ABSTRACT
The genetic variation in HIV-1 in patients is due to the high rate of viral replication, the high viral load, and the errors made during viral replication. Some of the mutations in reverse transcriptase (RT) that alter the deoxynucleoside triphosphate (dNTP)-binding pocket, including those that confer resistance to nucleoside/nucleotide analogs, affect dNTP selection during replication. The effects of mutations in RT on the spectrum (nature, position, and frequency) of errors made in vivo are poorly understood. We previously determined the mutation rate and the frequency of different types of mutations and identified hot spots for mutations in a lacZα (the α complementing region of lacZ) reporter gene carried by an HIV-1 vector that replicates using wild-type RT. We show here that four mutations (Y115F, M184V, M184I, and Q151M) in the dNTP-binding pocket of RT that had relatively small effects on the overall HIV-1 mutation rate (less than 3-fold compared to the wild type) significantly increased mutations at some specific positions in the lacZα reporter gene. We also show that changes in a sequence that flanks the reporter gene can affect the mutations that arise in the reporter. These data show that changes either in HIV-1 RT or in the sequence of the nucleic acid template can affect the spectrum of mutations made during viral replication. This could, by implication, affect the generation of drug-resistant mutants and immunological-escape mutants in patients.
IMPORTANCE RT is the viral enzyme that converts the RNA genome of HIV into DNA. Errors made during replication allow the virus to escape from the host's immune system and to develop resistance to the available anti-HIV drugs. We show that four different mutations in RT which are known to be associated with resistance to anti-RT drugs modestly increased the overall frequency of errors made during viral replication. However, the increased errors were not uniformly distributed; the additional errors occurred at a small number of positions (hot spots). Moreover, some of the RT mutations preferentially affected the nature of the errors that were made (some RT mutations caused an increase in insertion and deletion errors; others caused an increase in substitution errors). We also show that sequence changes in a region adjacent to a target gene can affect the errors made within the target gene.
INTRODUCTION
Although most HIV infections appear to be initiated by a single virus, enough mutations occur within a few years of infection to generate a swarm of related viruses with differing genomic sequences. These include immunological-escape mutants and drug-resistant mutants that arise during viral replication. Understanding the generation of mutations is critical to understanding both the course of the disease and the development of resistance to antiretroviral therapy. As pointed out by Coffin (1), there are multiple factors that contribute to the rapid genesis of mutant forms of HIV-1. Among the most important are the large population size and the rapid replication of the virus. However, all of the mutant viruses that are found in patients arise from errors made during viral replication. There are three polymerases that play a role in viral replication and can contribute to the mutations found in HIV: the replicative host DNA polymerase, host RNA polymerase II (Pol II), and the viral enzyme reverse transcriptase (RT). There are good reasons to believe that the fidelity of the human replicative DNA polymerase, together with its accompanying editing machinery, is significantly higher than that of either Pol II or RT (2, 3). HIV replicates rapidly in untreated patients. Most of the infected cells die before they can divide, and only Pol II and RT make a significant contribution to the errors made during viral replication in patients. Although all of the mutations that arise during HIV replication are commonly attributed to RT, the relative contributions of RT and Pol II to the overall error rate are not well defined. In the viral life cycle, the flow of genetic information is from the product of Pol II (RNA) to the product of RT (DNA). Theoretically, RT cannot be selected to have an error rate lower than that of Pol II. Therefore, RT probably makes at least half of the errors that arise during viral replication and it is possible that the fraction of the errors that are made by RT is considerably larger. If mutations in RT affect the nature of the errors and/or the rate at which the errors are made during viral DNA synthesis, these differences could affect the generation, and emergence, of immunological-escape mutants and drug-resistant mutants.
Attempts have been made to understand how mutations arise during HIV replication using purified RT. There are several problems with using this approach. First, it overlooks the contributions made by Pol II. Second, the mutation rates and the specific errors reported for wild-type (WT) and mutant forms of RT based on in vitro fidelity assays vary considerably in experiments done in different laboratories and, in some cases, in different experiments done in the same laboratory (discussed in more detail in a later section). Third, the fidelity of RT measured in vitro is approximately 20-fold lower than the fidelity of HIV replication measured using a viral vector carrying a reporter gene (4). Even if RT is responsible for the vast majority of the errors made during viral replication, its fidelity cannot be lower than the overall fidelity of viral replication. Finally, when the same target sequence was used for in vitro RT fidelity experiments and for viral replication fidelity experiments, the characteristics and sites at which mutations arose in the target gene did not match (see Results and Discussion).
For those reasons, we have used an HIV vector carrying the α complementing region of lacZ (lacZα) as the target to determine the nature and the frequency of the errors made during HIV replication (4). The vector is similar to lacZα-containing spleen necrosis virus (SNV) (5, 6) and HIV (7, 8) vectors developed by others. We previously described the errors that arose in the LacZα coding region in a single round of replication of an HIV-1 vector that replicates using WT RT. The mutations were divided into four classes: class 1, single nucleotide substitutions; class 2, single nucleotide frameshifts; class 3, multiple nucleotide substitutions; and class 4, insertions/deletions (indels). Class 1 single nucleotide substitution (missense) mutations were the most frequently detected, comprising 75% of the total. Approximately 85% of these missense mutations were transitions, with G-to-A mutations detected twice as often as C-to-T mutations. The most common transversions were T-to-A and C-to-A transversions (6.2% and 4.3%, respectively). These results suggested that RT and/or Pol II tends to insert an A in place of other nucleotides. There was a strong preference for missense mutations to arise at specific sites (hot spots). Most of the missense mutations were not associated with homopolymeric nucleotide runs and thus could not be explained by a misalignment/slippage mechanism. In contrast, the data obtained with the WT RT vector suggested that frameshift mutations do involve misalignment/slippage and that the majority of the single nucleotide frameshift errors made during replication of the WT and the mutant vectors were insertions rather than deletions.
We report here the effects of a set of four mutations (Y115F, M184I, M184V, and Q151M) in the polymerase active site (see Fig. 1) that alter the susceptibility of HIV-1 RT to nucleoside analogs (nucleoside RT inhibitors [NRTIs]) on the nature, frequency, and position of mutations that arise during viral replication. The majority of the errors made during HIV replication are missense mutations, which involve a failure in the ability of RT to appropriately discriminate between the correct incoming deoxynucleoside triphosphate (dNTP) and one of the other three dNTPs. It is possible that mutations in the polymerase active site that affect drug resistance, some of which are associated with a change in the ability of RT to discriminate between NRTI-TPs and normal dNTPs, also affect fidelity. Although we cannot say with confidence that any specific mutation in the lacZα reporter was made by RT or Pol II, any statistically significant change in the overall numbers of mutations, types of mutations, or sites at which the mutations arise (comparing the results obtained with the RT mutants to the WT results) can be attributed to the change in RT.
FIG 1.
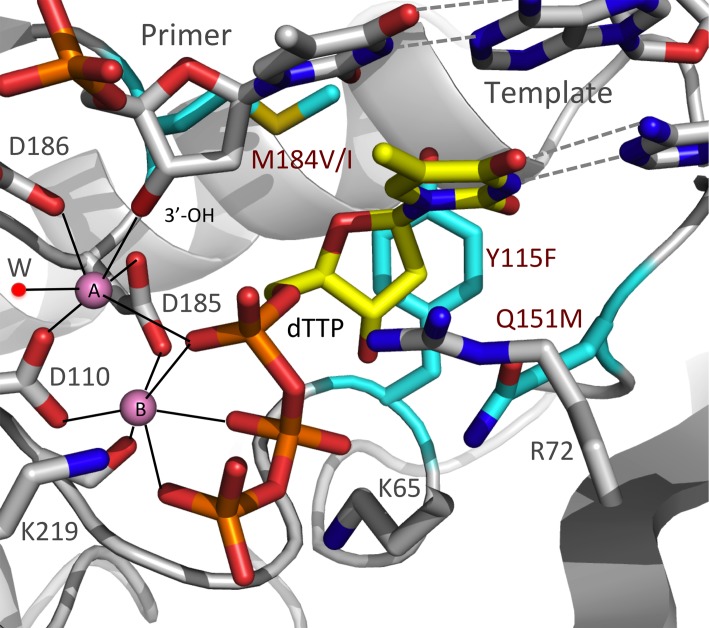
A model representing the binding of dTTP at the polymerase active site of RT prior to nucleotide incorporation. The model was built using structural information from structures 1RTD (20), 2IAJ (77), and 3V4I (78). The three amino acid residues that were mutated in the analysis, Y115, Q151, and M184, are shown in cyan. These residues form part of the polymerase active site that surrounds the bound dTTP (yellow). The coordination environments of the two catalytic Mg2+ ions (pink) are represented by dotted lines; a coordinating water molecule is labeled “W”.
In contrast to previous reports, we found that each of the RT mutants we tested caused a modest but statistically significant decrease in the fidelity of replication. The largest overall effect was seen with the Y115F mutation, which increased the overall error rate of viral replication by 2.6-fold. However, the mutations in RT had a much greater effect on the errors made at certain positions in the target gene than on the overall error rate. Y115F and M184I increased the frequency of specific missense mutations. Y115F and Q151M increased the frequency of specific single nucleotide frameshift mutations, most of which were insertions. The data show that relatively subtle changes in RT, for example, the loss of an OH group in the Y115F mutant or the small difference in M184I compared to M184V, can affect which mutations preferentially arise at specific sites. The data show that simply measuring the overall error rate is not a good way to monitor the impact of a mutation in RT on the errors made during HIV replication. Mutations in RT that have a relatively modest effect on the overall error rate can still have a significant impact on the mutations that arise at specific sites in a target gene and, by implication, on the generation of immunological-escape mutants and/or drug resistance mutations. We also found that sequence changes outside the essential LacZα coding region affected the sites at which missense (but not frameshift) mutations occur. This shows that the template sequence, and perhaps its structure, can affect the mutations that arise during viral replication.
MATERIALS AND METHODS
Plasmid construction.
We previously described a 4-vector system that can be used to generate virus stocks for the analysis of HIV-1 fidelity (4). This system is based on the use of four plasmids: pMDL-SH.IN-, pRSV-REV, pCMV-VSV-G, and pSICO-LZF. Both the pRSV-REV and pMDLg/pRRE viral vectors were obtained from Didier Trono (EPFL SV-DO, Lausanne, Switzerland [9]) through Addgene Inc. (Cambridge, MA). The vector used to produce viral RNA, pSICO-LZF, was derived from pSICO-XBX (10) by the introduction of a shuttle cassette (as a NotI-MluI fragment) containing the lacZα reporter gene in the forward orientation (lacZα-F). The pMDL-SH.IN- vector, which encodes HIV-1 Gag and Pol and contains a rev-response element (RRE), was derived from pMDLg/pRRE by replacing the gag and pol genes with the equivalent sequences from pHIV1-SH (11) and by the introduction of an active site mutation (D116N) in integrase (IN-) using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), according to manufacturer's instructions. Mutations were introduced into the pMDL-SH.IN- vector to produce viral vectors that had the following amino acid substitutions in RT: Y115F, Q151M, M184I, and M184V.
The shuttle cassette previously used to measure the fidelity of an HIV-1 vector that replicated using WT RT (4) contained a zeocin resistance gene with an upstream EM-7 promoter (EM-ZeoR), a 477-nucleotide (nt) region containing the lacZα sequence inserted in the same orientation as the viral genes (lacZα-F), and a ColE1 origin of replication (oriE). The 477-nt region comprised a regulatory region (108 nt), the first 5 codons of lacZα (15 nt), a multiple-cloning-site/polylinker region (180 nt), and a segment encoding the remaining 58 amino acids of LacZα (174 nt). We defined the total length of the mutational target of lacZα as 174 nt, beginning after the polylinker region (position 1) and continuing to the first TAA termination codon (position 174). Using this system, we showed that most mutations occurred within the first 120 nt of lacZα, consistent with the historical report that amino acids 3 to 41 of LacZ are the minimal required region for α-complementation (12). To eliminate extraneous sequences for the experiments reported here, we used overlap extension PCR to delete the polylinker region (180 nt). We then inserted 2 codons (6 nt) in place of the polylinker to align the first 5 codons of lacZα (15 nt) with the remaining 58 codons (174 nt), which generated a full-length 65-codon (195-nt) lacZα sequence consistent with that used by others (13). We also removed 42 nt (codons 52 to 65) from the 3′end of lacZα, reducing the overall size of the reporter from 195 to 153 nt (codons 1 to 51). In reporting the results obtained with the new pSICO-LZF vector, we have used a numbering convention for lacZα different from that used in our previous report but identical to that reported by previous researchers (8, 13, 14). For comparison purposes, nucleotide positions 22 to 150 of our newly modified lacZα sequence (a 129-nt region) match positions 4 to 132 of our previously published lacZα sequence (4) (see Fig. S1 in the supplemental material).
Cells, transfection, and infection.
HEK (human embryonic kidney) 293T and HOS (human osteosarcoma) cells were maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 5% (vol/vol) fetal bovine serum, 5% (vol/vol) newborn calf serum, 100 μg/ml penicillin G, and 100 μg/ml streptomycin (Quality Biological, Gaithersburg, MD). Virus stocks were generated by calcium phosphate-mediated cotransfection of 293T cells, seeded at 9 × 105 cells in 100-mm-diameter culture plates, with 10 μg pMDL-SH IN-, 5 μg pRSV-REV, 4 μg pCMV-VSV-G, and 15 μg pSICO-LZF. At 6 h posttransfection, vector DNA was thoroughly washed from the culture plates with phosphate-buffered saline (PBS) supplemented with 1% (vol/vol) fetal bovine serum. Virus-containing culture supernatants were harvested at 48 h posttransfection, clarified by filtration through Steriflip 0.22-μm-pore-size filter units (Millipore, Billerica, MA), and stored at −80°C until use. The amount of recombinant virus was determined using HIV-1 p24 antigen enzyme-linked immunosorbent assay kits (PerkinElmer, Boston, MA). On the day of infection, virus stocks were thawed and treated for 1 h at 37°C with 30 U/ml RNase-free DNase I (Roche, Indianapolis, IN) supplemented with 5 mM MgCl2 to remove residual vector DNA carried over from the transfection. Virus (equivalent to 500 ng of p24) and 8 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO) were then added to 150-mm-diameter culture plates followed by the addition of 2 × 106 HOS cells per plate.
Recovery of unintegrated HIV-lacZα plasmids.
Circularized unintegrated HIV-lacZα plasmids were recovered from infected HOS cells at 48 h postinfection using a modified Hirt extraction procedure, described previously (4). The circular viral DNAs were clonally amplified by electroporating 1 μl of the resuspended DNA directly into DH10B cells that express the ω-complementing segment of β-galactosidase (Invitrogen, Carlsbad, CA). Recipient Escherichia coli cells were subjected to a single 7.5-ms pulse (field strength, 1.5 kV/cm; capacitance, 25 μF; resistance, 186 Ω) using a 1-mm-gap E-Shot Standard electroporation cuvette at room temperature. The transformed E. coli bacteria were allowed to recover for 10 min (this recovery period is too brief to allow the bacteria to divide) prior to plating on 15-cm-diameter low-salt Luria-Bertani (LB) plates with 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 50 μg/ml zeocin, and 250 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (InVivoGen, San Diego, CA). After 24 h of growth at 37°C, the β-galactosidase activity in the E. coli colonies was monitored by color (white to light blue, inactive/reduced activity; blue, fully active). The color of the colonies was determined by visual inspection, and the data (colony counts) were recorded using an ECount electronic colony counter (Heathrow Scientific LLC, Vernon Hills, IL). Individual mutant colonies were inoculated into 96-well blocks containing 1.5 ml LB media containing 50 μg/ml zeocin and were grown for 48 to 72 h at 37°C. Plasmid DNA was extracted using a Biorobot 3000 workstation (Qiagen, Valencia, CA), and the lacZα-coding region was sequenced (Macrogen USA Inc., Rockville, MD).
Classification of mutations and compilation of lacZα mutation spectra.
Sequenced viral DNA products encoding the LacZα protein (153 nt) were compiled by alignment using Sequencher v4.1.4 software (Gene Codes Corporation, Ann Arbor, MI). Pairwise BLAST analysis of each recovered lacZα sequence and the reference wild-type sequence (153 nt) was conducted using the command line BLAST program blastall (http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/blastall/) installed on a computer with a Linux operating system. Only sequences with mutations in the 153-nucleotide region were counted and used in the analysis. A Perl script was written to parse the alignment and sort the lacZα sequences initially into 4 separate classes according to the types of mutations detected: class 1, single nucleotide substitutions; class 2, single nucleotide frameshifts (+1 or −1); class 3, multiple nucleotide substitutions, including spaced singlets and doublets (cutoff of two consecutive bases); and class 4, indel (insertion/deletion) mutations, including deletions, deletions with insertions, duplications, and multiples of single nucleotide substitutions plus frameshifts (cutoff of 3 or more consecutive bases). Mutation spectra were compiled from the sequences in each mutation class to summarize the total numbers, types, and positions of errors detected within the 153-nt lacZα target sequence. Transition-transversion substitutions were tabulated for class 1 and class 3.
Determination of mutation frequencies and rates.
Mutation frequencies were determined for each of the RT mutants by dividing the number of lacZα mutations by the total number of colonies screened. Because the data were obtained with a one-round vector, the rate was calculated as mutations/bp/generation. Mutation rates were determined by dividing mutation frequencies by the length of the lacZα target (153 nt). Because some of the missense mutations that arise in lacZα are silent, this strategy necessarily underestimates the actual mutation rate. A small number of mutations that did not alter the protein sequence (typically in the third base of a codon) were detected. These were invariably accompanied by another mutation that did alter the protein sequence. The fractions of the total represented by each of the four classes of mutations were determined by dividing the number of mutations in a given class by the total number of mutations. These fractions were graphically represented in the form of pie charts in which the overall size of each pie chart is proportional to the overall mutation rate. We counted the mutations using the same rules we had used previously (4): class 1 and 2 mutations were counted as independent mutations, while class 3 and 4 mutations were counted as dependent or single mutations.
Comparative analysis of the positions of errors.
The data were analyzed by log-linear categorical analysis, contingency table analysis, and related methods. The numbers of mutations in the 153-nt sequence of lacZα were detailed in 2-by-153 tables for each pairwise comparison of the lacZα mutational profiles. These tables were further collapsed to include only those sites which contained nonzero counts of mutations into smaller 2-by-n tables for which n was considerably less than 153. The 2-by-n tables were then collapsed into still smaller subtables, following the rules for partitioning of contingency tables, in conjunction with the corresponding partitioning of the likelihood-ratio chi-square statistic (G2) into orthogonal, additive components (15). Positions where frequent mutations (hot spots) occurred were thus identified through the application of rigorous statistical procedures that resulted in independent, nonoverlapping components. Prospective hot spots were then further subjected to global Fisher's exact tests. Compounding type I error rates were controlled through the use of the Benjamini-Hochberg correction. To that end, minimal criterion probability levels of at least 0.05 were obtained. The statistical significance of hot spot differences between the mutation spectra of WT and RT mutants is reported at the 0.05, 0.01, 0.001, and 0.0001 levels of significance.
Comparative analysis of the types of errors within and between mutation classes.
The mutation frequencies of each class of mutation, as generated and detected for each RT mutant, were compared to corresponding wild-type mutation frequencies using a global Fisher's exact test to determine statistical significance. Further detailed analyses included comparisons of ADAR (adenosine deaminases acting on RNA)-dependent versus non-ADAR-dependent mutations in class 3.
RESULTS
Flanking sequences can affect the missense mutations that arise in a target gene carried by an HIV vector.
We previously described a one-round HIV vector that carries the α complementing region of the lacZ gene (lacZα) (4). The vector also carries both a plasmid origin of replication and a selectable marker (zeocin resistance) such that circular forms of the viral DNA can be selected in E. coli. The vector plasmid is cotransfected into 293T cells together with the plasmids that express VSV-G, Gag-Pol (encoding a WT or mutant form of RT), and Rev (see Materials and Methods). The Gag-Pol plasmid has a mutation that inactivates integrase, thus increasing the amounts of the circular forms of the viral DNA that are produced in infected cells. We constructed a version of the Gag-Pol expression plasmid that encodes the WT RT and versions that encode four RT mutants (Y115F, M184I, M184V, and Q151M). Virions were recovered, treated with DNase to reduce the amount of carryover of vector DNA, and used to infect HOS cells. Circular forms of the viral DNA were recovered from infected cells by Hirt fractionation and electroporated into E. coli cells expressing the complementary segment of lacZ (ω). Errors in the lacZα gene were generated either when it was transcribed into viral genomic RNA by the host Pol II or when the viral RNA was converted into DNA by RT. Errors that affected the ability of the lacZα gene to complement the ω segment were detected by growing the E. coli transformants on selective media in the presence of X-gal. Light-blue and white colonies were picked, and the viral DNAs were sequenced. This method detects only mutations that affect LacZα activity, which means that we missed some of the mutations that arose in the experiments. However, it allows us to accurately compare the frequencies at which mutations are generated during the replication of a vector that carries WT RT and of vectors that carry mutant RTs (see Discussion).
In the experiments reported here, we modified the original lacZα vector, removing extraneous sequences adjacent to the essential 153-bp lacZα coding region. This resulted in a change in the numbering system for the sites in lacZα (see Fig. S1 in the supplemental material). Position 1 then corresponded to the first nucleotide of the initiator ATG, and the fidelity data we present here are based on an analysis of a 153-bp region that encodes the portion of LacZα essential for α-complementation (see Materials and Methods). To demonstrate that the changes we made in the vector did not have a profound effect on the errors that arose during HIV replication, we compared the mutations obtained using the new lacZα vector and the old lacZα vector that carries WT RT. The overall distribution of the mutations into the four classes was unaffected. There were no statistically significant differences in three of the four classes of mutations, but there were significant differences in the class 1 mutations.
Although the overall patterns of missense mutations were quite similar for the two vectors, there were three positions (positions 36, 52, and 62) in lacZα where there were significant differences (see Fig. S1 in the supplemental material). At position 36, more T-to-C transitions were obtained using the new lacZα vector, while at positions 52 and 62, more G-to-A transitions were obtained using the old lacZα vector. The old and new lacZα vectors differ over a stretch of 22 nucleotides immediately downstream of the initiator ATG in the new lacZα vector (see Fig. S1 in the supplemental material). Positions 36, 52, and 62 were, respectively, 14, 30, and 40 bp away from the nearest part of the divergent sequence. Pol II uses a double-stranded DNA (dsDNA) substrate, and it is not immediately obvious how its fidelity would be affected by a change in the sequence of the DNA template 40 bp away from the active site. In contrast, RT uses single-stranded RNA as the template for the synthesis of the first strand of viral DNA. The RNA template is highly structured, and, although RNA folding analyses failed to uncover any linkage between class 1 hot spots and the secondary structure of the template RNA, we think it is likely that RT is responsible for the differences in the errors made at positions 14, 30, and 40 that were seen in the assays done with the two vectors (see Discussion).
Mutations in RT affect the frequency and position of errors made in lacZα.
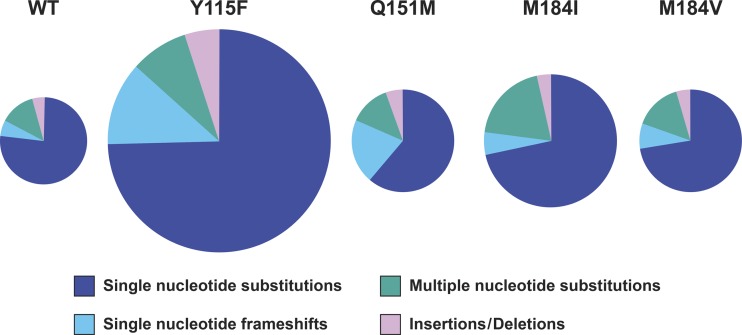
We tested the effects of four mutations in RT (Y115F, Q151M, M184I, and M184V) on the fidelity of HIV-1 replication. The positions of the mutations relative to the RT polymerase active site are shown in Fig. 1. The impact of these RT mutations on the overall mutation rate, and the distribution of mutations into the four classes of mutations, is shown graphically in Fig. 2. The size of each of the pie charts reflects the overall mutation rate in a single cycle of viral replication, while the sections of the pie charts represent the proportions associated with the four mutant classes. Although each of the RT mutations caused a statistically significant increase in the overall number of mutations (Table 1), the greatest increase was only 2.6-fold (Y115F). Some of the RT mutations also affected the distribution of the mutations into the four classes. For example, Q151M caused a significant increase in the proportion of frameshift mutations. More importantly, the mutations in RT also affected the specific sites at which either missense or frameshift mutations occurred in lacZα. In considering the data, several things should be kept in mind. Although we cannot tell whether any given mutation was made by RNA Pol II or by RT, these enzymes use different substrates and are likely to have different mutational hot spots. More importantly, the nature, position, and frequency of mutations made by RNA Pol II remain constant, as shown by comparisons of the mutations that arise in vectors that replicate using WT RT and the mutant RTs. If there is a significant difference in the number of errors at a given position, that difference can be attributed to the effects of the mutation in RT on the fidelity of HIV replication. Finally, although the majority of mutations are deleterious, on average, frameshift mutations are more deleterious than missense mutations. For this reason, there is probably strong selective pressure on the structure and function of DNA polymerases, including RT, to keep the number of frameshift mutations to a minimum.
FIG 2.
Graphical depiction of the effects of the RT mutants on the frequency of lacZα mutations and on their distribution into the four classes that are described in the text. The overall sizes of the pies are proportional to the mutation rates for WT RT and each of the RT mutants; the segments of the pies are proportional to the contribution of each of the four classes to the total mutation rate (see also Table 1).
TABLE 1.
Comparison of RT mutation frequencies versus WT: all classes of mutant lacZα sequencesc
| Enzyme ID | Mutation frequencya | Mutation rateb | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|---|
| WT | 451/205,171 | 1.4 × 10−5 | |||
| Y115F | 524/93,725 | 3.7 × 10−5 | <0.0001 | <0.0001 | **** |
| Q151M | 537/209,064 | 1.7 × 10−5 | 0.0154 | 0.0177 | * |
| M184I | 479/150,110 | 2.1 × 10−5 | <0.0001 | <0.0001 | **** |
| M184V | 466/172,601 | 1.8 × 10−5 | 0.0020 | 0.0027 | ** |
Mutation frequency data represent the number of mutant lacZα sequences divided by the total number of recovered clones.
Mutation rate data represent the mutation frequency divided by the size of the lacZα target sequence. Because the data were obtained with a one-round vector, the rate data were calculated as mutations/bp/generation.
Raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fischer's exact test (FET). To control for compounding type I error in these multiple comparisons, the Benjamini-Hochberg (BH) false-discovery-rate (FDR) corrections to significance calculations were applied to the raw (FET) P values where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less. ID, identifier.
Mutants. (i) Y115F.
Although Y115F is not as strongly associated with drug resistance as the other three RT mutants, it occurs in approximately 10% of patients receiving abacavir (ABC) alone and in 1% of patients treated with combination therapies. It has been reported to be occasionally associated with the K65R, L74V, and M184V mutations (16–18). Y115F causes a reduction in susceptibility to ABC and has a smaller effect on susceptibility to tenofovir disoproxil fumarate (TDF) (16, 19). In WT HIV-1 RT, Y115 forms part of the dNTP-binding pocket (20). The aromatic side chain of Y115 is positioned directly “under” the deoxyribose ring of the incoming dNTP (Fig. 1). The presence of the phenyl ring of tyrosine at this position helps HIV-1 RT discriminate between dNTPs and NTPs by forming a steric gate with Q151 (21). In MLV RT and hepatitis B virus polymerase, there is an F at the equivalent position that plays a similar role (22, 23). The polymerase defects associated with other amino acid substitutions at this position (22, 24, 25) highlight the critical importance of having an amino acid that has a phenyl ring in the side chain at this position. Although the exact mechanism by which Y115F causes ABC resistance is unclear, this mutation is likely to induce small changes in the hydrophobic interactions in the RT active site that lead to changes in the biological and enzymatic behavior of the polymerase (26).
The Y115F mutation has been reported to have a positive effect on both HIV-1 titer (27) and RT polymerase activity (28), but most of the available reports suggest that the Y115F mutation decreases the fidelity of HIV-1 RT. In murine leukemia virus (MLV) and SNV vector-based systems, the converse mutation at the equivalent position (F155Y) did not measurably affect the fidelity of replication (29). In HIV-1 RT-based in vitro-assays, the Y115F mutation has been reported to cause a 1.6-fold decrease in mutation frequency using a lacZα template and a 2.6-fold increase in mutation frequency in a mismatch extension assay (30). Other mutations at position 115, such as Y115A, were reported to cause 4-fold and 2.3-fold increases in the mutation frequency using a lacZα template in an in vitro assay and a viral vector-based assay, respectively (31, 32). Of the RT mutations we tested, the Y115F mutation had the greatest impact on the overall frequency of mutations in lacZα (2.6-fold higher than the WT) (Table 1 and Fig. 2).
At the sites where class 1 missense mutations preferentially occurred, there were two sites (positions 44 and 76) at which there was a highly significant increase and four other sites (positions 32, 95, 83, and 101) where there were moderately significant increases in the frequency of mutations compared to the WT results (Table 2; see also Fig. S2 in the supplemental material). The mutations at positions 44 and 76 were G-to-A and A-to-G transitions, respectively. The same mutations were also seen in the lacZα sequence when it was replicated using WT RT. Most of the additional mutations at positions 32, 83, and 101 were T-to-C transitions, while those at position 95 were C-to-T transitions. There were no missense mutations found at position 95 using a vector that was replicated using WT RT. However, a small number of T-to-A transversions (2 to 4 total) were found at positions 32, 83, and 101 in the lacZα sequence replicated using WT RT. Overall, the Y115F mutation in RT did not simply cause a uniform increase in the number of errors at the sites where WT RT tends to make errors; rather, the Y115F mutation increased the fraction of specific transition mutations at a small number of sites.
TABLE 2.
Sites in LacZα at which the RT mutations caused significant increases in the number of class 1 single nucleotide substitution (missense) mutationsa
| Enzyme ID | Site no. | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|
| Y115F | 32 | 0.0138 | 0.0193 | * |
| Y115F | 44 | 0.0002 | 0.0006 | *** |
| Y115F | 76 | <0.0001 | <0.0001 | **** |
| Y115F | 83 | 0.0125 | 0.0193 | * |
| Y115F | 95 | 0.0097 | 0.0193 | * |
| Y115F | 101 | 0.0165 | 0.0193 | * |
| Q151M | ||||
| M184I | 52 | <0.0001 | <0.0001 | **** |
| M184I | 76 | <0.0001 | 0.0001 | **** |
| M184I | 109 | 0.0057 | 0.0152 | * |
| M184V | 52 | 0.0005 | 0.0045 | ** |
| M184V | 109 | 0.0042 | 0.0187 | * |
Site number data refer to the position in LacZα, and raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fischer's exact test. To control for compounding type I error in these multiple comparisons, Benjamini-Hochberg (BH)/false-discovery-rate (FDR) corrections to significance were applied to each enzyme ID where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less.
The decrease in missense fidelity caused by the Y115F mutation may have been due to increased flexibility in the dNTP-binding pocket. The structure of the RT-template/primer-dNTP ternary complex shows that Y115 contacts the base of the nucleotide at the end of the primer strand on the top and is supported by P137 at the bottom, thus participating in dNTP binding and incorporation (20). This interaction, which involves the OH of Y115, would tend to stabilize the relative positions of Y115 and the end of the primer. The Y115F mutation leads to an increase in local hydrophobicity and structural adaptability. These changes may increase the flexibility of the dNTP-binding pocket, which could potentially increase missense mutations.
We also looked at the class 2 frameshift mutations. In contrast to missense mutations, the majority of the frameshift mutations made during the replication of an HIV vector carrying WT RT were insertions in homopolymeric runs of nucleotides, and the majority of the frameshift mutations were insertions (4). Because it is not possible to be certain where in a run a frameshift mutation occurred, frameshifts are shown at the end of nucleotide runs. There were three sites at which the Y115F mutation caused a moderately significant (position 53) or highly significant (positions 94 and 99) increase in the frequency of frameshift mutations (Table 3; see also Fig. S3 in the supplemental material). At all three positions, the majority of the additional mutations involved single nucleotide insertions: an A in a run of four As (position 53), a C in a run of five Cs (position 84), and a T in a run of three Ts (position 99). These same frameshift mutations were also seen, at a lower frequency, in the lacZα sequences that were replicated using WT RT. Thus, the Y115F mutation did not change the sites at which frameshift errors preferentially arise but rather caused RT to make additional errors at sites where WT RT (or possibly Pol II) makes frameshift errors. The preference for RT to make insertions rather than deletions may be due to the extensive protein-nucleus contacts that involve the template strand near the polymerase active site. The primer strand is more exposed to solvent and may more easily accommodate an unpaired or extrahelical base. Because more insertions were detected in the lacZα sequences that were replicated using the Y115F RT, this RT mutant may be more tolerant of an unpaired base in the primer strand than WT RT.
TABLE 3.
Sites in LacZα at which the RT mutations caused significant increases in the number of class 2 single nucleotide frameshift mutationsa
| Enzyme ID | Site no. | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|
| Y115F | 53 | 0.0064 | 0.0064 | ** |
| Y115F | 94 | <0.0001 | <0.0001 | **** |
| Y115F | 99 | <0.0001 | <0.0001 | **** |
| Q151M | 32 | 0.0005 | 0.0005 | *** |
| Q151M | 94 | <0.0001 | 0.0001 | **** |
| Q151M | 99 | <0.0001 | <0.0001 | **** |
| M184I | ||||
| M184V |
Site number data refer to the position in LacZα, and raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fischer's exact test. To control for compounding type I error in these multiple comparisons, Benjamini-Hochberg (BH)/false-discovery-rate (FDR) corrections to significance were applied to each enzyme ID independently of the others where where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less.
Given that the overall error rate of the WT vector was quite low (1.4 × 10−5), it was surprising that 12% of lacZα DNAs replicated by WT RT contained multiple missense errors (4; see also Fig. 2 and Table 4). Most of these class 3 multiple nucleotide substitutions involved A-to-G transitions (45 of the 55 lacZα DNAs), and a small fraction of the multiple mutations involved G-to-A transitions (4 of 55 lacZα DNAs). Based on an analysis of the sequence context, we previously reported that some or all of the multiple A-to-G mutations made by a vector that replicated using WT RT may have been caused by a host adenosine deaminase that acts on RNA (ADAR) (4). We also looked for multiple G-to-A mutations that matched the preferred sequence context of APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like, 3G) or APOBEC3F. Based on the nature of the mutation(s) and the sequence context, only a small fraction of the multiple mutations could have been caused by an APOBEC protein. This is presumably because the 293T cell line, like the kidney cells from which it is derived, expresses little or no APOBEC (33, 34). A small fraction of the class 3 multiple mutations (6 of 55 lacZα DNAs) could not be explained by the action of an ADAR or an APOBEC host enzyme (Table 5) (see Discussion) (35–37). We found that the Y115F mutation caused a small but statistically significant increase in the total number of number of class 3 mutations (Table 4).
TABLE 4.
Comparison of RT mutation frequencies versus WT: class 3 multiple nucleotide substitution mutations (total)d
| Enzyme ID | Mutation frequencya | Mutation rateb | Mutation fractionc (%) | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|---|---|
| WT | 55/205,171 | 1.8 × 10−6 | 12 | |||
| Y115F | 42/93,725 | 2.9 × 10−6 | 8 | 0.0155 | 0.0414 | * |
| Q151M | 69/209,064 | 2.2 × 10−6 | 13 | 0.2811 | 0.3213 | |
| M184I | 84/150,110 | 3.7 × 10−6 | 18 | <0.0001 | 0.0002 | *** |
| M184V | 68/172,601 | 2.6 × 10−6 | 15 | 0.0371 | 0.0741 |
Mutation frequency data represent the number of mutant lacZα sequences divided by the total number of recovered clones.
Mutation rate data represent the mutation frequency divided by the size of the lacZα target sequence. Because the data were obtained with a one-round vector, the rate data were calculated as mutations/bp/generation.
Mutation fraction data represent the number of class-specific mutant lacZα sequences divided by the total number of mutant lacZα sequences.
Raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fisher's exact test. To control for compounding type I error in these multiple comparisons, the Benjamini-Hochberg (BH)/false-discovery-rate (FDR) corrections to significance were applied to the significant P values where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less.
TABLE 5.
Comparison of RT mutation frequencies versus WT: class 3 multiple nucleotide substitution mutations (non-ADAR/non-APOBEC)d
| Enzyme ID | Mutation frequencya | Mutation rateb | Mutation fractionc (%) | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|---|---|
| WT | 6/205,171 | 1.9 × 10−7 | 1 | |||
| Y115F | 10/93,725 | 7.0 × 10−7 | 2 | 0.0125 | 0.0332 | * |
| Q151M | 5/209,064 | 1.6 × 10−7 | 1 | 0.7724 | 0.7724 | |
| M184I | 17/150,110 | 7.4 × 10−7 | 4 | 0.0026 | 0.0209 | * |
| M184V | 7/172,601 | 2.7 × 10−7 | 2 | 0.5885 | 0.6725 |
Mutation frequency data represent the number of mutant lacZα sequences divided by the total number of recovered clones.
Mutation rate data represent the mutation frequency divided by the size of the lacZα target sequence. Because the data were obtained with a one-round vector, the rate data were calculated as mutations/bp/generation.
Mutation fraction data represent the number of class-specific mutant lacZα sequences divided by the total number of mutant lacZα.
Raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fisher's exact test. To control for compounding type I error in these multiple comparisons, the Benjamini-Hochberg (BH)/false-discovery-rate (FDR) corrections to significance were applied where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less.
A small fraction (4%) of the lacZα mutations made during the replication of a vector that replicated using WT RT were indels (class 4) (Table 6). Nearly all (16 of 18) of these indel mutations involved large deletions (see Discussion). The indel mutations did not have an apparent preference for any specific sequence. The indel mutations generated during the replication of all four of the HIV-1 RT mutants were similar to the indel mutations generated in the lacZα genes that were replicated by WT RT. Y115F was the only HIV-1 RT mutant that showed a statistically significant increase in the total number of class 4 mutations (Table 6).
TABLE 6.
Comparison of RT mutation frequencies versus WT: class 4 indel mutationsd
| Enzyme ID | Mutation frequencya | Mutation rateb | Mutation fractionc (%) | Raw (FET) P value | BH (FDR)-adjusted P value | BH significance |
|---|---|---|---|---|---|---|
| WT | 18/205,171 | 5.7 × 10−7 | 4 | – | – | – |
| Y115F | 26/93,725 | 1.8 × 10−6 | 5 | 0.0002 | 0.0018 | ** |
| Q151M | 29/209,064 | 9.1 × 10−7 | 5 | 0.1447 | 0.5787 | |
| M184I | 17/150,110 | 7.4 × 10−7 | 4 | 0.4954 | 0.7927 | |
| M184V | 21/172,601 | 8.0 × 10−7 | 5 | 0.3370 | 0.6739 |
Mutation frequency data represent the number of mutant lacZα sequences divided by the total number of recovered clones.
Mutation rate data represent the mutation frequency divided by the size of the lacZα target sequence. Because the data were obtained with a one-round vector, the rate data were calculated as mutations/bp/generation.
Mutation fraction data represent the number of class-specific mutant lacZα sequences divided by the total number of mutant lacZα sequences.
Raw (FET) data represent P values of the significance of the results of comparisons to the WT by Fisher's exact test. To control for compounding type I error in these multiple comparisons, the Benjamini-Hochberg (BH)/false-discovery-rate (FDR) corrections to significance were applied where a single asterisk (*) represents 0.05 or less, two asterisks (**) represent 0.01 or less, three asterisks (***) represent 0.001 or less, and four asterisks (****) represent 0.0001 or less.
(ii) M184I and M184V.
M184 is part of the evolutionarily conserved YXDD motif at the polymerase active site. However, the amino acid at the X position, which is M in most retroviral RTs, is V in MLV RT (38). In HIV-1 RT, D185, D186, and D110 form the carboxylate triad that positions the two Mg2+ ions that catalyze DNA polymerization. The M184I and M184V mutations are selected by treatment with either lamivudine (3TC) or emtricitabine (FTC). In patients, M184I usually appears first and is subsequently replaced by M184V (39). In virus growth competition studies in cultured cells, M184V had a less deleterious effect on viral replication than M184I (40). The M184V mutation also had a less deleterious effect on the polymerase activity of RT than M184I (41, 42). Both 3TC and FTC have an oxathiolane ring in place of the normal deoxyribose, and the oxathiolane ring is in the l configuration as opposed to the normal d configuration. When the triphosphate form of either of these analogs is bound to the active site of RT, the l-oxathiolane ring projects toward the β-position of the side chain of residue 184. If methionine is present, the active site is large enough to accommodate either a normal dNTP or 3TCTP/FTCTP. However, the presence of a β-branched amino acid (isoleucine or valine) at position 184 causes steric hindrance that selectively interferes with the incorporation of the analogs (20, 43, 44).
The data in the published reports describing the effects of the M184V and M184I mutations on the fidelity of HIV-1 RT are complex and contradictory. In vitro misincorporation assays suggested that M184V increases RT fidelity by anywhere from 2.4-fold to 17.5-fold (45–48), whereas in similar assays, M184I has been reported to increase the fidelity of RT 2.6-fold (49, 50). On the basis of mismatch extension data, some groups have suggested that the M184V RT has much higher fidelity to the WT (48.6-fold) than M184I (6.1-fold) (51), while others have argued the opposite (52) or proposed that M184V decreases fidelity compared to the WT (3.5-fold) (53). In contrast, in RT-based in vitro assays using lacZα, the M184V and M184I mutations have been reported to cause 1.6-fold and 4-fold increases in fidelity, respectively (49, 50, 54). In a viral vector-based lacZα assay, M184V was reported to cause a very small (1.3-fold) decrease in the overall mutation rate of HIV-1 (55), and the analogous V223M mutation in MLV RT resulted in 1.8-fold increase in the mutation rate (29). However, it has been argued that M184V-M184I mutations do not significantly increase the overall fidelity of HIV-1 replication, based on the observation that the appearance of new drug resistance mutations is not delayed either in vivo or in vitro (56, 57).
We found that the M184V mutation caused a modest but statistically significant 1.3-fold increase in the frequency of mutations that arose in a single cycle of viral replication (Table 1). The distribution of mutations into the four major classes was not greatly affected by the M184V mutation (Fig. 2). When we looked at the sites at which missense mutations preferentially occurred, there were, compared to the WT, significant increases in the numbers of mutations at positions 52 and 109 (Table 2; see also Fig. S2 in the supplemental material). These were A-to-G transitions at position 52 and T-to-C transitions at position 109. In contrast, there were no significant differences in the frameshift errors made in the replication of the M184V mutant relative to the WT (Table 3; see also Fig. S3 in the supplemental material).
The M184I mutation caused a 1.5-fold increase in the overall frequency of mutations. There was a significant increase in missense mutations but no significant increase in frameshift mutations (Fig. 2). Compared to the WT, there were highly significant increases in the number of missense mutations at positions 52 and 76 and a moderate increase at position 109. This suggests that the two mutations at position 184 of HIV-1 RT have similar but not identical impacts on the nature and position of the errors made during viral replication. M184 interacts with the deoxyribose of the incoming dNTP. It seems reasonable to propose that at least some of the errors made at position 52 during the replication of the WT vector are made by RT and that the mutations at M184 make RT less precise, thus enhancing its propensity to make nucleotide substitution errors. However, WT RT did not make any errors at position 109 in the lacZα gene. In comparing the M184V and M184I mutants, it is not clear from the available data why an isoleucine side chain has a greater effect on RT than a valine side chain. However, the negative impact of these substitutions on the biochemical properties of the polymerase of RT parallels the impact on fidelity.
In analyzing the multiple mutations made by M184V, we focused on those that did not appear to have been made either by APOBEC or by ADAR (Table 5). There was a statistically significant increase in the fraction multiple made by M184V that did not appear to be caused by either ADAR or APOBEC. However, both the total number of these mutations and the fraction of the total that they represent are small. M184I also showed a significant increase in the number of replicated vectors that had multiple point mutations in lacZα. We were surprised to see a statistically significant increase in the number of M184I multiple mutations that could be attributed to ADARs. It is possible that some of these “ADAR-like” mutations were made by the mutant HIV-1 RT and not ADAR. Alternatively, it could be a rare statistical anomaly (Table S1 in the supplemental material). As mentioned earlier, neither the M184V mutation nor the M184I mutation caused a statistically significant change in the number of indel mutations.
(iii) Q151M.
Q151 is well conserved in retroviral RTs (38). In HIV-1, the Q151M mutation causes a moderate loss of susceptibility to zidovudine (AZT), didanosine (ddI), stavudine (d4T), and ABC and has a smaller effect on susceptibility to TDF, 3TC, and FTC. Accessory mutations (A62V, V75I, F77L, and F116Y), which accumulate rapidly in patients, result in a decreased susceptibility to most NRTIs but not to TDF (58–60). In WT HIV-1 RT, Q151 is positioned between Y115 and R72 and forms a part of the dNTP-binding pocket. Q151 interacts with the guanidinium group of the highly conserved R72, which stacks with the base and interacts with the α-phosphate of the incoming dNTP (Fig. 1) (20, 61). Through this interaction network, Q151 helps position the sugar ring, enhances base stacking, and facilitates the interactions of R72 with the α-phosphate of an incoming dNTP. RTs carrying the Q151M mutation incorporate the triphosphate forms of NRTIs less efficiently than WT RT, leading to resistance (62–64).
The Q151M mutation has been reported to have a slight to modest impact on both the HIV-1 titer (27) and the in vitro biochemical properties of RT (58, 64–66). In RT-based in vitro assays, the Q151M mutation caused a 1.2-fold decrease in the mutation frequency in lacZα and a 6-fold decrease in the misinsertion frequency (63, 67). Using a viral vector-based lacZα assay, Q151M was reported to have 5.7-fold higher fidelity than the WT (32, 68). However, data generated using an MLV vector carrying the corresponding mutation (Q190M) suggested that this mutation has no significant effect on the fidelity of retroviral replication (29). In our experiments, the Q151M mutation caused a modest but significant 1.2-fold increase in the frequency of errors that arose in a single cycle of HIV-1 replication (Table 1).
A comparison of the lacZα positions where class 1 missense mutations preferentially occurred showed no significant differences from WT RT (Table 2; see also Fig. S2 in the supplemental material). This result makes sense from a structural standpoint. Although the Q151M mutation may weaken the interactions that would normally involve R72 and/or the 3′ OH of the incoming dNTP, it does not appear that this mutation would affect base pairing at the polymerase active site.
When we looked at the sites at which class 2 frameshift mutations preferentially occurred, there was a highly significant increase in the frequency of mutations compared to the WT at three sites (positions 32, 94, and 99) (Table 3; see also Fig. S3 in the supplemental material). At two of these positions, an increase in the number of single nucleotide insertions was seen: a C in a run of five Cs (position 94) and a T in a run of three Ts (position 99). At the third position, both deletions and insertions of a T were seen in a run of four Ts (position 32). The propensity of the Q151M RT mutant to make frameshift errors at position 32 differed from that of WT RT, which made no frameshift errors at this position. Y115F and Q151M increased the frameshift errors at similar positions. Two of the three sites where these RT mutants caused an increase in the frameshift errors, 94 and 99, are the same, but only Y115F had a significant effect on missense errors (see Discussion). If, as the frameshift data suggest, the Q151M mutation makes HIV-1 RT more tolerant of unpaired or extrahelical nucleotides in both the primer and the template strand, it does so without affecting the ability of RT to tolerate mispaired nucleotides at the active site. Because the side chains of the residues at both position 115 and position 151 are close to the incoming dNTP, the template nucleotide, and each other (Fig. 1), it seems reasonable to expect that the Y115F and Q151M mutations would have similar effects on fidelity. Q151M did not cause a statistically significant change in either the class 3 (multiple) or class 4 (indel) mutations (Table 4 and Table 6).
DISCUSSION
Errors made during HIV replication underlie the development of drug resistance and the ability of the virus to develop mutations that allow it escape from the host's immune system. Thus, it is important to understand the contribution that RT makes to the errors made during HIV replication and to understand how mutations in RT affect the errors that arise during HIV replication. Despite extensive work attempting to measure the fidelity of WT HIV-1 RT and the impact of mutations on the fidelity of RT using purified recombinant proteins, relatively little is known about how mutations in RT affect the number of errors, the nature of the errors, and the positions at which the errors arise during viral replication. All of the RT mutations we tested (Y115F, M184I, M184V, and Q151M), three of which have been reported to increase RT fidelity, caused a modest but significant increase in the overall error rate of HIV replication. This reduction in fidelity was a direct result of an increase in the errors made by the mutant RTs. The additional errors were not uniformly distributed but arose at specific positions (hot spots). Both the specific positions at which the errors arose (the hot spots) and the nature of the increased errors in the lacZα reporter gene (for example, missense versus frameshift errors) depended on the nature and the position of the mutation in RT.
Using the lacZα gene as the reporter underestimates the overall error rate (by perhaps 2- to 3-fold), because silent mutations are not scored, but that should not affect the ability of the assay to detect changes in the fidelity of HIV replication caused by mutations in RT. WT HIV replication (and, by implication, WT HIV-1 RT) has a level of fidelity in a single-round infection that is similar to that seen with other retroviruses (4). The mutation rate for HIV replication is due to the errors made by both the host RNA Pol II and RT. Even if we assume that all of the errors are made by RT, the fidelity of HIV-1 RT is approximately 1.4 × 10−5, which is reasonably good for an enzyme that lacks an editing function. Thus, HIV-1 RT is not a low-fidelity reverse transcriptase. As discussed earlier, there are reasons to believe that the fidelity of RNA Pol II places an upper limit on the fidelity of RT. If the fidelity of RT has been selected to be relatively high (as discussed below), then the levels of fidelity of the two enzymes would be fairly similar. If this idea is correct, then even a significant increase in the fidelity of RT would, at most, increase the fidelity of HIV replication by 2-fold.
Small changes in the polymerase active site of RT (for example, the Y115F mutation and the difference between M184I and M184V) that have only a modest effect on the overall fidelity of replication can still have a significant effect on the number and nature of mutations that arise at specific positions in the target gene lacZα. This implies that these mutations in RT could affect the mutations that arise at specific positions in the HIV genome, including mutations that are involved in escape from immune surveillance and drug resistance. The side chains of the amino acids at positions 115, 184, and 151 are involved in positioning the incoming dNTP. This in turn impacts base pairing and the interaction of the incoming dNTP with catalytic metals and thus would influence the selection of the correct incoming dNTP (see Fig. 1). The fact that all of the mutations we tested reduced the overall fidelity of viral replication supports the idea that the amino acids at the polymerase active site of HIV-1 RT have been selected to optimize the fidelity of HIV-1 RT.
We also show that changes in the sequence of the vector outside the reporter gene can affect the mutations that arise within the target gene. Based on the nature of the templates, it is likely that this is the result of differences that occurred during reverse transcription, where the first-strand template(s) is single-stranded RNA, and not during RNA synthesis, which involves a double-stranded DNA template. Although we were not been able to find a simple or direct correlation between the structure of the RNA template and the sites at which mutations arise (4), the fact that sequences 30 to 40 nucleotides from the site of the mutation can affect the outcome suggests that it is possible that the structure of the RNA template can affect the errors that are made by RT.
As was previously mentioned, there is not a good correlation between the data that were obtained in experiments designed to measure the fidelity of HIV-1 RT that were done with purified recombinant RT (14, 24, 30, 31, 46–54, 67, 69–73) and the results that we obtained measuring the fidelity of HIV-1 replication. Given that viral replication involves the errors made by both RNA Pol II and RT, one might have expected that the error rate measured for purified RT would be lower than the error rate measured for viral replication. That, however, was not the case. The error rate measured for RT in most assays was approximately 20-fold higher than the error rate for viral replication (4). Moreover, in experiments in which the same lacZα sequence was copied in the two systems, the characteristics and positions of the errors were quite different. Part of the problem may be the absence, in the in vitro experiments, of viral and host cell factors that could provide a more favorable environment for RT to carry out accurate DNA synthesis. Given these important differences, it is not surprising that the data we obtained on the effects of specific mutations in RT on the fidelity of HIV replication contradict much of the published literature (14, 24, 30, 31, 46–54, 67, 69–74).
Our results are more similar to the data that were previously reported for the effects of RT mutations on the fidelity of HIV replication, which were based on the inactivation of a reporter gene carried by an HIV vector (32, 75). However, there are also some significant differences. We used a form of the viral vector that allowed us to show that the viral DNAs we analyzed had gone through the viral life cycle, and we cloned and sequenced each of the mutant viral DNAs. This approach allowed us to discard false positives and false negatives that would have skewed the results. In contrast, the previous reports of the effects of mutations in RT relied on determining the fraction of eukaryotic cells, or bacterial colonies, in which a reporter gene carried by a vector was scored as being either functional or inactive (32, 75).
More importantly, the assay we use makes it possible to determine both the overall error rate and the precise nature of the underlying mutations. Simply measuring the effects of an RT mutation on the overall error rate does not accurately measure its full impact on fidelity. For WT HIV-1 RT, the vast majority of both the missense and frameshift mutations occur at a subset of the available mutable sites. Mutations in RT that have a very modest effect on the overall mutation rate can still significantly affect the nature of missense and frameshift mutations and the positions at which they arise in the viral genome. Thus, the effects of the RT mutations on fidelity are themselves specific; mutations in RT cause significant changes in the errors that are made at a limited subset of mutable sites in the lacZα gene. This highlights the importance of using an assay that not only measures the overall mutation rate but also defines the nature of the errors and the positions at which they are made. Some of the sites in lacZα where mutations in RT caused a significant increase in the errors were hot spots for WT RT. This suggests that the mutations we analyzed in RT do not simply cause a monotonic increase in the probability that WT RT will make a missense or frameshift error every time it inserts a nucleotide. The effects of the RT mutations are more subtle and more specific.
Three of the four mutations we tested (M184V, M184I, and Q151M) have a strong impact on NRTI resistance. The fourth (Y115F) has a more modest effect. Nonetheless, the Y115F mutation had the greatest impact on fidelity. This shows that there is not a direct correlation between the ability of a mutant enzyme to discriminate between a normal dNTP and an NRTI-TP and its impact on the fidelity of HIV replication. However, all of the mutations we tested, including M184V, which is commonly found in patients who are treated with 3TC/FTC, affected the nature and the spectrum of mutations that arise during HIV replication. It is therefore likely that mutations in RT associated with NRTI resistance can affect the evolution of the virus and its ability to evade the immune system and/or develop new resistance mutations. Obvious similarities and substantial differences were seen in a comparison of the sites with increased mutations in lacZα for the four RT mutants. Some RT mutations affected its propensity to make missense versus frameshift errors, and some of the RT mutations caused an increase in the errors that were made at exactly the same positions in the lacZα target. For example, Q151M did not cause an increase in the number of missense mutations but did cause an increase in the frameshift mutations. In contrast, M184I and M184V caused an increase in missense mutations but not in frameshift mutations. Not surprisingly, the increased missense mutations made by M184I and M184V, while not identical, were more similar than the missense mutations made by Y115F. However, while both the M184I and M184V mutants showed a significant increase in missense mutations at positions 52 and 109, Y115F and M184I (but not M184V) showed a significant increase in missense mutations at position 76. Both Y115F and Q151M caused an increase in the frameshift mutations at positions 94 and 99.
When we analyzed the mutations made during the replication of a vector carrying WT RT, we were surprised, given the relatively low overall error rate of HIV replication, to find lacZα sequences that contained multiple missense mutations. The nature of the errors, and the sequence context in which they arose, suggested that many of these mutations could be explained by the action of ADAR (4). Because the majority of the multiple mutations were apparently made by a host enzyme, the data we obtained with the RT mutants were, in general, similar to the data obtained with WT RT. Some of these multiple mutations could be explained by the action of ADAR, and a small number of the multiple mutations could be explained by the action of an APOBEC. Although the number of multiple mutations was relatively small, there was, in the M184V data set, a highly significant increase in the overall number of multiple mutations that we cannot explain. There was a smaller but still significant increase in the multiple mutations that occurred in the Y115F data set. If we separate out the mutations that could not be attributed to either ADAR or APOBEC, there were small increases in the fractions of the multiple mutations in the Y115F and M184I data sets, but the total numbers were low.
Template switching is frequent during reverse transcription and has been implicated in the generation of indel mutations (6). Nonhomologous strand transfer(s) between two RNA templates during minus-strand DNA synthesis can result in either deletions or insertions. Theoretically, deletions should occur twice as often as insertions because, for a deletion, there are two RNA strands to which minus-strand DNA could be transferred. In contrast, for an insertion, there is only a single RNA to which minus-strand DNA could be transferred. It is possible that the nonhomologous strand transfers that give rise to deletions are directed specifically to the RNA strand which was being copied into DNA. However, this seems unlikely, given the ease with which homologous transfers occur between templates, creating recombinant retroviral genomes (6, 76). Alternatively, indels may arise primarily at positions where both RNA strands are broken at the same, or nearly the same, position. Having both RNA strands broken at the same position would prevent normal homologous recombination during minus-strand DNA synthesis, and continued minus-strand DNA synthesis would necessarily involve a nonhomologous strand transfer event. However, a stand transfer that generates an insertion would place the growing end of the minus-strand DNA 3′ of the breaks in the two template RNAs, which would not solve the problem of getting DNA synthesis past the breaks. Conversely, a transfer event 5′ of the break would allow minus-strand DNA synthesis to continue and would create a deletion. If indels are associated with an attempt to copy genomic RNAs broken at the same, or nearly the same, site, then a strand transfer event that created a deletion would more likely lead to the synthesis of a nearly complete viral DNA genome than would a strand transfer event that generated an insertion.
Only one of the RT mutations we tested, Y115F, increased the number of indel mutations. This could have been due to the Y115F mutation decreasing the polymerase activity of RT relative to the RNase H activity, which is known to increase recombination (27). Increasing the number of times that the nascent minus-strand DNA is released from the RNA template by RNase H degradation would increase recombination and the probability that a dissociation event would occur opposite a break in the second RNA genome. If the break in the second RNA strand prevented the nascent DNA strand from making a homologous strand transfer to the second RNA template, then minus-strand DNA synthesis could be continued only through the generation of an indel, which, as discussed above, is more likely to be a deletion than an insertion.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and an R37 MERIT Award (AI 27690) to E.A.
We are grateful to Allen Kane for help in preparing the figures and to Teresa Burdette and Carolyn Crisp for help in preparing the manuscript.
Footnotes
Published ahead of print 23 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00302-14.
REFERENCES
- 1.Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489. 10.1126/science.7824947 [DOI] [PubMed] [Google Scholar]
- 2.Wabl M, Burrows PD, von Gabain A, Steinberg C. 1985. Hypermutation at the immunoglobulin heavy chain locus in a pre-B-cell line. Proc. Natl. Acad. Sci. U. S. A. 82:479–482. 10.1073/pnas.82.2.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunkel TA, Bebenek K, Roberts JD, Fitzgerald MP, Thomas DC. 1989. Analysis of fidelity mechanisms with eukaryotic DNA-replication and repair proteins. Genome 31:100–103. 10.1139/g89-019 [DOI] [PubMed] [Google Scholar]
- 4.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 21 July 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 10.1128/JVI.00915-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns DP, Temin HM. 1994. High rates of frameshift mutations within homo-oligomeric runs during a single cycle of retroviral replication. J. Virol. 68:4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pathak VK, Temin HM. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. U. S. A. 87:6019–6023. 10.1073/pnas.87.16.6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansky LM. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retroviruses 12:307–314. 10.1089/aid.1996.12.307 [DOI] [PubMed] [Google Scholar]
- 8.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. U. S. A. 101:10380–10385. 10.1073/pnas.0403954101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, Preston BD. 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem. 277:38053–38061. 10.1074/jbc.M204774200 [DOI] [PubMed] [Google Scholar]
- 12.Langley KE, Villarejo MR, Fowler AV, Zamenhof PJ, Zabin I. 1975. Molecular basis of beta-galactosidase alpha-complementation. Proc. Natl. Acad. Sci. U. S. A. 72:1254–1257. 10.1073/pnas.72.4.1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel TA, Alexander PS. 1986. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J. Biol. Chem. 261:160–166 [PubMed] [Google Scholar]
- 14.Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948–16956 [PubMed] [Google Scholar]
- 15.Agresti A, Mehta CR, Patel NR. 1990. Exact inference for contingency-tables with ordered categories. J. Am. Stat Assoc. 85:453–458. 10.1080/01621459.1990.10476220 [DOI] [Google Scholar]
- 16.Miller V, Ait-Khaled M, Stone C, Griffin P, Mesogiti D, Cutrell A, Harrigan R, Staszewski S, Katlama C, Pearce G, Tisdale M. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:163–171. 10.1097/00002030-200001280-00012 [DOI] [PubMed] [Google Scholar]
- 17.Stone C, Ait-Khaled M, Craig C, Griffin P, Tisdale M. 2004. Human immunodeficiency virus type 1 reverse transcriptase mutation selection during in vitro exposure to tenofovir alone or combined with abacavir or lamivudine. Antimicrob. Agents Chemother. 48:1413–1415. 10.1128/AAC.48.4.1413-1415.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tisdale M, Alnadaf T, Cousens D. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrigan PR, Stone C, Griffin P, Najera I, Bloor S, Kemp S, Tisdale M, Larder B. 2000. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J. Infect. Dis. 181:912–920 [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Chopra R, Verdine GL, Harrison SC. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669–1675. 10.1126/science.282.5394.1669 [DOI] [PubMed] [Google Scholar]
- 21.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2000. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 97:3056–3061. 10.1073/pnas.97.7.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao G, Goff SP. 1998. Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J. Virol. 72:5905–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, Arnold E. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771–4779. 10.1128/JVI.75.10.4771-4779.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer PL, Hughes SH. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 74:6494–6500. 10.1128/JVI.74.14.6494-6500.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cases-Gonzalez CE, Gutierrez-Rivas M, Ménendez-Arias L. 2000. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:19759–19767 [DOI] [PubMed] [Google Scholar]
- 26.Ray AS, Basavapathruni A, Anderson KS. 2002. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J. Biol. Chem. 277:40479–40490. 10.1074/jbc.M205303200 [DOI] [PubMed] [Google Scholar]
- 27.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761–8770. 10.1128/JVI.78.16.8761-8770.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris D, Kaushik N, Pandey PK, Yadav PN, Pandey VN. 1998. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 273:33624–33634. 10.1074/jbc.273.50.33624 [DOI] [PubMed] [Google Scholar]
- 29.Halvas EK, Svarovskaia ES, Pathak VK. 2000. Role of murine leukemia virus reverse transcriptase deoxyribonucleoside triphosphate-binding site in retroviral replication and in vivo fidelity. J. Virol. 74:10349–10358. 10.1128/JVI.74.22.10349-10358.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martín-Hernández AM, Gutiérrez-Rivas M, Domingo E, Menéndez-Arias L. 1997. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 25:1383–1389. 10.1093/nar/25.7.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonckheere H, De Clercq E, Anne J. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 267:2658–2665. 10.1046/j.1432-1327.2000.01272.x [DOI] [PubMed] [Google Scholar]
- 32.Mansky LM, Le Rouzic E, Benichou S, Gajary LC. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 77:2071–2080. 10.1128/JVI.77.3.2071-2080.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296. 10.1006/geno.2002.6718 [DOI] [PubMed] [Google Scholar]
- 34.Piroozmand A, Yamamoto Y, Khamsri B, Fujita M, Uchiyama T, Adachi A. 2007. Generation and characterization of APOBEC3G-positive 293T cells for HIV-1 Vif study. J. Med. Invest. 54:154–158. 10.2152/jmi.54.154 [DOI] [PubMed] [Google Scholar]
- 35.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- 36.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. 10.1126/science.1083338 [DOI] [PubMed] [Google Scholar]
- 37.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. 10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- 38.Barber AM, Hizi A, Maizel JV, Jr, Hughes SH. 1990. HIV-1 reverse transcriptase: structure predictions for the polymerase domain. AIDS Res. Hum. Retroviruses 6:1061–1072. 10.1089/aid.1990.6.1061 [DOI] [PubMed] [Google Scholar]
- 39.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner SA, Mulder J, Loveday C, Christopherson C, et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411–1419. 10.1093/infdis/171.6.1411 [DOI] [PubMed] [Google Scholar]
- 40.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, Svarovskaia E, Miller MD, White KL. 2012. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J. Acquir. Immune Defic. Syndr. 59:47–54. 10.1097/QAI.0b013e31823aca74 [DOI] [PubMed] [Google Scholar]
- 41.Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040–4049 [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer PL, Hughes SH. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624–1628. 10.1128/AAC.39.7.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. U. S. A. 96:10027–10032. 10.1073/pnas.96.18.10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao HQ, Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300:403–418. 10.1006/jmbi.2000.3823 [DOI] [PubMed] [Google Scholar]
- 45.Bakhanashvili M, Avidan O, Hizi A. 1996. Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett. 391:257–262. 10.1016/0014-5793(96)00747-8 [DOI] [PubMed] [Google Scholar]
- 46.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad VR. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282–1285. 10.1126/science.271.5253.1282 [DOI] [PubMed] [Google Scholar]
- 47.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. 1996. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry 35:2168–2179. 10.1021/bi9516642 [DOI] [PubMed] [Google Scholar]
- 48.Feng JY, Anderson KS. 1999. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry 38:9440–9448. 10.1021/bi990709m [DOI] [PubMed] [Google Scholar]
- 49.Rezende LF, Drosopoulos WC, Prasad VR. 1998. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res. 26:3066–3072. 10.1093/nar/26.12.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oude Essink BB, Berkhout B. 1999. The fidelity of reverse transcription differs in reactions primed with RNA versus DNA primers. J. Biomed. Sci. 6:121–132. 10.1007/BF02256443 [DOI] [PubMed] [Google Scholar]
- 51.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad VR, Wainberg MA. 1997. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 25:4532–4536. 10.1093/nar/25.22.4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oude Essink BB, Back NK, Berkhout B. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212–3217. 10.1093/nar/25.16.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamburgh ME, Drosopoulos WC, Prasad VR. 1998. The influence of 3TC-resistance mutations E89G and M184V in the human immunodeficiency virus reverse transcriptase on mispair extension efficiency. Nucleic Acids Res. 26:4389–4394. 10.1093/nar/26.19.4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drosopoulos WC, Prasad VR. 1998. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J. Virol. 72:4224–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansky LM, Bernard LC. 2000. 3′-Azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 74:9532–9539. 10.1128/JVI.74.20.9532-9539.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keulen W, Nijhuis M, Schuurman R, Berkhout B, Boucher C. 1997. Reverse transcriptase fidelity and HIV-1 variation. Science 275:229; author reply 230–231 [PubMed] [Google Scholar]
- 57.Balzarini J, Pelemans H, Karlsson A, De Clerc QE, Kleim JP. 1996. Concomitant combination therapy for HIV infection preferable over sequential therapy with 3TC and non-nucleoside reverse transcriptase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 93:13152–13157. 10.1073/pnas.93.23.13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer PL, Clark PK, Hughes SH. 2012. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J. Virol. 86:5885–5894. 10.1128/JVI.06597-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsumi S, Kosalaraksa P, Tsang H, Kavlick MF, Harada S, Mitsuya H. 2003. Pathways for the emergence of multi-dideoxynucleoside-resistant HIV-1 variants. AIDS 17:1127–1137. 10.1097/00002030-200305230-00003 [DOI] [PubMed] [Google Scholar]
- 60.Iversen AK, Shafer RW, Wehrly K, Winters MA, Mullins JI, Chesebro B, Merigan TC. 1996. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J. Virol. 70:1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss KK, Isaacs SJ, Tran NH, Adman ET, Kim B. 2000. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry 39:10684–10694. 10.1021/bi000788y [DOI] [PubMed] [Google Scholar]
- 62.Ueno T, Shirasaka T, Mitsuya H. 1995. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J. Biol. Chem. 270:23605–23611. 10.1074/jbc.270.40.23605 [DOI] [PubMed] [Google Scholar]
- 63.Kaushik N, Talele TT, Pandey PK, Harris D, Yadav PN, Pandey VN. 2000. Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry 39:2912–2920. 10.1021/bi991376w [DOI] [PubMed] [Google Scholar]
- 64.Deval J, Selmi B, Boretto J, Egloff MP, Guerreiro C, Sarfati S, Canard B. 2002. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using alpha-boranophosphate nucleotide analogues. J. Biol. Chem. 277:42097–42104. 10.1074/jbc.M206725200 [DOI] [PubMed] [Google Scholar]
- 65.Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. 2006. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2:e10. 10.1371/journal.ppat.0020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarafianos SG, Das K, Hughes SH, Arnold E. 2004. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 14:716–730. 10.1016/j.sbi.2004.10.013 [DOI] [PubMed] [Google Scholar]
- 67.Rezende LF, Curr K, Ueno T, Mitsuya H, Prasad VR. 1998. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J. Virol. 72:2890–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss KK, Chen R, Skasko M, Reynolds HM, Lee K, Bambara RA, Mansky LM, Kim B. 2004. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry 43:4490–4500. 10.1021/bi035258r [DOI] [PubMed] [Google Scholar]
- 69.Boyer PL, Stenbak CR, Hoberman D, Linial ML, Hughes SH. 2007. In vitro fidelity of the prototype primate foamy virus (PFV) RT compared to HIV-1 RT. Virology 367:253–264. 10.1016/j.virol.2007.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji J, Loeb LA. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323–330. 10.1006/viro.1994.1130 [DOI] [PubMed] [Google Scholar]
- 71.Ji JP, Loeb LA. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954–958. 10.1021/bi00119a002 [DOI] [PubMed] [Google Scholar]
- 72.Roberts JD, Bebenek K, Kunkel TA. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171–1173. 10.1126/science.2460925 [DOI] [PubMed] [Google Scholar]
- 73.Stuke AW, Ahmad-Omar O, Hoefer K, Hunsmann G, Jentsch KD. 1997. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse transcriptases and DNA polymerases. Arch. Virol. 142:1139–1154. 10.1007/s007050050148 [DOI] [PubMed] [Google Scholar]
- 74.Hamburgh ME, Curr KA, Monaghan M, Rao VR, Tripathi S, Preston BD, Sarafianos S, Arnold E, Darden T, Prasad VR. 2006. Structural determinants of slippage-mediated mutations by human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 281:7421–7428. 10.1074/jbc.M511380200 [DOI] [PubMed] [Google Scholar]
- 75.Mansky LM, Pearl DK, Gajary LC. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J. Virol. 76:9253–9259. 10.1128/JVI.76.18.9253-9259.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen J, Powell D, Hu WS. 2006. High frequency of genetic recombination is a common feature of primate lentivirus replication. J. Virol. 80:9651–9658. 10.1128/JVI.00936-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Das K, Sarafianos SG, Clark AD, Jr, Boyer PL, Hughes SH, Arnold E. 2007. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J. Mol. Biol. 365:77–89. 10.1016/j.jmb.2006.08.097 [DOI] [PubMed] [Google Scholar]
- 78.Das K, Martinez SE, Bauman JD, Arnold E. 2012. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 19:253–259. 10.1038/nsmb.2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.