ABSTRACT
Hypoxia-inducible factor 1α (HIF-1α) has been frequently implicated in many cancers as well as viral pathogenesis. Kaposi's sarcoma-associated herpesvirus (KSHV) is linked to several human malignancies. It can stabilize HIF-1α during latent infection and undergoes lytic replication in response to hypoxic stress. However, the mechanism by which KSHV controls its latent and lytic life cycle through the deregulation of HIF-1α is not fully understood. Our previous studies showed that the hypoxia-sensitive chromatin remodeler KAP1 was targeted by the KSHV-encoded latency-associated nuclear antigen (LANA) to repress expression of the major lytic replication and transcriptional activator (RTA). Here we further report that an RNA interference-based knockdown of KAP1 in KSHV-infected primary effusion lymphoma (PEL) cells disrupted viral episome stability and abrogated sub-G1/G1 arrest of the cell cycle while increasing the efficiency of KSHV lytic reactivation by hypoxia or using the chemical 12-O-tetradecanoylphorbol-13-acetate (TPA) or sodium butyrate (NaB). Moreover, KSHV genome-wide screening revealed that four hypoxia-responsive clusters have a high concurrence of both RBP-Jκ and HIF-1α binding sites (RBS+HRE) within the same gene promoter and are tightly associated with KAP1. Inhibition of KAP1 greatly enhanced the association of RBP-Jκ with the HIF-1α complex for driving RTA expression not only in normoxia but also in hypoxia. These results suggest that both KAP1 and the concurrence of RBS+HRE within the RTA promoter are essential for KSHV latency and hypoxia-induced lytic reactivation.
IMPORTANCE Kaposi's sarcoma-associated herpesvirus (KSHV), a DNA tumor virus, is an etiological agent linked to several human malignancies, including Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL). HIF-1α, a key hypoxia-inducible factor, is frequently elevated in KSHV latently infected tumor cells and contributes to KSHV lytic replication in hypoxia. The molecular mechanisms of how KSHV controls the latent and lytic life cycle through deregulating HIF-1α remain unclear. In this study, we found that inhibition of hypoxia-sensitive chromatin remodeler KAP1 in KSHV-infected PEL cells leads to a loss of viral genome and increases its sensitivity to hypoxic stress, leading to KSHV lytic reactivation. Importantly, we also found that four hypoxia-responsive clusters within the KSHV genome contain a high concurrence of RBP-Jκ (a key cellular regulator involved in Notch signaling) and HIF-1α binding sites. These sites are also tightly associated with KAP1. This discovery implies that KAP1, RBP-Jκ, and HIF-1α play an essential role in KSHV pathogenesis through subtle cross talk which is dependent on the oxygen levels in the infected cells.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is a large DNA tumor virus implicated in the etiology of many human cancers: Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and a subset of multicentric Castleman's disease (MCD) (1, 2). Like all herpesviruses, KSHV establishes a similar paradigm for viral cancers, which results in a productive cycle or latent replication. During latency, the latency-associated nuclear antigen (LANA) encoded by open reading frame 73 (ORF73) is the dominant latent protein, with multiple functions for establishment and maintenance of latent infection (3–5). The majority of the viral genes are silenced, and the viral genome persists in host cells as an episome with a condensed chromatin state. In lytic replication, the replication and transcriptional activator (RTA) encoded by ORF50 is essential for initiation of viral lytic replication (6–9). Reactivation of KSHV from latency leads to increased risk of developing KS and PEL (6, 8). The switch from KSHV latent to lytic replication can be initiated by specific intracellular signals or extracellular stimuli, including hypoxia (10, 11), 12-O-tetradecanoylphorbol-13-acetate (TPA,) and sodium butyrate (NaB) (12); the viral episome gradually relaxes its compact chromatin structure, leading to expression of all viral genes and lytic replication (13, 14). These findings indicate that this transition of KSHV life cycle involves viral chromatin remodeling from the heterochromatin to the euchromatin state.
KAP1 (also known as TRIM28 or TIF1β), initially identified as a universal transcriptional corepressor, is a member of the family of KRAB (Kruppel-associated box) domain-containing zinc finger proteins (13, 15, 16). KAP1 can coordinate the assembly of several chromatin-remodeling proteins, such as histone deacetylase complex (HDAC), the histone methyltransferases (e.g., SETDB1), and HP1, to create an epigenetically stable and heritable heterochromatin (17, 18). However, neither KAP1 nor any of the above-mentioned interacting partners have DNA binding domains (17, 18). Therefore, other protein partners are required for recruitment of KAP1 to the genome. This suggests that the major role of KAP1 may lie outside its documented role in transcriptional regulation. In addition to acting as a transcriptional corepressor, KAP1 is also involved more broadly in various nuclear functions, including DNA damage response and p53-mediated apoptosis (19, 20). Emerging evidence suggests that posttranslational modifications, such as phosphorylation and SUMOylation, are important for KAP1 to condense or relax chromatin (21). Phosphorylation of KAP1 at Ser824 is critical to chromatin relaxation in response to genotoxic stress. While antagonistic to phosphorylation of Ser824, SUMOylation of KAP1 at Lys554, Lys779, and Lys804 generates binding platforms for SETDB1 and HDAC1 to condense chromatin (21). In regard to the role of KAP1 in controlling the chromatin state, it has been demonstrated that KAP1 can function as a chromatin remodeler and is widely targeted by a number of viruses, such as Epstein-Barr virus (EBV), HIV, and other retroviruses, for regulating cellular and viral gene expression (22–25). For KSHV, we and other groups have found that KAP1 plays a role in remodeling KSHV chromatin from latency to lytic replication (14, 26, 27). However, additional transcriptional factors are also important for gene activation of viral lytic replication. Particularly, the transcriptional factors related to KSHV reactivation with chromatin remodeling under hypoxic stress are not well understood.
Hypoxia as a common feature of KS tumors is physiologically linked with the development of KSHV-associated cancers as well as their malignant progression (28). We and others have found that hypoxia-inducible factor 1α (HIF-1α) is aberrantly accumulated in both KSHV latently infected cells and KS patient tissues in normoxia (29, 30). Meanwhile, hypoxic stress in vitro reactivates KSHV lytic replication through the hypoxia-responsive elements (HRE; 5′-RCGTGC-3′) within the RTA gene promoter (10, 11). More recently, we also found that LANA (the major latent antigen) plays a dual role in controlling HIF-1α (a key hypoxia responder) transcriptional activity via LANASIM (the SUMO2-interacting motif of LANA)-mediated KAP1 in both normoxia and hypoxia (26). However, how KAP1 coordinates with HIF-1α to regulate KSHV latency and reactivation in the hypoxic microenvironment remains unclear.
In this study, we further demonstrated that inhibition of KAP1 reduces the copy number of KSHV episomes and abrogates hypoxia-mediated sub-G1/G1 arrest of the cell cycle while facilitating KSHV reactivation induced by hypoxia. Strikingly, genome-wide screening analysis revealed a high concurrence of RBP-Jκ and HIF-1α binding sites on the KSHV genome. Particularly, we found that inhibition of KAP1 dramatically enhanced the association of RBP-Jκ with HIF-1α-containing complexes at the RTA promoter for regulating gene transcription. This report describes the first mechanism by which two main cellular transcription factors, RBP-Jκ and HIF-1α, can coordinate with KAP1 to remodel viral chromatin to regulate KSHV latent and lytic replication.
MATERIALS AND METHODS
Antibodies.
KAP1 (20C1) antibodies were purchased from Abcam (Cambridge, MA). HIF-1α antibodies were from BD Transduction Laboratory (San Jose, CA). Sin3A (AK-11) and PARP1 (F2) were purchased from Santa Cruz Biotech. Inc. (Santa Cruz, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; G8140-01) antibodies were from United States Biological Inc. (Swampascott, MA). Mouse monoclonal antibodies against LANA, RTA, and RBP-Jκ (BWH39) were used as described previously (31).
Cell culture and hypoxic incubation.
KSHV-positive (BC3 and BCBL1) B lymphoma cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS; HyClone). HEK293 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS (HyClone). Cells were grown in a humidified atmosphere at 37°C and gas tensions of 21% O2–5% CO2 for normoxic incubation and 1% O2–5% CO2 for hypoxic incubation as described previously (32).
Stable RNAi-expressing cell line production and transduction.
The KAP1 short hairpin RNA (shRNA) sequence (5′-GCATGAACCCCTTGTGCTG-3′), Sin3A shRNA sequence (5′-CAACTGCTGAGAAGGTTGATTCTGT-3′), and control sequence (5′-TGCGTTGCTAGTACCAAC-3′; nontargeting sequence) were individually inserted into the pGIPz vector according to the manufacturer's instructions (Clonetech). The pGIPz vector containing shRNA sequence was cotransfected with lentivrus packaging plasmids (Rev, vesicular stomatitis virus G protein [VSVG], and gp) into CoreT cells by the calcium phosphate method to generate virus. The packaged viruses were used to individually transduce target cells (BC3 and BCBL1) and selected using 2 μg/ml of puromycin. The RNA interference (RNAi) efficiency was assessed by Western blot analysis with specific KAP1 or Sin3A antibodies.
Flow cytometry of cell cycle.
Cells were harvested, washed in ice-cold phosphate-buffered saline (PBS), and fixed in cold methanol-acetone (50/50). Cells were stained with PBS containing 40 μg/ml of propidium iodide (PI), 200 μg/ml of RNase A (Sigma), and 0.05% Triton X-100 for 1 h at room temperature in the dark. Cell cycle profiles of stained cells were analyzed using FACScan (BD Biosciences, Foster, CA) and FlowJo software.
Quantitative PCR.
Total RNA from cells was extracted using TRIzol, and cDNA was made with a Superscript II reverse transcription kit (Invitrogen, Inc., Carlsbad, CA). The primers used for real-time PCR are shown in Table 1. The cDNA was amplified using 10 μl of master mix from the DyNAmo SYBR green quantitative real-time PCR kit (MJ Research, Inc.), a 1 μM concentration of each primer, and 2 μl of the cDNA product in a 20-μl total volume. Thirty cycles of 1 min at 94°C, 30 s at 55°C, and 40 s at 72°C were followed by 10 min at 72°C in a Step-One thermocycler (Applied Biosystems Inc.). A melting-curve analysis was performed to verify the specificities of the amplified products. The values for the relative levels of change were calculated by the threshold cycle (ΔΔCT) method, and samples were tested in triplicates.
TABLE 1.
Primers used for qPCR

Extraction and quantitation of KSHV episome DNA.
Total DNA was extracted by lysis buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM EDTA, 1% SDS), followed by proteinase K digestion. Relative numbers of KSHV episomal copies were calculated by quantitative PCR (qPCR) amplification of the terminal repeats (TR) or K8 gene primers as previously described (33). For intracellular episome DNA, GAPDH content in each sample was first amplified as an internal control to normalize DNA input for TR amplification. For extracellular viral DNA, the viral particles were obtained by following the protocol for the virion purification. Bac36 DNA was used as a standard control.
KSHV virion purification and primary infection.
The equal input amount of PEL cells was subjected to induction for KSHV reactivation. After induction, the supernatant of culture medium was collected and filtered through a 0.45-μm filter, and viral particles were spun down at 25,000 rpm for 2 h at 4°C. The concentrated virus was collected and used for infection or viral DNA quantitation. For primary infection, 2 × 106 peripheral blood mononuclear cells (PBMCs) were incubated with virus suspension in 1 ml of RPMI 1640 (10% FBS) medium in the presence of 5 μg/ml of Polybrene (Sigma, Marlborough, MA) and incubated for 4 h at 37°C. Cells were centrifuged for 5 min at 1,500 rpm, the supernatant was discarded, and pelleted cells were resuspended in fresh RPMI 1640 (10% FBS) medium in 6-well plates and cultured with 5% CO2 in a 37°C humidified incubator. The infection was checked by visualization of LANA staining using fluorescence microscopy.
Immunofluorescence and immunoblotting.
Immunofluorescence and immunoblotting assays were performed as described previously (10). Briefly, for immunofluorescence, cells were washed with ice-cold PBS, incubated on polylysine-treated coverslips for 20 min, and then fixed in 3% paraformaldehyde for 20 min at room temperature. After fixation, cells were washed three times in PBS and permeabilized in PBS containing 0.2% fish skin gelatin (G-7765; Sigma, Chicago, IL) and 0.2% Triton X-100 for 5 min, followed by primary and secondary antibody staining. DNA was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and coverslips were mounted with p-phenylenediamine. Cells were visualized with a Fluoview FV300 (Olympus Inc., Melville, NY) confocal microscope.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were done as previously described (34, 35), with some modifications. Cells (3 × 108) were cross-linked with 1.1% (vol/vol) formaldehyde, 100 mM NaCl, 0.5 mM EGTA, and 50 mM Tris-HCl (pH 8.0) in growth medium at 37°C for 10 min and then at 4°C for 50 min. Formaldehyde was quenched by adding 0.05 volume of 2.5 M glycine. Fixed cells were washed with PBS and incubated for 15 min in 15 ml of 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5 mM EGTA, and 0.25% (vol/vol) Triton X-100, followed by 15 min in 15 ml of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, and 200 mM NaCl, and finally sonicated in 1 ml of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% (wt/vol) SDS plus 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 μg/ml of aprotinin, leupeptin, and pepstatin to an average fragment size of 300 to 500 bp. Solubilized chromatin extracts were clarified by centrifugation at 12,000 × g and diluted to 6 optical density units (optical density at 260 nm [OD260])/ml in IP buffer (140 mM NaCl, 1% [wt/vol] Triton X-100, 0.1% [wt/vol] sodium deoxycholate, 1 mM PMSF, 100 μg/ml of salmon sperm DNA, and 100 μg/ml of bovine serum albumin [BSA]), preincubated for 1 h at 4°C with 10 μl/ml of 50% (vol/vol) protein A-agarose with normal mouse or rabbit serum, reconstituted in PBS, and washed several times in IP buffer. Aliquots (600 μl) were incubated with 20 μg of each specific antibody or IgG control overnight at 4°C. Immune complexes were separated into bound and unbound complexes with protein A-agarose, and cross-links were reversed by treatment at 65°C overnight. After treatment with RNase A and proteinase K, samples were extracted once with phenol-chloroform, and the DNA was precipitated with 2 volumes of ethanol plus 10 μg of glycogen as a carrier (Roche). Precipitated DNA was pelleted, washed once with 70% ethanol, dried, and resuspended in 100 μl of water. The ChIP DNA and 10% input were amplified by qPCR using specific primers with amplicons of 150 to 200 bp.
Preparation of nuclear extracts.
Cells were resuspended in 4 times the volume of the pellet in NE buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2 with protease inhibitors) after a cold PBS wash, incubated on ice for 1 h, and then transferred to a prechilled Dounce homogenizer and homogenized with 25 strokes. Homogenized samples were transferred to Eppendorf tubes and spun at 2,000 rpm for 5 min at 4°C. The supernatant was aspirated and the pellet resuspended in 2 times the volume of NE buffer B (20 mM HEPES [pH 7.9], 10% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA with protease inhibitors), followed by incubation on ice for 30 min and centrifugation at 13,000 rpm for 20 min at 4°C. The supernatants were transferred to fresh Eppendorf tubes, and an equal volume of NE buffer C (20 mM HEPES [pH 7.9], 30% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA with protease inhibitors) was added. This was then aliquoted, and the samples were snap-frozen at −80°C before use.
In vitro DNA binding affinity assay.
Two complementary 5′-biotinylated oligonucleotides with or without the HIF-1α binding site (wild type, 5′-GGATTCCAAACGTGCCCAGCGGT-3′; mutant, 5′-GGATTCCAATTAATCCCAGCGGT-3′; underline indicates HIF-1α binding site) were annealed to be double-stranded DNA and coupled to streptavidin-conjugated agarose beads. Per sample, 3 μg of biotinylated oligonucleotide was incubated with 40 μl of 50% streptavidin-conjugated agarose bead slurry in a total volume of 100 μl of a lysis buffer comprised of 50 mM Tris-HCl (pH 8.0), 15 mM NaCl, 0.1 mM EDTA, 10% glycerol, 10 mM N-ethylmaleimide (NEM), 1 mM PMSF, 1 mM dithiothreitol (DTT), 1 μg/ml of aprotinin, 1 μg/ml of leupeptin, and 1 μg/ml of pepstatin for 2 h at 4°C. Cell nuclear extracts (400 μg) were incubated with 40 μl of DNA-coupled agarose beads in lysis buffer at a total volume of 500 μl for 3 h at 4°C. The precipitated complexes were washed three times with lysis buffer. Purified DNA-binding proteins were boiled in SDS sample buffer and analyzed by SDS-PAGE and immunoblotting. A 5% input amount of biotinylated DNA oligonucleotide was verified by dot blotting.
Dual-luciferase reporter assay.
Firefly luciferase reporter plasmid pRta-luc with HRE or RBS+HRE was used to detect the effect of KAP1 knockdown on RTA gene expression in normoxia and hypoxia. The luciferase reporter assays were performed as described previously (36). Thymidine kinase (TK) promoter-driven Renilla luciferase was used as a control to normalize the transfection efficiency.
Statistical analysis.
Statistical significance of differences between means of at least 3 experiments was determined using Student's t test (P values indicated accordingly in figure legends or text below). Error bars represent positive and negative standard errors of the means (SEM).
RESULTS
Establishment of PEL cells with constitutive knockdown of KAP1.
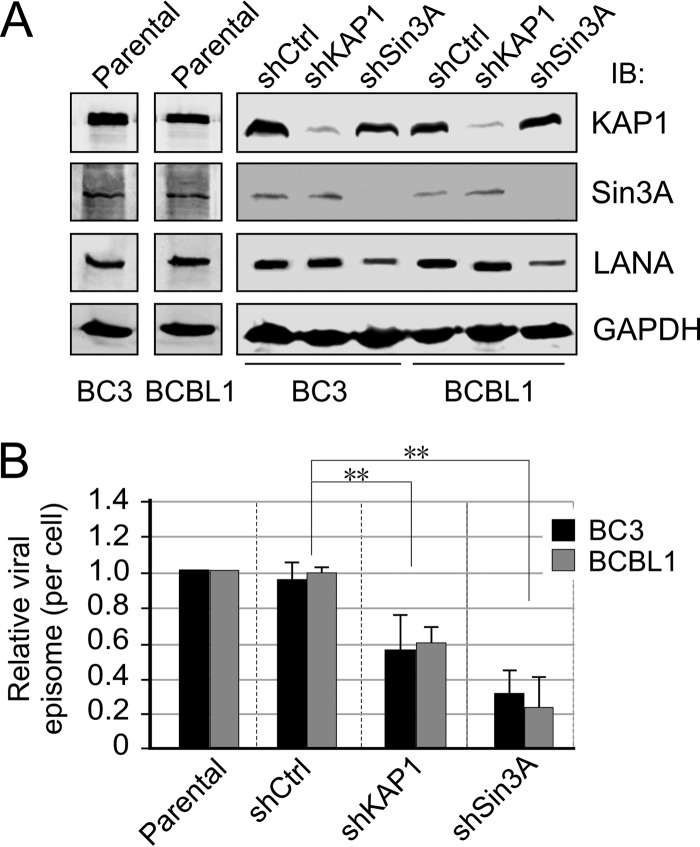
Previous studies have shown that LANA associated with KAP1 and Sin3A through its SUMO2-interacting motif (LANASIM) to maintain KSHV latency (26). To further determine if inhibition of KAP1 or Sin3A has effects similar to those of the LANASIM deletion on the long-term maintenance of the KSHV episome during latency, we individually generated 3 clones each of KSHV-infected PEL cell lines BC3 and BCBL1 with constitutive knockdown of KAP1 or Sin3A or a nonspecific control by using lentivirus carrying shRNA. The results of immunoblotting analyses showed that there was a high efficiency of KAP1 or Sin3A knockdown in both BC3 and BCBL1 cells (Fig. 1A). Interestingly, we observed that inhibition of Sin3A but not KAP1 caused an approximately 60% reduction of LANA expression in both PEL cell lines compared with the parental or knockdown control (Fig. 1A).
FIG 1.
Establishment of PEL stable cell lines with KAP1 or Sin3A knockdown. (A) Constitutive knockdown of KAP1 and Sin3A in PEL cells. PEL cell lines BC3 and BCBL1, parental and with KAP1 (shKAP1) or Sin3A (shSin3A) knockdown or scramble control (shCtrl), were individually generated by lentivirus-mediated transduction followed by selection with 2 μg/ml of selection. IB, immunoblotting. (B) Lower KSHV episome copy number in PEL cells with KAP1 or Sin3A constitutive knockdown. The genome DNA extracted from PEL cell lines BC3 and BCBL1 with shKAP1 or shSin3A or scramble control shRNA (shCtrl) from panel A were used to detect the relative intracellular viral episome copy number by quantitative PCR with TR as a target. GAPDH was used as an internal control. The results are presented as the average relative fold compared with parental cells from 3 independent knockdown clones from panel A. Double asterisks indicate a P value less than 0.05.
Inhibition of KAP1 reduced KSHV episomal copy number but facilitated lytic reactivation induced by TPA-NaB or hypoxia.
To investigate whether the inhibition of KAP1 or Sin3A disrupts KSHV episome stability, we checked the copy number of KSHV episomal DNAs in these KAP1 or Sin3A knocked-down cell lines by quantitative PCR analysis. The results showed that both KAP1 and Sin3A knockdown markedly reduced the viral episome copy number compared with parental or knockdown control (Fig. 1B). In view of the fact that both KAP1 and Sin3A are critical chromatin silencers for KSHV latency, we wanted to determine if the reduced expression of KAP1 or Sin3A would enhance the relative efficiency of viral reactivation induced by hypoxia, although constitutive knockdown of KAP1 or Sin3A resulted in a lower viral episomal DNA copy number in the parental PEL cells. To do so, we exposed KAP1 or Sin3A knockdown cell lines to hypoxia at different time points (0, 18, and 36 h), followed by quantitation of progeny virion production. The chemical stimuli TPA and NaB were used as a positive control. The results revealed that KAP1 or Sin3A knockdown in PEL cells consistently led to a relatively higher response to hypoxic stress within 36 h of exposure compared to parental or control knockdown (Fig. 2A, right side; similar results were obtained with BCBL1 knockdown cells). Intriguingly, a similar pattern was observed in the KAP1 or Sin3A knockdown cells in response to TPA-NaB treatment, although there was a slightly higher response in the TPA-NaB group than that in the hypoxia group after 18 h treatment (Fig. 2A; similar results were obtained for the BCBL1 knockdown cells).
FIG 2.
Loss of KAP1 or Sin3A dramatically enhances the relative efficiency of KSHV virion induction by TPA-NaB or hypoxia. (A) Equal amounts of parental BC3 cells with constitutive knockdown of KAP1 (shKAP1) or Sin3A (shSin3A) or scramble control (shCtrl) were cultured and exposed to TPA and butyrate (NaB) or 1% oxygen treatment for 0, 18, and 36 h. The supernatant of culture medium was subjected to quantitative PCR with TR or K8 as a target for virion production. The efficiency of extracellular KSHV DNA (virion) induction by TPA-NaB or hypoxia was detected and is presented as relative fold compared with mock treatment before induction. (B) A representative image of human PBMC infection with 36-h hypoxia induction virion from panel A at day 4 postinfection by LANA and DAPI nuclear staining.
To determine if the higher response to stimuli leads to more infectious virion production of KAP1 or Sin3A knockdown cells, we performed PBMC primary infection in vitro using purified virions from the same amount of parentally derived PEL cells induced for 36 h. The results consistently showed that about 87.5% of the PBMCs were infected by virions from the KAP1 knockdown cells, compared to 42% and 40.1% from parental and the control knockdown cells, respectively (Fig. 2B). In contrast, although Sin3A knockdown cells showed a relatively higher sensitivity to lytic replication stimuli, there was much less production of infectious virions (17.3%) due to a smaller amount of viral episomes retained. This indicates that KAP1 is involved in both TPA-NaB- and hypoxia-mediated signaling pathways.
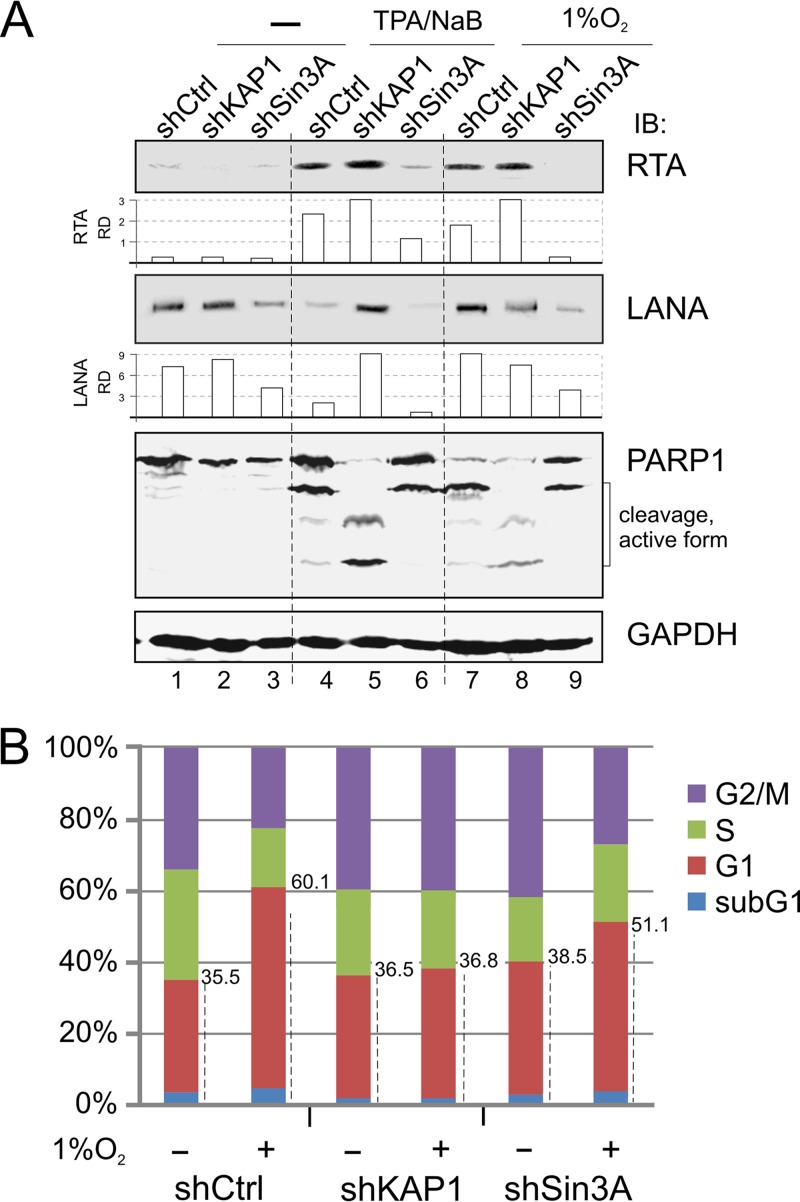
To further address the effects of KAP1 and Sin3A inhibition on TPA-NaB- and hypoxia-mediated lytic reactivation, we determined the protein levels of LANA and RTA expression by Western blot analysis. Unexpectedly, we observed that TPA-NaB treatment led to enhanced RTA expression along with reduced LANA expression, whereas hypoxia treatment not only enhanced RTA expression but also moderately enhanced LANA expression (Fig. 3A, compare lanes 1, 4, and 7). In contrast, KAP1 knockdown efficiently blocked these effects on LANA levels induced by TPA-NaB or hypoxia, while Sin3A knockdown showed similarly less LANA expression and almost undetectable levels for RTA after TPA-NaB or hypoxia treatment (Fig. 3A, compare lanes 4 and 7 with 5 and 8). Meanwhile, in monitoring the apoptotic marker PARP1, the results showed more cleavage to active forms of PARP1 in the KAP1 knockdown cells than that seen in control cells, as well as the Sin3A knockdown cells treated with TPA-NaB or hypoxia (Fig. 3A, compare lanes 5 and 8 with lanes 4 and 6 plus lanes 7 and 9). This further suggests that less KAP1 may result in more sensitivity to extracellular stresses during TPA-NaB and hypoxia treatment.
FIG 3.
Inhibition of KAP1 or Sin3A alters sensitivity of PEL cells to different extracellular stress. (A) Inhibition of KAP1 or Sin3A alters the sensitivity of PEL cells in response to lytic replication stimuli. BC3 cells with constitutively knocked down KAP1 or Sin3A were individually subjected to treatment with hypoxia (1% O2) or TPA-NaB (20 ng/ml, 1.5 mM) for 36 h. Cell lysates were subjected to immunoblotting as indicated. The relative density (RD) of RTA and LANA was quantified and is shown in the middle. (B) BC3 cells with constitutive KAP1 or Sin3A knockdown were individually subjected to treatment with or without 1% hypoxia overnight, followed by analysis of cell cycle profile. The average percentage of different phases (sub-G1, G1, S, G2/M) from three repeats is presented in a histogram.
To determine if KAP1 knockdown in PEL cells causes a less apoptotic response than Sin3A or nonspecific knockdown, we determined the cell cycle profile of those knockdown PEL cells with or without hypoxia treatment. Consistently, the data in Fig. 3B showed that the sub-G1/G1 population of KAP1 knockdown cells after treatment (36.5% to 36.8%) presented a much lower response to hypoxia than that seen with Sin3A (38.5% to 51.1%) or nonspecific control (35.5% to 60.1%) knockdown cells. The combined evidence above showed that KAP1, collaborating with Sin3A, is critical for silencing replication of the KSHV genome and is a key sensor involved in both TPA-NaB- and hypoxia-mediated signaling pathways.
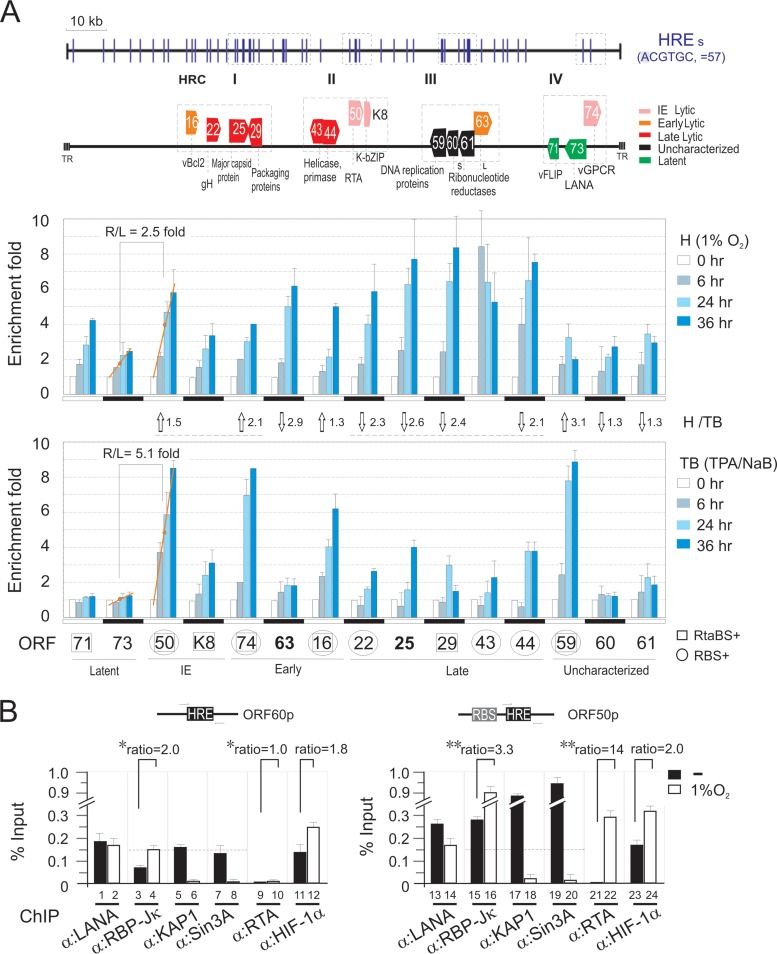
High concurrence of RBP-Jκ and HIF-1α binding sites within KSHV promoters associated with KAP1 and Sin3A.
Previous studies have shown that the transcription factor RBP-Jκ is targeted by KSHV for regulation of LANA and RTA expression and is important for induction of the switch from latency to lytic replication upon treatment with TPA-NaB (37–39). Interestingly, inhibition of KAP1 enhances KSHV reactivation not only in hypoxia but also upon TPA-NaB treatment. Therefore, we looked at the DNA binding sites of RBP-Jκ, HIF-1α, KAP1, and Sin3A, along with RTA and LANA, across the KSHV genome in normoxia and hypoxia by chromatin immunoprecipitation (ChIP) assays. Strikingly, the results showed that the major HIF-1α binding sites across the whole KSHV genome (about 46%) showed a high degree of overlap with KAP1 and Sin3A in the presence of RBP-Jκ and LANA in normoxia (Fig. 4A and B). However, the association of KAP1 and Sin3A within the KSHV genome was significantly decreased in hypoxia (Fig. 4A and B). No dramatic change in the protein levels of both KAP1 and Sin3A were observed in normoxia and hypoxia in our previous study (26). Interestingly, analysis of the major HIF-1α binding sites under hypoxic stress identified four clusters, HRC I, II, III, and IV, with a relative higher degree of the conserved HRE (ACGTGC) sequence within these KSHV genomic regions (compare Fig. 4A with Fig. 5A, bottom). Further, there were several important latent and lytic genes, including those for RTA and LANA, located in HRC II and IV, respectively (Fig. 5A, top). To further confirm whether the viral genes within these four HRC regions respond to hypoxic stress, the transcriptional levels of each gene in PEL cells with 1% oxygen treatment at different time points (0, 6, 24, and 36 h) were determined by quantitative PCR. The TPA-NaB-treated cells were used as a parallel control. We did observe that most of the 15 genes were gradually enhanced with hypoxic treatment within a 36-h period (Fig. 5A, middle). Consistent with the results in Fig. 3A, we saw that the expression of LANA was greatly induced by hypoxia but not TPA-NaB within 36 h of treatment. In contrast, the transcription levels of RTA after induction were relatively higher (2.5-fold by hypoxia and 5.1-fold by TPA-NaB) than those of LANA (Fig. 5A, middle and bottom). Interestingly, immediate early and early viral genes (ORF50 and ORF74) were higher in response to TPA-NaB than hypoxia, while late viral genes (ORF22, ORF25, ORF29, and ORF44) in general showed an opposite response (Fig. 5A, middle and bottom). In addition, upon TPA-NaB induction, we also observed that the gene promoters of ORF50, ORF74, ORF16, and ORF59, containing both the RBP-Jκ and RTA binding sites (RBS+RtaBS) along with HRE, presented consistently higher transcriptional levels after induction (Fig. 5A, bottom). This suggested that the transcriptional factors RBP-Jκ and HIF-1α are targeted by KAP1 for gene silencing and argues in support of a scenario in which expression of KSHV-associated hypoxia-responsive genes is dependent on the transcriptional activity of RTA during hypoxia. By analyzing the ORF50 and ORF60 gene promoters, which individually contained both the HRE-RBS and HRE alone, the results of BC3 ChIP assays with or without hypoxia treatment with antibodies against KAP1, Sin3A, RBP-Jκ, HIF-1α, RTA, or LANA revealed that the concurrence of RBS with HRE at the same promoter not only enhanced KAP1 and Sin3A association with the HRE gene promoters in normoxia (Fig. 5B, compare lanes 5 and 7 with lanes 17 and 19) but also increased the association of RTA and RBP-Jκ in hypoxia 14-fold and 3.3-fold, respectively (Fig. 5B, compare lanes 15 and 21 with lanes 16 and 22). In contrast to the HRE promoter alone, the association of RTA and RBP-Jκ in hypoxia increased only 2.0-fold and 1-fold, respectively (Fig. 5B, compare lanes 3 and 9 with lanes 4 and 10).
FIG 4.
Genome-wide screening of LANA, KAP1, Sin3A, HIF-1α, RTA, and RBP-Jκ binding sites within the KSHV genome. (A) (ChIP) assay was done by real-time PCR using a genome-wide array of 100 pair primers spaced across the KSHV genome using specific antibody as indicated. The enrichment of specific antibody bound to DNA was calculated by compared with input DNA and verified by an affinity more than 2-fold higher than for the nonspecific IgG parallel control. Graphs represent means ± standard deviations of two independent experiments. Chromatin DNA were prepared from KSHV-positive cell lines BC3 and BCBL1 with 21% (normoxia) or 1% (hypoxia) oxygen treatment for overnight before harvest. The results reveal four hypoxia-responsive clusters (HRC I, II, III, and IV) within the KSHV genome based on the HIF-1α–DNA binding region during hypoxia and indicated in light green. The main genes within these four clusters are shown in Fig. 5A. (B) Summary of KSHV whole-genome peak analysis for LANA, KAP1, Sin3A, HIF-1α, RBP-Jκ, and RTA binding chromatin from BC3 and BCBL1 cells under normoxic and hypoxic conditions. The amount of each protein binding site on KSHV genome is summarized as fold more than 2 from panel A, in parentheses.
FIG 5.
RBP-Jκ is involved in hypoxia-induced transcription of KSHV lytic genes. (A) Quantitative PCR of transcriptional levels of HRE-containing viral genes from BC3 cells induction by hypoxia or TPA-NaB at different time points. The viral genes within the four hypoxia-responsive clusters (HRC) are presented at the top. The gene promoters containing HRE with an RTA (RtaBS) or RBP-Jκ (RBS) binding site are individually indicated by squares and circles. The relative ratio of each gene stimulated by hypoxia (H) and TPA-NaB (TB) was calculated as average value of induction at three time points. R/L, ratio of RTA with LANA. (B) ChIP assay of the HRE or HRE+RBS (RBP-Jκ binding site)-containing promoter. Chromatin DNA prepared from KSHV-positive cell line BC3 with 21% (−) or 1% oxygen (O2) treatment overnight before harvest was subjected to ChIP assays by antibodies as indicated. The DNA binding level of each protein was detected by qPCR, and the results are presented as percentages compared with input. Asterisks indicate P values as follows: *, P > 0.05, and **, P < 0.01.
Inhibition of KAP1 enhanced the association of RBP-Jκ with HIF-1α in both normoxia and hypoxia.
To investigate whether RBP-Jκ is involved or is a component of the LANASIM-mediated HIF-1α complex, including KAP1 in hypoxia, we determined the levels of endogenous RBP-Jκ associated with a HIF-1α binding site oligonucleotide (HRE) in vitro in PEL cells with or without KAP1 knockdown under normoxic and hypoxic conditions. The results showed that the level of RBP-Jκ associated with HIF-1α was greatly enhanced in hypoxia, with a 4.5-fold increase for nonspecific knockdown control and a 6-fold increase for KAP1 knockdown (Fig. 6A). Strikingly, as shown in Fig. 6B, the results from luciferase reporter assays of the RTA promoter containing HRE and RBS (RBP-Jκ binding site) or HRE alone in the presence or absence of KAP1 suggested that in addition to enhancing RBP-Jκ-binding activity, the RBP-Jκ binding sites along with HRE further enhanced the transcriptional activity of the RTA promoter in hypoxia, from 2- to 6.2-fold in the knockdown control group (Fig. 6B, compare lanes 1 and 2 with lanes 5 and 6) and from 2.5- to 3.1-fold in the KAP1 knockdown group (Fig. 6B, compare lanes 3 and 4 with lanes 7 and 8). We also observed that the presence of RBS did reduce the transcriptional level of the RTA promoter in the nonspecific control instead of the KAP1 knockdown group under the normoxic condition (Fig. 6B, compare lanes 1 and 3 with lanes 5 and 7). These data support the hypothesis that the concurrence of RBS with HRE could be targeted by KSHV for selectively regulating specific gene promoters with HRE during normoxia and/or hypoxia.
FIG 6.
The transcription activities of gene promoters containing HREs in hypoxia are impaired by both KAP1 and RBP-Jκ. (A) Inhibition of KAP1 enhances the association of RBP-Jκ with HIF complex in hypoxia. In vitro DNA-binding assays of RBP-Jκ were performed under normoxic and hypoxic conditions. Nuclear cell extracts from BC3/shCtrl or BC3/shKAP1 with 21% or 1% oxygen treatment overnight were individually incubated with wild-type (Wt) or mutant (Mut) HIF-1α-binding DNA oligonucleotide followed by three washes, and the precipitations were Western blotted with RBP-Jκ antibodies. The relative density (RD) of RBP-Jκ binding affinity is presented. (B) Inhibition of KAP1 dramatically enhances the transcriptional level of the RTA promoter with a RBP-Jκ binding site in hypoxia. 293 cells were transfected with the luciferase reporter of the RTA promoter (pRta-Luc) with RBS and HRE or HRE alone in the presence or absence of KAP1 knockdown (shKAP1). At 36 h posttransfection, cells were subjected to treatment with or without 1% oxygen for 12 h before harvest. The results are shown as the RLU (relative luciferase unit) fold induction of pRta-Luc compared with pGL-3 vector alone. Data are presented as means ± standard deviations of three independent experiments. (C) Percentages of HREs and RBP-Jκ binding sites (RBS) within the same gene promoter of total HREs in host cellular and KSHV genomes. The data are summarized from Table 2. The sequence logo of conserved RBS used for analysis is shown at the bottom.
To further answer whether there is a significant difference in the concurrence rates of RBS and HREs within the same gene promoters in the host cellular and KSHV genome in order for the virus to selectively regulate cellular and viral genes, we statistically analyzed the conserved RBS and HRE sites of each gene promoter length within 500 bp across both the host cellular and KSHV genomes with a stringent P value cutoff. Intriguingly, the results showed that the concurrence rate of RBS and HRE was significantly higher than that seen in the human genome (46.8% versus 34.1% as shown in Fig. 6C; raw data in Table 2). This indicated that RBP-Jκ could be a second subtle regulator along with KAP1 which is targeted by KSHV to differentiate between its own and the host cellular gene promoters containing HRE in response to the accumulated HIF-1α in normoxia and hypoxia.
TABLE 2.
Analysis of HRE and RBS within the gene promoter from host cellular and KSHV (GenBank accession number NC_009333) genome sequencea
| Genome parameter | Host cellular sequence | KSHV sequence |
|---|---|---|
| Promoter length (bp) | 500 | 500 |
| Conservation P value cutoff | 0.05 | 0.05 |
| TFBS scan P value cutoff | 1.00E−03 | 1.00E−03 |
| Total gene no. | 34,306 | 86 |
| No. of genes with RBS (TTCCCAC) | 5,394 | 7 |
| No. of genes with HRE (ACGTGC) | 9,236 | 14 |
| No. of genes with both RBS and HRE (ACGTGC) | 3,053 | 6 |
| Concurrence P value | <1.70E−308 | 4.02E−05 |
| No. of genes with HRE (GCGTGC) | 6,941 | 13 |
| No. of genes with both RBS and HRE (GCGTGC) | 2,508 | 7 |
| Concurrence P value | <1.70E−308 | 3.19E−07 |
| No. of genes with HRE (both HRE) | 10,851 | 16 |
| No. of genes with both RBS and HRE (both HRE) | 3,587 | 7 |
| Concurrence P value | <1.70E−308 | 2.13E−06 |
Conservation P value is obtained by a permutation test. It means that the probability of getting a sequence with the conservation randomly is less than 0.05. TFBS scan P value shows the probability of getting a sequence with the same similarity between the detected TFBS sequence and those in the motif database randomly is less than 0.001. Concurrence P value is obtained by hypergeometric test. It indicates the significance of the concurrence between the genes with HRE and those with RBS. For cross-species conservation, the human host cellular genome is compared with that of the squirrel monkey, while KSHV is compared with saimirine herpesvirus 2.
DISCUSSION
Increasing evidence suggests that KAP1, a member of the tripartite motif (TRIM) protein family which displays antiviral properties in addition to its documented transcriptional corepressor function, is also involved in various biological functions (including complex function with HDACs leading to reversible acetylation of nonhistone proteins [such as p53 and E2F1]) and mediates inhibition of viral integration (40). The different molecular mechanisms of KAP1 restriction parallel its dual activities at the cellular level: as a transcription regulator functioning as a corepressor and as a regulator of protein activity through acetylation. Hence, the absence of KAP1 in transcriptional repression will disfavor a hypothetical role for KAP1 in viral persistent infection. Consistent with the fact that KAP1 and Sin3A are chromatin silencers, our finding showing that inhibition of KAP1 or Sin3A in PEL cells led to high sensitivity to hypoxic stress in terms of viral reactivation further supports this hypothesis.
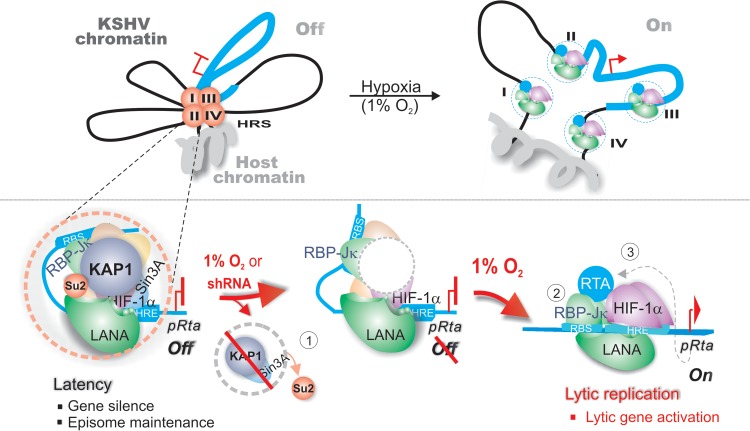
In regard to KSHV, it is well known that latency is the dominant form of KSHV persistent infection associated with host cell transformation. However, reactivation from latency is also important for the growth and dissemination of virus-associated tumor cells (41). KSHV reactivation by TPA-NaB has been extensively documented by directly or indirectly targeting transcriptional activation of the promoter regulating the lytic master regulator RTA (ORF50) (42). However, it is not fully understood how hypoxia induces reactivation from KSHV latency. To address this question, we and other groups have found that hypoxia not only induces the key latent antigen LANA encoded by KSHV but also cooperates with HIF-1α bound to HREs within the RTA promoter (10, 43) to directly induce lytic gene expression (i.e., ORF34-37) for lytic replication (44). Interestingly, we and other groups have found that HIF-1α is stabilized and accumulated in both PEL and KS cells in normoxia (29, 36). To further address how KSHV controls its own latent infection with HIF-1α accumulation while it induces lytic replication during hypoxia, we recently demonstrated that LANA can target the hypoxia-sensitive chromatin remodeler KAP1 through its SUMO2-interacting motif (LANASIM) (26). Deletion of LANASIM motif not only disrupts maintenance of the KSHV episome but also turns on lytic gene expression (26). However, the detailed mechanism of KAP1 involvement in the hypoxic response still remains unclear. In this report, we show that KAP1 knockdown not only led to a dramatic loss of KSHV episome copies but also enhanced the efficiency of reactivation in response to hypoxia. Importantly, we also reveal that there are at least four hypoxia-responsive sites within the KSHV genome and that the concurrence of the DNA binding site of both RBP-Jκ and HIF-1α (which is the downstream transcriptional effector of TPA-NaB and hypoxia stimuli) within the same gene promoter is high and tightly associated with KAP1. Thus, our studies indicate that RBP-Jκ is a second subtle regulator along with KAP1 which is targeted by KSHV for selectively silencing hypoxia-responsive viral and cellular gene expression in normoxia. RBP-Jκ coordinates with HIF-1α to initiate expression of HRE-containing lytic genes, including that for RTA in the absence of the inhibitory function of KAP1 during hypoxia. This production of RTA therefore functions as a third factor to further enhance its own expression and, directly or indirectly, that of other RTA-dependent genes (Fig. 7).
FIG 7.
Schematic illustrating the role of both KAP1 and RBP-Jκ in hypoxia-induced KSHV reactivation. There are at least four hypoxia-responsive clusters (HRC I, II, III, and IV) within KSHV chromatin. During latent infection, to efficiently silence HRE-responsive gene expression and maintain the viral episome, LANASIM-mediated chromatin silencer KAP1 targets the gene promoter of each HRC region with a high concurrence of RBP-Jκ binding site and HRE to inhibit viral gene expression (i.e., RTA) in normoxia. In contrast, each HRC region of KSHV chromatin in hypoxia (1% O2) undergoes three steps to reactivate lytic replication. In step 1, loss of SUMO-modified KAP1 induced by hypoxia (confirmed by shRNA-mediated inhibition of KAP1) remodels the chromatin structure of RBP-Jκ and HIF-1α binding sites; in step 2, the formation of RBP-Jκ–LANA–HIF-1α active complex in hypoxia greatly induces RTA expression; and in step 3, the expressed RTA will feed back to further globally activate KSHV lytic replication to produce virion progeny.
RBP-Jκ is a key responder to TPA-NaB stimulation for KSHV lytic replication, and TPA-NaB-induced lytic protein vPK (ORF36) can block the repressive activity of KAP1 (14). Our discovery further supports the notion that KAP1 is a repressor for KSHV lytic reactivation in response not only to hypoxia but also to TPA-NaB. However, in contrast, hypoxia but not TPA-NaB treatment greatly enhanced the expression of LANA, which is consistent with a recent report showing that hypoxia enhanced LANA expression (43). In addition, the role of KAP1 in response to extracellular stress like hypoxia or TPA-NaB treatment is not unique. Other groups and our current studies have shown that KAP1 responds to cells treated by serum starvation (data not shown) as well as DNA-damaging agents to arrest the cell cycle and apoptosis, respectively (45). These results indicate that KAP1 is a global transcriptional corepressor of KSHV lytic gene promoters through association with two key DNA-binding proteins, RBP-Jκ and HIF-1α, and can play a critical role in maintaining viral latency. A recent report showing that the KSHV-encoded latent protein kaposin B also deregulates KAP1-mediated inhibition of STAT3 further demonstrates an important role for KAP1 in KSHV latency infection (27).
How cellular signaling pathways cross talk is a rapidly emerging field in many branches of biology. The recent elucidation of the Notch-hypoxia intersection indicates the existence of cross talk between hypoxia and other key signaling (including Notch) mechanisms (46) and that interactions of two signaling pathways can occur at multiple steps in the signaling cascade (46). RBP-Jκ (also called CSL) is the major downstream transcription factor of the Notch signaling pathway and is a sequence-specific DNA binding factor which recruits corepressor or coactivator complexes to its cis-regulatory binding elements within the target gene promoters (47). It has recently been reported that RBP-Jκ deficiency triggers viral replication but attenuates reactivation by TPA-NaB for virion production in KSHV-infected B cells. This indicates that RBP-Jκ is required for both latency maintenance and lytic reactivation (48). In addition, RTA as the key master regulator of lytic replication has been demonstrated to either directly bind to DNA or use other DNA binding factors like RBP-Jκ as DNA adaptors. Consistent with previous reports (49–51), our studies also showed that there are a total of about 18 RTA binding sites within KSHV genome, which are tightly located in KSHV genes, including PAN RNAs, ORF16, ORF29, ORF45, ORF50, ORFK8, ORF59, ORF71/72, ORFK14/ORF74, and ORFK15. Also, most of the viral transcripts that encode structural proteins are expressed following transcripts for the replication proteins. Intriguingly, a KSHV nucleotide sequence search with well-known HRE and RBS sequences from cellular genes found 58 individual matches for HIF-1α and 43 for RBP-Jκ within the KSHV genome. Many of them are located at regions containing known or speculated lytic gene promoters (52). For example, ORF57, ORF59, and ORF47 activation by RTA is dependent on RBP-Jκ binding (38, 53). Since RBP-Jκ appears to be a signaling molecule controlling cellular survival and the viral life cycle of KSHV-infected cells, it is not surprising that RBP-Jκ is identified as a critical molecule engaged in response to hypoxic stress. What we observed about collaboration of HIF-1α and RBP-Jκ in KSHV suggests a cross talk between HIFα and Notch signaling occurring at the intersection between the oxygen-sensing and developmental pathways. Notch has been reported to potentiate HIF-1α-mediated transactivation of hypoxia-inducible genes in the context of both tumorigenesis and cellular differentiation (46, 54). Conversely, HIF-1α has also shown to enhance Notch signaling through several distinct mechanisms (46, 54–56). HIF-1α has been shown to stabilize Notch ICD in both normoxia and hypoxia in a manner independent of its transcriptional activity (46, 54), although the precise mechanism remains unclear. Meanwhile, HIF-1α interaction with Notch at the HEY2 and HES1 promoters has also been reported, and this association augments Notch-mediated transactivation under hypoxia in a manner that depends upon a functional CTAD but not DNA binding of HIF-1α (55, 56). This supports our observation that RBP-Jκ could bind to HIF-1α, and it requires further investigation.
In addition, to address the physiological relevance of KSHV to selectively activate gene expression via the virus-regulated HIF-1α, we have found that the transcription factor RBP-Jκ is also associated with the LANA-associated inhibitory complex and responds to hypoxic stress. The fact that a higher percentage of RBP-Jκ binding sites concurrently associates with HRE on the KSHV genome (particularly at the RTA promoter) than that seen on the human genome (i.e., VEGF promoter) may explain why KSHV can silence viral lytic gene expression and selectively activates cellular genes through accumulation of HIF-1α in normoxia. However, this accumulation strongly reactivates lytic replication in hypoxia.
In summary, our study provides new mechanistic insights on the programmed regulation of the KSHV genome during latent and lytic replication controlled by the major latent antigen LANA for gene silencing or activation based on KAP1. Particularly, the discovery that the LANASIM-associated KAP1 complex is highly sensitive to hypoxia (a physiologically relevant environmental condition) provides a potential therapeutic target against KSHV-associated cancers.
ACKNOWLEDGMENTS
We are grateful to the members of the Robertson laboratory and the Cai laboratory for critical discussions.
This work was supported by the Research and Innovation Key Project of the Shanghai Municipal Education (grant 13zz011), the Program for New Century Excellent Talents in University (grant NCET-14-0168), and funds from the National Key Basic Research “973” program (grant 2012CB519001) of China to Q.C., funds from the Natural Science Foundation of China (grant 81171649) to Y.G., and public health grants from the NIH (R01CA919972, R01DE017338, R01CA171979, and P01CA174439) to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print 2 April 2014
REFERENCES
- 1.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280 [PubMed] [Google Scholar]
- 2.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- 3.Cai Q, Verma SC, Lu J, Robertson ES. 2010. Molecular biology of Kaposi's sarcoma-associated herpesvirus and related oncogenesis. Adv. Virus Res. 78:87–142. 10.1016/B978-0-12-385032-4.00003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10:707–719. 10.1038/nrc2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma SC, Lan K, Robertson E. 2007. Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312:101–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukac DM, Renne R, Kirshner JR, Ganem D. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304–312. 10.1006/viro.1998.9486 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205–4220. 10.1128/JVI.77.7.4205-4220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866–10871. 10.1073/pnas.95.18.10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, AuCoin DP, Huete AR, Cei SA, Hanson LJ, Pari GS. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79:3479–3487. 10.1128/JVI.79.6.3479-3487.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Q, Lan K, Verma SC, Si H, Lin D, Robertson ES. 2006. Kaposi's sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1 alpha to upregulate RTA expression during hypoxia: latency control under low oxygen conditions. J. Virol. 80:7965–7975. 10.1128/JVI.00689-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis DA, Rinderknecht AS, Zoeteweij JP, Aoki Y, Read-Connole EL, Tosato G, Blauvelt A, Yarchoan R. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244–3250. 10.1182/blood.V97.10.3244 [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Black JB, Goldsmith CS, Browning PJ, Bhalla K, Offermann MK. 1999. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J. Gen. Virol. 80(Part 1):83–90 [DOI] [PubMed] [Google Scholar]
- 13.Iyengar S, Farnham PJ. 2011. KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 286:26267–26276. 10.1074/jbc.R111.252569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 69:5681–5689. 10.1158/0008-5472.CAN-08-4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. 1996. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701–6715 [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., III 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10:2067–2078. 10.1101/gad.10.16.2067 [DOI] [PubMed] [Google Scholar]
- 17.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., III 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19:4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, Peng H, Ivanov AV, Simpson AJ, Longley BJ. 2007. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 67:9954–9962. 10.1158/0008-5472.CAN-07-1478 [DOI] [PubMed] [Google Scholar]
- 20.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8:870–876. 10.1038/ncb1446 [DOI] [PubMed] [Google Scholar]
- 21.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. 2007. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282:36177–36189. 10.1074/jbc.M706912200 [DOI] [PubMed] [Google Scholar]
- 22.Liao G, Huang J, Fixman ED, Hayward SD. 2005. The Epstein-Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J. Virol. 79:245–256. 10.1128/JVI.79.1.245-256.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9:484–495. 10.1016/j.chom.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 24.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- 25.Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. 10.1016/j.cell.2007.07.026 [DOI] [PubMed] [Google Scholar]
- 26.Cai Q, Cai S, Zhu C, Verma SC, Choi J, Robertson ES. 2013. A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog. 9:e1003750. 10.1371/journal.ppat.1003750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King CA. 2013. Kaposi's sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J. Virol. 87:8779–8791. 10.1128/JVI.02976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Q, Cavallin LE, Yan B, Zhu S, Duran EM, Wang H, Hale LP, Dong C, Cesarman E, Mesri EA, Goldschmidt-Clermont PJ. 2009. Antitumorigenesis of antioxidants in a transgenic Rac1 model of Kaposi's sarcoma. Proc. Natl. Acad. Sci. U. S. A. 106:8683–8688. 10.1073/pnas.0812688106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Q, Murakami M, Si H, Robertson ES. 2007. A potential alpha-helix motif in the amino terminus of LANA encoded by Kaposi's sarcoma-associated herpesvirus is critical for nuclear accumulation of HIF-1alpha in normoxia. J. Virol. 81:10413–10423. 10.1128/JVI.00611-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll PA, Kenerson HL, Yeung RS, Lagunoff M. 2006. Latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J. Virol. 80:10802–10812. 10.1128/JVI.00673-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 79:7453–7465. 10.1128/JVI.79.12.7453-7465.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Q, Verma SC, Kumar P, Ma M, Robertson ES. 2010. Hypoxia inactivates the VHL tumor suppressor through PIASy-mediated SUMO modification. PLoS One 5:e9720. 10.1371/journal.pone.0009720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Si H, Verma SC, Lampson MA, Cai Q, Robertson ES. 2008. Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation. J. Virol. 82:6734–6746. 10.1128/JVI.00342-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei F, Zaprazna K, Wang J, Atchison ML. 2009. PU.1 can recruit BCL6 to DNA to repress gene expression in germinal center B cells. Mol. Cell. Biol. 29:4612–4622. 10.1128/MCB.00234-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Y, Srinivasan L, Perkins L, Atchison ML. 2005. Protein acetylation regulates both PU.1 transactivation and Ig kappa 3′ enhancer activity. J. Immunol. 175:5160–5169 [DOI] [PubMed] [Google Scholar]
- 36.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. 10.1371/journal.ppat.0020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, Verma SC, Cai Q, Saha A, Dzeng RK, Robertson ES. 2012. The RBP-Jkappa binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation. PLoS Pathog. 8:e1002479. 10.1371/journal.ppat.1002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977–1989. 10.1101/gad.996502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan K, Kuppers DA, Robertson ES. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468–3478. 10.1128/JVI.79.6.3468-3478.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F. 2009. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat. Cell Biol. 11:580–591. 10.1038/ncb1864 [DOI] [PubMed] [Google Scholar]
- 41.Ganem D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273–296. 10.1146/annurev.pathol.1.110304.100133 [DOI] [PubMed] [Google Scholar]
- 42.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veeranna RP, Haque M, Davis DA, Yang M, Yarchoan R. 2012. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induction by hypoxia and hypoxia-inducible factors. J. Virol. 86:1097–1108. 10.1128/JVI.05167-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haque M, Wang V, Davis DA, Zheng ZM, Yarchoan R. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated herpesvirus ORF34-37 gene cluster. J. Virol. 80:7037–7051. 10.1128/JVI.00553-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. 2011. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 12:R78. 10.1186/gb-2011-12-8-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greer SN, Metcalf JL, Wang Y, Ohh M. 2012. The updated biology of hypoxia-inducible factor. EMBO J. 31:2448–2460. 10.1038/emboj.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanigaki K, Honjo T. 2010. Two opposing roles of RBP-J. in Notch signaling. Curr. Top. Dev. Biol. 92:231–252. 10.1016/S0070-2153(10)92007-3 [DOI] [PubMed] [Google Scholar]
- 48.Scholz BA, Harth-Hertle ML, Malterer G, Haas J, Ellwart J, Schulz TF, Kempkes B. 2013. Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines. PLoS Pathog. 9:e1003336. 10.1371/journal.ppat.1003336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Ye F, Xie J, Kuhne K, Gao SJ. 2009. Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290–302. 10.1016/j.virol.2009.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paulose-Murphy M, Ha NK, Xiang C, Chen Y, Gillim L, Yarchoan R, Meltzer P, Bittner M, Trent J, Zeichner S. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843–4853. 10.1128/JVI.75.10.4843-4853.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891–902. 10.1128/JVI.75.2.891-902.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Persson LM, Wilson AC. 2010. Wide-scale use of Notch signaling factor CSL/RBP-Jkappa in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes. J. Virol. 84:1334–1347. 10.1128/JVI.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Cao Y, Liang D, Gao Y, Xia T, Robertson ES, Lan K. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element. Virology 380:264–275. 10.1016/j.virol.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lendahl U, Lee KL, Yang H, Poellinger L. 2009. Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 10:821–832. 10.1038/nrg2665 [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Imanaka N, Griffin JD. 2010. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br. J. Cancer 102:351–360. 10.1038/sj.bjc.6605486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. 2005. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9:617–628. 10.1016/j.devcel.2005.09.010 [DOI] [PubMed] [Google Scholar]