ABSTRACT
It is accepted that an effective prophylactic HIV-1 vaccine is likely to have the greatest impact on viral transmission rates. As previous reports have implicated DNA-priming, protein boost regimens to be efficient activators of humoral responses, we sought to optimize this regimen to further augment vaccine immunogenicity. Here we evaluated single versus concurrent intradermal (i.d.) and intramuscular (i.m.) vaccinations as a DNA-priming strategy for their abilities to elicit humoral and cellular responses against a model HIV-1 vaccine antigen, CN54-gp140. To further augment vaccine-elicited T and B cell responses, we enhanced cellular transfection with electroporation and then boosted the DNA-primed responses with homologous protein delivered subcutaneously (s.c.), intranasally (i.n.), i.m., or transcutaneously (t.c.). In mice, the concurrent priming regimen resulted in significantly elevated gamma interferon T cell responses and high-avidity antigen-specific IgG B cell responses, a hallmark of B cell maturation. Protein boosting of the concurrent DNA strategy further enhanced IgG concentrations but had little impact on T cell reactivity. Interestingly protein boosting by the subcutaneous route increased antibody avidity to a greater extent than protein boosting by either the i.m., i.n., or t.c. route, suggesting that this route may be preferential for driving B cell maturation. Using an alternative and larger animal model, the rabbit, we found the concurrent DNA-priming strategy followed by s.c. protein boosting to again be capable of eliciting high-avidity humoral responses and to also be able to neutralize HIV-1 pseudoviruses from diverse clades (clades A, B, and C). Taken together, we show that concurrent multiple-route DNA vaccinations induce strong cellular immunity, in addition to potent and high-avidity humoral immune responses.
IMPORTANCE The route of vaccination has profound effects on prevailing immune responses. Due to the insufficient immunogenicity and protection of current DNA delivery strategies, we evaluated concurrent DNA delivery via simultaneous administration of plasmid DNA by the i.m. and i.d. routes. The rationale behind this study was to provide clear evidence of the utility of concurrent vaccinations for an upcoming human clinical trial. Furthermore, this work will guide future preclinical studies by evaluating the use of model antigens and plasmids for prime-boost strategies. This paper will be of interest not only to virologists and vaccinologists working in the HIV field but also to researchers working in other viral vaccine settings and, critically, to the wider field of vaccine delivery.
INTRODUCTION
To date, most licensed vaccines are based on the generation of neutralizing antibodies which are effective against invariant antigen-bearing pathogens. However, as antigenic variability increases, the number of licensed vaccines that are effective dramatically decreases (1). As a consequence, HIV-1, a retrovirus with exceptionally high antigenic variability, may require a completely novel vaccination strategy. Hence, in an attempt to augment vaccine-induced anti-HIV-1 T helper and antibody responses, we utilized three distinct concepts to formulate a novel immune-priming paradigm to precede protein boost vaccination. Specifically, we utilized (i) a DNA plasmid vector called Auxo-GTU, previously described to induce strong and durable T cell responses, in combination with (ii) electroporation (EP) and (iii) concurrent intradermal (i.d.) and intramuscular (i.m.) vaccinations.
The Auxo-GTU technology is a nonreplicating plasmid vector which utilizes the bovine papilloma virus type 1 (BPV1) transcription activator, the segregation/partitioning factor E2 protein, and its multimeric binding sites (2, 3). This has been shown to result in the enhanced transcriptional activity of the transgenes along with the potential for increasing the number of cells expressing the transgene (3). Furthermore, it has previously been utilized in clinical and preclinical studies and has been shown to display a good safety profile (4). DNA-based vaccination is an attractive mode of vaccine delivery. DNA vaccines utilize the host for in vivo biosynthesis of transgene products (5), hence imitating infectious pathways, and through host cell posttranslational modifications, the transgene products more accurately represent the conformation of naturally expressed viral antigens (6). Despite the many advantages, most conventional DNA vaccination strategies appear to be poorly immunogenic. Therefore, DNA vaccines have failed to translate from earlier murine studies to humans, leading to poor efficacy in human clinical trials (7, 8), and as a consequence, no prophylactic DNA vaccine is clinically approved for use in humans (5). To enhance the immunogenicity of DNA vaccines, strategies such as promoter selection, codon optimization, and different routes of administration have been employed (5). However, the delivery of DNA in association with EP has been shown to dramatically increase gene expression and vaccine-induced responses over and above those that have been obtained by the use of most existing adjuvant technologies (9–12).
Other aspects may play a significant role in the as yet limited immunogenicity of DNA vaccines. For instance, while most vaccinations, including DNA, are delivered via the i.m. route (13), the lower number of antigen-presenting cells (APCs) within muscle tissues has been suggested to be a factor contributing to reduced efficacy (2, 5, 14). i.m. vaccination has been described to result in poor antigen-dependent T cell activation owing to the lack of APCs in muscle tissue (14). Certainly, previous studies have shown that i.d. vaccination increases the magnitude of polyfunctional CD4+ T cell responses compared to that achieved with i.m. vaccination (15). Therefore, despite myocytes being good transfection candidates (5), the location may be suboptimal for DNA-based immune activation. As opposed to previously published findings from studies evaluating single routes of DNA administration (16), here we report on the use of concurrent multiple-route vaccination and inoculation i.d., a route that allows the vaccine to reach a site with a plentiful and diverse number of APCs (17), combined with concurrent i.m. vaccination. We hypothesized that this dual-route vaccination may result in enhanced immune responses to the in vivo-expressed plasmid DNA vaccine transgenes. Here we report that combination of the concurrent DNA vaccination strategy (i.d. [EP] and i.m. [EP]) with a homologous protein boost vaccination regimen enhances the levels of production of antibody with a higher antigen-specific binding avidity. Furthermore, we show that concurrent DNA priming can augment the vaccine-elicited T cell response, a factor critical for providing the necessary T cell help to B cells. Finally, we demonstrate that the optimized DNA-priming, protein boost regimen can translate to a larger animal model and drive the production of neutralizing antibodies against different HIV-1 strains. To our knowledge, this is the first report of an optimized concurrent parenteral approach utilizing DNA EP in a prime-boost regimen to achieve enhanced immune responses against the HIV-1 envelope.
MATERIALS AND METHODS
Plasmids, protein, and peptides.
The HIV-1 vaccine construct was developed and prepared using the Auxo-GTU vector technology. The HIV-1 CN54-gp140 clade C/B gene (codon optimized) was inserted into the Auxo-GTU vector backbone and DNA vaccine produced by Fit Biotech, Finland. The recombinant CN54-gp140 protein was purchased from Polymun Scientific (Austria). The gp140-stimulatory peptides, 15-mers overlapping by 11 amino acids, were devised, synthesized (Insight Biotechnology, United Kingdom), and split into two pools.
Immunizations.
Six- to-8-week old female BALB/c mice (n = 8; Harlan, United Kingdom) were handled and procedures were performed in accordance with the terms of a project license granted under the United Kingdom Home Office Animals (Scientific Procedures) Act 1986. Mice received three or four plasmid DNA (GTU-CN54-gp140) immunizations at 3-week intervals as either i.d. (10 μg DNA in the lower back), i.m. (50 μg in the quadriceps), or i.d. and i.m. combination vaccinations. Injection sites then received EP using either 5-mm or tweezer electrodes using an ECM 830 square-wave electroporation system (BTX). Pulses consisted of 100 V of positive and negative polarity at a rate of 1 pulse/s, with each pulse lasting 50 ms. Protein boost studies involved either two subcutaneous (s.c.), intranasal (i.n.), transcutaneous (t.c.), or i.m. protein boosts (20 μg) using the CN54-gp140 protein in the absence of adjuvant. New Zealand White rabbits (n = 8; Harlan, United Kingdom) were handled and procedures were performed in accordance with the terms of a project license granted under the United Kingdom Home Office Animals (Scientific Procedures) Act of 1986. Rabbits were placed into three groups. Groups 1 and 2 received three i.d. (EP) and i.m. (EP) DNA-priming vaccinations, while groups 1 and 3 received two s.c. recombinant gp140 protein boost vaccinations. For i.d. and i.m. vaccinations, 20 and 100 μg of DNA were used, while 40 μg protein was used for s.c. protein boosts.
Sampling and processing.
Blood was collected and centrifuged at 1,000 × g for 20 min. The serum was harvested and stored at −20°C. To assess gamma interferon (IFN-γ) T cell responses, cultures of lymphocytes from spleens were prepared as described previously (18, 19).
Bioluminescence imaging.
Real-time bioimaging of i.d. and i.m. DNA vaccinations was performed using a multispectral Carestream in vivo FX Pro system. Briefly, either 50 μg (i.m.) or 30 μg (i.d.) GTU-Luc-GFP plasmid was administered with or without EP to anesthetized BALB/c mice. Luciferase activity was measured 15 min after i.p. administration of d-luciferin (150 mg/kg of body weight; Promega, United Kingdom). Bioluminescence images were acquired and analyzed using Carestream software and 60-s exposures.
Immunoglobulin ELISA and avidity assay.
A quantitative immunoglobulin enzyme-linked immunosorbent assay (ELISA) protocol described previously (18, 19) was followed. Briefly, 5 μg/ml gp140-coated ELISA plates were blocked with 200 μl/well 1% bovine serum albumin and 0.05% phosphate-buffered saline (PBS)–Tween 20 (PBST). After washing, samples were diluted and added to the plates for 1 h before washing and addition of a 1:4,000 dilution of either anti-mouse IgG–horseradish peroxidase (HRP), IgG1–HRP, IgG2a–HRP, or IgA–HRP (Southern Biotech). Samples were developed using tetramethylbenzidine (TMB), and the reaction was stopped using Stop solution (Insight Biotechnologies, United Kingdom). Standards consisted of coating with anti-mouse kappa (1:3,200) and lambda (1:3,200) light chains (Serotec, United Kingdom), blocking as described above, and then addition of purified IgG (Southern Biotech, United Kingdom) or IgA (Southern Biotech, United Kingdom) starting at 200 ng/ml. The absorbance was read on a spectrophotometer (VersaMax; Molecular Devices) using SoftMax Pro GxP (v5) software. For the rabbit antigen-specific IgG ELISA, a protocol similar to that described above was followed. For standards, plates were initially coated with a 1:1,250 dilution of 2 mg/ml goat anti-rabbit IgG (Millipore, United Kingdom), followed by 1/5 dilutions of purified rabbit IgG (1,000 ng/ml; Serotec, United Kingdom), and rabbit IgG was detected using monoclonal anti-rabbit gamma chain-specific IgG (clone RG-96; Sigma, United Kingdom).
The avidity of serum immunoglobulin was determined by the antibody-antigen binding resistance to 8 M urea. Samples were prediluted to give an optical density (OD) at 450 nm (OD450) readout of between 1.0 and 1.5 and were added to gp140-coated plates. The plates were then washed three times with either PBST or 8 M urea in PBST, before incubating with anti-mouse IgG–HRP. Samples were developed with TMB as described above. The avidity index (in percent) was calculated as the OD450 of urea-treated samples/OD450 of PBST-treated samples. Antisera with index values exceeding 50% were ascribed a high avidity, those with index values of 30 to 50% were ascribed intermediate avidity, and those with index values of <30% were ascribed a low avidity.
IFN-γ antigen-specific ELISpot assays.
IFN-γ enzyme-linked immunosorbent spot (ELISpot) assays (Mabtech, United Kingdom) were carried out on splenocytes per the manufacturer's instructions and as described previously (18, 19). Plates were blocked for 30 min using complete RPMI before addition of 5 × 106 cells/ml. Next, 5 μg/ml of gp140 peptide pools 1 and 2 was used to stimulate cells for 16 h to assess the responses. Unstimulated and 5 μg/ml phytohemagglutinin (Sigma, United Kingdom)-stimulated cells served as controls. To detect spots, biotinylated anti-IFN-γ antibody was added at 1 μg/ml for 2 h before washing and incubating with streptavidin-HRP for 1 h. Plates were washed as described above, and 100 μl/well of TMB substrate was added.
Virus neutralization assay.
HIV-1 serum neutralization assays were performed using a luciferase-based assay in TZM.bl cells as described previously (20). Briefly, 3-fold serial dilutions of serum samples in 10% Dulbecco modified Eagle growth medium (100 μl/well) were performed in duplicate in 96-well plates. A 50-μl volume of virus was added to each well, and the plates were incubated for 1 h at 37°C. TZM.bl cells in growth medium (1 × 104 cells/well per 100-μl volume) containing 11 μg/ml DEAE-dextran (Sigma) were then added to each well. The neutralization assay used replicate wells of TZM.bl cells alone (cell control) and TZM.bl cells with virus (virus control). Following a 48-h incubation at 37°C, a 150-μl volume of assay medium was removed from each well and 100 μl of Bright-Glo luciferase reagent (Promega) was added. The cells were lysed for 2 min, before a 150-μl volume of cell lysate was transferred to a 96-well black solid plate, and luminescence was measured using a Victor 3 luminometer (PerkinElmer). The 50% inhibitory dose (ID50) titer was calculated as the serum dilution that caused a 50% reduction in relative luminescence units (RLUs) compared to that for virus control wells after subtraction of cell control RLUs.
Statistical analysis.
Statistical analysis of the data was carried out using a Mann-Whitney nonparametric t test and GraphPad Prism software.
RESULTS
Concurrent i.d. and i.m. vaccination with EP enhances cellular responses as well as alters antibody avidity.
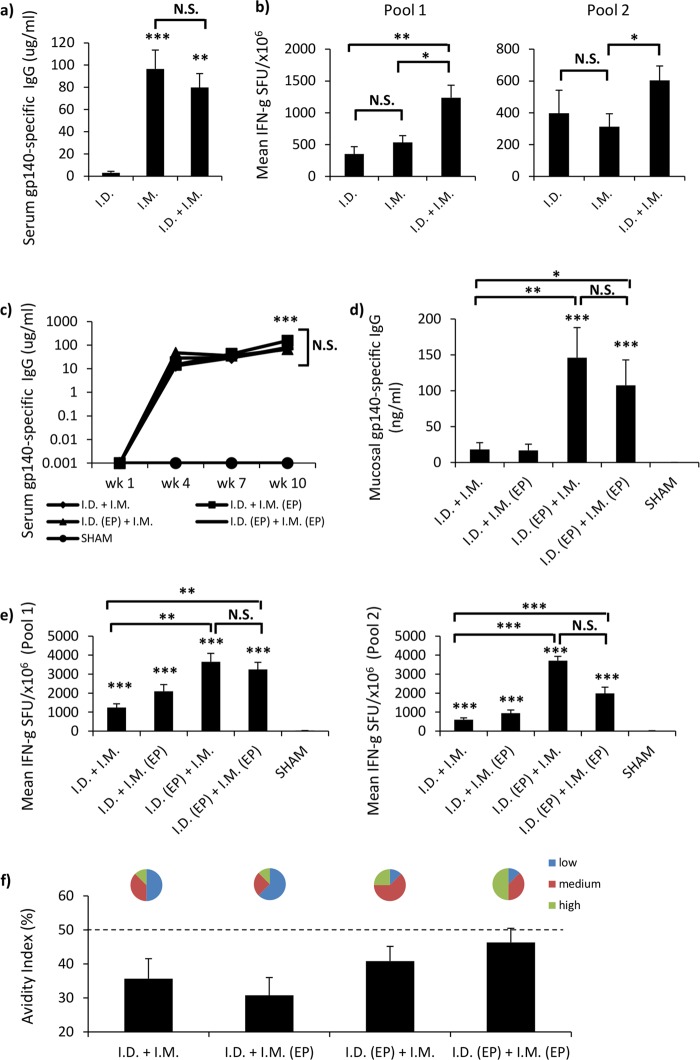
i.m. vaccination is the most common route for DNA vaccination. However, even though myocytes are highly susceptible to transfection, the response obtained by the targeting of muscle tissue for vaccine gene expression may be limited because of the low number of local APCs (21). This is in stark contrast to the response obtained by the targeting of skin, which is notable for its abundance of APCs. We therefore hypothesized that combining i.m. vaccination with concurrent i.d. vaccination might enhance vaccine-mediated humoral and cellular immune responses over and above the response normally seen by either route alone. Initially, we compared single-site i.d. or i.m. vaccination with concurrent i.d. and i.m. vaccination without EP (see Table S1 in the supplemental material). i.m. vaccination resulted in significantly (P = 0.0009) elevated levels of antibody in serum compared to those achieved by i.d. vaccination (Fig. 1a). However, both i.d. and i.m. vaccinations individually induced nonstatistically distinct IFN-γ spot-forming unit (SFU) responses (Fig. 1b), suggesting that individual i.m. vaccinations were equivalent to i.d. vaccinations for eliciting cellular responses. When comparing concurrent i.d. and i.m. vaccination with individual i.d. and i.m. vaccinations, the concurrent i.d. and i.m. vaccination regimen generated specific IgG concentrations similar to those generated by i.m. vaccination, and the specific IgG concentrations were significantly elevated compared to those achieved with i.d. vaccination (P = 0.0019) (Fig. 1a); however, the concurrent regimen increased antibody avidity compared to that achieved by i.m. vaccination (see Fig. S1a in the supplemental material). Furthermore, concurrent i.d. and i.m. vaccination significantly enhanced the T cell responses against peptide pool 1 (P = 0.003 and P = 0.014 compared to the responses for the individual i.m. and i.d. vaccination groups, respectively) and peptide pool 2 (P = 0.0379 compared to the responses for both the individual i.m. and i.d. vaccination groups) (Fig. 1b) relative to the responses seen for the individual i.m. or i.d. vaccination group, indicating that concurrent i.d. and i.m. vaccination is superior to i.m. vaccination with respect to avidity and the magnitude of cellular responses.
FIG 1.
Concurrent priming vaccination with EP elicits stronger cellular and higher-avidity antibody responses. (a and b) Single i.d. (10 μg) or i.m. (50 μg) vaccination was compared to concurrent i.d. and i.m. (10 μg and 50 μg) vaccination (n = 8 mice per group), in the absence of EP, for the elicited antigen-specific serum IgG antibody (a) and cellular (b) responses. Humoral and cellular responses were evaluated 1 week after the fourth and final vaccination by antigen-specific IgG ELISA and IFN-γ ELISpot assay and are shown as group means (±SEM). (c to e) Comparison of the ability of different concurrent vaccination regimens, in the presence or absence of EP, to elicit antigen-specific serum (μg/ml) (c) and mucosal (ng/ml) (d) IgG responses and to stimulate systemic cellular immunity (numbers of SFU per million antigen-stimulated cells [±SEM]) (e). Mice (n = 8 per group) were vaccinated 4 times at 3-week intervals, with bleeds occurring 1 week after each vaccination. Mucosal sampling occurred during study week 10, 1 week after the final vaccination. Statistical significance was assessed using the Mann-Whitney U test. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005; N.S., not significant. (f) To assess antibody avidity, concurrent vaccination serum samples were prediluted to give an optical density of between 1 and 1.5 using an in-house antigen-specific ELISA. Samples were then titrated in an endpoint ELISA in duplicate using nonreducing (PBS) and reducing (8 M urea) washes after sample addition. Results are shown as the percent change in binding [(reducing OD/nonreducing OD) × 100].
Next, we evaluated the impact of EP on concurrent i.d. and i.m. vaccination and the relative contribution that EP made to the cellular and humoral responses when applied to each vaccination site. All concurrent vaccination combinations generated equivalent and significant concentrations of serum IgG compared to those for the controls (P ≥ 0.0005) (Fig. 1c). As all the tested concurrent vaccination regimens elicited similar antibody titers, we utilized an avidity assay as a means to differentiate and qualitatively evaluate the humoral responses. Here we show that the i.d. (EP) and i.m. group and the i.d. (EP) and i.m. (EP) group generated the highest proportions of medium- and high-avidity responses in serum (Fig. 1f). Strikingly, the i.d. (EP) and i.m. (EP) regimen generated 50% more mice that seroconverted with a high-avidity response than the i.d. (EP) and i.m. regimen. This clearly demonstrates that the different concurrent vaccination regimens had different impacts on B cell maturation.
Interestingly, significant differences in mucosal IgG responses were observed. The i.d. and i.m. and the i.d. and i.m. (EP) regimens failed to generate significant quantities of IgG (Fig. 1d). However, the i.d. (EP) and i.m. (146 ± 42 ng/ml, P = 0.0006) and the i.d. (EP) and i.m. (EP) (107 ± 35.4 ng/ml, P = 0.0006) regimens elicited highly significant IgG responses in the vagina compared to those in the control and the i.d. and i.m. groups (P = 0.0024 and P = 0.0165, respectively). No significant difference was observed between the i.d. (EP) and i.m. and the i.d. (EP) and i.m. (EP) groups (Fig. 1d), suggesting that administration of EP via the i.d. route facilitates antibody localization in mucosal secretions. Evaluation of cellular responses revealed that all concurrent regimens elicited significant T cell responses (P ≥ 0.0005) to peptide pools 1 and 2 compared to the response by the controls (Fig. 1e). Furthermore, the i.d. (EP) and i.m. and the i.d. (EP) and i.m. (EP) regimens elicited significantly elevated T cell responses compared to the i.d. and i.m. and the i.d. and i.m. (EP) regimens for both peptide pool 1 (P = 0.003 and 0.001, respectively) and peptide pool 2 (P = 0.002 and 0.003, respectively). No significant differences in T cell responses were recorded between the i.d. (EP) and i.m. and the i.d. (EP) and i.m. (EP) groups (Fig. 1e). Taken together, these results show the concurrent i.d. (EP) and i.m. (EP) treatment regimen to be superior for eliciting strong humoral and cellular responses.
In addition to increasing transgene expression levels, EP has been described to act as an adjuvant through as yet undefined mechanisms (22–24). Therefore, using the empty vector, we compared the ability of EP delivered via the i.m. route (see Fig. S2a in the supplemental material) and EP delivered via the i.d. route (see Fig. S2b in the supplemental material) to alter antigen-specific immune responses. Using the empty vector, electroporation via the i.m. route either augmented or significantly increased cellular immune responses to the vaccine transgene delivered via the i.d. route. In contrast, this adjuvant activity was not observed when EP and empty vector were delivered via the i.d. route. Thus, suggesting that the route of EP delivery can significantly affect the ensuing cellular immune response.
Concurrent DNA-priming (EP) and protein boost vaccinations enhance humoral responses.
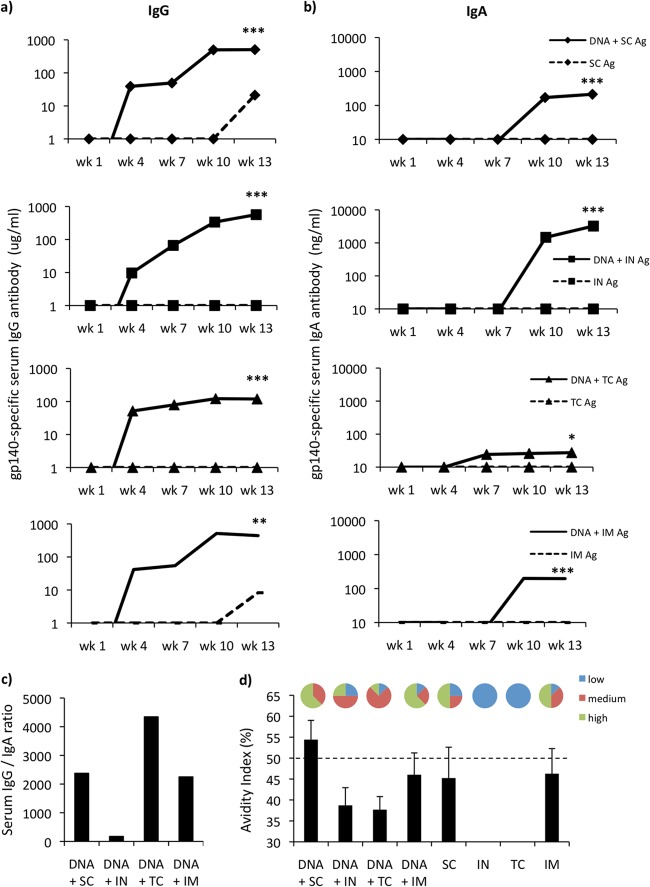
DNA priming and subsequent protein boosting have been shown to significantly amplify humoral responses. Therefore, we assessed the ability of protein boost vaccination to augment the antibody responses elicited. We evaluated the concurrent i.d. (EP) and i.m. (EP) DNA-priming strategy (here referred to as simply DNA) in combination with two subsequent protein boost vaccinations via the s.c., i.n., t.c., and i.m. routes (see Table S2 in the supplemental material). All prime-boost regimens generated significantly elevated IgG levels in the systemic compartment (Fig. 2a). The DNA vaccinations efficiently primed the immune responses, resulting in elevated IgG levels in all groups by the end of the 3rd priming (week 6). These responses were significantly amplified by all routes of protein boosting. DNA priming boosted by the s.c. (507 ± 60.5 μg/ml, P = 0.0003) and i.n. (566 ± 78.6 μg/ml, P = 0.0003) routes generated the highest IgG responses, closely followed by i.m. boosting (442 ± 62 μg/ml, P = 0.0047). Although the response was highly significant (119 ± 30 μg/ml, P = 0.0003), the DNA and t.c. protein boost regimen resulted in the lowest IgG profile. Serum IgA responses for all boost regimens were also significantly elevated compared to those for the protein controls (Fig. 2b). The DNA and i.n. route of protein boost vaccination was clearly superior to the other boost regimens for the production of antigen-specific IgA, resulting in highly significant serum IgA concentrations being recorded during study week 13 (3,107 ± 1.1 ng/ml, P = 0.0002). The DNA and s.c. (213 ± 83 ng/ml, P = 0.0002) and DNA and i.m. (196 ± 56.7 ng/ml, P = 0.0002) routes elicited similar levels of serum IgA; however, the DNA and t.c. (P = 0.029) regimen resulted in the least antigen-specific IgA being generated (Fig. 2b). Interestingly, however, the DNA and t.c. route was the only one that elicited elevated antigen-specific IgA responses after the final DNA-priming vaccination. Assessment of the serum antigen-specific IgG/IgA ratios revealed that the i.n. boosting route could dramatically alter the ratio of IgG/IgA, while the t.c. boosting regimen promoted IgG production over IgA production (Fig. 2c.). The i.m. and s.c. routes produced almost equivalent ratios of IgG/IgA (Fig. 2c.).
FIG 2.
The route of administration of protein boost impacts the ensuing serum humoral responses. (a and b) Antigen (Ag)-specific IgG (a) and IgA (b) from serum were assessed (n = 8 per group) 1 week after three concurrent DNA-priming vaccinations (i.d. [10 μg] and i.m. [50 μg]) and two protein boosts (20 μg/dose). Protein boosts were delivered by either the s.c., i.n., t.c., or i.m. route. Antibody results are expressed as group means (μg/ml or ng/ml). Statistical significance was assessed using the Mann-Whitney U test. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005. (c) To assess antibody avidity, vaccine serum samples were prediluted to give an optical density between 1 and 1.5 using an in-house antigen-specific ELISA. Samples were then titrated in an endpoint ELISA in duplicate using nonreducing (PBS) and reducing (8 M urea) washes after sample addition. Results are shown as the percent change in binding [(reducing OD/nonreducing OD) × 100].
The impact of different protein boost vaccination routes on IgG avidity was assessed in order to ascertain the optimal prime-boost regimen. Interestingly, the DNA and s.c. route of boost vaccination outperformed all other boost regimens, generating the highest-avidity antibody responses (54.4%) (Fig. 2d). Furthermore, 62.5% of vaccinated animals in the DNA and s.c. group displayed high-avidity antibody binding, while the remaining animals displayed medium-avidity antibody binding. Interestingly, while the DNA and s.c. and the DNA and i.n. regimens generated similar IgG levels (Fig. 2a), the avidities of the elicited antibodies were markedly different (Fig. 2d). A noticeable reduction (15.7%) in avidity for the DNA and i.n. regimen compared to that for the DNA and s.c. regimen was recorded, clearly suggesting that the route of protein boost greatly impacts the quality of the antibody elicited. Furthermore, it is interesting to note that the DNA and i.m. (46%) regimen elicited antibody that had an avidity similar to that of antibody from the non-DNA-primed (46.2%) vaccinated controls, suggesting that although it is capable of immunologically priming for large quantities of antibody, DNA priming via the i.m. route had a minimal impact on B cell maturation (Fig. 2d). Collectively, these results show that the s.c. route of protein boosting is preferential for the generation of high levels of serum antibodies with high avidity. The presence of protective antibody in external mucosal secretions is reasoned to add a significant layer of protection against HIV-1 infections occurring through susceptible mucosal surfaces. Therefore, we assessed the impact of the different protein boost vaccination regimens on vaginal antibody responses. All groups receiving DNA priming and protein boosting generated significant quantities of IgG in the vaginal vault compared to the quantities for the controls receiving protein only (see Fig. S3a in the supplemental material). In terms of the elicited vaginal IgA responses, only the DNA and i.n. protein boost vaccination group generated statistically significant responses (60 ± 28 ng/ml, P = 0.0025) (see Fig. S3b in the supplemental material). Hence, taken together, these data show that s.c., i.n., t.c., and i.m. protein boosting significantly augments mucosal IgG antibody responses, while only the i.n. route significantly increases the presence of mucosal IgA within external secretions.
The route of protein boost vaccination does not significantly alter the mean quantity of the vaccine-elicited cellular response.
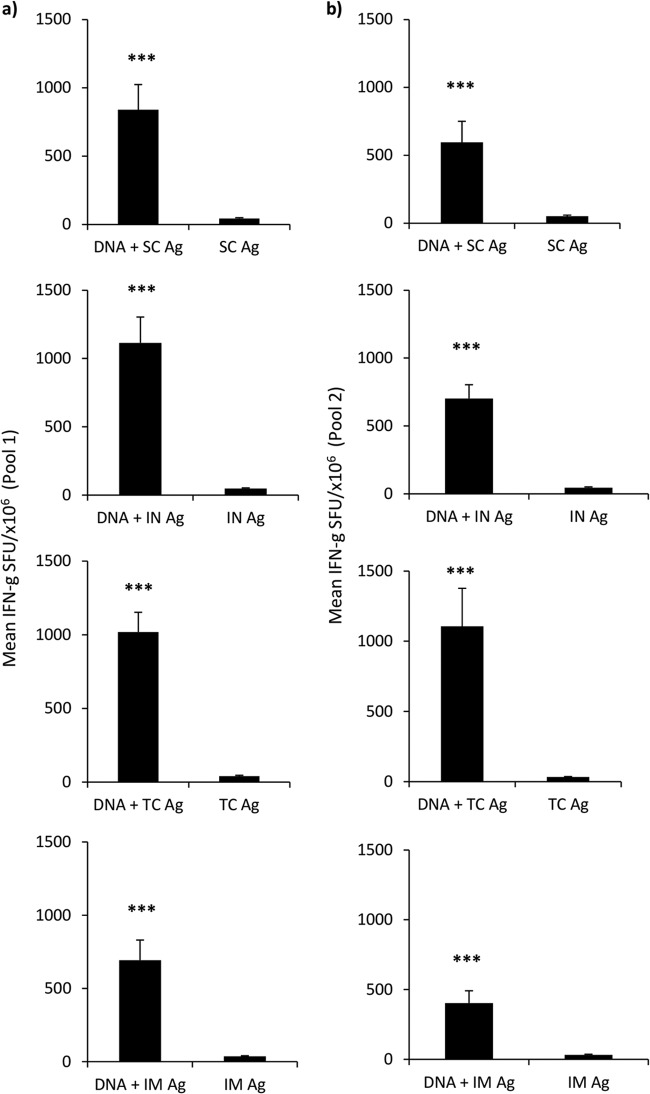
Next, we evaluated the impact of various routes of protein boost vaccination on the cellular response of DNA-primed mice. Here we show that all vaccine regimens which involved prior DNA priming, DNA and s.c. (840 ± 184 ng/ml, P = 0.0002), DNA and i.n. (1,113 ± 188 ng/ml, P = 0.0002), DNA and t.c. (1,018 ± 133 ng/ml, P = 0.0002), and DNA and i.m. (692 ± 137 ng/ml, P = 0.0002), generated highly significant cellular responses against peptide pool 1 compared to the responses for their respective controls (Fig. 3a). This was also seen for Env peptide pool 2, when the results obtained by the DNA and s.c. (595 ± 154 ng/ml, P = 0.0009), DNA and i.n. (702 ± 102 ng/ml, P = 0.0002), DNA and t.c. (1106 ± 271 ng/ml, P = 0.0002), and DNA and i.m. (403 ± 88 ng/ml, P = 0.0002) regimens were compared to those for the controls (Fig. 3b). Comparisons between the regimens revealed no statistically significant difference between groups and, therefore, no advantage of one protein boost regimen over any other for augmenting total cellular immune responses.
FIG 3.
The route of protein boost vaccination does not significantly affect the magnitude of the cellular response. Vaccinated mice (n = 8 per group) were sacrificed 1 week after three concurrent DNA-priming vaccinations (i.d. [10 μg] and i.m. [50 μg]) and two protein boosts (20 μg/dose). Protein boosts were delivered by either the s.c., i.n., t.c., or i.m. route. Splenocytes were assessed by IFN-γ ELISpot assay for antigen-reactive T cells using two sets of peptide pools consisting of 15-mers overlapping by 11 amino acids (a and b). Graphs are expressed as group means (numbers of SFU per million antigen-stimulated cells [±SEM]). Statistical significance was assessed using the Mann-Whitney U test. ***, P ≤ 0.0005.
Concurrent DNA (EP) and s.c. protein boost vaccinations elicit strong humoral responses and increase antibody avidity and HIV-1 neutralization in rabbits.
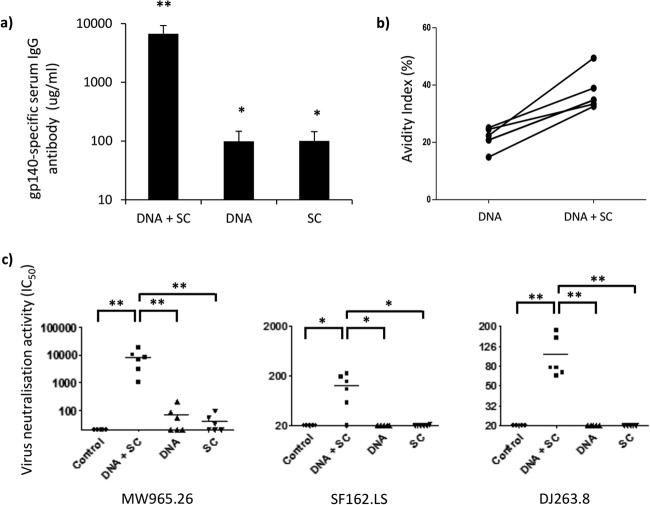
The murine model is suboptimal for the assessment of neutralizing antibody responses due to high background levels of nonspecific inhibitor activity. Therefore, we extended our murine experiments into the larger rabbit model to test the antibody responses elicited in HIV-1 neutralization assays. New Zealand White rabbits were vaccinated using a triple DNA-priming (i.d. [EP] and i.m. [EP]) regimen followed by a double s.c. protein boost regimen (see Table S3 in the supplemental material). This was chosen on the basis of the observation that it induced the highest-avidity antibodies in the murine model. DNA priming utilized concurrent i.d. (20 μg) and i.m. (100 μg) vaccination and EP followed by homologous s.c. protein boosting (40 μg). The DNA and s.c. regimen (6,697.6 ± 2,547 μg/ml) elicited higher IgG responses than DNA (99.2 μg/ml ± 47.6) or protein alone (100.4 ± 44.1 μg/ml) (Fig. 4a), although all three vaccine regimens, DNA and s.c. (P = 0.002), DNA (P = 0. 0167), and s.c. (P = 0.0167), elicited significant IgG responses compared to that in baseline control serum. Interestingly, triple i.d. (EP) and i.m. (EP) DNA vaccination induced antigen-specific antibody levels similar to those induced by the two s.c. protein vaccinations.
FIG 4.
Concurrent DNA priming (EP) and recombinant protein boost elicits strong humoral responses with virus-neutralizing activity. (a) Antigen-specific IgG from serum was assessed (n = 6 NWZ rabbits per group) by an in-house ELISA 1 week after either three concurrent DNA-priming vaccinations (i.d. [20 μg] and i.m. [100 μg]) and two protein boosts (40 μg/dose), three concurrent DNA-priming vaccinations (i.d. [20 μg] and i.m. [100 μg]), or two protein boosts. Antibody results are expressed as group means (±SEM) (μg/ml). (b) To assess antibody avidity, rabbit serum samples were prediluted immediately after DNA priming and after two protein boost vaccinations to give an optical density of between 1 and 1.5 using an in-house antigen-specific ELISA. Samples were then titrated in an endpoint ELISA in duplicate using nonreducing (PBS) and reducing (8 M urea) washes after sample addition. Results are shown as the percent change in binding [(reducing OD/nonreducing OD) × 100[. (c) TZM.bl virus neutralization assays were performed on week 13 serum samples using tier 1 pseudoviruses from clades A (DJ263.8), B (SF162.LS), and C (MW965.26) and MuLV as a control. The ID50 titer was calculated as the serum dilution that caused a 50% reduction in the numbers of RLUs compared to those for the virus control wells (TZM.bl cells with virus) after subtraction of the number of RLUs for the cell control (TZM.bl cells alone). Statistical significance was assessed using the Mann-Whitney U test. *, P ≤ 0.05; **, P ≤ 0.005.
To better understand the elicited humoral response, we evaluated the avidity of serum from vaccinated rabbits. Here we found that after two protein boost vaccinations, the avidity index increased for all rabbits that had received prior DNA immunizations (Fig. 4b), suggesting that B cell maturation had occurred in rabbits vaccinated with DNA and s.c. Importantly, this is in agreement with bilayer interferometry data showing increased binding and reduced dissociation kinetics (see Fig. S4a in the supplemental material). Here we found that the first protein boost vaccination (3.55 × 10−3 nm s−1) had a 1.45-fold lower rate of change than the second protein boost vaccination (5.16 × 10−3 nm s−1), demonstrating the increased binding kinetics of the serum after each protein boost. Evaluation of the dissociation rates (Kds) revealed that after each protein boost vaccination, antibody dissociated from antigen at a lower rate, indicating increased antibody binding and, therefore, increased B cell maturation (see Fig. S4b in the supplemental material).
Next, we evaluated the neutralizing activity of serum from vaccinated rabbits in a TZM.bl assay using a variety of tier 1 pseudoviruses from clades A, B, and C. Here we found that the DNA and s.c. vaccination regimen elicited significant neutralizing activity against clade C MW965.26 (ID50 titer, 7,887.7 ± 2,525.6; P = 0.0037), clade B SF162.LS (ID50 titer, 126.5 ± 32.2; P = 0.014), and clade A DJ263.8 (ID50 titer, 104 ± 21; P = 0.0037) pseudoviruses compared to that against the murine leukemia virus (MuLV) negative control (Fig. 4c). Furthermore, the DNA and s.c. regimen elicited significant neutralizing activity compared to that for the regimens of DNA alone and s.c. alone for the MW965.26 (P = 0.0022 and P = 0.0022, respectively), SF162.LS (P = 0.014 and P = 0.0167, respectively), and DJ263.8 (P = 0.005 and P = 0.005, respectively) pseudoviruses (Fig. 4c). Therefore, these results demonstrate the cross-clade neutralizing activity of the DNA and s.c. vaccination regimen in rabbits.
DISCUSSION
Despite the overwhelming success of vaccines, there is a clear requirement for the immediate development of prophylactic interventions against a number of disease-causing organisms. This is particularly true for HIV-1, where vaccines are not available and current vaccine concepts have failed. Hence, novel vaccine technologies or paradigms will undoubtedly play an important role in protecting against HIV-1 acquisition. In this study, we used a DNA plasmid vector expressing gp140 as part of a preclinical study to deliver HIV-1 Env as a priming vaccine immunogen to two different sites (by the i.d. and i.m. routes) concurrently. The aim was to elicit through rational vaccine design high-titer antibody and cellular responses over and above what is seen through conventional systemic single-site vaccination. While humoral antibody responses were the principal focus of this study, we also evaluated gp140-specific cellular responses, as CD4+ T cell help is critical for not only the development of cytotoxic CD8 responses but also the development of Env-specific high-affinity neutralizing antibody responses. With that said, the results presented here clearly demonstrate that concurrent vaccinations are highly effective activators of both cellular and humoral responses. Of particular note is the i.d. (EP) and i.m. (EP) combination, which generated strong T and B cell responses, with the antibody elicited by this regimen demonstrating a higher avidity than antibody elicited by all other combinations tested. To our knowledge, this is the first time that a preclinical vaccination study has evaluated concurrent plasmid DNA vaccination as part of a prime-boost strategy, and this study suggests that concurrent DNA vaccine delivery may offer a new approach in HIV-1 prime-boost vaccine delivery research.
During each stage of evaluation, we selected the best vaccine combination on the basis of the elicited humoral and cellular responses. The optimized DNA regimen was then tested in a DNA-priming, protein boost protocol for any elicited virus-neutralizing activity. Prime-boost vaccination has previously been used to synergistically amplify memory T cell responses to vaccinating antigens (25) by invoking CD4+ T cells and corresponding T cell-dependent antibody responses (26). Furthermore, it is believed that DNA priming can facilitate memory B cell development, which can be selectively amplified upon recombinant protein boost vaccination and enhance antibody avidity (26). In this study, we detected a robust expansion in elicited antibody responses upon protein boosting in DNA-primed animals, with maximal affinity maturation being associated with the s.c. route. Hence, i.d. (EP) and i.m. (EP) vaccination with s.c. protein boosting was found to be the optimal vaccination regimen with respect to the magnitude and avidity of the systemic IgG responses. Curiously, while the s.c., i.n., and i.m. routes of protein boost vaccination elicited similar antigen-specific IgG concentrations in serum, the i.n. route elicited IgG responses which were the lowest in avidity (Fig. 2a and c), suggesting a lack of affinity maturation. This may be due to suboptimal stimulation of the ensuing immune responses through a combination of low antigen availability and a lack of Toll-like receptor activation. It is feasible to assume that i.n. protein boosting could result in the stimulation of germinal centers in an alternative repertoire of draining lymph nodes not normally associated with s.c., i.m., and t.c. vaccination. This could result in the de novo generation of antibody responses which have had less time for somatic hypermutation and affinity maturation. Hence, future studies evaluating the elicited T and B cell responses for affinity maturation and subsequent epitope breadth, as well as the impact of various coadministered adjuvants, could prove crucial. Using the rabbit as a second animal model, we demonstrate increased antibody affinity with each subsequent s.c. protein boost following optimized DNA priming. This confirmed our earlier murine studies demonstrating that protein boosting augmented antibody avidity following prior DNA priming. Interestingly, previous studies in HIV infection models have correlated increased antibody avidity with lower viral titers at peak viremia (27), hence suggesting that B cell antibody maturation can play a protective role during HIV-1 infection. However, it is important to note that in previous studies, avidity maturation was ascribed to the use of adjuvants, such as granulocyte-macrophage colony-stimulating factor (27). Therefore, as we did not use any immunomodulatory adjuvant, we associate our increased avidity maturation to the optimized vaccination protocol.
The production of a neutralizing antibody response has been the goal for most prophylactic HIV-1 vaccines. This is especially true since a number of neutralizing antibody-passive infusion studies provided a proof of concept regarding protection against infection in the macaque model (28). Furthermore, the appearance of increased polyclonal serum avidity has been shown to coincide with the appearance of neutralizing antibody responses (29). However, the broadly cross-neutralizing antibody response thought to be required for protection against diverse strains of HIV-1 can take years to develop, if it develops at all (30). Thus, any vaccination strategy that preferentially drives the development of affinity maturation or at least speeds up the process may induce the production of protective neutralizing antibodies. By employing a sequential experimental process based on avidity, the humoral response, and cellular concentrations, we sought to harness the elusive vaccine-mediated neutralizing antibody response. Consequently, critical to our data was the ability of our vaccination regimen to elicit neutralizing antibody responses. The serum from the DNA-priming, protein boost group was able to neutralize viruses in both an intraclade and an interclade fashion, indicating that the elicited neutralizing antibody response had a degree of breadth. Currently, there is only one published phase 1 human clinical trial employing DNA-priming, protein boost vaccination (31). Within this particular study, the elicited antibody was capable of neutralizing tier 1 autologous pseudoviruses (31), hence providing compelling evidence that this type of vaccination regimen can translate effectively to humans. This is further supported by DNA-priming, protein boost vaccine regimens that elicited antibody responses capable of neutralizing homologous and heterologous viral strains in rhesus macaques (32). More recently, a coimmunization study was evaluated in mice and macaques (33). Here, the authors used a DNA and protein covaccination procedure and plasmids containing a mixture of Env and Gag DNA delivered by the i.m. (EP) route. The authors also administered gp120 protein formulated in EM-005 (glucopyranosyl lipid adjuvant-stable emulsion) to the same site. Interestingly, the authors reported that the concurrent regimen enhanced antibody responses and T cell polyfunctionality and had a broader cross-clade neutralizing potential (33).
Our current study did not use molecular adjuvants in order to clearly differentiate the impacts of the different components of the concurrent regimens. However, it would be interesting to evaluate the impact of adjuvants on the DNA-priming process as well as the protein boost to measure their impact on avidity and neutralizing potential. This may provide further enhancement, as adjuvants have been described to drive affinity maturation and promote antibody avidity against viral antigens (27, 34). As elicitation of HIV-1 broadly neutralizing antibody responses has been shown to be coincident with antibody affinity maturation as well as to possibly be driven by the presence of diverse simultaneously circulating forms of Env (35), it would be interesting to test the concurrent DNA-priming (i.d. [EP] and i.m. [EP]) and s.c. protein boost regimen with a multiclade vaccine formulation. Therefore, as two previous human clinical trials (VAX003 and VAX004) involving bivalent recombinant gp120s adjuvanted with alum have been shown to elicit tier 1 neutralizing antibody (36, 37), the adjuvanation of our concurrent DNA regimen with s.c. protein boost presents an intriguing avenue of future investigation. Of particular note is the use of clade C recombinant gp140 in our studies, while the VAX003 and VAX004 clinical trials utilized AIDSVAX B/B′ and AIDSVAX B/E. Thus, our preclinical studies demonstrate the activation of tier 1 neutralization activity against a different model of Env. Furthermore, as our study was designed with the augmentation of humoral responses as its principal focus, it would be interesting to combine this optimized regimen with a dedicated T cell vaccine strategy. Collectively, the data presented here support the concept for the use of concurrent DNA-priming vaccination combinations as mediators of robust T and B cell responses, with protein boost immunizations serving to efficiently amplify antibody levels and augment affinity maturation and neutralization.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by CUT'HIVAC under a European Commission FP7 award (grant number 241904). We are also grateful to the Fondation Dormeur for provision of an equipment grant. Thanks go to Béhazine Combadière and Louis Chonco (INSERM, FRA) for training provisions. The neutralization studies were supported by the Bill and Melinda Gates Foundation (Comprehensive Antibody Vaccine Immune Monitoring Consortium, grant 1032144, to M. S. Seaman).
Special thanks go to Ralf Wagner for the provision of the codon-optimized CN54-gp140 plasmid (University of Regensburg, Germany) and to Andres Männik, Ioana Stanescu, Mart Ustav, and Fit Biotech (Finland) for the provision of the GTU-Luc-GFP and GTU-CN54-gp140 plasmids used in these studies. We also thank Dietmar Katinger (Polymun Scientific, Austria) for the provision of the CN54-gp140 antigen.
Footnotes
Published ahead of print 9 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00183-14.
REFERENCES
- 1.Rappuoli R. 2007. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 25:1361–1366. 10.1038/nbt1207-1361 [DOI] [PubMed] [Google Scholar]
- 2.Blazevic V, Mannik A, Malm M, Sikut R, Valtavaara M, Toots U, Ustav M, Krohn K. 2006. Induction of human immunodeficiency virus type-1-specific immunity with a novel gene transport unit (GTU)-multiHIV DNA vaccine. AIDS Res. Hum. Retroviruses 22:667–677. 10.1089/aid.2006.22.667 [DOI] [PubMed] [Google Scholar]
- 3.Martinon F, Kaldma K, Sikut R, Culina S, Romain G, Tuomela M, Adojaan M, Mannik A, Toots U, Kivisild T, Morin J, Brochard P, Delache B, Tripiciano A, Ensoli F, Stanescu I, Le Grand R, Ustav M. 2009. Persistent immune responses induced by a human immunodeficiency virus DNA vaccine delivered in association with electroporation in the skin of nonhuman primates. Hum. Gene Ther. 20:1291–1307. 10.1089/hum.2009.044 [DOI] [PubMed] [Google Scholar]
- 4.Tuomela M, Malm M, Wallen M, Stanescu I, Krohn K, Peterson P. 2005. Biodistribution and general safety of a naked DNA plasmid, GTU-multiHIV, in a rat, using a quantitative PCR method. Vaccine 23:890–896. 10.1016/j.vaccine.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 5.Kutzler MA, Weiner DB. 2008. DNA vaccines: ready for prime time? Nat. Rev. Genet. 9:776–788. 10.1038/nrg2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shedlock DJ, Weiner DB. 2000. DNA vaccination: antigen presentation and the induction of immunity. J. Leukoc. Biol. 68:793–806. 10.1189/jlb.1938-3673 [DOI] [PubMed] [Google Scholar]
- 7.Klavinskis LS, Gao L, Barnfield C, Lehner T, Parker S. 1997. Mucosal immunization with DNA-liposome complexes. Vaccine 15:818–820. 10.1016/S0264-410X(96)00278-2 [DOI] [PubMed] [Google Scholar]
- 8.Torrieri-Dramard L, Lambrecht B, Ferreira HL, Van den Berg T, Klatzmann D, Bellier B. 2011. Intranasal DNA vaccination induces potent mucosal and systemic immune responses and cross-protective immunity against influenza viruses. Mol. Ther. 19:602–611. 10.1038/mt.2010.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallengard D, Haller BK, Maltais AK, Gelius E, Nihlmark K, Wahren B, Brave A. 2011. Comparison of plasmid vaccine immunization schedules using intradermal in vivo electroporation. Clin. Vaccine Immunol. 18:1577–1581. 10.1128/CVI.05045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez LA, Arango T, Boyer J. 2013. Therapeutic and prophylactic DNA vaccines for HIV-1. Expert Opin. Biol. Ther. 13:563–573. 10.1517/14712598.2013.758709 [DOI] [PubMed] [Google Scholar]
- 11.Sardesai NY, Weiner DB. 2011. Electroporation delivery of DNA vaccines: prospects for success. Curr. Opin. Immunol. 23:421–429. 10.1016/j.coi.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin J, Dai A, Lecureux J, Arango T, Kutzler MA, Yan J, Lewis MG, Khan A, Sardesai NY, Montefiore D, Ruprecht R, Weiner DB, Boyer JD. 2011. High antibody and cellular responses induced to HIV-1 clade C envelope following DNA vaccines delivered by electroporation. Vaccine 29:6763–6770. 10.1016/j.vaccine.2010.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combadiere B, Liard C. 2011. Transcutaneous and intradermal vaccination. Hum. Vaccin. 7:811–827. 10.4161/hv.7.8.16274 [DOI] [PubMed] [Google Scholar]
- 14.Koutsonanos DG, Vassilieva EV, Stavropoulou A, Zarnitsyn VG, Esser ES, Taherbhai MT, Prausnitz MR, Compans RW, Skountzou I. 2012. Delivery of subunit influenza vaccine to skin with microneedles improves immunogenicity and long-lived protection. Sci. Rep. 2:357. 10.1038/srep00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brave A, Nystrom S, Roos AK, Applequist SE. 2011. Plasmid DNA vaccination using skin electroporation promotes poly-functional CD4 T-cell responses. Immunol. Cell Biol. 89:492–496. 10.1038/icb.2010.109 [DOI] [PubMed] [Google Scholar]
- 16.Kulkarni V, Rosati M, Bear J, Pilkington GR, Jalah R, Bergamaschi C, Singh AK, Alicea C, Chowdhury B, Zhang GM, Zhang GM, Kim EY, Wolinsky SM, Huang W, Guan Y, LaBranche C, Montefiori DC, Broderick KE, Sardesai NY, Valentin A, Felber BK, Pavlakis GN. 2013. Comparison of intradermal and intramuscular delivery followed by in vivo electroporation of SIV Env DNA in macaques. Hum. Vaccin. Immunother. 9:2081–2094. 10.4161/hv.25473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liard C, Munier S, Joulin-Giet A, Bonduelle O, Hadam S, Duffy D, Vogt A, Verrier B, Combadiere B. 2012. Intradermal immunization triggers epidermal Langerhans cell mobilization required for CD8 T-cell immune responses. J. Invest. Dermatol. 132:615–625. 10.1038/jid.2011.346 [DOI] [PubMed] [Google Scholar]
- 18.Mann JF, McKay PF, Arokiasamy S, Patel RK, Klein K, Shattock RJ. 2013. Pulmonary delivery of DNA vaccine constructs using deacylated PEI elicits immune responses and protects against viral challenge infection. J. Control. Release 170:452–459. 10.1016/j.jconrel.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann JF, McKay PF, Arokiasamy S, Patel RK, Tregoning JS, Shattock RJ. 2013. Mucosal application of gp140 encoding DNA polyplexes to different tissues results in altered immunological outcomes in mice. PLoS One 8:e67412. 10.1371/journal.pone.0067412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12:Unit 12.11. 10.1002/0471142735.im1211s64 [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Kjeken R, Mathiesen I, Barouch DH. 2008. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J. Virol. 82:5643–5649. 10.1128/JVI.02564-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiarella P, Massi E, De Robertis M, Sibilio A, Parrella P, Fazio VM, Signori E. 2008. Electroporation of skeletal muscle induces danger signal release and antigen-presenting cell recruitment independently of DNA vaccine administration. Expert Opin. Biol. Ther. 8:1645–1657. 10.1517/14712598.8.11.1645 [DOI] [PubMed] [Google Scholar]
- 23.Daftarian P, Chowdhury R, Ames P, Wei C, King AD, de Rivero Vaccari JP, Dillon L, Price J, Leung H, Ashlock B, Mesri E, Perez V, Züchner S, Reiser J, Lemmon V, Keane RW. 2011. In vivo electroporation and non-protein based screening assays to identify antibodies against native protein conformations. Hybridoma (Larchmt.) 30:409–418. 10.1089/hyb.2010.0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roos AK, Eriksson F, Timmons JA, Gerhardt J, Nyman U, Gudmundsdotter L, Brave A, Wahren B, Pisa P. 2009. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS One 4:e7226. 10.1371/journal.pone.0007226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodland DL. 2004. Jump-starting the immune system: prime-boosting comes of age. Trends Immunol. 25:98–104. 10.1016/j.it.2003.11.009 [DOI] [PubMed] [Google Scholar]
- 26.Lu S. 2006. Combination DNA plus protein HIV vaccines. Springer Semin. Immunopathol. 28:255–265. 10.1007/s00281-006-0028-1 [DOI] [PubMed] [Google Scholar]
- 27.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, Chennareddi L, Yu T, Kannanganat S, Ofielu L, Villinger F, Wyatt LS, Moss B, Amara RR, Robinson HL. 2007. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology 369:153–167. 10.1016/j.virol.2007.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210. 10.1038/72318 [DOI] [PubMed] [Google Scholar]
- 29.Richmond JF, Lu S, Santoro JC, Weng J, Hu SL, Montefiori DC, Robinson HL. 1998. Studies of the neutralizing activity and avidity of anti-human immunodeficiency virus type 1 Env antibody elicited by DNA priming and protein boosting. J. Virol. 72:9092–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 18:1688–1692. 10.1038/nm.2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, Shen S, Green S, Rothman AL, Ennis FA, Arthos J, Pal R, Markham P, Lu S. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 26:1098–1110. 10.1016/j.vaccine.2007.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal R, Wang S, Kalyanaraman VS, Nair BC, Whitney S, Keen T, Hocker L, Hudacik L, Rose N, Cristillo A, Mboudjeka I, Shen S, Wu-Chou TH, Montefiori D, Mascola J, Lu S, Markham P. 2005. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J. Med. Primatol. 34:226–236. 10.1111/j.1600-0684.2005.00120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Valentin A, Kulkarni V, Rosati M, Beach RK, Alicea C, Hannaman D, Reed SG, Felber BK, Pavlakis GN. 2013. HIV/SIV DNA vaccine combined with protein in a co-immunization protocol elicits highest humoral responses to envelope in mice and macaques. Vaccine 31:3747–3755. 10.1016/j.vaccine.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, Del Giudice G, Rappuoli R, Golding H. 2011. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci. Transl. Med. 3:85ra48. 10.1126/scitranslmed.3002336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, NISC Comparative Sequencing Program. Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. 10.1038/nature12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D'Souza P, Rodriguez-Chavez IR, DeCamp A, Giganti M, Berman PW, Self SG, Montefiori DC. 2010. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J. Infect. Dis. 202:595–605. 10.1086/654816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. 2012. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 206:431–441. 10.1093/infdis/jis367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.