ABSTRACT
Phosphatidylinositol 4-kinase IIIβ (PI4KB) is a host factor required for the replication of certain picornavirus genomes. We previously showed that nonstructural proteins 2B, 2BC, 2C, 3A, and 3AB of Aichi virus (AiV), a picornavirus, interact with the Golgi protein, acyl-coenzyme A binding domain containing 3 (ACBD3), which interacts with PI4KB. These five viral proteins, ACBD3, PI4KB, and the PI4KB product phosphatidylinositol 4-phosphate (PI4P) colocalize to the AiV RNA replication sites (J. Sasaki et al., EMBO J. 31:754–766, 2012). We here examined the roles of these viral and cellular molecules in the formation of AiV replication complexes. Immunofluorescence microscopy revealed that treatment of AiV polyprotein-expressing cells with a small interfering RNA targeting ACBD3 abolished colocalization of the viral 2B, 2C, and 3A proteins with PI4KB. A PI4KB-specific inhibitor also prevented their colocalization. Virus RNA replication increased the level of cellular PI4P without affecting that of PI4KB, and individual expression of 2B, 2BC, 2C, 3A, or 3AB stimulated PI4P generation. These results suggest that the viral protein/ACBD3/PI4KB complex plays an important role in forming the functional replication complex by enhancing PI4P synthesis. Of the viral proteins, 3A and 3AB were shown to stimulate the in vitro kinase activity of PI4KB through forming a 3A or 3AB/ACBD3/PI4KB complex, whereas the ACBD3-mediated PI4KB activation by 2B and 2C remains to be demonstrated.
IMPORTANCE The phosphatidylinositol 4-kinase PI4KB is a host factor required for the replication of certain picornavirus genomes. Aichi virus, a picornavirus belonging to the genus Kobuvirus, forms a complex comprising one of the viral nonstructural proteins 2B, 2BC, 2C, 3A, and 3AB, the Golgi protein ACBD3, and PI4KB to synthesize PI4P at the sites for viral RNA replication. However, the roles of this protein complex in forming the replication complex are unknown. This study showed that virus RNA replication and individual viral proteins enhance the level of cellular PI4P, and suggested that the viral protein/ACBD3/PI4KB complex plays an important role in forming a functional replication complex. Thus, the present study provides a new example of modulation of cellular lipid metabolism by viruses to support the replication of their genomes.
INTRODUCTION
The family Picornaviridae comprises a large group of nonenveloped, single-stranded positive-sense RNA viruses, which are classified into 17 genera, as well as many recently discovered unassigned viruses (http://www.picornaviridae.com). Picornaviruses infect mammals and birds and include important human and animal pathogens such as poliovirus (PV), enterovirus 71, coxsackieviruses, hepatitis A virus, and foot-and-mouth disease virus. The 7,100- to 8,500-nucleotide (nt) picornavirus genome contains a single long open reading frame (ORF) that encodes a polyprotein. This polyprotein is then cleaved into 11 to 12 proteins by virus-encoded proteases. After uncoating, the positive-sense RNA genome is first translated to produce viral proteins and then transcribed into negative-strand RNA that serves as a template for the synthesis of positive-strand RNA. The newly generated positive-strand RNA is packaged into capsid proteins to form the virion.
Positive-strand RNA viruses, including picornaviruses, utilize intracellular membranes for genome replication. The membranes of various intracellular organelles, e.g., endoplasmic reticulum (ER), ER-Golgi intermediate compartment, Golgi apparatus, mitochondria, lysosomes, or endosomes, are rearranged, often in association with vesicle formation, during viral infection to form replication complexes in which viral genome RNA replicates. Viral RNA and viral and host proteins interact with one another in a replication complex. For some picornaviruses, it is reported that some or all of 2B, 2C, and 3A, and cleavage intermediates 2BC and 3AB, which are membrane-associated nonstructural proteins, are involved in membrane rearrangement (1–9). Evidence indicates that the formation of a membranous replication complex increases the local concentrations of components required for replication, provides a scaffold for anchoring the replication complexes, confines the process of RNA replication to a specific cytoplasmic location, prevents the activation of certain host defense mechanisms, and provides certain lipids required for genome synthesis (reviewed in reference 10).
Phosphatidylinositol 4-phosphate (PI4P) is a minor component of cellular membranes. PI4P is synthesized by PI 4-kinases by phosphorylation of the 4 position of the inositol ring of phosphatidylinositol (PI). In mammals, there are four PI 4-kinases: PI4KIIα, PI4KIIβ, PI4KIIIα (also known as PI4KA), and PI4KIIIβ (also known as PI4KB). PI4P recruits certain effector proteins onto membranes during vesicular trafficking and lipid transport (reviewed in references 11, 12, and 13). Recent studies show that PI4P is important for the replication of certain positive-strand RNA viruses and that the viruses utilize various strategies to generate PI4P-enriched environments at genome replication sites. The nonstructural protein NS5A of hepatitis C virus (HCV), a member of the genus Hepacivirus of the family Flaviviridae, interacts with ER-resident PI4KA and stimulates its activity, leading to the accumulation of PI4P at viral replication sites where it is required for the structural and functional integrity of the membranous replication complex (14–18). For PV and coxsackievirus B3 (CVB3), which are members of the genus Enterovirus, a model is proposed, where the membrane-associated viral nonstructural protein 3A binds Golgi apparatus-specific brefeldin A resistance guanine nucleotide exchange factor 1 (GBF1)/ADP-ribosylation factor 1 (Arf1) to enhance the recruitment of PI4KB to the Golgi/trans-Golgi network, leading to PI4P accumulation at viral RNA replication sites (19). Moreover, CVB3 infection increases cellular level of PI4P. PI4P binds to viral RNA polymerase 3D to facilitate viral RNA synthesis. PI4KB activity is required for the replication of the genomes of other enteroviruses such as enterovirus 71 and human rhinovirus (20, 21).
In addition, we recently found that Aichi virus (AiV), a member of the genus Kobuvirus, recruits PI4KB to the viral RNA replication sites through a different mechanism. AiV was first isolated in 1989 from a patient in Japan during an outbreak of acute gastroenteritis associated with consumption of raw oysters (22). AiV was subsequently detected in Asian countries, Brazil, Europe, and Africa, and its association with acute gastroenteritis suggests that it is the causative agent of this disease (23–31). We found that the AiV nonstructural proteins—2B, 2BC, 2C, 3A, and 3AB—bind to the Golgi protein, acyl-coenzyme A binding domain containing 3 (ACBD3), which interacts with PI4KB to form a viral protein/ACBD3/PI4KB complex that synthesizes PI4P at sites of AiV RNA replication (32). Further, the 3A protein of some picornaviruses, including bovine kobuvirus, PV, CVB2, B3, and B5, and human rhinovirus 14, forms a complex with ACBD3 and PI4KB (33).
In the present study, we assessed the roles of AiV proteins (2B, 2BC, 2C, 3A, and 3AB), ACBD3, and PI4KB in the formation of the viral replication complexes. Immunofluorescence microscopy revealed that treatment of cells expressing the AiV polyprotein with a small interfering RNA (siRNA) targeted to the mRNA encoding ACBD3 abolished the colocalization of the viral proteins 2B, 2C, and 3A with PI4KB. Moreover, a PI4KB-specific inhibitor also disrupted colocalization of these same viral proteins with ACBD3 and PI4KB in polyprotein-expressing cells. Viral genomic RNA replication increased the level of cellular PI4P without affecting that of PI4KB, and the expression of the viral proteins, 2B, 2BC, 2C, 3A, and 3AB, also stimulated PI4P synthesis. These results suggest that the viral protein/ACBD3/PI4KB complex plays an important role in forming the functional replication complex by enhancing PI4P synthesis. Of the viral proteins, 3A and 3AB were shown to stimulate the in vitro kinase activity of PI4KB through forming a 3A or 3AB/ACBD3/PI4KB complex, whereas the ACBD3-mediated PI4KB activation by 2B and 2C remains to be demonstrated in vitro.
MATERIALS AND METHODS
Plasmids.
The plasmid pAV-FL-Luc-5′rzm encodes a full-length AiV cDNA in which the capsid-coding region is replaced by a firefly luciferase gene (34, 35). The DNA fragment encoding 2B, 2BC, 2C, 3A, or 3AB was amplified from pAV-FL-Luc-5′rzm by PCR using a specific primer pair (Sal-ATG-2B-P), annealing to the 5′ end of 2B (5′-GTCGACGTCATGGGTCTCCTCACCCTCTCT-3′); Mlu-2B-Stop-M, annealing to the 3′-end of 2B (5′-ACGCGTTATTGAGGTTCAAGGGTTGCCCGT-3′); Sal-ATG-2C-P, annealing to the 5′-end of 2C (5′-GTCGACGTCATGGGGCTCAAAGACTTCAAC-3′); Mlu-2C-Stop-M, annealing to the 3′ end of 2C (5′-ACGCGTTACTGGCGTCTGATGAGGGAGGC-3′); Sal-MET-5700-P, annealing to the 5′-end of 3A (36); Pst-3A-M, annealing to the 3′ end of 3A (5′-AAACTGCAGTTATTGAGGTTCCTGTGGCGTG-3′); or Mlu-Stop-6065M, annealing to the 3′-end of 3B (5′-AAACGCGTTTACTGGCGCTGGATGTGACGA-3′) and cloned into the SalI-SmaI sites of the pCI vector (Promega). Then, EcoRI-NotI fragments of these plasmids were cloned into the same sites of the pcDNA4T/O vector (Invitrogen), generating pcDNA-2B, -2BC, -2C, -3A, and -3AB. The sequence encoding the leader (L) protein was amplified from pAV-FL-Luc-5′rzm by PCR using primers (Sal-ATG-L [5′-TATGTCGACGTCATGGCTGCAACACGGGTT-3′] and L-3′ [5′-AGCGGCCGCTTATTGCCGTTGGAGGTTAGTGGTA-3′]), and the amplicon was digested with SalI and then ligated to the XhoI site and the blunt-ended XbaI site of pcDNA4T/O, yielding pcDNA-L.
To express the AiV polyprotein under the control of the cytomegalovirus (CMV) promoter in mammalian cells, we constructed the plasmid designated pCMV-polyprotein. To exclude the possibility that transcripts from the plasmid replicate in transfected cells, the AiV cDNA sequence lacking the 3′ untranslated region (3′UTR) was cloned into the pcDNA4T/O vector, as follows. The KpnI-HindIII (blunt-ended) fragment (nt 662 to the 3′ end of the AiV genome) of pAV-FL-Luc-5′rzm was cloned into the KpnI-EcoRV sites of pcDNA4T/O. Next, the 5′-terminal region of the genome was amplified from pAV-FL-Luc-5′rzm by PCR using the primers M-13 RV and 752M (5′-GCAGCCATGACTCCGACATAAAAGG-3′), digested with KpnI, and then cloned into the plasmid containing nt 662 to the 3′ end of the AiV genome. The PstI-HindIII fragment (nt 6771 to the 3′ end) of pAV-FL-Luc-5′rzm was subcloned into pUC118. This subclone served as a template for PCR using the primers EcoRV-polyA-P (5′-AGATTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGCTTGGCACT-3′) and Mlu-ORF3′-M (5′-AACGCGTCAGGCAGCCAGCAGATTTAGC-3′) to delete the 3′UTR. The PCR product was self-ligated, and the Pst/HindIII (blunt-ended) fragment was ligated to the PstI/NotI (blunt-ended) sites of pcDNA4T/O containing the AiV cDNA to generate the plasmid pCMV-polyprotein.
The plasmids pMAL-2B, -2C, -3A, and -3AB were described previously (6, 32, 36). For construction of pMAL-3A(R3A) and pMAL-3A(E11A), first, the 3A sequence was amplified from pAV-FL-Luc-5′rzm by PCR using the primers Eco-3A-P (5′-GAATTCGGCAACCGGGTCATCGACGCGGAA-3′) and Pst-3A-M and cloned into pGEM-T vector (Promega) to generate pG-Eco3APst. PCR was performed with primers Eco3A(R3A)-P (5′-GCAATTCGGCAACGCGGTCATCGACGCGGAA-3′) and Pst-3A-M using pG-Eco3APst as a template, and the amplicon was cloned into pGEM-T vector. The insert of the resultant plasmid was sequenced, excised with EcoRI and PstI, and ligated into pMAL-cX2 (New England BioLabs) to generate pMAL-3A(R3A). On the other hand, PCR was performed with the primers 3A(E11A)-P (5′-GAATCCCCCTGGAATACGCGGAT-3′) and 3A-5730M (5′-CCTGGGTTCCGCGTCGATGA-3′) using pG-Eco3APst as a template, and the amplicon was self-ligated. After the sequence was confirmed, the EcoRI/PstI fragment of this plasmid was ligated into pMAL-cX2 to generate pMAL-3A(E11A). Construction of the plasmid pCI-FLAG-ACBD3 was described previously (32). The plasmid pCI-FLAG-PI4KB was constructed by replacing the HA-tag sequence inserted into the NheI/MluI sites of pCI-HA-PI4KB (32) with the FLAG tag sequence, which was obtained by annealing the oligonucleotides 5′-CTAGCCACCATGGATTACAAGGATGACGACGATAAGA-3′ and 5′-CGCGTCTTATCGTCGTCATCCTTGTAATCCATGGTGG-3′.
Electroporation and DNA transfection.
Replicon RNA was transcribed from HindIII-linearized pAV-FL-Luc-5′rzm using a T7 RiboMAX Express large-scale RNA production system (Promega). Electroporation of Vero cells with replicon RNA was performed as described previously (37). Plasmid DNA transfection to cells was performed using TransIT 2020 (Mirus Bio) or FuGENE HD transfection reagent (Roche) according to the manufacturer's protocol.
Antibodies and reagents.
The preparation of rabbit and guinea pig polyclonal antibodies against 2B, 2C, and 3A was described previously (32). Antibodies were as follows (source): rabbit polyclonals ACBD3 and FLAG and mouse monoclonal α-tubulin (Sigma-Aldrich), rabbit polyclonal PI4KB (Millipore), mouse monoclonal PI4KB (BD Biosciences), mouse monoclonal IgM PI4P (Echelon Biosciences), Alexa Fluor 488-conjugated anti-rabbit IgG and anti-mouse IgM and Alexa Fluor 594-conjugated anti-rabbit IgG and anti-guinea pig IgG (Molecular Probes), AMCA-conjugated anti-guinea pig IgG (Millipore), horseradish peroxidase (HRP)-conjugated anti-rabbit (Invitrogen), and HRP-conjugated anti-mouse IgG (Zymed). T00127-HEV, a PI4KB-specific inhibitor, was kindly provided by M. Arita, National Institute of Infectious Diseases, Tokyo, Japan (20).
Immunofluorescence microscopy.
Vero cells that were mock electroporated or electroporated with replicon RNA were cultured on glass slides for 4 h, and Vero cells transfected with pCMV-polyprotein were grown in medium with or without 5 μM T00127-HEV1 on glass slides for 24 h. The cells were fixed with 4% paraformaldehyde and then permeabilized with 0.5% Triton X-100 or 20 μM digitonin as described previously (32). After fixation and permeabilization, the cells were washed three times with phosphate-buffered saline (PBS) and incubated with appropriate primary antibodies (dilutions) as follows: anti-2B and anti-2C (1:2,000), anti-3A (1:200), anti-ACBD3 (1:500), anti-PI4KB (1:400), and anti-PI4P (1:300) diluted in PBS containing 3% bovine serum albumin (BSA) or Can Get Signal Immunostain Solution A (Toyobo) for 1 h at room temperature or overnight at 4°C. The cells were washed three times with PBS and then incubated with appropriate secondary antibodies (dilutions) as follows: Alexa Fluor-conjugated anti-mouse, anti-rabbit, or anti-guinea pig (1:500) and AMCA-conjugated anti-guinea pig (1:100) diluted in PBS containing 3% BSA for 1 h at room temperature. After three washes with PBS, the slides were treated with Fluoromount Plus (Diagnostic BioSystems). Images were acquired by using a BZ-8000 fluorescence microscope (Keyence).
Immunoblotting.
Cells were washed three times with PBS and then lysed in 1× passive lysis buffer (Promega). Cell lysates were centrifuged at 15,000 rpm for 2 min. Proteins in the supernatants were separated using 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 2% ECL Advance blocking reagent (Amersham Biosciences) and probed with appropriate primary antibodies (anti-PI4KB and anti-α-tubulin diluted 1:10,000, anti-2B and anti-3A diluted 1:40,000, anti-2C diluted 1:8,000, and anti-FLAG diluted 1:20,000) in Can Get Signal Immunoreaction enhancer solution (Toyobo). After being washed with PBS containing 0.1% Tween 20, the membranes were incubated with HRP-conjugated secondary antibodies in Can Get Signal Immunoreaction enhancer solution. Chemiluminescence was visualized using ECL Advance reagents (Amersham Biosciences) and a LAS-4000 UV Mini-System (Fujifilm).
ACBD3 expression knockdown using siRNA.
ACBD3 expression knockdown using siRNA was performed as described previously (32). Vero cells were transfected with siRNA against ACBD3 or control siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions, cultured for 72 h, and then transfected with pCMV-polyprotein.
Lipid extraction and PI4P detection.
Vero cells that were mock electroporated or electroporated with viral replicon RNA were cultured on 10-cm dishes for 6 h. Vero cells transfected with pcDNA4T/O or a pcDNA construct encoding a viral protein were grown on 6-cm dishes for 24 h (for L, 2B, 2C, and 3A) or 48 h (for 2BC and 3AB) with or without 10 μM T00127-HEV1.
Extraction of lipids from the cells was performed using a PI4P Mass Strip kit (Echelon Bioscience) with modifications. All samples were centrifuged at 15,000 rpm for 2 min, except to harvest cells (6,000 rpm, 2 min) or for phase separation (9,400 rpm, 2 min). The lipid extracts were dissolved in CHCl3-methanol-H2O (1:2:0.8 [vol/vol/vol]) and then spotted onto a PI4P strip. PI4P was detected using the PI4P Mass Strip kit, and visualized using ECL Advance reagents (Amersham Biosciences) and the LAS 4000 UV Mini-System. The PI4P levels were determined using the Multi-Gauge v3.0 software.
Protein expression and purification.
Escherichia coli cells were transformed with pMAL-c2X or a pMAL construct, and maltose-binding protein (MBP) or MBP fusion proteins were expressed and bound to an amylose resin as described previously (32). After the resin was washed three times with column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA [pH 7.5]) and two times with kinase buffer (40 mM Tris-HCl, 20 mM MgCl2, 1 mM EGTA, 0.2% Triton X-100, 1 mM dithiothreitol [pH 7.5]) (38), the proteins were eluted with kinase buffer containing 10 mM maltose. The pCI-FLAG-ACBD3 or pCI-FLAG-PI4KB vectors were used to transfect 293T cells, and 24 h later, the cells were lysed as described previously (32). The cell lysate was incubated with Sepharose CL-4B beads (Sigma-Aldrich) for 1 h at 4°C and incubated with an anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1.5 h at 4°C. The resin was washed four times with lysis buffer and two times with kinase buffer and then incubated in kinase buffer containing 100 ng of FLAG peptide (Sigma-Aldrich)/μl for 1.5 h on ice. Kinase buffer containing 0.1% BSA (wt/vol) was used to elute FLAG-PI4KB. After centrifugation, the supernatant was collected. Purified proteins were electrophoresed with known BSA amounts and quantified densitometrically.
In vitro kinase assay.
In vitro kinase assays were performed using an ADP-Glo kinase assay kit (Promega) according to the manufacturer's protocol and the study of Tai et al. (38), with modifications. In this system, ADP generated through the kinase reaction is again converted to ATP, which in turn is used for the luciferase assay, and thus the luminescence reflects the kinase activity. One kinase reaction mixture contained 2 ng of FLAG-PI4KB, the indicated amounts of FLAG-ACBD3 and one of the MBP-fused viral proteins, 0.1 mM ATP, 0.75 mM PI, and 0.1% BSA in 5 μl of kinase buffer and was incubated for 60 min at room temperature. T00127-HEV1 was added to the kinase reaction mixture to a final concentration of 5 μM. Five microliters of the ADP-Glo Reagent provided with the kit was added to the reaction mixture to deplete the residual ATP. After incubation for 45 min at room temperature, 10 μl of the kinase detection reagent provided with the kit was added to generate ATP as the substrate for the luciferase/luciferin reaction. After incubation for 60 min at room temperature, luciferase activity in each reaction mixture was measured by using a luminometer (Lumat LB9507; Berthold). Under these reaction conditions, the luciferase activity reached a plateau when >25 ng of PI4KB was used (data not shown).
MBP pulldown assay.
MBP pulldown assays were performed using MBP or MBP-fused viral proteins and FLAG-ACBD3 as described previously (32). The captured FLAG-ACBD3 was detected by using immunoblotting with rabbit anti-FLAG antibody.
Statistical analysis.
All data are presented as the means ± the standard deviation (SD). Statistical analyses were performed using paired, two-tailed Student t tests and Microsoft Excel. Statistical significance was defined as P < 0.05.
RESULTS
Construction of a plasmid expressing the AiV polyprotein.
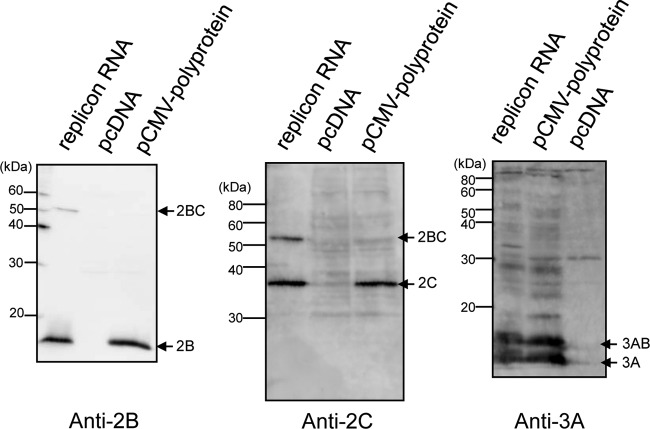
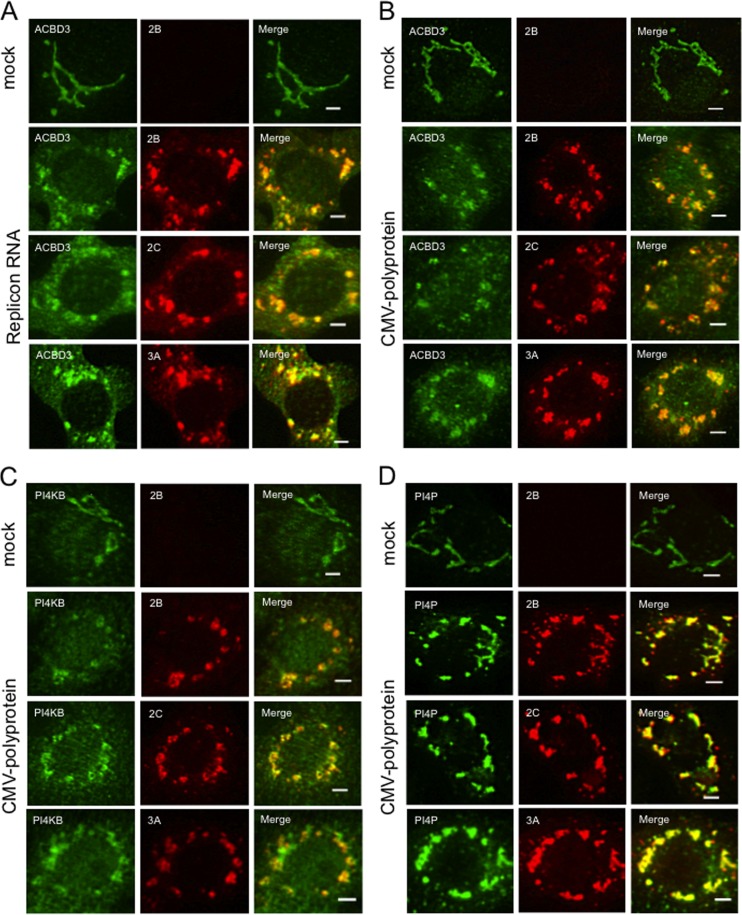
To examine the roles of ACBD3 and PI4KB in forming the AiV replication complex, our first strategy involved inhibiting ACBD3 expression using a specific siRNA and inhibiting PIK4B activity using a pharmacological inhibitor; however, this approach inhibited the replication of the AiV replicon RNA (32), resulting in the synthesis of insufficient amounts of viral proteins. Therefore, we constructed a vector (pCMV-polyprotein) to constitutively express the viral polyprotein at high levels. The vector was engineered to prevent the replication of viral RNA transcripts by deleting the 3′ UTR from the plasmid. Immunoblotting analyses with antibodies specific for viral proteins 2B, 2C, and 3A readily detected the expression and processing of the viral polyprotein in transfected cells after 24 h (Fig. 1); however, 2BC was only faintly detected. The intracellular localization of the viral proteins in cells transfected with pCMV-polyprotein was analyzed by immunofluorescence staining (Fig. 2). Viral proteins 2B, 2C, and 3A were detected as patchy structures in the perinuclear region, and colocalized with ACBD3 (Fig. 2B), PI4KB (Fig. 2C), or PI4P (Fig. 2D). These results are similar to those acquired from the analysis of viral replicon RNA-transfected cells (Fig. 2A) (32).
FIG 1.
Processing of the polyproteins expressed by AiV replicon RNA and pCMV-polyprotein. Vero cells were transfected with AiV replicon RNA, pCMV-polyprotein, or pcDNA, and cell lysates prepared 5 and 24 h after transfection were subjected to immunoblotting with anti-2B, anti-2C, or anti-3A antibodies.
FIG 2.
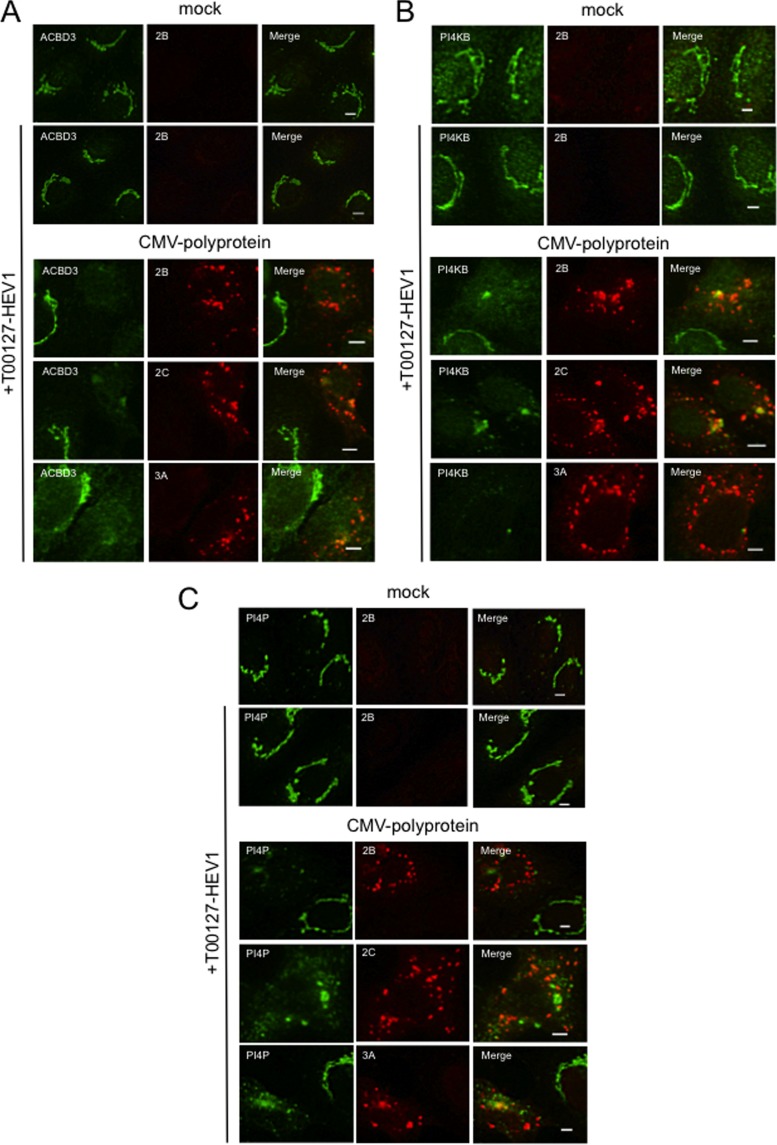
Transient expression of the AiV polyprotein induces the formation of the membrane structures containing viral proteins, ACBD3, PI4KB, and PI4P similar to those present in cells with replicating AiV RNA genomes. (A) Vero cells were mock electroporated or electroporated with replicon RNA. Four hours after electroporation, the cells were fixed and double stained with antibodies against ACBD3 (green) and 2B, 2C, or 3A (red). (B, C, and D) Vero cells were transfected with a plasmid (pCMV-polyprotein) that expresses the full-length polyprotein. After 24 h, the cells were immunostained with antibodies against ACBD3 (B), PI4KB (C), PI4P (D) (green), and 2B, 2C, or 3A (red). Scale bars, 20 μm.
PI4KB does not colocalize with the viral proteins when ACBD3 expression is inhibited.
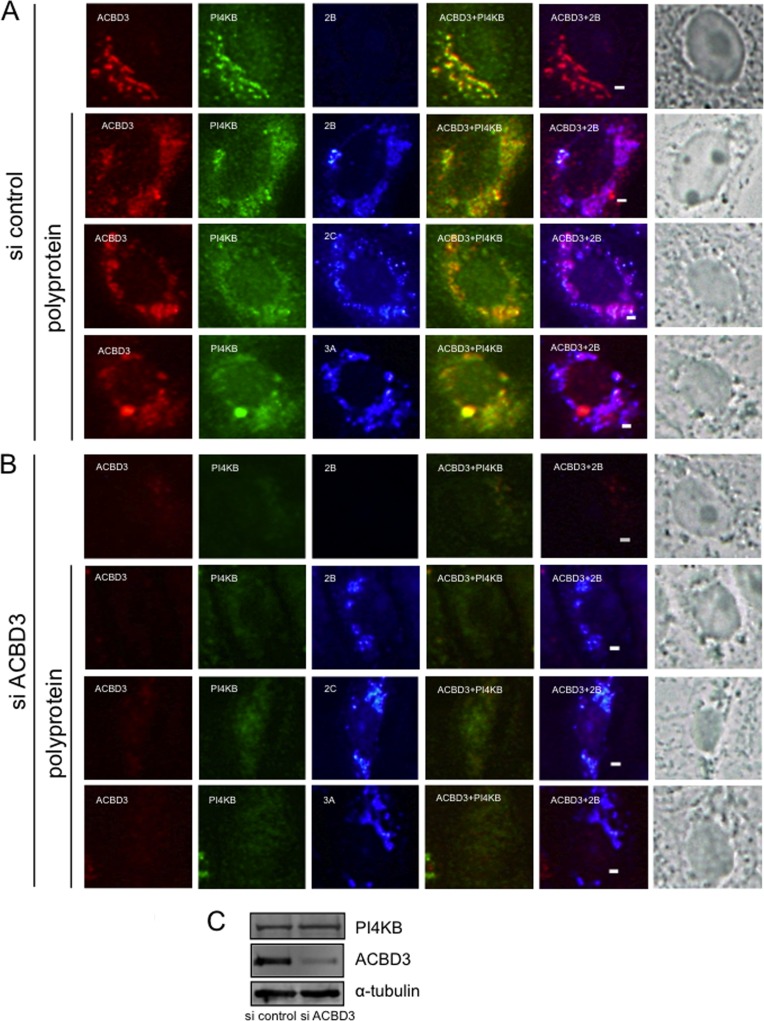
To determine the role of ACBD3 in the formation of the characteristic ACBD3-, PI4KB-, and viral protein-positive structures during viral RNA replication, Vero cells were transfected with ACBD3-specific or control siRNAs and transfected with pCMV-polyprotein 72 h later. After 24 h, protein expression and subcellular localization was determined by using immunofluorescence analysis. In cells transfected with ACBD3-siRNA, but not transfected with pCMV-polyprotein, PI4KB staining was dispersed throughout the cytoplasm, in contrast to its typical Golgi localization (Fig. 3A), suggesting a role of ACBD3 in Golgi localization of PI4KB in normal cells. When the viral polyprotein was expressed in ACBD3 knockdown cells, the viral proteins 2B, 2C, and 3A formed small clusters in the cytoplasm (Fig. 3B). However, unlike cells treated with the control siRNA (Fig. 3A), they did not colocalize with PI4KB, which was dispersed throughout the cytoplasm (Fig. 3B). The PI4KB expression level was not decreased in ACBD3 knockdown cells (Fig. 3C). These results suggest that ACBD3 plays an important role in the formation of the ACBD3-, PI4KB-, and viral protein-positive clusters during viral RNA replication.
FIG 3.
ACBD3 knockdown abolishes the colocalization of PI4KB with viral proteins. (A and B) Vero cells were transfected with control siRNA (A) or ACBD3-siRNA (B) and cultured for 72 h before transfection with pCMV-polyprotein. After 24 h, the cells were fixed and triple stained with antibodies against ACBD3 (red), PI4KB (green), and 2B, 2C, or 3A (blue). The panels in the column on the far right show visible light micrographs of cells. Asterisks indicate cells with decreased ACBD3 expression. Scale bars, 20 μm. (C) Western blotting shows that decreased ACBD3 expression did not affect the levels of PI4KB or α-tubulin.
Inhibiting PI4KB activity alters the distribution of viral proteins, ACBD3, PI4KB, and PI4P in cells expressing the viral polyprotein.
Next, we examined the role of PI4KB activity in the characteristic subcellular localization of the viral proteins, ACBD3, and PI4KB during viral RNA replication. Vero cells were transfected with pCMV-polyprotein and, at the same time, treated with T00127-HEV1, a PI4KB-specific inhibitor (20). Treatment of this almost completely inhibited viral RNA replication in Vero cells (32) and PI4KB activity in vitro (see Fig. 6B). When mock-transfected cells were treated with T00127-HEV1, no significant differences were observed in the subcellular distribution and signal intensities of ACBD3, PI4KB, and PI4P (Fig. 4A, B, and C, upper panels). Because PI4KB activity was inhibited by treatment with T00127-HEV1, PI4P in the Golgi apparatus was likely synthesized by other PI 4-kinases such as PI4KIIα. In the cytoplasm of cells transfected with pCMV-polyprotein and treated with T00127-HEV1, 2B, 2C, and 3A formed small dot-like structures (Fig. 4, lower panels), ACBD3 and PI4KB were dispersed (Fig. 4A and B, respectively), and PI4P formed small dots (Fig. 4C). Although ACBD3 colocalized to some extent with 2C or 3A and PI4KB colocalized with 2B or 2C at the large clusters, which may represent incompletely dispersed Golgi structures, ACBD3, PI4KB, or PI4P did not detectably colocalize with the viral proteins at most small clusters (Fig. 4C). These results suggest that the PI4KB activity is essential for the formation of the viral protein-, ACBD3-, PI4KB-, and PI4P-positive structures observed during viral RNA replication.
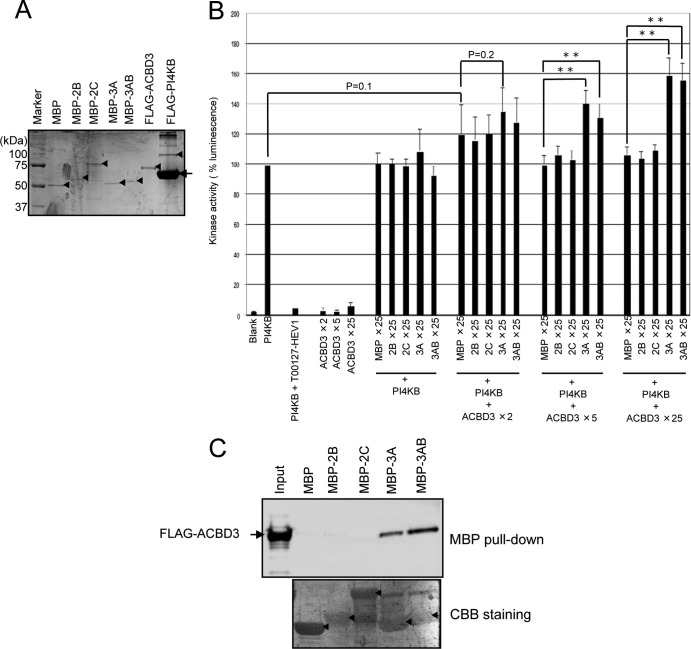
FIG 6.
Viral proteins 3A and 3AB stimulate PI4KB activity in the presence of ACBD3. (A) Purified proteins were electrophoresed through a 10% SDS-polyacrylamide gel and stained with Coomassie brilliant blue. Arrowheads and an arrow indicate each purified protein and BSA contained in the PI4KB elution buffer, respectively. (B) In vitro kinase assays. The kinase activity of PI4KB (2 ng) was assayed in the presence or absence of the indicated molar excess of ACBD3 and MBP or MBP-fused viral proteins compared to PI4KB. The concentration of T00127-HEV1 in the reaction mixtures was 5 μM. Luminescence generated by PI4KB alone was defined as 100%. The experiments were repeated at least three times, and the data are means ± the SD. **, P < 0.001. (C) MBP pulldown assay. MBP or MBP-fused viral proteins immobilized on amylose resins were incubated with purified FLAG-tagged ACBD3, and proteins binding to the resin were analyzed using SDS-PAGE, followed by immunoblotting with anti-FLAG antibody (upper panel). Proteins bound to the membrane were stained with Coomassie brilliant blue (lower panel). Asterisks indicate MBP or MBP-fused viral proteins.
FIG 4.
Inhibiting PI4KB activity disrupts PI4KB-, ACBD3-, and viral protein-positive membrane structures. (A, B, and C) Vero cells were transfected with pCMV-polyprotein and then cultured in medium containing 5 μM T00127-HEV1, a PI4KB-specific inhibitor. After 24 h, the cells were fixed and double stained with antibodies against ACBD3 (A), PI4KB (B), or PI4P (C) (green), and 2B, 2C, or 3A (red). Scale bars, 20 μm.
Virus replication and expression of the viral proteins stimulate PI4P synthesis in Vero cells.
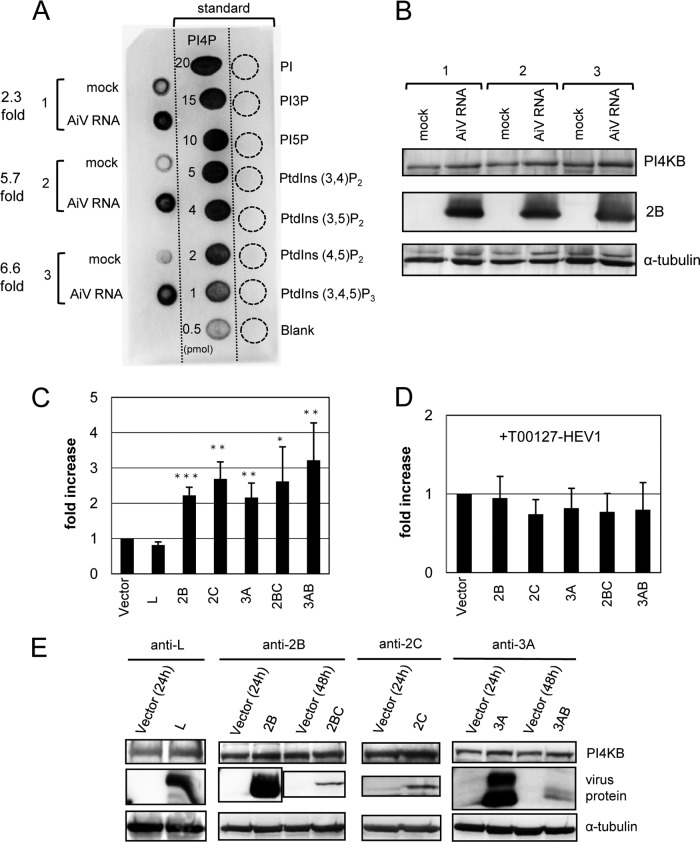
We examined whether the synthesis of PI4P is stimulated in viral RNA-replicating cells. Lipids were extracted from replicon RNA- or mock-transfected cells at 6 h after transfection, and PI4P were detected using a PI(4)P Mass Strip kit. The results showed that PI4P levels increased by factors of 2.3, 5.7, and 6.6 in three independent analyses of cells transfected with the viral replicon RNA compared to mock-transfected cells (Fig. 5A). Immunoblotting analysis showed that the PI4KB amount in these cells was not significantly different from that in mock-transfected cells (Fig. 5B).
FIG 5.
AiV replication and AiV membrane protein expression increase cellular PI4P levels. (A) Vero cells were mock electroporated or electroporated with replicon RNA. PI4P levels were determined 6 h after electroporation. Blots from three independent experiments (left blots 1 to 3) are shown with 0.5- to 20-pmol PI4P standards (middle blots) and 20-pmol PIPn controls (right blots). (B) Lysates prepared from the cells used in panel A were subjected to immunoblotting with antibodies against PI4KB, 2B, or α-tubulin. (C) Vero cells were transfected with pcDNA or the indicated constructs encoding a viral protein. At 24 h (for L, 2B, 2C, or 3A) or 48 h (for 2BC or 3AB) after transfection, each PI4P level was measured as described in panel A and expressed as the value relative to that for pcDNA. (D) Experiments similar to those described in panel C were performed by adding T00127-HEV1 to the cultured medium. The data are means ± the SD for at least six independent experiments. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (E) Western blotting of cell lysates from panel C.
To determine which of the viral proteins, 2B, 2BC, 2C, 3A, and 3AB, has the ability to stimulate PI4P synthesis, each protein was individually expressed in the Vero cells (Fig. 5C). The effect of the nonstructural viral protein L, which does not interact with ACBD3 (32), was also assessed. PI4P synthesis was stimulated in cells transfected with 2B, 2BC, 2C, 3A, or 3AB but not L (Fig. 5C), and the PI4KB level was not significantly altered (Fig. 5E). To examine whether other PI-4 kinases contributed to the increase in PI4P synthesis, similar experiments were performed using T00127-HEV1. The results showed that the expression of the viral proteins did not enhance PI4P synthesis (Fig. 5D). This suggests that PI4KB is mainly involved in the increase in PI4P synthesis in cells expressing 2B, 2BC, 2C, 3A, or 3AB. T00127-HEV1 treatment did not significantly change the PI4KB amount (data not shown). These results suggest that in AiV-infected cells, 2B, 2BC, 2C, 3A, and 3AB mediate an increase in PI4P synthesis by enhancing PI4KB activity.
Formation of the 3A/ACBD3/PI4KB complex stimulates PI4KB activity.
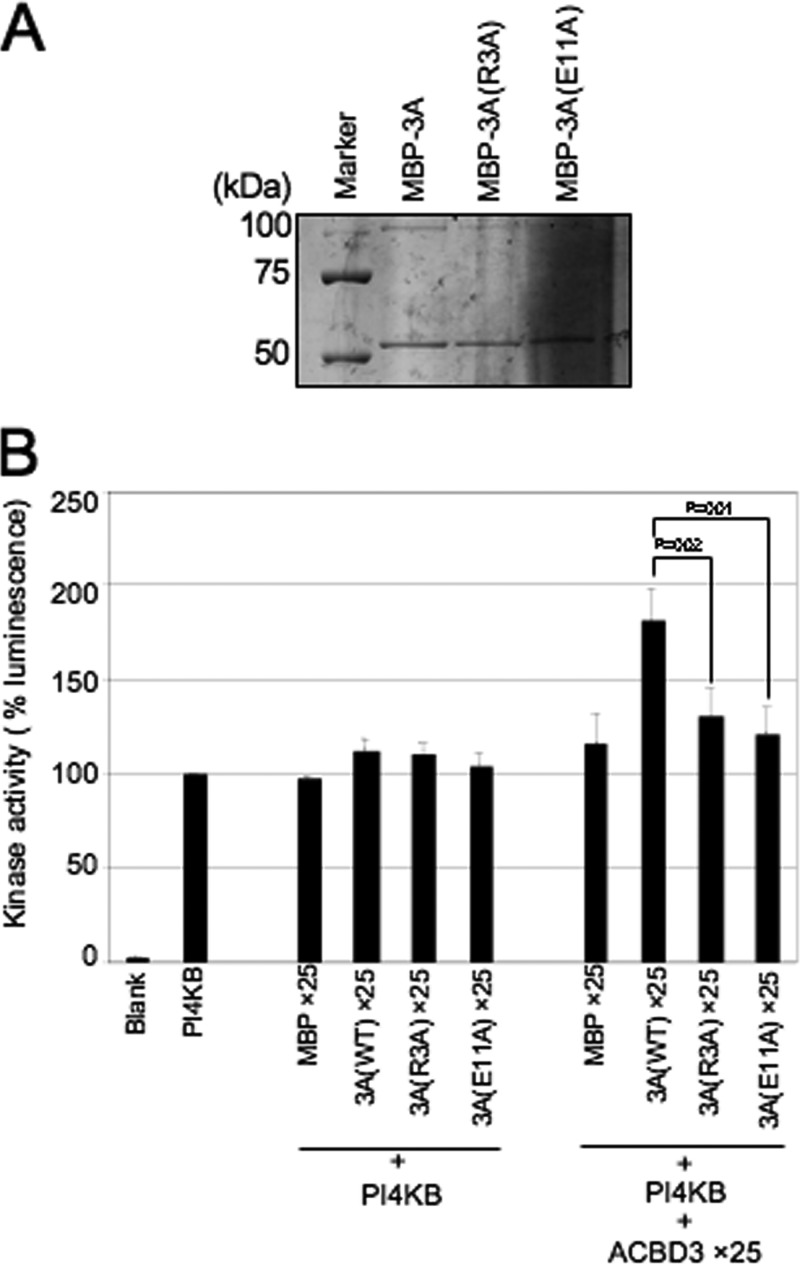
The 2B, 2BC, 2C, 3A, and 3AB proteins do not bind to PI4KB directly. Instead, they bind to ACBD3, which interacts with PI4KB (32). To determine whether the formation of the viral protein/ACBD3/PI4KB complex is involved in the stimulation of PI4KB activity, we conducted in vitro kinase assays. FLAG-tagged PI4KB and ACBD3 were individually expressed in 293T cells and then purified. The MBP-fused 2B, 2C, 3A, and 3AB proteins were expressed in E. coli and purified (Fig. 6A). For 2BC, we were unable to purify sufficient amount of the MBP-2BC protein, as described previously (32).
Assays included 2 ng of PI4KB in the absence or presence of a 25-fold molar excess of MBP or MBP-fused viral proteins and in 2-, 5-, or 25-fold molar excess of ACBD3 compared to PI4KB (Fig. 6B). PI4KB activity was stimulated by 3A and 3AB in the presence of a >5-fold molar excess of ACBD3. In the presence of a 5- or 25-fold molar excess of ACBD3, 3A or 3AB enhanced PI4KB activity by approximately 1.3- and 1.5-fold, respectively, compared to MBP. In the absence of ACBD3 or in the presence of a 2-fold molar excess of ACBD3, 3A and 3AB did not significantly stimulate PI4KB activity. In contrast, 2B and 2C did not stimulate PI4KB activity with or without ACBD3. ACBD3 did not enhance PI4KB activity in the absence of the viral proteins.
Since MBP-fused viral proteins expressed in E coli were used in this experiment, we used an MBP pulldown assay to analyze the interaction of the viral proteins with FLAG-ACBD3, which was expressed in 293T cells. ACBD3 interacted strongly with 3A or 3AB but not with 2B or 2C (Fig. 6C). The reason for the failure of 2B and 2C to interact with ACBD3 is unknown but may be explained by steric interference by the MBP sequence or improper folding of 2B and 2C when expressed in E. coli.
We investigated whether a 3A/ACBD3/PI4KB complex is required for the stimulation of PI4KB activity using two 3A mutants designated 3A(R3A) and 3A(E11A). The R3A mutation inhibits the interaction between 3A and ACBD3, and the E11A mutation does not, but inhibits the interaction of the 3A(E11A)/ACBD3 complex with PI4KB (33). The results of in vitro kinase assays showed that the ability of the two mutants to stimulate PI4KB activity was decreased (Fig. 7B), suggesting that 3A and 3AB stimulate PI4KB activity through the formation of a 3A or 3AB/ACBD3/PI4KB complex.
FIG 7.
In vitro kinase assay of two 3A mutants. (A) Purified MBP-3A and its MBP-3A mutant fusion proteins were electrophoresed through a 10% SDS-polyacrylamide gel and stained with Coomassie brilliant blue. (B) In vitro kinase assay to examine the ability of the 3A mutants to stimulate PI4KB. The data are presented as described in Fig. 6B.
DISCUSSION
We recently reported that AiV genome replication requires the formation of a complex comprising PI4KB, ACBD3, and one of the AiV nonstructural proteins—2B, 2BC, 2C, 3A, and 3AB—and the resultant synthesis of PI4P lipids at the replication sites (32). In the present study, we examined the roles of this protein complex and PI4KB activity in AiV genome replication. We showed that ACBD3 knockdown in polyprotein-expressing cells abolished colocalization of the viral proteins, 2B, 2C, and 3A, with PI4KB, and that inhibition of PI4KB activity in polyprotein-expressing cells led to disruption of the viral protein-, ACBD3-, PI4KB-, and PI4P-positive structures. Further, PI4P levels in cells were increased by AiV replication or the expression of 2B, 2BC, 2C, 3A, or 3AB, and the experiments using PI4KB-specific inhibitor suggested that PI4KB is the main factor responsible for the increase in PI4P levels. These results suggest that the viral protein/ACBD3/PI4KB complex plays an important role in forming the functional replication complex by enhancing PI4P synthesis. Of the viral proteins, 3A and 3AB were shown to stimulate the in vitro kinase activity of PI4KB through forming a 3A or 3AB/ACBD3/PI4KB complex, whereas the ACBD3-mediated PI4KB activation by 2B and 2C remains to be demonstrated.
Inhibition of ACBD3 expression by transfecting cells with an ACBD3-siRNA abolished colocalization of the viral 2B, 2C, and 3A proteins with PI4KB. Further, when PI4KB activity was inhibited in polyprotein-expressing cells, the viral proteins no longer colocalized with ACBD3 and PI4KB. Thus, the viral proteins formed small dot-like structures at the cytoplasm. In contrast, ACBD3 and PI4KB were dispersed throughout the cytoplasm. PI4P did not colocalize with the viral proteins at small clusters in cells treated with the PI4KB-specific inhibitor (Fig. 4). Since knockdown of ACBD3 or inhibition of PI4KB activity inhibited AiV replication (32), AiV replication would not occur on small clusters containing viral proteins but lacking ACBD3, PI4KB, and PI4KB-generated PI4P. Because these host and viral proteins and PI4P lipids were colocalized at the viral RNA replication sites (32), ACBD3-mediated recruitment of PI4KB by the viral proteins and the ensuring synthesis of PI4P are likely required for the formation of the functional replication complex.
After treatment of cells with the PI4KB-specific inhibitor, a comparative amount of PI4P, which is probably synthesized mainly by another Golgi apparatus-located PI4-kinase, PI4KIIα, was still observed at the Golgi apparatus on fluorescence microscopy (Fig. 4). This suggests that the formation of the functional replication complex requires PI4P that is newly synthesized through the formation of the viral protein/ACBD3/PI4KB complex and that the preexisting PI4P at the Golgi apparatus prior to viral genome replication is not utilized for it. PI4KA expression knockdown inhibits HCV genome replication and causes the formation of a morphologically abnormal membranous web (14, 15, 17, 18). Regarding the point that PI4P synthesized at the replication site is required for the formation of the functional AiV membranous replication complex, the role of PI4P in AiV replication presented here is similar to that seen in HCV replication. On the other hand, for enteroviruses, PI4P synthesized at the replication sites recruits viral RNA polymerase 3D to initiate and facilitate viral RNA synthesis (19). We are now performing experiments to detect whether PI4P interacts with AiV RNA polymerase 3D.
AiV replication in Vero cells increased the level of PI4P without increasing that of PI4KB level (Fig. 5A and B). It is also reported that the PI4P levels increase in cells infected with CVB3 and HCV (15, 17, 19). Moreover, we showed that the PI4P levels were increased in cells individually transfected with plasmids expressing 2B, 2BC, 2C, 3A, or 3AB (Fig. 5C). This result suggests that these AiV proteins not only recruit PI4KB by recruiting ACBD3 to viral RNA replication sites but also enhance PI4KB activity.
We examined further the ability of 2B, 2C, 3A, or 3AB to stimulate PI4KB activity by in vitro kinase assays. The results showed that 3A and 3AB enhanced PI4KB activity only in the presence of ACBD3 (Fig. 6B), indicating that the formation of the 3A or 3AB/ACBD3/PI4KB complex is required for the stimulation of PI4KB activity. This assumption is supported by our demonstration that two 3A mutants, which are unable to form the 3A/ACBD3/PI4KB complex, failed to stimulate PI4KB activity compared to wild-type 3A (Fig. 7B). The HCV NS5A protein stimulates PI4KA activity (15, 17) through a direct interaction (15–18), whereas AiV 3A interacts with PI4KB through ACBD3 (32). ACBD3 solely can interact with PI4KB (32) but could not enhance PI4KB activity (Fig. 6B). These results suggest that binding of 3A triggers a conformational change of ACBD3, which in turn induces a conformational change of PI4KB, resulting in enhancement of the PI4KB activity. This hypothesis is supported by the findings that the R3A mutation of the 3A protein inhibits the interaction between 3A and ACBD3, but the E11A mutation does not, although this mutation inhibits the interaction between the 3A(E11A)/ACBD3 complex and PI4KB (33). Further work is required to define exactly how 3A stimulates PI4KB.
Although 3A and 3AB enhanced the PI4KB activity in the presence of ACBD3 in vitro by a factor of ∼1.6, this level of enhancement is modest compared to that detected in cells expressing 3A or 3AB (2- to 3-fold). It seems likely that infected cells support kinase activity to a greater extent than in vitro reaction conditions because of the use of FLAG-tagged or MBP fusion proteins purified from E. coli or because of the composition of the reaction mixture.
The mechanism that regulates PI4P levels in CVB3-infected cells is unknown. CVB3 3A can form the 3A/GBF1/Arf1/PI4KB (19) and 3A/ACBD3/PI4KB complexes (33). Therefore, it will be interesting to determine whether these protein complexes stimulate PI4KB activity.
PI4KB activity was not stimulated by 2B or 2C with or without ACBD3 in the in vitro kinase assay (Fig. 6B). For this experiment, MBP-fused 2B and 2C expressed in E. coli were used, and these fusion proteins did not interact with FLAG-tagged ACBD3 in vitro (Fig. 6C). The reason for the failure of 2B and 2C to interact with ACBD3 is unknown but may be explained by steric interference by the MBP sequence or improper folding of 2B and 2C when expressed in E. coli. Therefore, we could not determine whether the increase in the PI4P level detected in cells expressing 2B or 2C was due to the stimulation of PI4KB activity through the formation of the 2B or 2C/ACBD3/PI4KB complex.
The detailed role of PI4P in the formation of the AiV replication complex is unknown. Because changes in the lipid headgroup size affect membrane curvature (39), it is possible that the increase in the local concentration of PI4P at the Golgi body caused by AiV infection may induce a membrane rearrangement required for the formation of the replication complex. Further, it is likely that PI4P is required to anchor PI4P-binding proteins that induce membrane rearrangement. Proteins that bind to PI4P within the Golgi apparatus include oxysterol-binding protein-1 (OSBP1), ceramide transfer protein (CERT) or four phosphate adaptor protein 1 (FAPP1), FAPP2, Golgi phosphoprotein 3 (GOLPH3) (13), and Arfaptin1 and Arfaptin2 (40). FAPP1 and FAPP2 regulate lipid transport (41, 42) and Golgi-to-plasma membrane transport (43, 44) and induce membrane tubulation (45, 46). Membrane tubules are observed in CVB3 replication structures (47). Recently, it was reported that minor enviroxime-like compounds, PV replication inhibitors, target OSBP (48). Therefore, it will be necessary to examine the involvement of the PI4P-binding proteins in regulating AiV replication.
ACKNOWLEDGMENTS
We are grateful to Minetaro Arita for providing T00127-HEV1. We thank Moeka Sasaki for excellent technical assistance.
This study was supported in part by a Grant-in-Aid for Scientific Research (C), Strategic Research Base Development Program for Private Universities, and a Grant-in-Aid from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Aldabe R, Barco A, Carrasco L. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134–23137. 10.1074/jbc.271.38.23134 [DOI] [PubMed] [Google Scholar]
- 2.Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129–145. 10.1006/viro.1994.1329 [DOI] [PubMed] [Google Scholar]
- 3.Egger D, Teterina N, Ehrenfeld E, Bienz K. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570–6580. 10.1128/JVI.74.14.6570-6580.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knox C, Moffat K, Ali S, Ryan M, Wileman T. 2005. Foot-and-mouth disease virus replication sites form next to the nucleus and close to the Golgi apparatus, but exclude marker proteins associated with host membrane compartments. J. Gen. Virol. 86:687–696. 10.1099/vir.0.80208-0 [DOI] [PubMed] [Google Scholar]
- 5.Krogerus C, Samuilova O, Pöyry T, Jokitalo E, Hyypiä T. 2007. Intracellular localization and effects of individually expressed human parechovirus 1 nonstructural proteins. J. Gen. Virol. 88:831–841. 10.1099/vir.0.82201-0 [DOI] [PubMed] [Google Scholar]
- 6.Moffat K, Howell G, Knox C, Belsham GJ, Monaghan P, Ryan MD, Wileman T. 2005. Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J. Virol. 79:4382–4395. 10.1128/JVI.79.7.4382-4395.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suhy DA, Giddings TH, Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965. 10.1128/JVI.74.19.8953-8965.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towner JS, Ho TV, Semler BL. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810–26818. 10.1074/jbc.271.43.26810 [DOI] [PubMed] [Google Scholar]
- 10.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balla A, Balla T. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351–361. 10.1016/j.tcb.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 12.D'Angelo G, Vicinanza M, Di Campli A, De Matteis MA. 2008. The multiple roles of PtdIns(4)P—not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 121:1955–1963. 10.1242/jcs.023630 [DOI] [PubMed] [Google Scholar]
- 13.Graham TR, Burd CG. 2011. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 21:113–121. 10.1016/j.tcb.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582. 10.1073/pnas.0902693106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase IIIα-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J. Virol. 85:8870–8883. 10.1128/JVI.00059-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim YS, Hwang SB. 2011. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIα and regulates viral propagation. J. Biol. Chem. 286:11290–11298. 10.1074/jbc.M110.194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. 10.1016/j.chom.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tai AW, Salloum S. 2011. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One 6:e26300. 10.1371/journal.pone.0026300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJM, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arita M, Kojima H, Nagano T, Okabe T, Wakita T, Shimizu H. 2011. Phosphatidylinositol 4-kinase III beta is a target of enviroxime-like compounds for antipoliovirus activity. J. Virol. 85:2364–2372. 10.1128/JVI.02249-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spickler C, Lippens J, Laberge MK, Desmeules S, Bellavance E, Garneau M, Guo T, Hucke O, Leyssen P, Neyts J, Vaillancourt FH, Décor A, O'Meara J, Franti M, Gauthier A. 2013. Phosphatidylinositol 4-kinase III beta is essential for replication of human rhinovirus and its inhibition causes a lethal phenotype in vivo. Antimicrob. Agents Chemother. 57:3358–3368. 10.1128/AAC.00303-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954–957. 10.1093/infdis/164.5.954 [DOI] [PubMed] [Google Scholar]
- 23.Ambert-Balay K, Lorro M, Bon F, Giraudon H, Kaplon J, Wolfer M, Lebon P, Gendrel D, Pothier P. 2008. Prevalence and genetic diversity of Aichi virus in community and hospitalized patients. J. Clin. Microbiol. 46:1252–1258. 10.1128/JCM.02140-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goyer M, Aho LS, Bour JB, Ambert-Balay K, Pothier P. 2008. Seroprevalence distribution of Aichi virus among a French population in 2006-2007. Arch. Virol. 153:1171–1174. 10.1007/s00705-008-0091-0 [DOI] [PubMed] [Google Scholar]
- 25.Oh DY, Silva PA, Hauroeder B, Diedrich S, Cardoso DDP, Schreier E. 2006. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 151:1199–1206. 10.1007/s00705-005-0706-7 [DOI] [PubMed] [Google Scholar]
- 26.Pham NTK, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Ushijima H. 2007. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J. Clin. Microbiol. 45:2287–2288. 10.1128/JCM.00525-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter G, Boldizsår Å, Papp G, Pankovics P. 2009. Detection of Aichi virus shedding in a child with enteric and extraintestinal symptoms in Hungary. Arch. Virol. 154:1529–1532. 10.1007/s00705-009-0473-y [DOI] [PubMed] [Google Scholar]
- 28.Sdiri-Loulizi K, Gharbi-Khélifi H, de Rougemont A, Slaheddine C, Sakly N, Ambert-Balay K, Hassine M, Neji Guédiche M, Aouni M, Pothier P. 2008. Acute infantile gastroenteritis associated with human enteric viruses in Tunisia. J. Clin. Microbiol. 46:1349–1355. 10.1128/JCM.02438-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita T, Sakae K, Kobayashi S, Ishihara Y, Miyake T, Mubina A, Isomura S. 1995. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol. Immunol. 39:433–435. 10.1111/j.1348-0421.1995.tb02225.x [DOI] [PubMed] [Google Scholar]
- 30.Yamashita T, Sugiyama M, Tsuzuki H, Sakae K, Suzuki Y, Miyazaki Y. 2000. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 38:2955–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Zhang W, Shen Q, Yang Z, Zhu J, Cui L, Hua X. 2009. Aichi virus strains in children with gastroenteritis, China. Emerg. Infect. Dis. 15:1703–1705. 10.3201/eid1510.090522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31:754–766. 10.1038/emboj.2011.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greninger AL, Knudsen GM, Betegon M, Burlingame AL, DeRisi JL. 2012. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIβ. J. Virol. 86:3605–3616. 10.1128/JVI.06778-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagashima S, Sasaki J, Taniguchi K. 2003. Functional analysis of the stem-loop structures at the 5′ end of the Aichi virus genome. Virology 313:56–65. 10.1016/S0042-6822(03)00346-5 [DOI] [PubMed] [Google Scholar]
- 35.Nagashima S, Sasaki J, Taniguchi K. 2005. The 5′-terminal region of the Aichi virus genome encodes cis-acting replication elements required for positive- and negative-strand RNA synthesis. J. Virol. 79:6918–6931. 10.1128/JVI.79.11.6918-6931.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagashima S, Sasaki J, Taniguchi K. 2008. Interaction between polypeptide 3ABC and the 5′-terminal structural elements of the genome of Aichi virus: implication for negative-strand RNA synthesis. J. Virol. 82:6161–6171. 10.1128/JVI.02151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki J, Kusuhara Y, Maeno Y, Kobayashi N, Yamashita T, Sakae K, Takeda N, Taniguchi K. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021–8030. 10.1128/JVI.75.17.8021-8030.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai AW, Bojjireddy N, Balla T. 2011. A homogeneous and nonisotopic assay for phosphatidylinositol 4-kinases. Anal. Biochem. 417:97–102. 10.1016/j.ab.2011.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature 438:590–596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- 40.Cruz-Garcia D, Ortega-Bellido M, Scarpa M, Villeneuve J, Jovic M, Porzner M, Balla T, Seufferlein T, Malhotra V. 2013. Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. 32:1717–1729. 10.1038/emboj.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. 2007. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 449:62–67. 10.1038/nature06097 [DOI] [PubMed] [Google Scholar]
- 42.Halter D, Neumann S, van Dijk SM, Wolthoorn J, de Mazière AM, Vieira OV, Mattjus P, Klumperman J, van Meer G, Sprong H. 2007. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 179:101–115. 10.1083/jcb.200704091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393–404. 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- 44.Vieira OV, Verkade P, Manninen A, Simons K. 2005. FAPP2 is involved in the transport of apical cargo in polarized MDCK cells. J. Cell Biol. 170:521–526. 10.1083/jcb.200503078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao X, Coskun U, Rössle M, Buschhorn SB, Grzybek M, Dafforn TR, Lenoir M, Overduin M, Simons K. 2009. Golgi protein FAPP2 tubulates membranes. Proc. Natl. Acad. Sci. U. S. A. 106:21121–21125. 10.1073/pnas.0911789106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. 2011. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J. Biol. Chem. 286:18650–18657. 10.1074/jbc.M111.233015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Bárcena M. 2011. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio 2:e00166-11. 10.1128/mBio.00166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arita M, Kojima H, Nagano T, Okabe T, Wakita T, Shimizu H. 2013. Oxysterol-binding protein family I is the target of minor enviroxime-like compounds. J. Virol. 87:4252–4260. 10.1128/JVI.03546-12 [DOI] [PMC free article] [PubMed] [Google Scholar]