ABSTRACT
Gammaherpesviruses are ubiquitous pathogens that establish a lifelong infection and are associated with cancer. In spite of the high seroprevalence of infection, the risk factors that predispose the host toward gammaherpesvirus-induced malignancies are still poorly understood. Interferon (IFN) regulatory factor 1 (IRF-1) is a tumor suppressor that is also involved in the regulation of innate and adaptive immune responses. On the basis of its biology, IRF-1 represents a plausible host factor to attenuate gammaherpesvirus infection and tumorigenesis. In this study, we show that IRF-1 restricts gammaherpesvirus replication in primary macrophages, a physiologically relevant immune cell type. In spite of the known role of IRF-1 in stimulating type I IFN expression, induction of a global type I IFN response was similar in IRF-1-deficient and -proficient macrophages during gammaherpesvirus infection. However, IRF-1 was required for optimal expression of cholesterol-25-hydroxylase, a host enzyme that restricted gammaherpesvirus replication in primary macrophages and contributed to the antiviral effects of IRF-1. In summary, the current study provides an insight into the mechanism by which IRF-1 attenuates gammaherpesvirus replication in primary immune cells, a mechanism that is likely to contribute to the antiviral effects of IRF-1 in other virus systems.
IMPORTANCE Interferon regulatory factor 1 (IRF-1) is a transcription factor that regulates innate and adaptive immune responses and functions as a tumor suppressor. IRF-1 restricts the replication of diverse viruses; however, the mechanisms responsible for the antiviral effects of IRF-1 are still poorly understood. Gammaherpesviruses are ubiquitous pathogens that are associated with the induction of several malignancies. Here we show that IRF-1 expression attenuates gammaherpesvirus replication in primary macrophages, in part by increasing expression of cholesterol-25-hydroxylase (CH25H). CH25H and its product, 25-hydroxycholesterol, restrict replication of diverse virus families. Thus, our findings offer an insight into the mechanism by which IRF-1 attenuates the replication of gammaherpesviruses, a mechanism that is likely to be applicable to other virus systems.
INTRODUCTION
Gammaherpesviruses are ubiquitous pathogens that establish a lifelong infection and are associated with cancer. Specifically, two known human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), are associated with lymphoproliferative diseases, including those in immunocompromised patients (1). Given the high prevalence of EBV and KSHV infections, it is important to identify host factors that control gammaherpesvirus infection and pathogenesis, a quest that is hindered by the exquisite species specificity of human gammaherpesviruses. Murine gammaherpesvirus 68 (MHV68) is genetically and biologically related to the human gammaherpesviruses and offers an experimental system that overcomes the challenges associated with studies of EBV and KSHV infection (2, 3).
Interferon (IFN) regulatory factor 1 (IRF-1) is the founding member of a family of transcription factors that orchestrate type I IFN expression following virus infection (4–6). IRF-1 restricts replication of flavi-, toga-, and retroviruses (7–9); however, the mechanism by which IRF-1 controls viral replication is poorly understood. Furthermore, IRF-1 is capable of inducing antiviral genes independently of type I IFN (10), suggesting that some of the IRF-1 antiviral functions may be IFN independent. In addition to its role in the innate immune response, IRF-1 suppresses cellular transformation in vitro and in vivo (11, 12). Specifically, IRF-1 is required for apoptosis and cell cycle arrest of irradiated T cells (11, 13). Not surprisingly, IRF-1 expression is decreased in a variety of human cancers (14–16).
The antiviral and tumor suppressor roles of IRF-1 pose it as a likely regulator of gammaherpesvirus infection and pathogenesis. Importantly, little is known about the regulation of gammaherpesvirus infection by IRF-1. KSHV encodes a multifunctional viral IRF-1 protein which shares limited homology with the DNA binding domain of cellular IRF-1 and inhibits expression of IRF-1-driven genes in a reporter-based system (17). However, it is not clear whether IRF-1 regulates KSHV replication or latency. In contrast, EBV usurps IRF-1 to facilitate viral gene transcription in latently infected cell lines (18, 19); it is not known if EBV inhibits IRF-1-driven induction of antiviral genes or its tumor-suppressive functions. MHV68 replication is increased in IRF-1-deficient myeloid dendritic cells in vitro and during acute infection of IRF-1-deficient mice (20). Importantly, the mechanism by which IRF-1 suppresses viral replication is unknown.
Here we show that IRF-1 attenuates MHV68 replication in primary bone marrow-derived macrophages, an immune cell type that is biologically relevant for MHV68 replication and latency (21, 22). The extent of type I IFN signaling was similar in IRF-1-deficient and wild-type (wt) MHV68-infected macrophages; further, genetic evidence suggested that IRF-1 functions downstream of type I IFN expression to restrict MHV68 replication in primary macrophages. Importantly, IRF-1 was required for optimal induction of cholesterol-25-hydroxylase (CH25H), a host enzyme that attenuated MHV68 replication. Because 25-hydroxycholesterol (25HC), a product of CH25H activity, is associated with broad antiviral effects (23, 24), the results of our study provide an important insight into the mechanism of IRF-1-dependent antiviral functions.
MATERIALS AND METHODS
Animals and primary cell cultures.
C57BL/6J (BL6), IRF-1−/− (25), and CH25H−/− (26) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). IFNAR1−/− mice (27) were obtained from Mitchell Grayson. IRF-1−/− and IFNAR1−/− mice were crossed to generate an IRF-1−/− IFNAR1−/− (double-knockout [DKO]) mouse strain. Mice were bred and housed in a specific-pathogen-free barrier facility in accordance with institutional and federal guidelines. All experimental manipulations of mice were approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin. Bone marrow was harvested from mice between 3 and 10 weeks of age. Primary bone marrow-derived macrophages were generated as previously described (21).
Viral DNA quantitation.
Infected cells were washed with phosphate-buffered saline (PBS) and lysed in a buffer containing 10 mM Tris-HCl, 1 mM EDTA, 8% SDS, and 20 μg/ml of proteinase K (Sigma-Aldrich, St. Louis, MO). Following overnight protein digestion at 56°C, DNA was extracted with phenol-chloroform and precipitated using standard sodium acetate-ethanol treatment. The DNA pellet was resuspended in water, viral DNA was measured by real-time PCR using core gene 50 promoter primers, and the viral DNA level was normalized to the corresponding GAPDH (glyceraldehyde-3-phosphate dehydrogenase) level as previously described (28).
mRNA measurement.
Total RNA was harvested from cells, DNase treated, and reverse transcribed as previously described (29). cDNA was assessed in triplicate, along with corresponding negative reverse transcription (RT) reactions, by real-time PCR using an iCycler (Bio-Rad, Hercules, CA) (29, 30). The relative abundance of each cDNA was normalized to the corresponding GAPDH levels using the ΔCT threshold cycle (CT) method. Primers for the MHV68 DNA synthesis genes, IRF-1, universal alpha IFN (IFN-α), IFN-β, GAPDH (29, 31, 32), and CH25H (26) were described previously. The primers used to measure the levels of mRNA for MHV68-targeting antiviral genes are listed in Table 1.
TABLE 1.
RT-PCR primer sequences for MHV68-targeted genes
| Primer | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| MNDA | GGAGTGGACAATGGCACAACATCA | TGACGAAACTGTGGTCTCCACACA |
| MITD1 | AGTAGTGGCCTGGAAGAAATAAA | GTGGGATGCAGAAGGAAGAA |
| Mx2 | AGCAGAGTGACACAAGCGAGAAGA | AGCCCTTCTGTCCCTGAATCACAA |
| IFIH | CGATCCGAATGATTGATGCA | AGTTGGTCATTGCAACTGCT |
| ADAR1 | GTGCAAAGCCCTGCATAGA | AGGAGGCAGTAGCCATTAGA |
| SPRY2 | GCACTGTTCGTAGAGGGTTAG | TAGGAGTGTTGGCTGCTTTAG |
Western analysis.
Cell lysates were collected and analyzed as previously described (29, 30). The antibodies used were anti-β-actin (1:20,000; Novus Biological, Littleton, CO), anti-total Stat1 (1:3,000; Santa Cruz Biotechnology, Santa Cruz, CA), anti-IRF-1 (1:1,000; Cell Signaling Technology, Danvers, MA), anti-ssDBP (1:1,000 [30]), anti-ISG15 (1:4,000; a gift of Debbie Lenschow), and a secondary goat antimouse or antirabbit horseradish peroxidase-conjugated secondary antibody (1:25,000; Jackson ImmunoResearch, West Grove, PA).
Interferon treatment.
Macrophages were treated with recombinant mouse IFN-β (BioLegend, San Diego, CA) or IFN-γ (R&D Systems, Minneapolis, MN) diluted in tissue culture medium. Cells were treated with 10 U/ml for 3 h prior to lysis.
Viral stock preparation and infections.
N36S virus mutant (21, 33) and wt MHV68 stocks were prepared and titers were determined on NIH 3T12 cells as previously described (28). Bone marrow-derived macrophages were infected with MHV68 for 1 h at 37°C in 5% CO2 to allow adsorption and washed twice with PBS prior to medium replenishment.
Interferon bioassay.
Conditioned medium (CM) was collected from bone marrow-derived macrophages which were mock infected or infected with MHV68. The amount of antiviral activity in CM was determined as previously described using an encephalomyocarditis virus (EMCV) bioassay (32). Briefly, monolayers of L929 cells were overlaid with UV-irradiated CM or diluted IFN-β standards and incubated overnight at 37°C in 5% CO2. Subsequently, monolayers were infected with EMCV (B strain) at a multiplicity of infection (MOI) of 5 PFU/cell, and cell survival was determined after an additional 24 h.
Measurement of 25-hydroxycholesterol.
Bone marrow macrophages derived from BL6, IRF-1−/−, and CH25H−/− mice were mock infected or infected with wt MV68 at an MOI of 10 PFU/cell for 24 h. Conditioned medium was removed, and the cells were washed twice with cold PBS and subsequently scraped into fresh PBS. Cells were treated with 0.01% butyrated hydroxytoluene (BHT; Fisher Science Education, Nazareth, PA) to prevent auto-oxidation, after which 60 ng of 24(R/S)-hydroxycholesterol-d7 (24-RS-HC; Avanti Polar Lipids Inc., Alabaster AB), 3 μg 5-α-cholestane (Sigma-Aldrich, St. Louis, MO), and 10 M NaOH were added. Oxygen was decreased by blowing 100% nitrogen onto the samples, after which the samples were placed in a shaking water bath at 60°C for 2 h. Sterols and oxysterols were sequentially extracted 3 times with hexanes and concentrated under 100% nitrogen. Samples were redissolved in 100% ethanol and adjusted to 70% ethanol, after which the oxysterols were separated from cholesterol using a certified Sep-Pak C18 cartridge (Waters, Milford, MA). Sterols and cholesterol were eluted using 70% and 100% ethanol, respectively. Eluates were concentrated under 100% nitrogen, derivatized using a 99:1 solution of N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA/TMCS) and trimethylchlorosilane (TMCS), and dried under 100% nitrogen. Samples were resuspended in ethyl acetate and analyzed using gas chromatography-mass spectroscopy (GC-MS; Thermo Finnigan DSQ; Thermo Fisher Scientific, Madison, WI) using the conditions described previously (34). 25HC had an m/z of 131.0 with a retention time of 6.26 min, and 24-RS-HC had an m/z of 413.0 and a retention time of 17.80 min; these values were comparable to those reported previously (35).
Treatment of infected macrophages with 25-hydroxycholesterol.
Macrophages derived from BL6, IRF-1−/−, and CH25H−/− mice were infected with wt MHV68 at an MOI of 0.1 PFU/cell. After 30 min of adsorption, the inoculum was removed and the cells were washed three times with PBS and incubated in medium containing either 2 μM 25-hydroxycholestrol, 7-ketone (Steraloids, Inc., Newport, RI), or 0.5% ethanol (carrier) throughout the infection. Medium and cells were collected at the time points indicated below to determine the total virus yield.
Statistical analysis.
Student's t test or the Mann-Whitney test (GraphPad Prism, La Jolla, CA) was used to measure statistical significance, and the α value was 0.05.
RESULTS
IRF-1 reduces MHV68 replication in primary macrophages.
The broad antiviral activities of IRF-1 attenuate the replication of several viruses, including hepatitis C virus, West Nile virus (WNV), and EMCV (6, 7, 36). Further, overexpression of IRF-1 in transformed neuronal human cells (the 293T cell line [37]) reduces subsequent infection and replication of MHV68 (38). Importantly, the mechanism by which IRF-1 restricts viral replication remains poorly understood. To determine the extent to which IRF-1 regulates MHV68 replication in primary murine macrophages, a physiologically relevant cell type that supports MHV68 replication and latency (21, 22), the virus yield in bone marrow-derived macrophages generated from BL6 and IRF-1−/− mice was measured. In the absence of IRF-1 expression, MHV68 replication was increased 5- to 10-fold at all time points examined under conditions of both high (Fig. 1A) and low (Fig. 1B) MOIs. Thus, IRF-1 restricted MHV68 replication in primary macrophages.
FIG 1.
IRF-1 is induced by type I IFN signaling and restricts MHV68 replication in primary macrophages. Bone marrow-derived macrophages from control (BL6) and IRF-1−/− mice were infected with MHV68 at an MOI of 5 (A) or 0.01 (B) PFU/cell. The total virus yield was determined by plaque assay on NIH 3T12 fibroblasts. (C to E) BL6 or IFNAR1−/− macrophages were mock infected or infected at an MOI of 5 PFU/cell with wt MHV68 or an orf36-null virus mutant (N36S). (C) The levels of IRF-1 mRNA were measured by quantitative RT-PCR at the indicated times postinfection and normalized to the corresponding GAPDH mRNA levels. (D, E) Protein levels of IRF-1, ssDBP, and β-actin were measured at the indicated times postinfection. Additional controls for panel E included uninfected primary macrophages treated with recombinant IFN-β or IFN-γ. The mRNA expression data in panel C were pooled from three independent experiments, with two replicates within each experiment. Error bars represent SEMs. *, P < 0.05.
MHV68 infection induces IRF-1 expression in a type I IFN-dependent but viral DNA damage response-independent manner.
IRF-1 is a short-lived transcription factor that is constitutively expressed at low levels in a variety of tissues, including spleen (39). Importantly, transcription of IRF-1 is significantly increased by type I and type II IFNs (38) and in the context of RNA virus infections (9, 40). Additionally, induction of the DNA damage response increases both transcription and the protein stability of IRF-1 in an ATM-dependent manner (41). Because MHV68 infection of primary macrophages induces both type I IFN and a DNA damage response (21, 32), we aimed to define the relative contribution of these two host signaling pathways to the expression of IRF-1 in MHV68-infected macrophages.
MHV68 infection increased IRF-1 transcription, with peak levels of IRF-1 mRNA being observed at 8 h postinfection and a subsequent decrease in IRF-1 mRNA levels being observed by 24 h postinfection (Fig. 1C). Changes in IRF-1 mRNA levels were mirrored by the changes in protein levels (Fig. 1D), suggesting that during MHV68 infection, IRF-1 expression is regulated at the level of transcription. Expression of MHV68 protein kinase orf36 is necessary and sufficient to induce a DNA damage response in infected macrophages (21). IRF-1 mRNA and protein levels were not decreased in macrophages infected with the orf36-null MHV68 mutant N36S (Fig. 1C and D), suggesting that the virus-induced DNA damage response does not increase IRF-1 expression. In contrast, expression of type I IFN receptor 1 (IFNAR1) was required to increase IRF-1 mRNA and protein levels in MHV68-infected macrophages (Fig. 1E; data not shown). Thus, MHV68 infection of primary macrophages increased the expression of IRF-1 in a type I IFN-dependent manner.
IRF-1 restricts MHV68 replication downstream of IE viral gene expression.
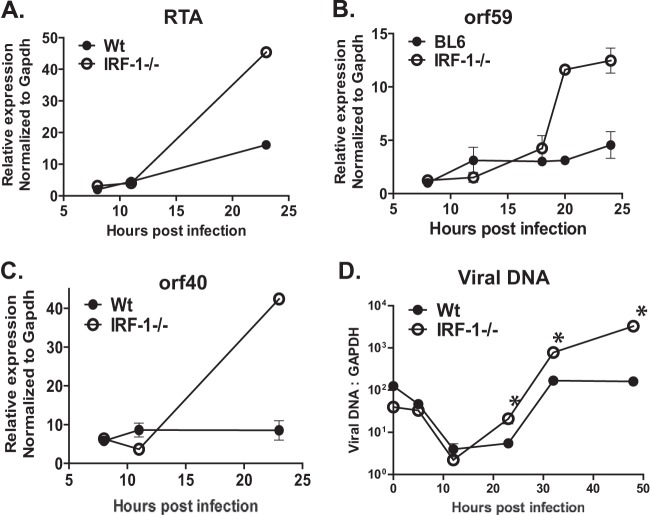
Having observed increased MHV68 replication in IRF-1−/− macrophages (Fig. 1), we sought to determine the stage of viral replication cycle at which IRF-1 imposes a restriction. Expression of immediate early (IE) genes is one of the earliest processes that take place following virus entry. MHV68 RTA is an immediate early viral protein that is essential for gammaherpesvirus lytic replication and reactivation from latency (42–46). Because peak IRF-1 protein levels were observed at 8 h postinfection (Fig. 1), coincident with the known timing of RTA expression (28), we hypothesized that IRF-1 attenuates the expression of immediate early MHV68 genes. Surprisingly, RTA mRNA levels were indistinguishable in control and IRF-1−/− primary macrophages as late as 12 h postinfection (Fig. 2A).
FIG 2.
IRF-1 restricts MHV68 replication downstream of immediate early viral gene expression. Bone marrow-derived macrophages from control (BL6) and IRF-1−/− mice were infected with MHV68 at an MOI of 5 PFU/cell. (A to C) RNA was isolated at the indicated times postinfection, and the levels of RTA, orf59, and orf40 mRNA were measured by quantitative RT-PCR and subsequently normalized to the corresponding GAPDH mRNA levels. (D) Total DNA was isolated at the indicated times postinfection, and the levels of viral DNA were quantified by real-time PCR and subsequently normalized to the corresponding cellular DNA levels. The data shown are representative of those from at least three independent experiments, with each data point being derived from 2 to 3 independent biological repeats within each experiment. Error bars represent SEMs. *, P < 0.05.
Interestingly, by 24 h postinfection, RTA mRNA levels were significantly higher in IRF-1−/− macrophages than control macrophages (Fig. 2A). A similar increase in viral gene expression at 24 h but not 12 h postinfection was seen for every MHV68 gene that encodes critical components of the viral DNA synthesis machinery (orf59, orf56, orf40, orf44, orf9, orf6; Fig. 1E and 2B and C; data not shown). Consistent with the increased expression of viral DNA synthesis genes, increased levels of viral DNA were observed in IRF-1−/− macrophages (Fig. 2D). Thus, IRF-1 attenuated MHV68 replication downstream of immediate early gene expression.
Type I IFN signaling is similar in IRF-1-deficient and -proficient MHV68-infected primary macrophages.
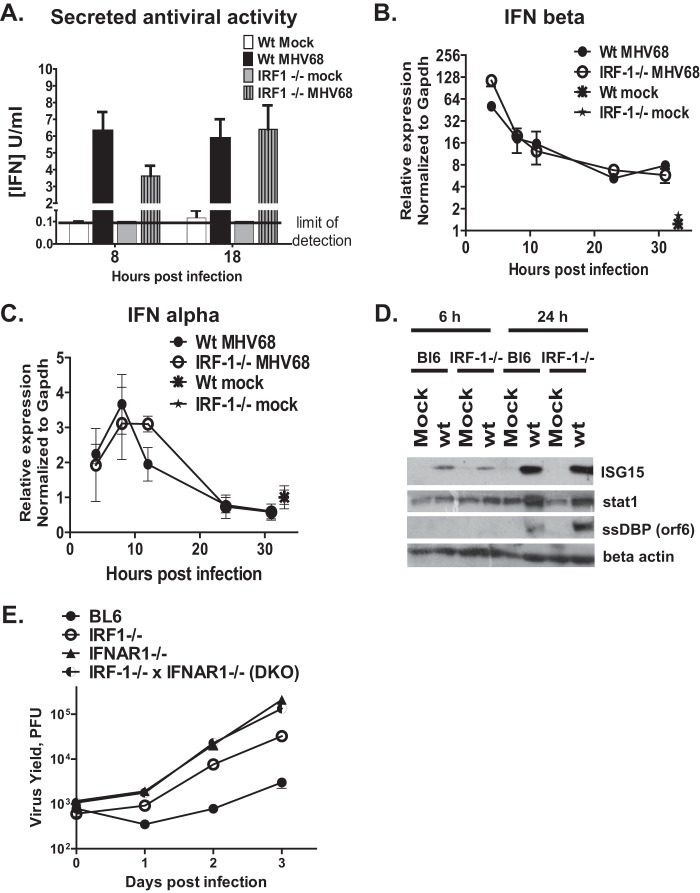
IRF-1 can induce expression of IFN-β, a member of an extensive family of cytokines that engage a unique type I IFN receptor. Receptor engagement stimulates signaling that culminates in the transcriptional induction of hundreds of antiviral interferon-stimulated genes (ISGs) (47). We previously showed that interferon regulatory factor 3 (IRF-3) is required for type I IFN expression in MHV68-infected primary macrophages (32). However, it was not clear whether IRF-1 contributed to the induction of a type I IFN response, which is known to restrict MHV68 replication in vivo and in primary macrophages in vitro (32, 33, 48). To comprehensively assess the antiviral activity produced in an IRF-1-dependent manner, control or IRF-1−/− macrophages were mock treated or infected with MHV68, and the antiviral activity of the conditioned tissue culture medium was measured at several times postinfection using an EMCV bioassay. MHV68 infection of control macrophages resulted in a robust increase in secreted antiviral activity at 8 and 18 h postinfection (Fig. 3A). While IRF-1−/− MHV68-infected macrophages demonstrated a modest decrease in secreted antiviral activity at 8 h postinfection compared to control infections (Fig. 3A), antiviral activity was similar in MHV68-infected cultures of both genotypes at 18 h postinfection. Furthermore, IFN-β and IFN-α mRNA levels were similar in wt and IRF-1−/− macrophages throughout the infection (Fig. 3B and C).
FIG 3.
IRF-1 functions downstream of the type I IFN receptor to attenuate MHV68 replication in primary macrophages. Bone marrow-derived macrophages from control (BL6) and IRF-1−/− mice were mock infected or infected with MHV68 at an MOI of 10 PFU/cell. (A) Antiviral activity in the culture supernatants was analyzed at the indicated times postinfection using an EMCV bioassay and expressed as the number of units of type I IFN based on the standard curve generated with recombinant IFN-β in the same assay. (B, C) Total RNA was harvested at the indicated times postinfection, and the levels of IFN-β and IFN-α mRNA were measured by quantitative RT-PCR; data from 2 to 3 independent experiments were pooled, each data point was derived from 2 to 3 replicates within each experiment, and error bars represent SEMs. (D) Protein levels of ISG15, total Stat1, MHV68 ssDBP, and β-actin were measured at the indicated times postinfection. (E) Macrophages of the indicated genotypes were infected with MHV68 at an MOI of 1 PFU/cell. The virus yield in triplicate cultures was measured at the indicated times postinfection. Data are representative of those from two independent experiments, and error bars represent SEMs.
To determine the extent to which IRF-1 regulates global type I IFN signaling downstream of the type I IFN receptor, expression of ISG15 and Stat1 was measured in control and IRF-1−/− macrophages following MHV68 infection. The levels of the ISG15 and Stat1 proteins were similarly increased in macrophages of both genotypes (Fig. 3D), indicating that signaling downstream of the type I interferon receptor occurred at similar levels in IRF-1−/− and wild-type macrophages in response to MHV68 infection.
IRF-1 functions downstream of the type I IFN receptor to attenuate MHV68 replication.
IRF-1 was constitutively expressed at low levels in naive primary macrophages (Fig. 1D). Furthermore, IRF-1 can induce antiviral gene expression independently of type I IFN (10), suggesting that at least some antiviral functions of IRF-1 may be IFN independent. To test the extent to which IFN signaling was required for the ability of IRF-1 to attenuate MHV68 replication, the growth of MHV68 was assessed in macrophages isolated from mice with a single or combined (DKO) deficiency in IRF-1 and IFNAR1. Replication of MHV68 was increased in IRF-1−/− macrophages and was further potentiated in the absence of the type I IFN receptor (IFNAR1−/−; Fig. 3E). In contrast, the replication of MHV68 was similar in macrophages with a single deficiency of IFNAR1 (IFNAR1−/−) and combined deficiencies of IRF-1 and IFNAR1 (DKO) (Fig. 3E). Thus, in the context of primary macrophages, IRF-1 acted downstream of the type I IFN receptor to restrict MHV68 replication.
IRF-1 facilitates expression of CH25H in MHV68-infected primary macrophages.
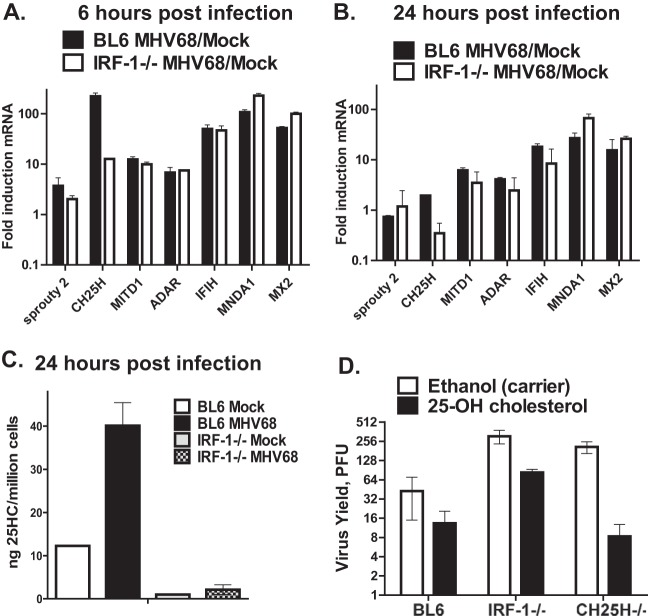
Having determined that IRF-1 acts downstream of the type I IFN receptor to attenuate MHV68 replication, we wanted to test the hypothesis that IRF-1 is required for expression of genes that interfere with MHV68 replication. A subset of such genes was recently identified in a screen performed by Liu et al. (2012), where individual genes induced by type I or type II IFN were overexpressed in the 293T cell line prior to MHV68 infection (38); interestingly, IRF-1 was identified as one of the MHV68-restricting genes in this screen. To determine the extent to which IRF-1 regulated the transcription of MHV68-restricting genes identified by Liu et al. (38), expression of these genes was compared in wt and IRF-1−/− MHV68-infected primary macrophages. Expression of all examined MHV68-targeting ISGs was induced at 6 h postinfection (Fig. 4A) and either remained induced or returned to baseline levels by 24 h postinfection (Fig. 4B). Intriguingly, out of seven examined mRNAs, CH25H mRNA was the only mRNA whose levels were significantly different between wt and IRF-1−/− macrophages. Specifically, at 6 h postinfection, wild-type macrophages displayed an ∼230-fold increase in CH25H relative mRNA levels compared to those in the mock-infected controls (Fig. 4A). In contrast, only a 12-fold increase in CH25H mRNA levels was observed in MHV68-infected IRF-1−/− macrophages compared to the levels in the uninfected cultures (Fig. 4A). By 24 h postinfection, the relative mRNA levels of CH25H decreased to levels near or below those at the baseline (Fig. 4B), consistent with the corresponding decrease in IRF-1 protein and mRNA levels (Fig. 1C and D). Thus, IRF-1 was required for optimal expression of CH25H in MHV68-infected primary macrophages.
FIG 4.
IRF-1 facilitates the CH25H expression that contributes to IRF-1-dependent attenuation of MHV68 replication. (A, B) Control and IRF-1−/− macrophages were mock infected or infected with MHV68 at an MOI of 10 PFU/cell. At 6 and 24 h postinfection, the mRNA levels of the indicated genes were measured by quantitative RT-PCR, normalized to the GAPDH mRNA levels, and subsequently expressed as the fold induction over the levels measured in corresponding mock-infected macrophages. (C) Cell-associated 25HC levels in macrophages of the indicated genotypes were measured at 24 h following mock or MHV68 infection (MOI = 5 PFU/cell). Data are representative of those from two independent experiments. (D) Primary macrophages of the indicated genotypes were infected with MHV68 (MOI = 0.1 PFU/cell) and treated with ethanol or 2 μM 25HC (from an ethanol-based stock) immediately following viral absorption for the duration of infection. The virus yield was measured at 48 h postinfection.
CH25H is an endoplasmic reticulum-associated enzyme that converts cholesterol into 25HC. Along with other oxysterols, 25HC is a signaling moiety that regulates several biological pathways, including cholesterol synthesis (49), liver X receptor-mediated transcription (50, 51), and B cell migration (52). Significantly, two independent groups recently demonstrated that 25HC has broad antiviral activity, including activity against MHV68 (23, 24). Specifically, 25HC pretreatment restricted MHV68 spread in primary mouse embryo fibroblasts (23); furthermore, MHV68 gene expression was increased during acute infection of CH25H−/− mice (24). Having observed lower CH25H mRNA levels in IRF-1−/− MHV68-infected macrophages (Fig. 4A), accumulation of its enzymatic product, 25HC, was examined. As expected, 25HC levels were increased in MHV68-infected BL6 macrophages compared to those in the mock-infected BL6 controls (Fig. 4C). In contrast, 25HC levels were reduced in IRF-1−/− macrophages, regardless of infection status. Thus, IRF-1 facilitated the expression of CH25H and 25HC, its enzymatic product, in MHV68-infected primary macrophages.
25HC contributes to IRF-1-dependent attenuation of MHV68 replication in primary macrophages.
Because of the broad antiviral activity of 25HC (23, 24), we examined its relative contribution to IRF-1-mediated restriction of MHV68 replication. Treatment of control BL6 macrophages with exogenous 25HC produced a modest decrease in MHV68 replication, consistent with the endogenous production of 25HC by these macrophages (Fig. 4D). Increased levels of MHV68 replication were observed in carrier-treated CH25H−/− macrophages compared to BL6 cultures (also see the results presented in Fig. 5). Exogenous addition of 25HC to CH25H−/− macrophages decreased MHV68 replication to the levels observed in 25HC-treated BL6 macrophages, indicating that the increased MHV68 replication observed in CH25H−/− carrier-treated cultures was due to the lack of 25HC. In contrast, while exogenous 25HC treatment of IRF-1−/− macrophages decreased the level of MHV68 replication compared to that for the carrier-treated control, the levels of viral replication after this decrease failed to reach the levels observed in 25HC-treated BL6 and CH25H−/− macrophages. Thus, the production of 25HC by CH25H contributed to but was not exclusively responsible for the IRF-1-mediated attenuation of MHV68 replication.
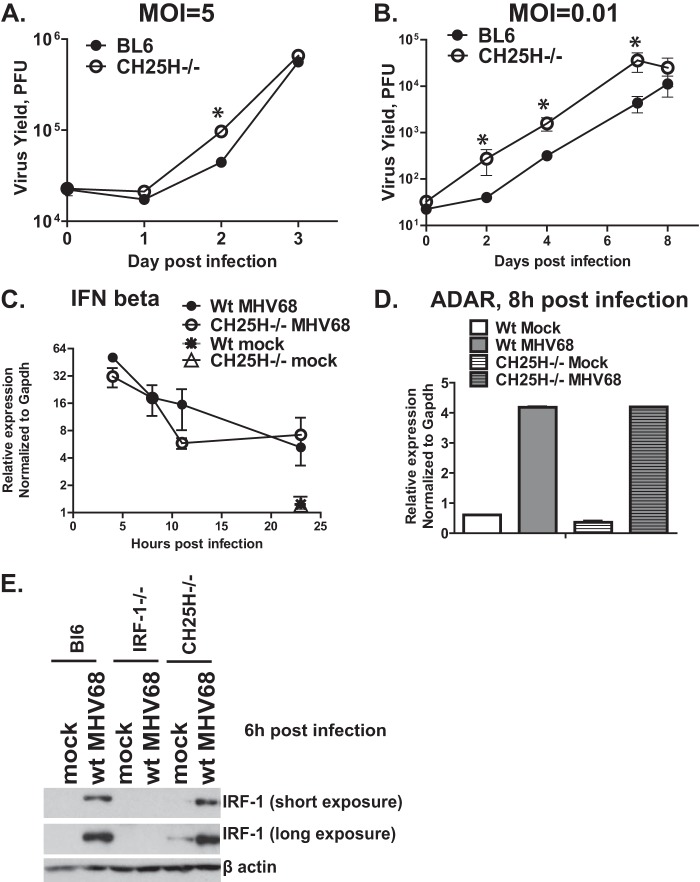
FIG 5.
CH25H attenuates MHV68 replication in primary macrophages. (A, B) BL6 or CH25H−/− macrophages were infected with MHV68 at an MOI of 5 or 0.01 PFU/cell. The virus yield was measured at the indicated times postinfection. (C, D) IFN-β or ADAR mRNA levels were measured by quantitative RT-PCR and normalized to the corresponding GAPDH mRNA levels. (E) Macrophages of the indicated genotypes were infected with MHV68 at an MOI of 5 PFU/cell. Protein levels of IRF-1 and β-actin were measured at 6 h postinfection. *, P < 0.05.
CH25H expression attenuates MHV68 replication.
In agreement with the broad antiviral activity of CH25H (23, 24), higher levels of MHV68 replication were observed in CH25H−/− primary macrophages than the BL6 controls at both high and low MOIs (Fig. 5A and B), confirming that CH25H attenuated MHV68 replication in these immune cells. Similar to the observations made in IRF-1−/− macrophages (Fig. 3), the induction of IFN-β mRNA was comparable in control and CH25H−/− macrophages (Fig. 5C). Furthermore, the mRNA levels of ADAR, an MHV68-restricting ISG, and the protein levels of IRF-1 were similar in control and CH25H−/− macrophages following MHV68 infection (Fig. 5D and E). Thus, CH25H expression restricted MHV68 replication in primary macrophages but did not affect global type I IFN expression or signaling in infected cells.
CH25H restricts late stages of MHV68 gene expression and viral DNA synthesis in primary macrophages.
In spite of the broad antiviral activity of CH25H, the antiviral mechanism remains controversial and poorly understood. Restriction of viral entry is one proposed CH25H-dependent antiviral mechanism, especially in the case of enveloped viruses (24). Surprisingly, the mRNA levels of MHV68 RTA, orf59, and orf40 immediate early/early genes were similar in control and CH25H−/− macrophages during the first 24 h of infection (Fig. 6A to C), suggesting that CH25H expression does not attenuate MHV68 entry or the initial stages of lytic replication in primary macrophages. This observation was in contrast to the increased mRNA levels of the same MHV68 genes observed in IRF-1−/− macrophages by 24 h postinfection (Fig. 2A to C), supporting the existence of additional, CH25H-independent but IRF-1-dependent antiviral mechanisms that restrict MHV68 replication.
FIG 6.
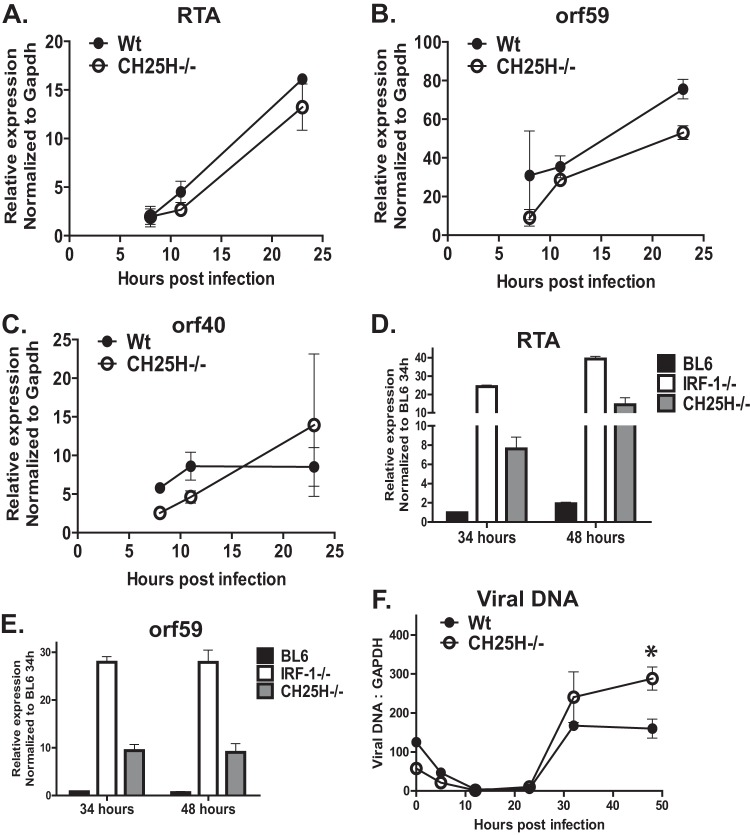
CH25H restricts late stages of MHV68 gene expression and viral DNA synthesis in primary macrophages. BL6, CH25H−/−, or IRF-1−/− macrophages were infected with MHV68 at an MOI of 5 PFU/cell. (A to E) RNA was isolated at the indicated times postinfection, and the levels of RTA, orf59, and orf40 mRNA were measured by quantitative RT-PCR and subsequently normalized to the corresponding GAPDH mRNA levels. (F) Total DNA was isolated at the indicated times postinfection, and the levels of viral DNA were quantified by real-time PCR and subsequently normalized to the corresponding cellular DNA levels (measured using GAPDH-specific primers). *, P < 0.05.
In contrast to the levels in the first 24 h of infection, the mRNA levels of RTA and viral DNA synthesis genes were increased in CH25H−/− macrophages at 34 and 48 h postinfection (Fig. 6D and E; data not shown). However, the levels of RTA and orf59 mRNA in CH25H−/− macrophages remained below the corresponding mRNA levels in IRF-1−/− macrophages (Fig. 6D and E). Similar to what was observed in IRF-1−/− macrophages, CH25H−/− macrophages displayed higher levels of viral DNA accumulation (Fig. 6F); however, these increased viral DNA levels appeared with delayed kinetics in CH25H−/− cells (at 30 to 48 h postinfection versus 18 to 24 h postinfection in IRF-1−/− macrophages; compare Fig. 6F and 2D). In conclusion, the IRF-1-imposed attenuation of the MHV68 lytic life cycle was more profound than that imposed by CH25H, supporting the existence of several IRF-1-dependent antiviral mechanisms that restrict MHV68 replication.
DISCUSSION
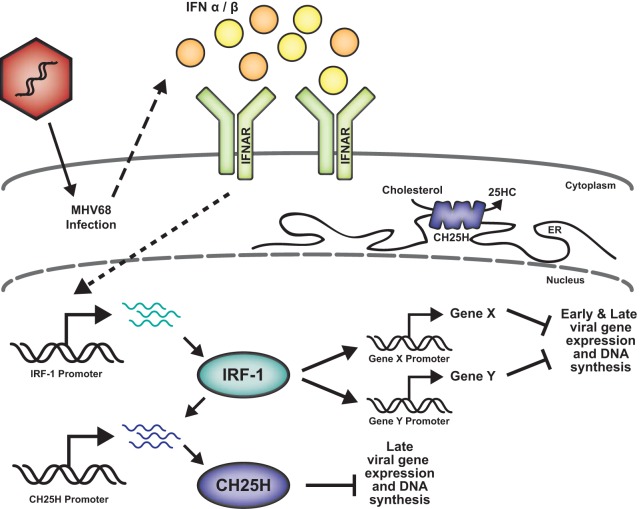
In this study, we show that IRF-1 attenuates gammaherpesvirus replication in primary macrophages and offer an insight into the mechanism by which IRF-1 exerts its antiviral effects. Based on the results of the current study, we propose the following working model (Fig. 7). Constitutive expression of IRF-1 in primary macrophages is further stimulated by the type I IFN that is induced following MHV68 infection. While the global type I IFN response appears to be similar in IRF-1-deficient and wild-type MHV68-infected macrophages, IRF-1 transcriptionally induces a number of cellular genes, including the gene for CH25H. These IRF-1-induced genes interfere with MHV68 gene expression and DNA synthesis, thus restricting viral replication.
FIG 7.
IRF-1-driven restriction of MHV68 replication. Type I IFN expression in MHV68-infected macrophages results in the transcriptional induction of IRF-1. Subsequently, IRF-1 increases the expression of several host genes, including the CH25H gene, that attenuate MHV68 replication. While CH25H inhibits late stages of viral DNA synthesis, other, yet unidentified IRF-1-induced cellular proteins attenuate viral gene expression and viral DNA replication at earlier stages of infection. ER, endoplasmic reticulum.
Antiviral functions of IRF-1.
IRF-1 has broad antiviral activities that restrict the replication of several families of viruses, including flavi-, toga-, and retroviruses (7–9); importantly, the mechanism by which IRF-1 exerts its antiviral functions remains poorly understood. Replication of West Nile virus (WNV) was restricted by IRF-1 in primary macrophages but not fibroblasts, indicating that IRF-1 antiviral activities may be further modified in a cell type-specific manner (7). Furthermore, IRF-1−/− mice demonstrated higher levels of WNV replication in vivo (7). Interestingly, similar to our observations (Fig. 3), the global type I IFN response was not attenuated in the absence of IRF-1 following either in vivo or in vitro WNV infection.
On the basis of genetic evidence presented in this study (Fig. 3E), IRF-1 functions downstream of the type I IFN receptor to attenuate MHV68 replication in primary macrophages. Interestingly, IRF-1 expression was required for IFN-γ-mediated suppression of mouse norovirus (MNV) replication in primary macrophages in vitro (53). A similar phenotype was seen with vaccinia virus (VACV), where IRF-1 was required for IFN-γ-mediated suppression of VACV replication in mouse fibroblasts (54). Because IFN-γ is not expressed in primary macrophages infected with MHV68 in vitro (our unpublished observations), it is possible that IRF-1 has additional, type I IFN-independent functions that attenuate MHV68 infection, especially in vivo where IFN-γ is present (55). Identification of such additional IRF-1 functions is the focus of ongoing studies.
The exact stages of the viral life cycle that are inhibited by IRF-1 are mostly unknown. In this study, IRF-1 suppressed viral gene expression starting between 12 and 24 h postinfection and attenuated the accumulation of MHV68 DNA (Fig. 2). Interestingly, the timing of IRF-1-dependent antiviral effects in our study lagged behind the time of peak IRF-1 expression (Fig. 1C and D), suggesting that IRF-1 primarily exerts its antiviral functions by inducing expression of host genes, such as the CH25H gene. However, at this time we cannot exclude the possibility of potential direct effects of IRF-1 on the regulation of viral gene expression, as IRF-1 protein levels still remain above those at the baseline at 24 h postinfection (Fig. 1D), when the antiviral effects are observed.
Finally, IRF-1 antiviral effects may be further regulated by a yet-to-be-identified MHV68 protein, similar to the postulated regulation of IRF-1 by KSHV-encoded viral IRF-1 in the context of infection (17). It is clear, however, that the inhibition of IRF-1 in the context of MHV68 infection is incomplete, as evidenced by increased viral replication in IRF-1−/− macrophages (Fig. 1). Instead, MHV68 may usurp IRF-1 to mediate putative proviral functions, similar to the positive effect of IRF-1 on the transcription of the EBV latent gene in vitro (18, 19) and suppression of the MHV68 M2 gene by a related IRF, IRF-2, in vivo (56). Thus, the identification of a putative IRF-1 MHV68 regulator will be an important focus of future studies.
Antiviral functions of CH25H.
In this study, we show that IRF-1 facilitates transcription of the gene for CH25H (Fig. 4A), an activity that contributes to IRF-1-mediated attenuation of MHV68 (Fig. 4D). CH25H expression can be induced by Stat1 (23, 57), and IRF-1 cooperates with Stat1 in the induction of select cytokine genes (58). Thus, it is possible that IRF-1 cooperates with Stat1 to directly induce CH25H expression in MHV68-infected macrophages. Alternatively, another transcriptional factor induced or activated by IRF-1 can be directly responsible for the differential expression of CH25H in IRF-1-deficient and -proficient macrophages. Importantly, it is clear that increased CH25H expression is not sufficient to account for all the antiviral effects of IRF-1 in the context of MHV68 infection. Thus, a comprehensive analysis of IRF-1-dependent gene expression in the context of MHV68 infection is a critical future direction for research that will allow identification of additional cellular genes that restrict MHV68 replication.
CH25H is an endoplasmic reticulum-associated enzyme that modifies cholesterol to generate 25HC, an oxysterol with multiple roles in diverse cell signaling pathways. Interestingly, 25HC is the predominant oxysterol produced by IFN-treated or mouse cytomegalovirus-infected primary macrophages (23, 26). In addition to the well-characterized roles of CH25H in the regulation of cholesterol biosynthesis and liver X receptor-mediated transcription (49–51), CH25H and its product, 25HC, were recently shown to restrict the replication of diverse viruses, including MHV68 (23, 24). However, the exact mechanism by which CH25H limits virus replication remains controversial and is likely to be further modified in the context of individual virus families. In this study, CH25H expression attenuated viral gene transcription and MHV68 DNA synthesis during late stages of replication, making it unlikely that the endoplasmic reticulum-associated enzyme itself directly suppresses these viral processes. Thus, an important focus of future studies will be identification of the exact mechanism by which CH25H and its product, 25HC, suppress viral gene expression and DNA replication.
Finally, identification of the roles of IRF-1 and CH25H during chronic gammaherpesvirus infection and virus-driven lymphomagenesis is an important and immediate future direction of research, especially given the tumor suppressor functions of IRF-1. The analysis of in vivo phenotypes is further complicated by the functions of IRF-1 and CH25H in the innate and adaptive immune response (26, 39, 59, 60) and IRF-1 involvement in the DNA damage response, which is known to regulate chronic MHV68 infection (61, 62). Dissection of the antiviral mechanism by which IRF-1 suppresses gammaherpesvirus replication and chronic infection is likely to benefit understanding of IRF-1 antiviral functions in the context of other virus families.
ACKNOWLEDGMENTS
Expert technical assistance was provided by Chelsea Spurley. We are grateful to Debbie Lenschow for the generous gift of anti-ISG15 antibody. We thank John Corbett and William Jackson for helpful discussions.
This study was funded by American Cancer Society research scholar grant RSG-12-174-01-MPC and a grant from Advancing Healthier Wisconsin (to V.L.T.) and by a grant from the Biomedical Laboratory Research and Development Program, U.S. Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (to S.B.P.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 305:163–174. 10.1016/j.canlet.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virgin HW, IV, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. 1990. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J. Gen. Virol. 71:1365–1372. 10.1099/0022-1317-71-6-1365 [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 6.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, Taniguchi T. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921–1924. 10.1126/science.8009222 [DOI] [PubMed] [Google Scholar]
- 7.Brien JD, Daffis S, Lazear HM, Cho H, Suthar MS, Gale M, Jr, Diamond MS. 2011. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8(+) T cell immune responses against West Nile virus infection. PLoS Pathog. 7:e1002230. 10.1371/journal.ppat.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pflugheber J, Fredericksen B, Sumpter R, Jr, Wang C, Ware F, Sodora DL, Gale M., Jr 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:4650–4655. 10.1073/pnas.062055699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stirnweiss A, Ksienzyk A, Klages K, Rand U, Grashoff M, Hauser H, Kroger A. 2010. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 184:5179–5185. 10.4049/jimmunol.0902264 [DOI] [PubMed] [Google Scholar]
- 11.Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. 1996. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature 382:816–818. 10.1038/382816a0 [DOI] [PubMed] [Google Scholar]
- 12.Nozawa H, Oda E, Nakao K, Ishihara M, Ueda S, Yokochi T, Ogasawara K, Nakatsuru Y, Shimizu S, Ohira Y, Hioki K, Aizawa S, Ishikawa T, Katsuki M, Muto T, Taniguchi T, Tanaka N. 1999. Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes Dev. 13:1240–1245. 10.1101/gad.13.10.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak TW, Taki S, Taniguchi T. 1995. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature 376:596–599. 10.1038/376596a0 [DOI] [PubMed] [Google Scholar]
- 14.Connett JM, Badri L, Giordano TJ, Connett WC, Doherty GM. 2005. Interferon regulatory factor 1 (IRF-1) and IRF-2 expression in breast cancer tissue microarrays. J. Interferon Cytokine Res. 25:587–594. 10.1089/jir.2005.25.587 [DOI] [PubMed] [Google Scholar]
- 15.Kuroboshi H, Okubo T, Kitaya K, Nakayama T, Daikoku N, Fushiki S, Honjo H. 2003. Interferon regulatory factor-1 expression in human uterine endometrial carcinoma. Gynecol. Oncol. 91:354–358. 10.1016/S0090-8258(03)00515-8 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Liu DP, Chen PP, Koeffler HP, Tong XJ, Xie D. 2007. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 67:2535–2543. 10.1158/0008-5472.CAN-06-3530 [DOI] [PubMed] [Google Scholar]
- 17.Zimring JC, Goodbourn S, Offermann MK. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J. Virol. 72:701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nonkwelo C, Ruf IK, Sample J. 1997. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J. Virol. 71:6887–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer BC, Paulson E, Strominger JL, Speck SH. 1997. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol. Cell. Biol. 17:873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz F, Heit A, Guggemoos S, Krug A, Mages J, Schiemann M, Adler H, Drexler I, Haas T, Lang R, Wagner H. 2007. Interferon-regulatory-factor 1 controls Toll-like receptor 9-mediated IFN-beta production in myeloid dendritic cells. Eur. J. Immunol. 37:315–327. 10.1002/eji.200636767 [DOI] [PubMed] [Google Scholar]
- 21.Tarakanova VL, Leung-Pineda V, Hwang S, Yang C-W, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW., IV 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275–286. 10.1016/j.chom.2007.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weck KE, Kim SS, Virgin HW, IV, Speck SH. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, Meljon A, Talbot S, Krishnan K, Covey DF, Wenk MR, Craigon M, Ruzsics Z, Haas J, Angulo A, Griffiths WJ, Glass CK, Wang Y, Ghazal P. 2013. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38:106–118. 10.1016/j.immuni.2012.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. 10.1016/j.immuni.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, Potter J, Furlonger CL, Narendran A, Suzuki H, Ohashi PS, Paige CJ, Taniguchi T, Mak TW. 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75:83–97. 10.1016/S0092-8674(05)80086-8 [DOI] [PubMed] [Google Scholar]
- 26.Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 2009. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl. Acad. Sci. U. S. A. 106:16764–16769. 10.1073/pnas.0909142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S, Zhang X, Tough DF, Sprent J. 1998. Type I interferon-mediated stimulation of T cells by CpG DNA. J. Exp. Med. 188:2335–2342. 10.1084/jem.188.12.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mounce BC, Tsan FC, Kohler S, Cirillo LA, Tarakanova VL. 2011. Dynamic association of gammaherpesvirus DNA with core histone during de novo lytic infection of primary cells. Virology 421:167–172. 10.1016/j.virol.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mboko WP, Mounce BC, Wood BM, Kulinski JM, Corbett JA, Tarakanova VL. 2012. Coordinate regulation of DNA damage and type I interferon responses imposes an antiviral state that attenuates mouse gammaherpesvirus type 68 replication in primary macrophages. J. Virol. 86:6899–6912. 10.1128/JVI.07119-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mounce BC, Tsan FC, Droit L, Kohler S, Cirillo LA, Tarakanova VL. 2011. Gammaherpesvirus gene expression and DNA synthesis are facilitated by viral protein kinase and histone variant H2AX. Virology 420:73–81. 10.1016/j.virol.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarakanova VL, Molleston JM, Goodwin M, Virgin HW., IV 2010. MHV68 complement regulatory protein facilitates MHV68 replication in primary macrophages in a complement independent manner. Virology 405:50–61. 10.1016/j.virol.2010.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood BM, Mboko WP, Mounce BC, Tarakanova VL. 2013. Mouse gammaherpesvirus-68 infection acts as a rheostat to set the level of type I interferon signaling in primary macrophages. Virology 443:123–133. 10.1016/j.virol.2013.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, Song MJ, Sanchez DJ, O'Connell RM, Cheng G, Sun R. 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5:166–178. 10.1016/j.chom.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solca C, Tint GS, Patel SB. 2013. Dietary xenosterols lead to infertility and loss of abdominal adipose tissue in sterolin-deficient mice. J. Lipid Res. 54:397–409. 10.1194/jlr.M031476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dzeletovic S, Breuer O, Lund E, Diczfalusy U. 1995. Determination of cholesterol oxidation products in human plasma by isotope dilution-mass spectrometry. Anal. Biochem. 225:73–80. 10.1006/abio.1995.1110 [DOI] [PubMed] [Google Scholar]
- 36.Kanazawa N, Kurosaki M, Sakamoto N, Enomoto N, Itsui Y, Yamashiro T, Tanabe Y, Maekawa S, Nakagawa M, Chen CH, Kakinuma S, Oshima S, Nakamura T, Kato T, Wakita T, Watanabe M. 2004. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J. Virol. 78:9713–9720. 10.1128/JVI.78.18.9713-9720.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw G, Morse S, Ararat M, Graham FL. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869–871. 10.1096/fj.01-0995fje [DOI] [PubMed] [Google Scholar]
- 38.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. 2012. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 109:4239–4244. 10.1073/pnas.1114981109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. 1988. Related expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54:903–913. 10.1016/S0092-8674(88)91307-4 [DOI] [PubMed] [Google Scholar]
- 40.Fujita T, Kimura Y, Miyamoto M, Barsoumian EL, Taniguchi T. 1989. Induction of endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature 337:270–272. 10.1038/337270a0 [DOI] [PubMed] [Google Scholar]
- 41.Pamment J, Ramsay E, Kelleher M, Dornan D, Ball KL. 2002. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene 21:7776–7785. 10.1038/sj.onc.1205981 [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Pavlova, Virgin HW, IV, Speck SH. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029–2037. 10.1128/JVI.74.4.2029-2037.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukac DM, Renne R, Kirshner JR, Ganem D. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304–312. 10.1006/viro.1998.9486 [DOI] [PubMed] [Google Scholar]
- 44.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu FX, Miller G. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866–10871. 10.1073/pnas.95.18.10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu TT, Usherwood EJ, Stewart JP, Nash AA, Sun R. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659–3667. 10.1128/JVI.74.8.3659-3667.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu TT, Tong L, Rickabaugh T, Speck S, Sun R. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262–9273. 10.1128/JVI.75.19.9262-9273.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375–386. 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 48.Barton ES, Lutzke ML, Rochford R, Virgin HW., IV 2005. Alpha/beta interferons regulate murine gammaherpesvirus latent gene expression and reactivation from latency. J. Virol. 79:14149–14160. 10.1128/JVI.79.22.14149-14160.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279:52772–52780. 10.1074/jbc.M410302200 [DOI] [PubMed] [Google Scholar]
- 50.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. 2007. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 5:73–79. 10.1016/j.cmet.2006.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383:728–731. 10.1038/383728a0 [DOI] [PubMed] [Google Scholar]
- 52.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, Knochenmuss R, Noel S, Gessier F, Kelly LM, Vanek M, Laurent S, Preuss I, Miault C, Christen I, Karuna R, Li W, Koo DI, Suply T, Schmedt C, Peters EC, Falchetto R, Katopodis A, Spanka C, Roy MO, Detheux M, Chen YA, Schultz PG, Cho CY, Seuwen K, Cyster JG, Sailer AW. 2011. Oxysterols direct immune cell migration via EBI2. Nature 475:524–527. 10.1038/nature10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maloney NS, Thackray LB, Goel G, Hwang S, Duan E, Vachharajani P, Xavier R, Virgin HW. 2012. Essential cell-autonomous role for interferon (IFN) regulatory factor 1 in IFN-gamma-mediated inhibition of norovirus replication in macrophages. J. Virol. 86:12655–12664. 10.1128/JVI.01564-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trilling M, Le VT, Zimmermann A, Ludwig H, Pfeffer K, Sutter G, Smith GL, Hengel H. 2009. Gamma interferon-induced interferon regulatory factor 1-dependent antiviral response inhibits vaccinia virus replication in mouse but not human fibroblasts. J. Virol. 83:3684–3695. 10.1128/JVI.02042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW., IV 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447:326–329. 10.1038/nature05762 [DOI] [PubMed] [Google Scholar]
- 56.Mandal P, Krueger BE, Oldenburg D, Andry KA, Beard RS, White DW, Barton ES. 2011. A gammaherpesvirus cooperates with interferon-alpha/beta-induced IRF2 to halt viral replication, control reactivation, and minimize host lethality. PLoS Pathog. 7:e1002371. 10.1371/journal.ppat.1002371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park K, Scott AL. 2010. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88:1081–1087. 10.1189/jlb.0610318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. 2013. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and Toll-like receptor signaling. Immunity 39:454–469. 10.1016/j.immuni.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, Mitsuyama M, Shin EH, Kojima S, Taniguchi T, Asano Y. 1997. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6:673–679. 10.1016/S1074-7613(00)80443-4 [DOI] [PubMed] [Google Scholar]
- 60.Yi T, Wang X, Kelly LM, An J, Xu Y, Sailer AW, Gustafsson JA, Russell DW, Cyster JG. 2012. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity 37:535–548. 10.1016/j.immuni.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulinski JM, Leonardo SM, Mounce BC, Malherbe LP, Gauld SB, Tarakanova VL. 2012. Ataxia-telangiectasia mutated kinase controls chronic gammaherpesvirus infection. J. Virol. 86:12826–12837. 10.1128/JVI.00917-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarakanova VL, Stanitsa E, Leonardo SM, Bigley TM, Gauld SB. 2010. Conserved gammaherpesvirus kinase and histone variant H2AX facilitate gammaherpesvirus latency in vivo. Virology 405:50–61. 10.1016/j.virol.2010.05.027 [DOI] [PubMed] [Google Scholar]