Abstract
Phage typing is used for the subtyping of clones of epidemic bacteria. In this study, we identified the outer membrane protein OmpW as the receptor for phage VP5, one of the typing phages for the Vibrio cholerae O1 El Tor biotype. A characteristic 11-bp deletion in ompW was observed in all epidemic strains resistant to VP5, suggesting that this mutation event can be used as a tracing marker in cholera surveillance.
TEXT
Seven cholera pandemics have been recorded historically. The first six pandemics were putatively caused by the classical biotype of Vibrio cholerae serogroup O1, whereas the ongoing seventh pandemic is caused by the O1 El Tor biotype (1). Subtyping of V. cholerae strains is useful in epidemiological and microbiological studies. A phage-biotyping scheme, in which five typing phages (VP1 to VP5) were included, was established for El Tor strain subtyping in the 1970s in China (2). During surveillance of V. cholerae strains, almost all El Tor strains from epidemics were found to be sensitive to all five of these typing phages and are designated phage type 1 (PT1) strains. However, during the 1998 cholera epidemic in Sichuan Province, some patient strains were resistant to phage VP5. These strains belong to the PT6 group and coexisted with PT1 strains. PT6 strains became predominant in 1999 and 2000 but disappeared after 2001 (3), which makes them unique among the epidemic strains.
Phages infect susceptible bacterial strains, beginning with binding to receptors on the surfaces of bacteria. OmpW is an outer membrane protein that serves as a receptor for Escherichia coli colicin S4 (4). The ompW gene has served as a species-specific gene of V. cholerae in many detection studies (5, 6). This gene was found to have an 11-bp deletion in the PT6 strains, but the deletion was not detected in any PT1 strain (3). Therefore, a role for OmpW in VP5 infection was suspected.
In this study, ompW genes from 44 strains isolated from 1998 to 2001, including 22 PT6 and 11 PT1 strains from Sichuan and 11 PT1 strains from other provinces, were sequenced. The same 11-bp deletion was in all PT6 strains (Fig. 1). The deletion corresponds to nucleotides (nt) 298 to 308 of ompW and causes a shift in the reading frame. A new stop codon is generated at 350 nt, ahead of the original stop codon (Fig. 1). All the PT1 strains had intact and identical ompW sequences.
FIG 1.

Alignment of the ompW gene of the PT6 and PT1 V. cholerae strains, showing the same 11-bp-deletion mutation in all PT6 strains. A new stop codon appeared at 350 nt, upstream of the original stop codon.
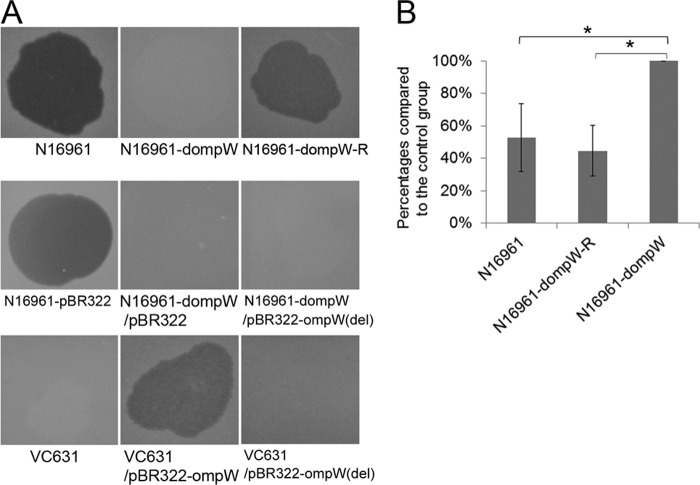
To assess the possible role of the ompW gene in VP5 infection, we constructed an ompW deletion mutant, designated N16961-dompW, with suicide plasmid pWM91 (7) from the VP5-sensitive strain N16961. The N16961-dompW mutant lost sensitivity to VP5 (Fig. 2A). This sensitivity was restored when it was complemented with plasmid pBR322-ompW (designated strain N16961-dompW-R) containing the intact ompW gene cloned from N16961 but not when it obtained pBR322-ompW(del) containing ompW with an 11-bp deletion cloned from the PT6 strain VC631. VC631 became sensitive to VP5 when it obtained plasmid pBR322-ompW but remained resistant when it obtained pBR322-ompW(del) (Fig. 2A), indicating that the intact ompW gene is needed for the sensitivity of V. cholerae El Tor to VP5.
FIG 2.
Experimental identification of the role of OmpW in VP5 infection. (A) Plaques formed by wild-type strains N16961 and VC631, deletion mutant N16961-dompW, and strains complemented with different plasmids. (B) VP5 adsorption by the wild-type strain N16961 and its ompW mutants. Plaques were counted in 10−5 dilutions in a double-layer agar plate containing the VP5-sensitive strain 2477c. The N16961-dompW mutant was used as the test and calculation control. Three independently prepared VP5 phages were used as the biological repeat tests. The numbers of plaques in the N16961- and N16961-dompW-R-treated groups were divided by the number of plaques in the N16961-dompW-treated group to produce plaque formation percentages to show adsorption ratios. Smaller values indicate that more phage particles were adsorbed. Statistically significant comparisons between the groups are marked with an asterisk (*) (analysis of variance [ANOVA] for randomized block design; P < 0.05). No difference was found between the N16961 and N16961-dompW-R groups.
The role of OmpW in infection with VP5 was further validated by strain adsorption tests. Phage VP5 (108 PFU/ml) was mixed equally with strains N16961, N16961-dompW, and N16961-dompW-R (108 CFU/ml of each). The mixture was centrifuged to remove the cells and the adsorbed phage particles, and the supernatants (containing the unadsorbed phages) were diluted 10 times serially and dropped onto a double-layer agar plate containing VP5-sensitive strain 2477c. The plaques of 10−5 dilutions of the supernatants were counted. When compared to that of the N16961-dompW mutant, the adsorption abilities of strains N16961 and N16961-dompW-R, which contain the intact ompW gene, were much stronger because there were fewer phage particles left in both supernatants, and the differences were statistically significant (Fig. 2B). Therefore, we identified OmpW as the receptor for VP5.
VP5 is one of the five typing phages in the phage-biotyping scheme. We have observed that almost all the toxigenic El Tor strains from the epidemics in China are VP5 sensitive, whereas some Sichuan strains from 1998 to 2000 with the ompW mutation are VP5 resistant. We searched the OmpW sequences in the GenBank protein database. Within the obtained 142 V. cholerae strains, 97 have the same OmpW sequence as N16961, whereas some strains have some amino acid residue substitutions (Table 1). Twenty-five strains possess two sequence types of the truncated OmpW. One type includes 20 strains from Haiti from 2010 and a Brazilian strain from 1978, and another type includes two strains which have the same sequence mutation as Sichuan PT6 strains. Our comparisons showed the divergence of OmpW in V. cholerae, and most likely VP5-resistant strains have also appeared in other areas.
TABLE 1.
OmpW mutant V. cholerae strains used in protein sequence comparisons and their OmpW mutations
| OmpW mutationa | Sequence changes | Strains and their sourcesb |
|---|---|---|
| Residue substitution | G142S, A147N, G148A | O395 (1964, India); 95412 (1997, Mexico); HE-40 (2010, Haiti); HE-46 (2010, Haiti); NHCC-008D (2010, Bangladesh); RC27 (1991, Indonesia); HE39 (2010, Haiti); HE48 (2010, Haiti); MZO-2 (2001, Bangladesh); MZO-3 (NA, Bangladesh); TMA 21 (1982, Brazil); V52 (1968, Sudan) |
| Residue substitution | A147N, G148A | 87395 (1983, Mexico); HE-09 (2010, Haiti) |
| Residue substitution | G142S, A147N, G148A, V180 M | EM-1676A (2011, Bangladesh); 1587 (1994, Peru) |
| Residue substitution | S15L, S16A, Q59H, A79R, F122N, G142S, A147N, G148A, S179Y, M210I | gi48376 (NA) |
| Residue substitution | G142S, A147N, G148E | V51 (NA) |
| Residue substitution | V45I, N47G, T48S, E92D, G94I, S96G, G100A, E101D, T145S, N146S, G148T, S150T, V164A, F166V | gi190689252 (2006, Laos) |
| Residue substitution | T121A | BJG-01 (NA) |
| Truncated | KLHHFL from residue 131 | 116063 (1978, Brazil); HC-02A1 (NA); HC-36A1 (NA); and the following 20 strains from Haiti, 2010: HC-02C1, HC-1A2, HC-41B1, HC-43B1, HC-44C1, HC-46B1, HC-50A1, HC-51A1, HC-52A1, HC-55A1, HC-55B2, HC-55C2, HC-56A1, HC-57A1, HC-59A1, HC-59B1, HC-60A1, HC-61A2, HC-78A1, HE-45 |
| Truncated | TFATYLYGPILLW from residue 100, same as PT6 strains | 2740-80 (1980, U.S. Gulf Coast); gi487821612 (NA) |
Compared to strain N16961.
NA, epidemic data not available.
Some biological effects of the inactivation of the ompW gene were detected in V. cholerae. A quantitative biofilm formation assay was performed using 96-well cell plates and crystal violet staining as described previously (8). The optical density at 570 nanometer (OD570)/OD600 values for N16961 were 1.53 ± 0.13, and those for the N16961-dompW mutant were 1.76 ± 0.22. Mild enhancement of biofilm formation was observed in the ompW mutant (P < 0.05). This may be a compensatory response, promoting environmental survival. In our colonization competition test of the wild-type and mutant strains in mice, a slight decrease in the concentration of the ompW mutant strain (C6706-dompW, constructed from the VP5-sensitive El Tor strain C6707) was observed (Fig. 3). This was consistent with the results of a previous study that showed the intestinal colonization activities of ompW-deficient mutants to be at only marginally lower levels than those of wild-type O1 strains (9). In this way, ompW mutants were shown to have less adaptability to their environments and to human hosts than their wild-type counterparts. In conclusion, OmpW may act as the receptor for typing phage VP5. Cholera phages may play a role in the emergence of new V. cholerae pandemic serogroups or clones, and phage-resistant strains may have a survival advantage (10). However, the PT6 strains disappeared while the PT1 strains continue to spread within the population. This suggests that the PT6 strain must have some deficiencies with respect to surviving in and outside of hosts. Nevertheless, when such strains contain the ompW mutation, they can be used as a tracer, allowing their paths through the environment and human populations to be tracked.
FIG 3.
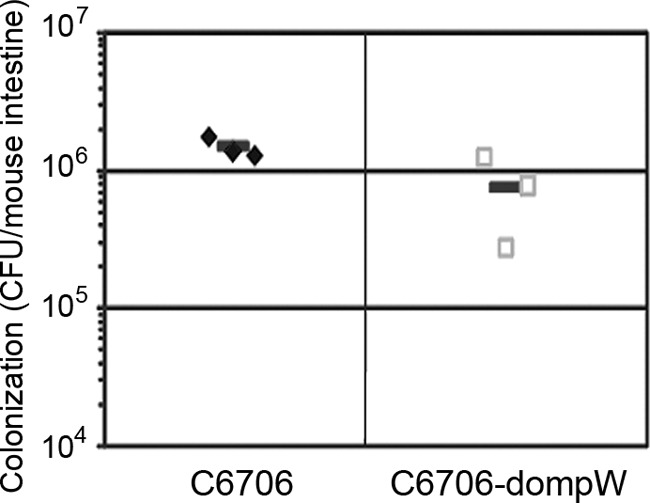
Colonization competition test of the wild-type strain C6706 and its ompW mutant strain in mice. Short lines indicate the average CFU values for the tests.
Nucleotide sequence accession numbers.
The ompW gene sequences of PT1 strain N16961 and PT6 strain VC631 have been deposited in GenBank under accession numbers KJ722608 and KJ722607.
ACKNOWLEDGMENTS
This study was supported by the Priority Project on Infectious Disease Control and Prevention (2012ZX10004215) from the Ministry of Health, China, and by the National Basic Research Priorities Program (2009CB522604) from the Ministry of Scientific Technology, China.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Feeley JC. 1965. Classification of Vibrio cholerae (Vibrio comma), including El Tor vibrios, by infrasubspecific characteristics. J. Bacteriol. 89:665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao S, Wu S, Liu B. 1984. Characteristics of typing phages of Vibrio cholerae biotype El Tor. Fu Huo Luan Zi Liao Hui Bian 4:237–245 (In Chinese.) [Google Scholar]
- 3.Xu DL, Wang HX, Diao BW, Liu HL, Xiong LF, Gao SY, Kan B. 2009. Molecular characterization of Vibrio cholerae phage-type 6b epidemic isolates from 1998 to 2001 in Sichuan province. Zhonghua Yu Fang Yi Xue Za Zhi 43:409–412 (In Chinese.) [PubMed] [Google Scholar]
- 4.Pilsl H, Smajs D, Braun V. 1999. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srisuk C, Chaivisuthangkura P, Rukpratanporn S, Longyant S, Sridulyakul P, Sithigorngul P. 2010. Rapid and sensitive detection of Vibrio cholerae by loop-mediated isothermal amplification targeted to the gene of outer membrane protein ompW. Lett. Appl. Microbiol. 50:36–42. 10.1111/j.1472-765X.2009.02749.x [DOI] [PubMed] [Google Scholar]
- 7.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. 10.1006/plas.1996.0001 [DOI] [PubMed] [Google Scholar]
- 8.Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nandi B, Nandy RK, Sarkar A, Ghose AC. 2005. Structural features, properties and regulation of the outer-membrane protein W (OmpW) of Vibrio cholerae. Microbiology 151:2975–2986. 10.1099/mic.0.27995-0 [DOI] [PubMed] [Google Scholar]
- 10.Faruque SM, Naser IB, Islam MJ, Faruque AS, Ghosh AN, Nair GB, Sack DA, Mekalanos JJ. 2005. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. U. S. A. 102:1702–1707. 10.1073/pnas.0408992102 [DOI] [PMC free article] [PubMed] [Google Scholar]