to immunoblotting analysis with the indicated antibodies. (B) HeLa cells were transfected with Pim-1 siRNA, AKT1/2 siRNA, or a nontargeting control siRNA for 72 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (C) Cell lines were treated with GNE-652 (1 μM) or AZD1208 (3 μM) for 3 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (D) Cell lysates from two independently isolated pairs of WT and Pim TKO MEF cells (left), TKO MEFs with empty vector (TKO-Vector) and TKO MEFs with a Pim-1-expressing construct (TKO-Pim-1) (middle), and WT MEFs and Pim-1 single-knockout MEFs (right) were subjected to Western blotting with the indicated antibodies. (E) HeLa cells were synchronized with a double-thymidine block protocol and simultaneously treated with Pim-1 siRNA. Cells were released into fresh medium and harvested at indicated times. Cell lysates were subjected to Western blotting with the indicated antibodies.
To determine whether the Pim-1 phosphorylation of elF4B occurs in tumor cells, we analyzed Pim-1 siRNA-transfected HeLa (Fig. 3B), PC3-LN4 (see Fig. S4B in the supplemental material), and BT474 (see Fig. S4C) cells. The knockdown of Pim-1 expression was associated with reduced phosphorylation of eIF4B S406 but did not affect phosphorylation on S422 in any of the three cell lines. Moreover, treatment of various benign or malignant cell lines, including BPH1, PC3-LN4, BT474, HeLa, 293T, MKN45, U2OS, and H1993, with either of two pan-Pim inhibitors, GNE-652 and AZD1208, reduced the phosphorylation of eIF4B S406, but only in one (HeLa) did it cause a reduction in the phosphorylation of S422 (Fig. 3C). Pim regulation of eIF4B phosphorylation also was found in triple-Pim knockout (TKO) MEFs. In these cells, the baseline level of eIF4B phosphorylation of S406 and S422 was significantly lower than that in wild-type MEFs (Fig. 3D, left). Expression of Pim-1 in the TKO cells partially restored the phosphorylation of eIF4B at S406 but did not affect phosphorylation at S422 (Fig. 3D, middle). Similarly, Pim-1-only (Pim-1−/−)-knockout MEFs displayed reduced levels of eIF4B S406 but not S422 compared to wild-type MEFs (Fig. 3D, right). Together, these data suggest that S406 is the major Pim-1-directed phosphorylation site on eIF4B. Because phosphorylation of S422 was reduced by Pim inhibitors or the absence of Pim kinases in some cellular contexts, regulation of this phosphorylation could be complex.
A natural model of altered levels of the Pim-1 protein kinase is the variation in the levels that occur during the cell cycle (16). To determine whether eIF4B S406 phosphorylation parallels the expression of Pim-1 during the cell cycle, HeLa cells were synchronized with a double thymidine block protocol and then released into normal medium. Both S406 and S422 phosphorylation of elF4B increased gradually and peaked as the cells entered mitosis, as evidenced by the increased expression of phosphohistone H3 and confirmed by the flow cytometry analyses (unpublished data), and then gradually decreased again (Fig. 3E). The elevation of eIF4B S406 phosphorylation during the cell cycle immediately followed the upregulation of Pim-1 protein levels (Fig. 3E). When the cells were treated with Pim-1 siRNA, the phosphorylation of eIF4B S406 during the cell cycle was reduced significantly whereas the phosphorylation of eIF4B S422 was less affected (Fig. 3E). Concomitantly, the levels of MET protein were reduced (Fig. 3E). This experiment confirms that the levels of Pim-1 are cell cycle regulated and parallel the levels of eIF4B S406 phosphorylation and MET expression during the cycle.
Pim-1 regulates eIF4B S406 phosphorylation in response to growth factors and serum.
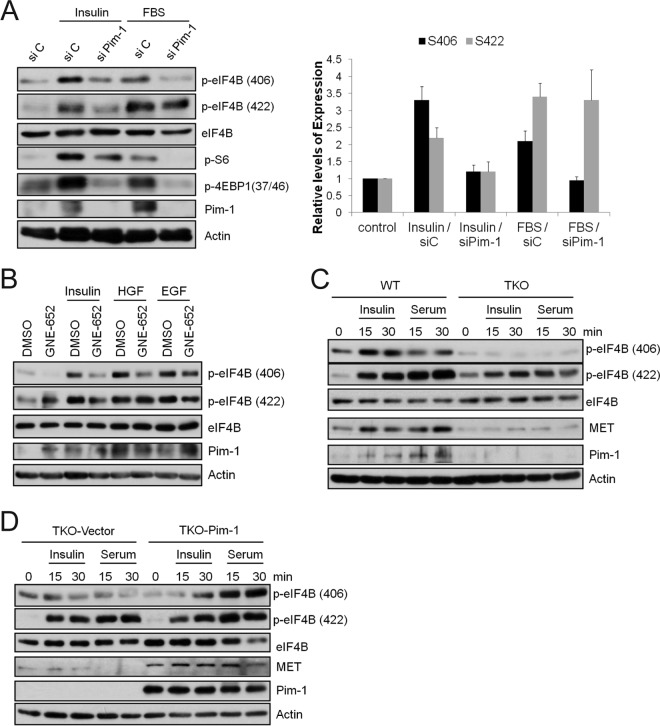
The eIF4B protein is phosphorylated and activated in response to insulin and serum stimulation (17, 26). Insulin and serum stimulation of serum-starved control tumor cells resulted in enhanced eIF4B phosphorylation at S406, and this effect was impaired in the Pim-1 siRNA-transfected HeLa (Fig. 4A), PC3-LN4 (see Fig. S4D in the supplemental material), and BT474 (see Fig. S4E) cells. Insulin and serum stimulation also resulted in enhanced eIF4B phosphorylation at S422. In siRNA-transfected tumor cells, a decrease in Pim-1 levels impaired the insulin-treated S422 phosphorylation, but it did not affect the serum-stimulated levels in all three cell lines (Fig. 4A; see also Fig. S4D and E). Interestingly, both insulin and serum increased the expression of Pim-1 protein in these cell lines (Fig. 4A; see also Fig. S4D and E), suggesting another level of regulation of eIF4B phosphorylation under these conditions. Furthermore, both HGF and EGF treatment induced the phosphorylation of both S406 and S422 in HeLa cells (Fig. 4B). In these cells, the phosphorylation of S406 and, to a lesser extent, S422 was blocked by the Pim inhibitor GNE-652 (Fig. 4B).
FIG 4.
Pim-1 regulates eIF4B S406 phosphorylation in response to growth factors and serum. (A) HeLa cells were treated with Pim-1 siRNA for 48 h, and then cells were serum starved for 24 h before stimulation with insulin (1 μg/ml) or 20% fetal bovine serum (FBS) for an additional 30 min. The levels of S406 and S422 phosphorylation were quantified by normalizing the levels to total eIF4B using ImageJ software. The average ± SD is shown. (B) HeLa cells were serum starved for 24 h and pretreated with GNE-652 (1 μM) for 3 h before stimulation with insulin (1 μg/ml), HGF (100 ng/ml), or EGF (100 ng/ml) for 30 min. Cell lysates were analyzed by immunoblot assays using the indicated antibodies. (C and D) MEFs as described for Fig. 3D were serum starved for 24 h and treated with insulin (1 μg/ml) or 20% FBS for 15 and 30 min. Cell lysates were analyzed by immunoblot assays using the indicated antibodies.
Additionally, treatment of wild-type MEFs with insulin or serum markedly increased the phosphorylation of eIF4B S406 whereas these stimuli failed to stimulate this phosphorylation in serum-starved TKO MEFs (Fig. 4C). In contrast, insulin or serum did stimulate S422 phosphorylation in the serum-starved TKO MEFs, although the level of phosphorylation was significantly lower than that induced in similarly treated wild-type MEFs (Fig. 4C). Reintroduction of Pim-1 into the TKO cells increased both the insulin- and serum-induced phosphorylation of eIF4B at S406 but did not affect the phosphorylation at S422 (Fig. 4D). Notably, the level of MET protein expression was lower in the TKO MEFs than in wild-type MEFs and could not be induced by stimulation with either insulin or serum (Fig. 4C) whereas its expression was enhanced on reexpression of Pim-1 (Fig. 4D). These data suggest that in MEFs the expression of MET is associated with the phosphorylation of eIF4B at S406 and that a single Pim kinase (Pim-1) is sufficient to control this expression.
Pim-1 controls eIF4B phosphorylation at S406 whereas PI3K/AKT/mTOR pathways control phosphorylation at S422.
It has been suggested that insulin-induced phosphorylation of eIF4B S406 is dependent on both MEK and mTOR activity (17). As described above, we found that treatment of various benign or malignant cell lines, including BPH1, PC3-LN4, BT474, HeLa, 293T, MKN45, U2OS, and H1993 (Fig. 3C), with Pim inhibitors markedly reduced the phosphorylation of eIF4B S406, and only in HeLa cells did it cause a reduction in the phosphorylation of S422. In contrast, treatment of these cell lines with the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathway inhibitor BEZ235, PP242, or AZD8055 blocked the phosphorylation of eIF4B S422 but had no effect on S406 phosphorylation (Fig. 5A). As the results could be affected by the experimental conditions, we further analyzed elF4B phosphorylation in HeLa, PC3-LN4, and BT474 cells that were grown under either serum-rich conditions (Fig. 5B; see also Fig. S5A and B in the supplemental material) or starved and insulin-stimulated conditions (Fig. 5C; see also Fig. S5C and D). Again, we found that phosphorylation of eIF4B S406 was not inhibited by small-molecule inhibitors that block the PI3K/AKT/mTOR pathway (MK2206 for AKT1,2,3; rapamycin for mTORC1; PP242 for mTORC1 and mTORC2; and BEZ25 for PI3K, mTORC1, and mTORC2) or the MEK pathway (U0126 for MEK1,2 and BI-D1870 for RSK1,2,3,4), when used alone or in combination. In marked contrast, the Pim inhibitors, GNE-652 and AZD1208, clearly blocked phosphorylation of eIF4B S406. However, PP242, BEZ235, and, to a lesser extent, MK2206 inhibited eIF4B S422 phosphorylation (Fig. 5B and C; see also Fig. S5). Meanwhile, although rapamycin alone did not inhibit the eIF4B S422 phosphorylation, its combination with U0126 or BI-D1870 did (Fig. 5B and C; see also Fig. S5). In addition, BI-D1870 alone also reduced insulin-induced eIF4B S422 phosphorylation (Fig. 5C; see also Fig. S5C and D). These results suggest that phosphorylation of eIF4B S422 is predominantly controlled by the PI3K/AKT/mTOR pathway. However, in agreement with other studies (17, 26), the MEK/ERK/RSK pathway also plays a role in phosphorylation of S422. Meanwhile, phosphorylation of eIF4B S406 is predominantly controlled by the Pim protein kinases. We have found previously that small-molecule AKT inhibitors can induce Pim-1 expression (27). Consistent with this observation, GSK690693, an AKT inhibitor, increased the phosphorylation of eIF4B S406 while reducing the levels of eIF4B S422 phosphorylation (Fig. 5D). This AKT inhibitor-induced increase in phosphorylation of eIF4B S406 was blocked by GNE-652, a Pim inhibitor, suggesting that under these circumstances the increased Pim-1 expression associated with inhibition of the AKT protein kinase plays a role in the phosphorylation of eIF4B S406.
FIG 5.
Pim and PI3K/AKT/mTOR pathways control phosphorylation of eIF4B S406 and S422, respectively. (A) Cell lines were treated with BEZ235 (0.5 μM), PP242 (1 μM), or AZD8055 (1 μM) for 3 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (B) HeLa cells were treated with MK2206 (1 μM), rapamycin (Rapa; 100 nM), PP242 (1 μM), BEZ235 (0.5 μM), U0126 (U0; 15 μM), BI-D1870 (BI; 10 μM), GNE-652 (1 μM), AZD1208 (3 μM), and the indicated inhibitor combinations for 3 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (C) HeLa cells were serum starved for 24 h, and then cells were treated with inhibitors as in panel B for 3 h before insulin (1 μg/ml) was added for an additional 30 min. Cell lysates were analyzed by immunoblot assays using the indicated antibodies. (D) PC3-LN4 cells were treated with GSK690693 and GNE-652 as indicated for 24 h. Cell lysates were analyzed by immunoblot assays using the indicated antibodies.
Phosphorylation of eIF4B regulates its association with the eIF3 translation initiation complex.
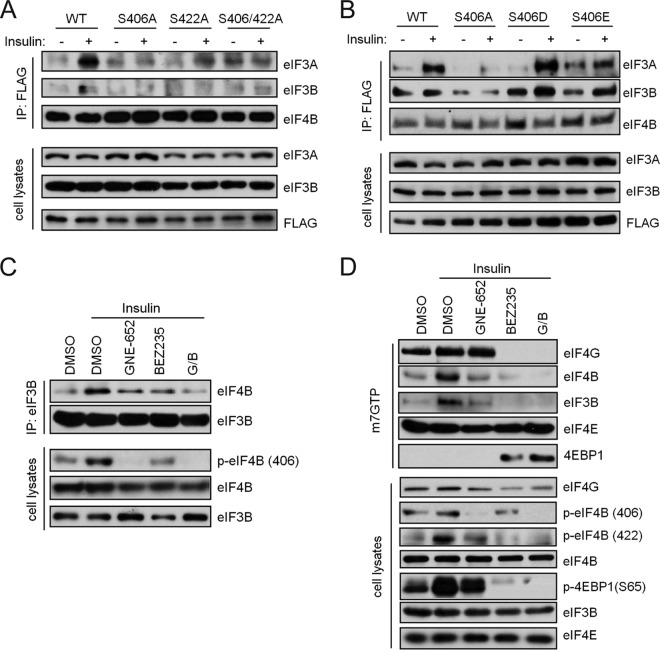
The protein translation initiation complex consists of several proteins in the eIF family. In yeast and mammalian cells, the binding of eIF4B to eIF3 enhances protein synthesis (28, 29). It has been suggested that phosphorylation of eIF4B S422 can regulate its interaction with eIF3 (26). To examine whether the association of eIF3 proteins with eIF4B is affected by eIF4B S406 phosphorylation, we carried out coimmunoprecipitation experiments. Cells were transfected with eIF4B phosphorylation-site mutation constructs S406A and S422A as well as S406D and S406E, which carry negative charges and thus mimic phosphorylation of eIF4B at S406. After transfection, cells were starved and then stimulated with insulin to promote the formation of the eIF3 complex. We find that insulin treatment stimulated an enhanced interaction of wild-type eIF4B with eIF3A and eIF3B and that this interaction with eIF3A was absent when the cells were transfected with the S422A or S406A/S422A mutants (Fig. 6A). This interaction also was abrogated when the cells were transfected with the S406A construct; moreover, the S406D and S406E mutant constructs formed a complex with eIF3A or eIF3B as efficiently as did wild-type eIF4B (Fig. 6B). Pretreatment of 293T cells with the pan-Pim inhibitor GNE-652 or the PI3K/mTOR inhibitor BEZ235, or both agents prior to insulin addition, blocked the ability of eIF4B to bind to eIF3B (Fig. 6C). Thus, elF4B S406 phosphorylation plays an essential role in the formation of the translation initiation complex.
FIG 6.
Phosphorylation of eIF4B S406 regulates its association with the eIF3 translational complex. (A and B) FLAG-tagged wild-type (WT) eIF4B and its mutants were expressed in 293T cells. Cells were starved and stimulated with insulin (1 μg/ml) for 15 min. Immunoprecipitates (IP) and cell lysates were immunoblotted as shown. (C) HeLa cells were serum starved and pretreated with GNE-652 (1 μM), BEZ235 (0.5 μM), or the combination (G/B) for 3 h before stimulation with insulin (1 μg/ml) for 15 min. Coimmunoprecipitation was carried out with anti-eIF3B antibody. The immunoprecipitates and cell lysates were immunoblotted as shown. (D) Cells were treated as in panel C. Cell lysates were incubated with m7-GTP-Sepharose, and elutes were analyzed by immunoblot assays using the indicated antibodies.
These data suggested that both the eIF4B S406 and S422 phosphorylation sites contribute to the binding of eIF4B to eIF3B and that inhibition of phosphorylation of both sites resulted in greater inhibition of binding than did inhibition of either site alone. We therefore extended the studies to determine whether the phosphorylation state of eIF4B affects the formation of translation initiation complex at the 5′ cap structure. The m7-GTP-Sepharose binding assay was used to identify the proteins that bound to the 5′ cap structure. Insulin treatment of starved HeLa cells enhanced the binding of both eIF4B and eIF3B to the m7-GTP beads (Fig. 6D). Treatment with the Pim inhibitor GNE-652 reduced binding of both elF4B and elF3B to the m7-GTP beads, as did treatment with BEZ235 (Fig. 6D). Concomitant treatment with both GNE-652 and BEZ235 resulted in the same or a better reduction in binding. The inhibitors did differ, however, in terms of their effects on the binding of other components of the translation complex: BEZ235 enhanced the interaction of 4EBP1 with the complex, but GNE-652 did not. Consistent with this observation, BEZ235 treatment also resulted in a decrease in the binding of eIF4G, whereas GNE-652 did not (Fig. 6D). These findings suggest that eIF4B S406 phosphorylation is an important regulatory element in controlling the protein binding of eIF4B to the eIF3 complex and that eIF4B S406 phosphorylation regulated by Pim kinases and S422 phosphorylation regulated by PI3K/AKT/mTOR pathway converge on eIF4B to control protein translation.
Phosphorylation of eIF4B S406 positively regulates MET expression.
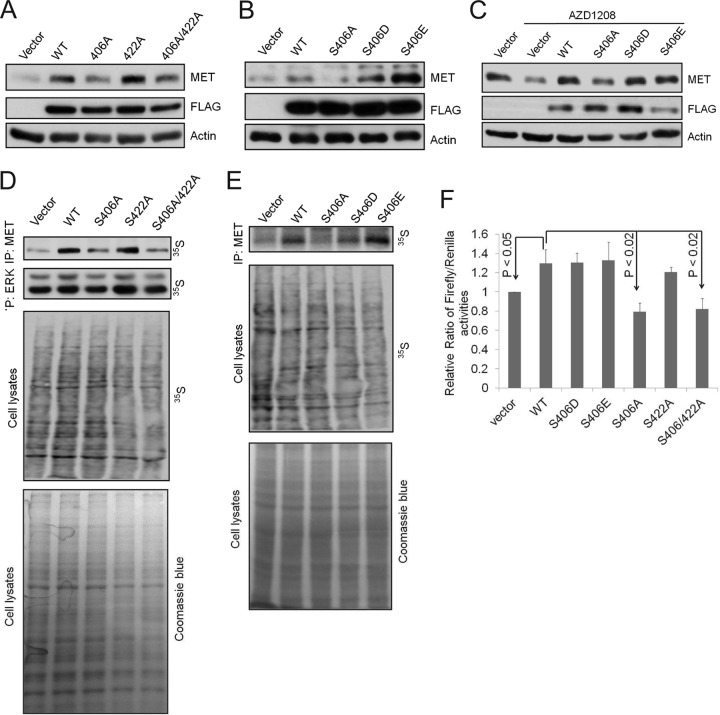
The experiments described above established that Pim-1 kinase activity may affect translational activity directly. Furthermore, we have previously demonstrated that Pim-1 does not affect the transcription of MET (13). To specifically determine the role of eIF4B S406 phosphorylation in the regulation of the translation of MET, we again utilized the eIF4B mutant constructs. First, the constructs were transfected into U2OS cells, an osteosarcoma cell line that expresses low levels of endogenous eIF4B (unpublished data). Transfection with the wild-type eIF4B construct increased the expression of MET compared to the vector control (Fig. 7A) whereas transfection with the S406A and S406A/S422A mutants failed to do so. The levels of MET protein were increased to the levels seen on transfection with wild-type eIF4B after transfection with eIF4B S422A (Fig. 7A) or S406D or S406E (Fig. 7B) mutants. Furthermore, treatment with the small-molecule Pim inhibitor AZD1208 caused downregulation of MET expression that was reversed by transfection with wild-type eIF4B or eIF4B S406D or S406E but not by eIF4B S406A (Fig. 7C). Thus, the eIF4B S406 phosphorylation site is critical for the regulation of MET expression in this cell line.
FIG 7.
eIF4B S406 phosphorylation positively regulates the MET expression. (A and B) U2OS cells were transfected with a control vector or plasmids expressing wild-type eIF4B (WT) and its mutants for 48 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (C) U2OS cells were transfected as in panels A and B. At 24 h after transfection, cells were treated with AZD1208 (3 μM) for an additional 24 h. Cell lysates were analyzed by immunoblot assays using indicated antibodies. (D and E) U2OS cells overexpressing eIF4B or its mutants S406A, S422A, S406/422A, S406D, and S406E were labeled with 35S for new protein synthesis. Newly synthesized MET and ERK were immunoprecipitated, separated by SDS-PAGE, and visualized by autoradiography. (F) HeLa cells were transfected with pR-MET-F construct together with eIF4B or its mutants. After 24 h, luciferase assays were performed. Relative ratios of firefly/Renilla luciferase activities are shown. The ratio for the vector control was set as 1. The averages ± SDs are shown. P < 0.05, WT versus vector; P < 0.02, WT versus S406A or S406/422A.
To determine the effects of eIF4B S406 phosphorylation on translation of MET protein, we monitored new protein synthesis rates by labeling cells with [35S]methionine. Treatment of both PC3-LN-4 and BT474 cells with the pan-Pim inhibitor AZD1208 but not the PI3K/mTOR inhibitor BEZ235 reduced MET synthesis (see Fig. S6A in the supplemental material). These data again suggest that eIF4B S406 but not S422 phosphorylation is important for translation of MET protein. Indeed, overexpression of wild-type eIF4B and S406D and S406E mutants but not the S406A protein increased the rate of MET protein synthesis in U2OS cells (Fig. 7D and E).
We then used a dicistronic luciferase construct containing the MET 5′ UTR inserted upstream of the firefly luciferase gene (13) to further examine the role of eIF4B in regulating MET translation. Transfection of HeLa (Fig. 7F) or U2OS (see Fig. S6B in the supplemental material) cells with the wild-type eIF4B, or either of the two phosphomimetic mutants (S406D or S406E), increased the ratio of firefly/Renilla luciferase activity whereas transfection with either the S406A or S406A/S422A constructs did not. In this assay, the activity of the S406/S422A construct was comparable to that of wild-type eIF4B (Fig. 7F; see also Fig. S6B). Taken together, these results indicate that Pim kinase phosphorylation of eIF4B S406 plays a crucial role in controlling the translation of MET.
Pim kinase inhibitors reduce eIF4B S406 phosphorylation and MET expression in human patient samples.
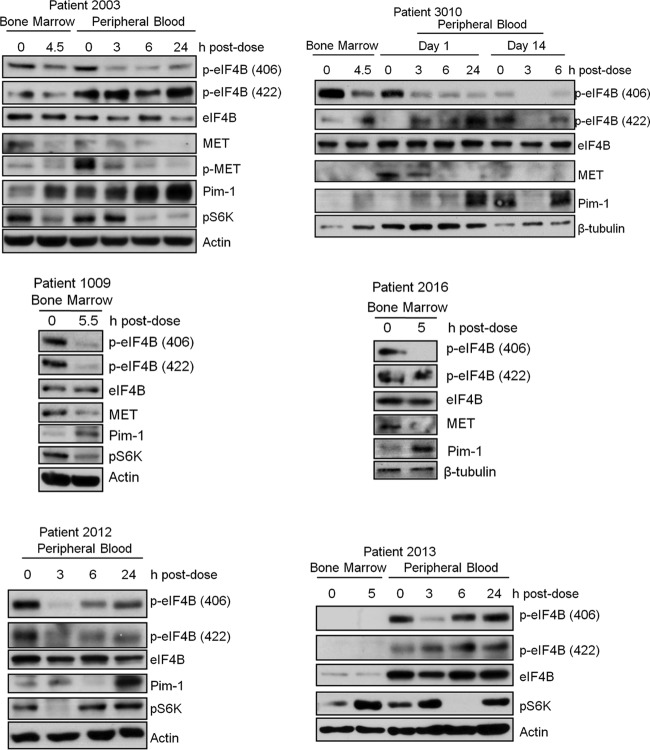
The above data were generated using cultured cells. To determine whether the results may be relevant clinically, we utilized samples from patients with AML. Pim-1 protein kinases are overexpressed in AML blasts and play a role in leukemogenesis (30), and recently, autocrine activation of MET has been demonstrated in human acute myeloid leukemia cells and samples from patients with AML (8). To determine whether inhibition of Pim kinases reduces eIF4B S406 phosphorylation and MET expression in human patient samples, we isolated bone marrow or peripheral blood mononuclear cells from the blood of patients with AML and treated these cells ex vivo with two different Pim inhibitors. Treatment with either AZD1208 or GNE-652 caused a dose-dependent downregulation of phosphorylation of eIF4B at S406 but hardly at S422 (Fig. 8). The Pim inhibitor treatment reduced the levels of MET protein in the samples from the three patients (patients 1, 2, and 3) in which MET was detectable, but the fourth patient (patient 4) did not express sufficient amounts of MET to make this protein evaluable.
FIG 8.
Pim inhibitors reduce eIF4B S406 phosphorylation and MET expression in cells derived from AML patients treated with Pim inhibitors ex vivo. Mononuclear cells freshly isolated from bone marrow and/or peripheral blood of four AML patients were treated with AZD1208 or GNE-652 for 24 h. Cell lysates were analyzed by immunoblot assays using the indicated antibodies.
In addition, we analyzed peripheral blood and/or bone marrow samples collected from AML patients in a phase I clinical trial of AZD1208 (Fig. 9; see also Table S1 in the supplemental material). In all evaluated patients, the infusion of the Pim inhibitor markedly increased Pim-1 protein levels and/or decreased S6K activity, suggesting that this small-molecule inhibitor was active in patients. Although MET was difficult to discern by Western blotting, analysis of blast samples from peripheral blood or bone marrow derived from these patients also demonstrated inhibition of eIF4B S406 phosphorylation and MET expression (Fig. 9). Analysis of the peripheral blood indicated inhibition of eIF4B phosphorylation, but the time course of inhibition (determined at 3, 6, and 24 h after infusion) differed among the patients, with prolonged inhibition seen in patients 2003 and 3010 but only transient changes visible in patients 2012 and 2013 (Fig. 9). These results could reflect the biology of the tumor or the individual drug pharmacokinetics. In patient 2003, MET was detected but phospho-MET could be more easily measured and was shown to decrease with Pim treatment. In patient 3010, MET could be measured in the peripheral blood and decreased with Pim treatment. For patients 1009 and 2016, peripheral blood was not available, but the bone marrow samples showed reduced MET protein. The MET was not detectable in samples from patients 2012 and 2013. These data suggest that inhibition of Pim regulates both the phosphorylation of eIF4B and the levels of MET protein in vivo.
FIG 9.
Pim inhibitors reduce eIF4B S406 phosphorylation and MET expression in samples obtained from patients with AML undergoing AZD1208 treatment. Cell lysates from mononuclear cells isolated from bone marrow and/or peripheral blood from six AML patients treated with AZD1208 in an ongoing phase I clinical trial were analyzed by immunoblot assays using the indicated antibodies.
DISCUSSION
Our present findings demonstrate that Pim-1 kinase activity plays a significant role in determining the levels of MET protein expression and that it does so by regulating protein translation. The regulation of MET expression by Pim-1 kinase activity has been generalized to multiple tumor types. Correlations between the levels of MET and Pim-1 were observed in normal tissues, in numerous tumor cell lines, and on analysis of patient-derived tissues, including analysis of primary prostate tissue microarrays and analysis of leukemic cells from patients with AML. Notably, in cell lines representing numerous cancer types, including prostate, breast, and lung cancer; leukemia; sarcoma; and cervical cancer, inhibition of the Pim activity with small molecules lowered MET expression significantly. Mutations and dysregulation of transcription in the MET-HGF axis are common in patients with certain cancers (renal papillary, hepatocellular, gastric, and esophageal cancer) and in some patients with lung cancer (4, 5). These results suggest that translational control of MET expression could be important in the control of this receptor in tumor cells.
The physiologic relevance of the Pim kinase regulation of the MET-HGF signaling axis is indicated by the observation that manipulation of Pim kinase activity had a significant effect on tumor cell behavior, including cell scattering, invasion, and migration, and that this was associated with the effects of the Pim kinase activity on MET expression. Furthermore, the induction of MET expression by both insulin and serum was blocked by inhibition or downregulation of Pim. This suggests that inhibiting Pim kinase activity blocks the proliferation and survival signals provided by hormones and growth factors that act by elevating MET expression.
In this paper, we demonstrate that Pim protein kinase phosphorylates eIF4B S406 and that this phosphorylation determines eIF4B binding to the eIF3 complex, thereby regulating the formation of the translation initiation apparatus. Our results demonstrate that Pim-1 modulation of this phosphorylation site occurs in cell lines and fresh tumor samples and in human leukemic samples taken from patients who have been infused with small-molecule Pim inhibitors. Previously, van Gorp and colleagues reported that insulin-induced phosphorylation of eIF4B S406 was dependent on both MEK and mTOR activity (17); however, we found that the phosphorylation of eIF4B S406 was not inhibited by MEK-ERK-RSK pathway inhibitors, PI3K/AKT/mTOR pathway inhibitors, or combinations of these inhibitors. In marked contrast, inhibitors of Pim kinase activity effectively blocked eIF4B S406 phosphorylation. This was found to be the case in several cell lines representing various tumor types and to be independent of the chemotype of the Pim kinase inhibitor. The use of various combinations of inhibitors further revealed that the Pim kinase pathway directly regulates S406 phosphorylation whereas the PI3K/AKT/mTOR pathways primarily regulate S422 phosphorylation. However, there did not appear to be a hierarchical relationship between S406 phosphorylation and S422 phosphorylation under the conditions used in these experiments. In multiple cell lines, blockading the phosphorylation of either site did not affect the phosphorylation of the other one. p70S6K did not appear to play a role in eIF4B S406 phosphorylation, as both Pim and PI3K/AKT/mTOR inhibitors blocked its activity, which is evidenced by the inhibition of ribosomal protein S6 phosphorylation, but only Pim inhibitors reduced eIF4B S406 phosphorylation. The difference in the primary eIF4B phosphorylation sites of the Pim kinase pathway and the PI3K/AKT/mTOR pathways suggests that these pathways can function in parallel to control protein synthesis. The convergence of the mechanisms of action on eIF4B phosphorylation suggests, however, that eIF4B may act as a nexus that integrates diverse signaling events. Our results do not rule out the potential for cross talk or overlap between these pathways. Indeed, we found that treatment of HeLa, PC3-LN4, and BT474 cells with Pim-1 siRNA reduced insulin- but not serum-stimulated eIF4B S422 phosphorylation, suggesting that in the case of insulin stimulation Pim-1 might influence the cross talk with the PI3K/AKT pathway. The ability to control eIF4B S406 phosphorylation may be related to our observation that both insulin and serum are found to elevate the protein expression of Pim-1. Furthermore, we observed that the reduced eIF4B S422 phosphorylation and impaired induction of eIF4B S422 phosphorylation by insulin in TKO MEFs compared to wild-type MEFs was consistent with our finding that Pim may play an important role in insulin signal transduction. Although serum treatment of starved cells elevated Pim-1 levels, blocking the activity of Pim-1 did not inhibit the ability of serum to control S422 phosphorylation. It is possible that serum, by strongly activating ERK, stimulates the phosphorylation of S422 through a p90RSK-dependent pathway and does not require the PI3K/AKT/mTOR pathway. Collectively, our data suggest a role for Pim-1 in the cell cycle and induced phosphorylation of eIF4B S406, together with an indirect context-dependent regulation of eIF4B S422 phosphorylation.
The importance of S406 in the control of translation is further demonstrated by our observation that an S406A mutant of eIF4B failed to bind to eIF3A and eIF3B in response to insulin treatment and that treatment with a Pim inhibitor reduced insulin-induced binding of eIF4B bound to eIF3B. Precipitation of the entire cap complex using m7-GTP-Sepharose beads after treatment with insulin together with a Pim inhibitor provided further evidence that eIF4B S406 phosphorylation is crucial for its interaction with eIF3 and thus the formation of a complete translation initiation complex. This assay also revealed that treatment with inhibitors of the PI3K/AKT pathway or Pim kinase had differential effects on the binding of eIF4G and 4EBP1. This suggests that although these pathways both control eIF4B phosphorylation, they can have very different effects on the structural interactions that drive formation of the translation complex.
The observation that Pim-1 may be a key regulator of MET-HGF signaling could have significant therapeutic relevance. MET inhibitors, including both antibodies and small molecules, are already in advanced-phase clinical trials (4). Pim protein kinase inhibitors, such as AZD1208, have now entered the clinic in phase I trials. The analysis of cells obtained from patients with AML treated with the Pim kinase inhibitor AZD1208 in a phase I clinical trial suggested that this compound inhibited eIF4B S406 phosphorylation and MET expression. While the number of patient samples is low, taken together with our analyses of tumor cell lines, these results suggest that eIF4B phosphorylation could function as a unique biomarker of Pim inhibitor activity in clinical trials and that exploration of a combination of MET and Pim inhibitors may be an effective therapeutic strategy in MET-driven cancers.
Although we have focused our investigations on MET, the mechanisms by which Pim-1 controls protein translation may also impact on the level of additional proteins, e.g., the insulin receptor (INSR) and c-Myc (see Fig. S7A and B in the supplemental material). In TKO compared to wild-type (WT) MEFs, we found that the MET and INSR mRNAs shifted from heavier to lighter polysomes, suggesting that Pim was needed for the translation of both of these proteins. In TKO cells, the defective binding of c-Myc mRNA to ribosomes across almost all polysome fractions was clearly seen (see Fig. S7C, bottom panel), again suggesting the importance of the Pim kinases to translational regulation. Furthermore, transfection of a wild-type eIF4B construct and the active S406D or S406E (see Fig. S7D) mutants increased, compared to the vector control (see Fig. S7D), the expression of not only MET but also the INSR and c-Myc, while in comparison, transfection with the S406A mutant failed to do so. Thus, the eIF4B S406 phosphorylation site is critical for the translation of not only MET but also the INSR and c-Myc proteins and potentially additional proteins that undergo translational control.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Hollings Cancer Center Cell Evaluation and Therapy Shared Resource and Biorepository and Tissue Analysis Shared Resource at MUSC for providing excellent support of this project. Paul J. Coffer (Department of Cell Biology, University Medical Center Utrecht, Utrecht, The Netherlands) kindly provided FLAG-tagged eIF4B expression plasmids. We thank Scott D. Cramer (University of Colorado School of Medicine) for sharing the mouse prostate epithelial cells expressing Pim-1. We are grateful to GlaxoSmithKline for supplying GSK690693. We thank Marina Konopleva and Juliana Benito (M. D. Anderson Cancer Center) for providing AML cell lines Molm-16 and OCI-AML2.
This work is supported by NIH grants 1K01DK085196 (to B.C.), 1R01CA173200 (to A.S.K.), and W81XWH-12-10560 (to A.S.K.) and the HCC support grant 5P30CA138313. This work was funded by a Conquer Cancer Foundation of ASCO Young Investigator Award (to A.M.).
Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the American Society of Clinical Oncology or the Conquer Cancer Foundation.
We declare no potential conflicts of interest.
Footnotes
Published ahead of print 28 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00147-14.
REFERENCES
- 1.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. 2003. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 4:915–925. 10.1038/nrm1261 [DOI] [PubMed] [Google Scholar]
- 2.Stoker M, Gherardi E, Perryman M, Gray J. 1987. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature 327:239–242. 10.1038/327239a0 [DOI] [PubMed] [Google Scholar]
- 3.Mazzone M, Comoglio PM. 2006. The Met pathway: master switch and drug target in cancer progression. FASEB J. 20:1611–1621. 10.1096/fj.06-5947rev [DOI] [PubMed] [Google Scholar]
- 4.Varkaris A, Corn PG, Gaur S, Dayyani F, Logothetis CJ, Gallick GE. 2011. The role of HGF/c-Met signaling in prostate cancer progression and c-Met inhibitors in clinical trials. Expert Opin. Invest. Drugs 20:1677–1684. 10.1517/13543784.2011.631523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benvenuti S, Comoglio PM. 2007. The MET receptor tyrosine kinase in invasion and metastasis. J. Cell. Physiol. 213:316–325. 10.1002/jcp.21183 [DOI] [PubMed] [Google Scholar]
- 6.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M, Allikmets R, Chidambaram A, Bergerheim UR, Feltis JT, Casadevall C, Zamarron A, Bernues M, Richard S, Lips CJ, Walther MM, Tsui LC, Geil L, Orcutt ML, Stackhouse T, Lipan J, Slife L, Brauch H, Decker J, Niehans G, Hughson MD, Moch H, Storkel S, Lerman MI, Linehan WM, Zbar B. 1997. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 16:68–73. 10.1038/ng0597-68 [DOI] [PubMed] [Google Scholar]
- 7.Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, Seiwert T, Christensen JG, Jagadeeswaran R, Krausz T, Vokes EE, Husain AN, Salgia R. 2008. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 47:1025–1037. 10.1002/gcc.20604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, Christie A, Valk PJ, Delwel R, Ngo V, Kutok JL, Dahlberg SE, Moreau LA, Byers RJ, Christensen JG, Vande Woude G, Licht JD, Kung AL, Staudt LM, Look AT. 2012. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat. Med. 18:1118–1122. 10.1038/nm.2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316:1039–1043. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 10.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. 2007. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 67:2081–2088. 10.1158/0008-5472.CAN-06-3495 [DOI] [PubMed] [Google Scholar]
- 11.Nawijn MC, Alendar A, Berns A. 2011. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer 11:23–34. 10.1038/nrc2986 [DOI] [PubMed] [Google Scholar]
- 12.Beharry Z, Zemskova M, Mahajan S, Zhang F, Ma J, Xia Z, Lilly M, Smith CD, Kraft AS. 2009. Novel benzylidene-thiazolidine-2,4-diones inhibit Pim protein kinase activity and induce cell cycle arrest in leukemia and prostate cancer cells. Mol. Cancer Ther. 8:1473–1483. 10.1158/1535-7163.MCT-08-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cen B, Mahajan S, Wang W, Kraft AS. 2013. Elevation of receptor tyrosine kinases by small molecule AKT inhibitors in prostate cancer is mediated by Pim-1. Cancer Res. 73:3402–3411. 10.1158/0008-5472.CAN-12-4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. 1996. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin. Cancer Res. 2:1627–1636 [PubMed] [Google Scholar]
- 15.Beharry Z, Mahajan S, Zemskova M, Lin YW, Tholanikunnel BG, Xia Z, Smith CD, Kraft AS. 2011. The Pim protein kinases regulate energy metabolism and cell growth. Proc. Natl. Acad. Sci. U. S. A. 108:528–533. 10.1073/pnas.1013214108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cen B, Mahajan S, Zemskova M, Beharry Z, Lin YW, Cramer SD, Lilly MB, Kraft AS. 2010. Regulation of Skp2 levels by the Pim-1 protein kinase. J. Biol. Chem. 285:29128–29137. 10.1074/jbc.M110.137240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Gorp AG, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F, Hershey JW, Burgering BM, Calkhoven CF, Coffer PJ. 2009. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 28:95–106. 10.1038/onc.2008.367 [DOI] [PubMed] [Google Scholar]
- 18.Merrick WC, Hensold JO. 2001. Analysis of eukaryotic translation in purified and semipurified systems. Curr. Protoc. Cell Biol. Chapter 11:Unit 11.9. 10.1002/0471143030.cb1109s08 [DOI] [PubMed] [Google Scholar]
- 19.Fuss IJ, Kanof ME, Smith PD, Zola H. 2009. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. Chapter 7:Unit 7.1. 10.1002/0471142735.im0701s85 [DOI] [PubMed] [Google Scholar]
- 20.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. 2012. Targeting MET in cancer: rationale and progress. Nat. Rev. Cancer 12:89–103. 10.1038/nrc3205 [DOI] [PubMed] [Google Scholar]
- 21.Fram ST, Wells CM, Jones GE. 2011. HGF-induced DU145 cell scatter assay. Methods Mol. Biol. 769:31–40. 10.1007/978-1-61779-207-6_3 [DOI] [PubMed] [Google Scholar]
- 22.Xia Z, Knaak C, Ma J, Beharry ZM, McInnes C, Wang W, Kraft AS, Smith CD. 2009. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J. Med. Chem. 52:74–86. 10.1021/jm800937p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambarotta G, Pistoi S, Giordano S, Comoglio PM, Santoro C. 1994. Structure and inducible regulation of the human MET promoter. J. Biol. Chem. 269:12852–12857 [PubMed] [Google Scholar]
- 24.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, Svitkin Y, Sonenberg N. 2010. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell. Biol. 30:1478–1485. 10.1128/MCB.01218-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 25:2781–2791. 10.1038/sj.emboj.7601166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cen B, Mahajan S, Wang W, Kraft AS. 2013. Elevation of receptor tyrosine kinases by small molecule AKT inhibitors in prostate cancer is mediated by Pim-1. Cancer Res. 73:3402–3411. 10.1158/0008-5472.CAN-12-4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vornlocher HP, Hanachi P, Ribeiro S, Hershey JW. 1999. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J. Biol. Chem. 274:16802–16812. 10.1074/jbc.274.24.16802 [DOI] [PubMed] [Google Scholar]
- 29.Methot N, Song MS, Sonenberg N. 1996. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16:5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L, Uzunov M, Ifrah N, Dreyfus F, Lacombe C, Mayeux P, Bouscary D. 2009. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 114:1618–1627. 10.1182/blood-2008-10-184515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.