ABSTRACT
Human papillomavirus type 6 (HPV6) is the major etiological agent of anogenital warts and laryngeal papillomas and has been included in both the quadrivalent and nonavalent prophylactic HPV vaccines. This study investigated the global genomic diversity of HPV6, using 724 isolates and 190 complete genomes from six continents, and the association of HPV6 genomic variants with geographical location, anatomical site of infection/disease, and gender. Initially, a 2,800-bp E5a-E5b-L1-LCR fragment was sequenced from 492/530 (92.8%) HPV6-positive samples collected for this study. Among them, 130 exhibited at least one single nucleotide polymorphism (SNP), indel, or amino acid change in the E5a-E5b-L1-LCR fragment and were sequenced in full. A global alignment and maximum likelihood tree of 190 complete HPV6 genomes (130 fully sequenced in this study and 60 obtained from sequence repositories) revealed two variant lineages, A and B, and five B sublineages: B1, B2, B3, B4, and B5. HPV6 (sub)lineage-specific SNPs and a 960-bp representative region for whole-genome-based phylogenetic clustering within the L2 open reading frame were identified. Multivariate logistic regression analysis revealed that lineage B predominated globally. Sublineage B3 was more common in Africa and North and South America, and lineage A was more common in Asia. Sublineages B1 and B3 were associated with anogenital infections, indicating a potential lesion-specific predilection of some HPV6 sublineages. Females had higher odds for infection with sublineage B3 than males. In conclusion, a global HPV6 phylogenetic analysis revealed the existence of two variant lineages and five sublineages, showing some degree of ethnogeographic, gender, and/or disease predilection in their distribution.
IMPORTANCE This study established the largest database of globally circulating HPV6 genomic variants and contributed a total of 130 new, complete HPV6 genome sequences to available sequence repositories. Two HPV6 variant lineages and five sublineages were identified and showed some degree of association with geographical location, anatomical site of infection/disease, and/or gender. We additionally identified several HPV6 lineage- and sublineage-specific SNPs to facilitate the identification of HPV6 variants and determined a representative region within the L2 gene that is suitable for HPV6 whole-genome-based phylogenetic analysis. This study complements and significantly expands the current knowledge of HPV6 genetic diversity and forms a comprehensive basis for future epidemiological, evolutionary, functional, pathogenicity, vaccination, and molecular assay development studies.
INTRODUCTION
Human papillomavirus 6 (HPV6) is classified taxonomically in the Alphapapillomavirus genus, as species Alpha-10 (1). HPV6 is considered a low-risk HPV, since it is rarely detected in invasive cervical cancer and other HPV-related anogenital cancers. It is the major etiological agent of anogenital warts and laryngeal papillomas, the most frequent benign tumors of the anogenital region and upper respiratory tract (2–5). In addition, HPV6 has been associated with Buschke-Löwenstein tumor (6) and with sporadic cases of anal (7), vulvar (8), penile (9), and head and neck (10) cancers, and it is the most prevalent low-risk type in HPV-positive women with normal cytology, with a global prevalence of 0.8% (11). Due to its clinical importance, HPV6 has been included in both the quadrivalent and nonavalent prophylactic HPV vaccines (12).
Although the genomic diversity of HPV6 has been investigated previously, only a limited number of HPV6 isolates from ethnogeographically closed cohorts were studied, focusing mostly on a single genomic region (13–16). Two recent Slovenian studies reported a substantial genomic variability across the entire HPV6 genome and identified several novel genomic variants (17, 18). The existence of two main HPV6 phylogenetic clusters and several smaller subclusters was identified recently in a study that included 43 complete HPV6 genome sequences (19). Two HPV6 variant groups were established: lineage A consisted of variants that were closely related to the HPV6b prototype sequence (accession no. X00203), while lineage B was further divided into three defined sublineages (B1, B2, and B3) (19). Apart from the preliminary observation of lineage B variant predominance in HPV6-related anogenital warts and laryngeal papillomas originating from Slovenia (17), as well as the recent description of sublineage B1 predominance in anogenital lesions originating from Australia (14), no other clinical correlations for HPV6 variant groups have been observed so far (13, 15, 16, 18).
The present comprehensive study, based on 724 HPV6 isolates and 190 complete genome sequences from six continents, was conducted to assess the global genomic diversity of HPV6 and to investigate possible ethnogeographical and clinical associations for HPV6 lineages and sublineages. To the best of our knowledge, the independent associations between specific HPV6 (sub)lineages and geographic location, anatomical location of infection, and gender were evaluated for the first time on more than 700 HPV6 sequences, using multivariate logistic regression.
MATERIALS AND METHODS
Study samples.
For the purpose of this study, a total of 530 HPV6 DNA-positive samples were obtained from 15 countries covering six continents: Argentina, Australia, Canada, Croatia, Czech Republic, Germany, Hong Kong, Japan, Lithuania, Malaysia, Slovenia, Switzerland, South Africa, United Kingdom, and United States. Samples were collected from the anogenital region (400 samples [75.5%]), the head and neck region (84 samples [15.8%]), or unspecified anatomical locations (46 samples [8.7%]). The majority of samples were cervical swabs (186 samples [35.1%]), followed by anogenital wart tissues (179 samples [33.8%]) and laryngeal papilloma tissues (83 samples [15.7%]) (Table 1).
TABLE 1.
Characteristics of 530 study samples containing HPV6
| Anatomical location and sample type | No. (%) of samples |
|---|---|
| Anogenital region | 400 (75.5) |
| Cervical swaba | 186 (46.5) |
| Normal cytology | 18 (9.7) |
| ASCUS | 23 (12.4) |
| LSIL | 30 (16.1) |
| HSIL | 1 (0.5) |
| CIN1 | 8 (4.3) |
| CIN1/2 | 10 (5.4) |
| ND | 96 (51.6) |
| Anogenital wart tissue | 179 (44.8) |
| Unspecified sample type | 35 (8.7) |
| Head and neck region | 84 (15.8) |
| Laryngeal papilloma tissue | 83 (98.8) |
| Nasal papilloma tissue | 1 (1.2) |
| Unspecified anatomical location | 46 (8.7) |
ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; CIN1, cervical intraepithelial neoplasia grade 1; CIN1/2, cervical intraepithelial neoplasia grade 1 or 2; ND, data not available.
The initial repertoire of HPV6 isolates for HPV6 genomic variant correlation analyses included 875 HPV6 sequences, as follows: (i) 130 complete genomes and 400 partial sequences, ranging from 839 to 6,664 bp, generated in this study; and (ii) 60 complete genomes and 285 partial sequences, ranging from 121 to 3,853 bp, obtained from the NCBI database, the PapillomaVirus Episteme database (PaVE), and various HPV6-related publications (data set collection closed June 2013). HPV6 sequences with a determined lineage/sublineage status and geographical location (724/875 sequences [82.7%]) were selected for further statistical analyses.
Whole-genome amplification and sequencing.
A fragment of 4,908 bp containing the complete E6, E5a, E5b, L2, L1, and LCR genomic regions was initially amplified using the HPV6-E5S and HPV6-E6R primers (17). HPV6 genomic variants that contained at least one unique single nucleotide polymorphism (SNP), amino acid change, or insertion/deletion (indel) in E5a, E5b, L1, and/or LCR were selected for complete genome sequencing. For this purpose, an additional overlapping DNA fragment of 4,511 bp, containing the complete E6, E7, E1, E2, E4, E5a, and E5b open reading frames (ORFs), was generated using the HPV6-E6S and HPV6-E5R primers (17).
Both PCRs were conducted using a Platinum Taq DNA Polymerase High Fidelity kit (Invitrogen, Carlsbad, CA), with 100 ng of DNA template, 2.5 μl of 10× High Fidelity PCR buffer, 0.1 μl of each primer (50 μM), 0.5 μl of deoxynucleoside triphosphates (dNTPs) (10 mM), 1 μl of MgSO4 (50 mM), and water up to 25 μl for each reaction mixture. The cycling conditions were as follows: DNA denaturation and polymerase activation at 94°C for 2 min, followed by 50 cycles of 30 s at 94°C, 30 s at 62 or 56°C, and 5 min at 68°C. The final extension step was done at 68°C for 7 min.
PCR products were visualized by gel electrophoresis, purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany), and sequenced at Macrogen Europe (Amsterdam, Netherlands) or in our in-house sequencing facility, using previously published primers (18). The in-house sequencing was performed using a BigDye Terminator sequencing kit (version 3.1; Applied Biosystems, Foster City, CA) and a sequencing protocol developed by Platt et al. (20). Sequencing reaction mixtures were purified with a BigDye XTerminator purification kit following the manufacturer's instructions (Applied Biosystems) and then run on a 3500 genetic analyzer (Applied Biosystems).
Nucleotide sequences were acquired in both directions and analyzed with CLC Main Workbench software, version 6.5 (CLC Bio, Denmark). The HPV6b reference genome (accession no. X00203), bracketed in the LCR genomic region and available in the PaVE database under PAVE ID HPV6REF, was used as a standard for comparisons and nucleotide position numbering in all analyses. Samples with mutations occurring only once or with wobble nucleotides were amplified and sequenced repeatedly.
Phylogenetic tree construction.
The global phylogenetic tree construction employed 130 complete HPV6 genomes determined in this study (accession no. HG793809 to HG793938 ) and 60 complete genomes available in GenBank (NCBI) (accession no. FR751320 to FR751338 [17], HE599226 to HE599246 and HE962026 to HE962032 [21], and JN252314 to JN252323 [22]), including the corrected HPV6b reference genome (PAVE ID HPV6REF) and nonprototypic HPV6a (accession no. L41216) and HPV6vc (accession no. AF092932) genomes. The complete genomes were linearized at the first ATG of the E6 ORF and aligned using MAFFT v6.846 software (23). Maximum likelihood trees were constructed using RAxML HPC2 v7.6.3, employing 1,000 bootstrap values (24). Bayesian trees were constructed using MrBayes v3.2.1 (25), with 10,000,000 cycles for Markov chain Monte Carlo (MCMC) runs; a general time-reversible model with gamma-distributed rate variation and a proportion of invariable sites (GTR+G+I) was used. Phylogenetic trees were visualized in FigTree v1.4.0 (26).
The tree construction processes were repeated for multiple HPV6 genomic regions to identify the most informative region(s) for whole-genome-based phylogenetic clustering.
Identification and naming of HPV6 variant lineages and sublineages.
Identification of HPV6 variant lineages and sublineages was based on 190 globally aligned HPV6 genomes (130 from our study and 60 obtained from NCBI and PaVE), using the classification and nomenclature system proposed recently (19): nucleotide differences of >1% to <10% and >0.5% to <1% of the complete genomes were used to define variant lineages and sublineages, respectively. Pairwise identities (p distances) were calculated in MEGA5 (27) and were used to construct a heat map scaled in color gradients, with blue indicating maximum (100%) pairwise identity and red indicating minimum (98.4%) pairwise identity.
Identification of lineage/sublineage-specific SNPs.
Out of 190 complete genomes (130 from our study and 60 from NCBI and PaVE), a representative complete genome with more than 0.05% pairwise dissimilarity to other isolates or with a unique variation pattern was selected for identification of lineage- and sublineage-specific SNPs with MacClade v4.08 (28). Based on the above-described criteria, a total of 48 complete genomes were selected, covering all six continents. All of the selected isolates clustered phylogenetically with one of the established HPV6 variant (sub)lineages.
Statistical methods.
Ethnogeographical and clinical associations for HPV6 variant lineages and sublineages were evaluated for 724 HPV6 samples, originating from 18 countries and covering 6 continents (see Data Set S1 in the supplemental material).
A contingency table (Table 2) was constructed to present the distribution of HPV6 lineage A and sublineages B according to geographical location (continent and country), anatomical location of infection (anogenital region or head and neck region), type of lesion (anogenital wart tissue or laryngeal papilloma tissue), and gender, using IBM SPSS 22.0 (released 2013; IBM Corp.). Pie charts representing global and continental distributions of HPV6 (sub)lineages were created in Excel 2013 (Microsoft Office; Microsoft Corporation, Santa Rosa, CA).
TABLE 2.
HPV6 lineage A and sublineage B distributions in 724 samples containing HPV6, according to geographical location (continent and country), anatomical location of infection, lesion type, and gender
| Variable | No. (%) of samples |
||||||
|---|---|---|---|---|---|---|---|
| A | B1 | B2 | B3 | B4 | B5 | Total | |
| Geographical location | |||||||
| Europe | |||||||
| Croatia | 6 (12.8) | 34 (72.3) | 2 (4.3) | 5 (10.6) | 0 (0) | 0 (0) | 47 (100) |
| Czech Republic | 17 (32.1) | 32 (60.4) | 2 (3.8) | 1 (1.9) | 1 (1.9) | 0 (0) | 53 (100) |
| Germany | 2 (4.2) | 26 (54.2) | 11 (22.9) | 9 (18.8) | 0 (0) | 0 (0) | 48 (100) |
| Lithuania | 6 (85.7) | 1 (14.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (100) |
| Serbia | 1 (50) | 0 (0) | 0 (0) | 1 (50) | 0 (0) | 0 (0) | 2 (100) |
| Slovenia | 16 (14.7) | 81 (74.3) | 11 (10.1) | 1 (0.9) | 0 (0) | 0 (0) | 109 (100) |
| Sweden | 2 (20) | 2 (20) | 5 (50) | 1 (10) | 0 (0) | 0 (0) | 10 (100) |
| Switzerland | 4 (7.8) | 27 (52.9) | 5 (9.8) | 12 (23.5) | 0 (0) | 3 (5.9) | 51 (100) |
| United Kingdom | 2 (7.1) | 17 (60.7) | 5 (17.9) | 2 (7.1) | 1 (3.6) | 1 (3.6) | 28 (100) |
| Asia | |||||||
| Hong Kong | 36 (69.2) | 14 (26.9) | 0 (0) | 1 (1.9) | 1 (1.9) | 0 (0) | 52 (100) |
| Japan | 16 (42.1) | 20 (52.6) | 0 (0) | 1 (2.6) | 1 (2.6) | 0 (0) | 38 (100) |
| Malaysia | 1 (33.3) | 2 (66.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (100) |
| North America | |||||||
| Canada | 7 (10.6) | 34 (51.5) | 7 (10.6) | 18 (27.3) | 0 (0) | 0 (0) | 66 (100) |
| USA | 3 (6.5) | 23 (50) | 1 (2.2) | 19 (41.3) | 0 (0) | 0 (0) | 46 (100) |
| South America | |||||||
| Argentina | 4 (7.3) | 29 (52.7) | 13 (23.6) | 9 (16.4) | 0 (0) | 0 (0) | 55 (100) |
| Brazil | 1 (5.6) | 12 (66.7) | 0 (0) | 5 (27.8) | 0 (0) | 0 (0) | 18 (100) |
| Australia | |||||||
| Australia | 5 (9.4) | 36 (67.9) | 10 (18.9) | 2 (3.8) | 0 (0) | 0 (0) | 53 (100) |
| Africa | |||||||
| South Africa | 2 (5.3) | 6 (15.8) | 2 (5.3) | 22 (57.9) | 0 (0) | 6 (15.8) | 38 (100) |
| Total | 131 (18.1) | 396 (54.7) | 74 (10.2) | 109 (15.1) | 4 (0.6) | 10 (1.4) | 724 (100) |
| Anatomical location | |||||||
| Anogenital region | 87 (17.4) | 287 (57.3) | 53 (10.6) | 68 (13.6) | 3 (0.6) | 3 (0.6) | 501 (100) |
| Head and neck region | 38 (21.5) | 84 (47.5) | 17 (9.6) | 30 (16.9) | 1 (0.6) | 7 (4.0) | 177 (100) |
| Total | 125 (18.4) | 371 (54.7) | 70 (10.3) | 98 (14.5) | 4 (0.6) | 10 (1.5) | 678 (100) |
| Lesion type | |||||||
| Anogenital wart tissue | 71 (26.2) | 165 (60.9) | 21 (7.7) | 10 (3.7) | 3 (1.1) | 1 (0.4) | 271 (100) |
| Laryngeal papilloma tissue | 38 (21.6) | 83 (47.2) | 17 (9.7) | 30 (17.0) | 1 (0.6) | 7 (4.0) | 176 (100) |
| Total | 109 (24.4) | 248 (55.5) | 38 (8.5) | 40 (8.9) | 4 (0.9) | 8 (1.8) | 447 (100) |
| Gender | |||||||
| Female | 44 (14.1) | 169 (54.0) | 33 (10.5) | 62 (19.8) | 1 (0.3) | 4 (1.3) | 313 (100) |
| Male | 62 (23.9) | 149 (57.5) | 26 (10.0) | 19 (7.3) | 2 (0.8) | 1 (0.4) | 259 (100) |
| Total | 106 (18.5) | 318 (55.6) | 59 (10.3) | 81 (14.2) | 3 (0.5) | 5 (0.9) | 572 (100) |
Bivariate associations of characteristics represented in the contingency table were then determined, comparing lineages A and B and sublineages B1 and B3 versus lineage A. Sublineages B2, B4, and B5 were excluded from the analysis since they did not meet the criteria for statistical analysis. Odds ratios (ORs) were calculated, and the significance level was set at an α value of 0.05. All tests were two-sided.
To evaluate the independence of associations among the above-mentioned characteristics, multivariate logistic regression was performed; depending on the number of categories of the dependent variable, binary or multinomial logistic regression was used. All analyses were performed in R software, version 2.12.0 (Free Software Foundation, Boston, MA), and IBM SPSS 22.0 (IBM Corp.).
Nucleotide sequence accession numbers.
The 130 complete HPV6 genome sequences determined in this study were deposited in the EMBL-EBI database under accession numbers HG793809 to HG793938.
RESULTS
A complete, approximately 2,800-bp fragment containing the E5a-E5b-L1-LCR genomic region was successfully amplified and sequenced from 492/530 (92.8%) HPV6 DNA-positive samples collected for this study. Among the 492 sequenced isolates, 130 (26.4%) exhibited at least one SNP, indel, or amino acid change in the E5a, E5b, L1, and/or LCR genomic region and were completely sequenced.
The minimum and maximum lengths of 190 globally aligned HPV6 genomes were 7,954 and 8,051 bp, respectively. In total, 471 (5.9%) variable nucleotide positions and 165 (6.6%) variable amino acid positions were identified. The genomic variabilities of 190 HPV6 isolates in nine ORFs and four noncoding genomic regions are summarized in Table 3. Briefly, the maximum frequencies of nucleotide changes in coding regions varied from 3.4% in the E7 ORF to 13.2% in the E5b ORF. The frequencies of amino acid changes varied from 2.0% in the E6 ORF to 27.4% in the E5b ORF. A maximum of 16 indels were detected across all compared sequences. Indels were detected in LCR and noncoding region 3 (NCR3; between the E5b and L2 ORFs). One in-frame deletion of 42 bp was detected in the E2 ORF of one isolate. The maximum pairwise nucleotide difference between the two most variable genomes was 1.6%.
TABLE 3.
Comparison of nucleotide and amino acid sequence variabilities within 190 complete HPV6 genomes
| ORF/noncoding regiona | Maximum % nt pairwise differenceb | No. of nt | No. (%) of variable nt positionsc | No. of variable nt in each codon position |
Maximum % aa pairwise differenced | No. of aa | No. (%) of variable aa positions | ||
|---|---|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | |||||||
| E6 | 1.99 | 453 | 19 (4.19) | 1 | 1 | 17 | 1.34 | 151 | 3 (1.99) |
| E7 | 1.68 | 297 | 10 (3.37) | 2 | 3 | 5 | 4.17 | 99 | 4 (4.04) |
| NCR1 | 20.00 | 5 | 1 (20.00) | ||||||
| E1 | 1.44 | 1,950 | 82 (4.21) | 21 | 10 | 51 | 1.71 | 650 | 30 (4.62) |
| E2 | 2.26 | 1,107 | 60 (5.42) | 22 | 13 | 25 | 5.03 | 396 | 38 (10.30) |
| E4 | 3.04 | 330 | 19 (5.76) | 6 | 5 | 8 | 6.70 | 110 | 9 (8.18) |
| NCR2 | 3.51 | 57 | 4 (7.02) | ||||||
| E5a | 5.43 | 276 | 28 (10.14) | 7 | 6 | 15 | 9.31 | 92 | 13 (14.13) |
| E5b | 5.94 | 219 | 29 (13.24) | 9 | 9 | 11 | 13.35 | 73 | 20 (27.40) |
| NCR3 | 9.09 | 45 | 7 (15.56) | ||||||
| L2 | 1.52 | 1,380 | 82 (5.94) | 18 | 8 | 56 | 1.98 | 460 | 28 (6.09) |
| L1 | 1.20 | 1,503 | 74 (4.92) | 13 | 8 | 53 | 0.80 | 501 | 20 (3.99) |
| LCR | 2.61 | 862 | 79 (9.16) | ||||||
| CGe | 1.58 | 8,052 | 471 (5.85) | 2,505 | 165 (6.59) | ||||
ORF, open reading frame (E6, E7, E1, E2, E4, E5a, E5b, L2, and L1); NCR1, noncoding region 1 (between E7 and E1 ORFs); NCR2, noncoding region 2 (between E2 and E5a ORFs); NCR3, noncoding region 3 (between E5b and L2 ORFs); LCR, long control region; CG, complete genome.
Calculated from the global alignment.
Nucleotide variations included SNPs. Indels and wobble nucleotides were excluded from the count.
Calculated from the global alignment.
A single genome size was inferred from the nucleotide alignment. Each nucleotide position was counted once. The total number and percentage of variable nucleotides included SNPs; indels and wobble nucleotides were excluded from the count. The cumulative number of amino acids was taken from 9 ORFs (E6, E7, E1, E2, E4, E5a, E5b, L2, and L1), with overlapping ORFs counted separately.
Among 130 newly sequenced complete genomes, we inspected double-peak reads for 15/130 (11.5%) HPV6 isolates. These double-peak nucleotide positions were marked as wobble nucleotides in the final complete genome sequences.
Lineage and sublineage identification.
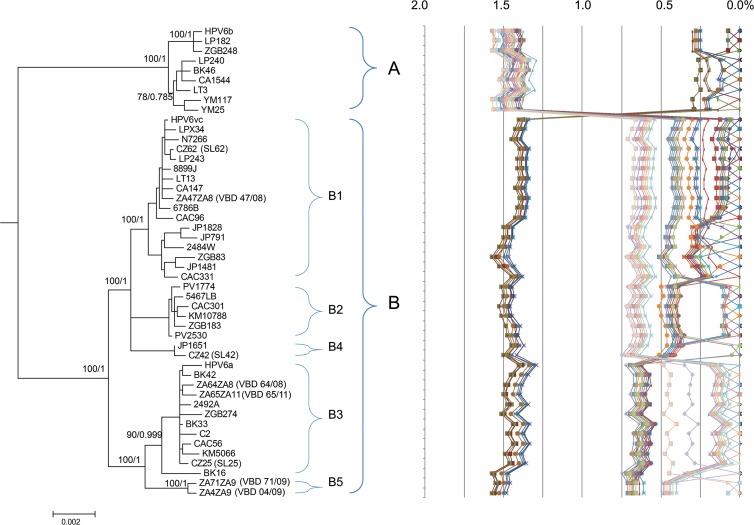
Phylogenetic analyses of the 190 complete HPV6 genomes confirmed the presence of two distinct HPV6 variant lineages, A and B, and revealed five variant sublineages of lineage B: B1, B2, B3, B4, and B5 (see Fig. S1 in the supplemental material). Lineage A included variants related to the HPV6b prototype sequence (HPV6REF), while two major sublineages in lineage B contained variants closely related to HPV6a (sublineage B3) and HPV6vc (sublineage B1). Sublineage B1 included the largest number of sequences (89/190 sequences [46.8%]), followed by lineage A (47/190 sequences [24.7%]), sublineage B3 (30/190 sequences [15.8%]), sublineage B2 (19/190 sequences [10.0%]), sublineage B4 (3/190 sequences [1.6%]), and sublineage B5 (2/190 sequences [1.1%]) (see Fig. S1). A phylogenetic tree of the 48 most divergent complete HPV6 genomes (>0.05% pairwise dissimilarity) clearly confirmed the clustering of variants into the lineages and sublineages established based on 190 complete HPV6 genomes (Fig. 1).
FIG 1.
HPV6 variant tree topology and pairwise comparisons of the 48 most variable complete HPV6 genomes. RAxML and Bayesian trees were inferred from the global alignment of the 48 most variable complete HPV6 genome sequences (the RAxML tree is shown). Numbers alongside branches indicate support scores in the following order: bootstrap percentages obtained by RAxML and Bayesian credibility values obtained by MrBayes 3.2.1. The percent nucleotide sequence differences were calculated for each isolate compared to all other isolates and are shown in the right panel. Values for each comparison for a given isolate are connected by lines, and the comparison to self is indicated as a 0% difference point.
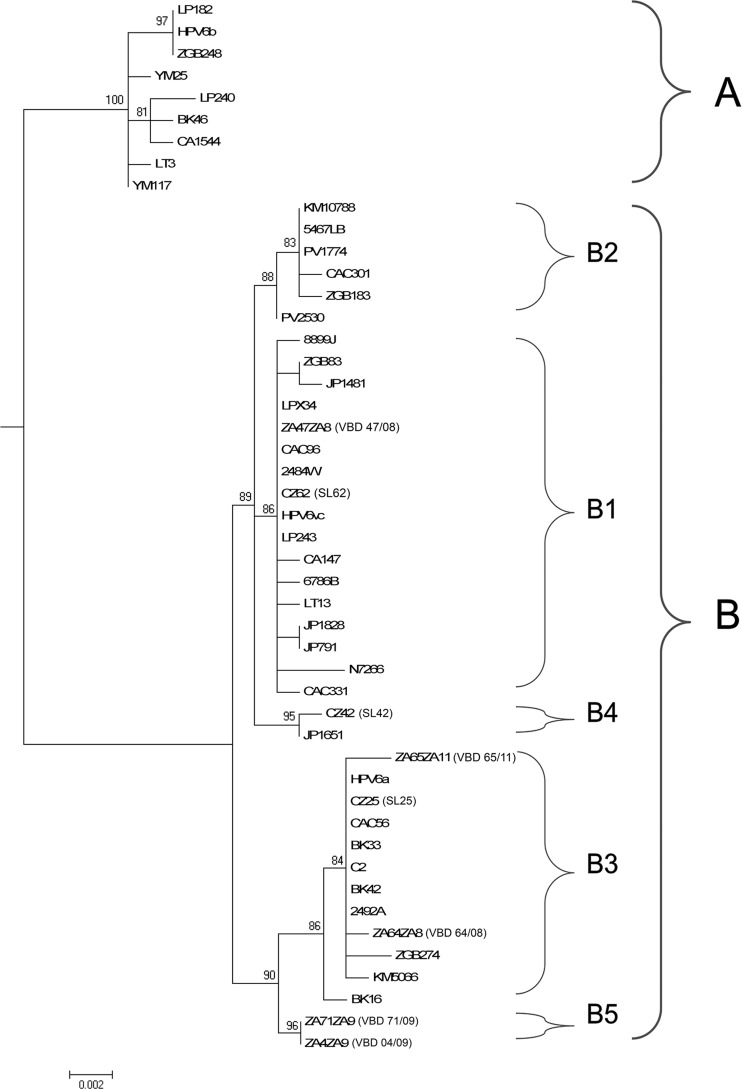
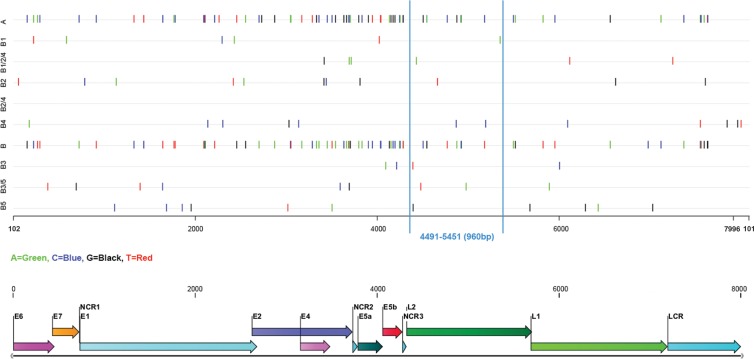
Among all HPV6 genomic regions, only the E1, E2, and L2 ORFs produced trees that were congruent with the whole-genome tree topology. Of these, the L2 ORF showed the optimal discriminatory power for total reproduction of the whole-genome phylogenetic tree topology. In addition, a 960-bp fragment at nucleotide positions 4,491 to 5,451 within the L2 ORF relative to the corrected prototype HPV6b genome (HPV6REF) could be used as a representative region for whole-genome-based phylogenetic clustering (Fig. 2). The 960-bp region contained at least one SNP specific for each lineage and sublineage (Fig. 3). Similarly, the concatenated E5a, E5b, L1, and LCR sequence of approximately 2,800 bp displayed a tree topology that was identical to that of the complete genome sequences.
FIG 2.
Maximum likelihood tree based on a 960-bp region within the L2 ORF for the 48 most variable HPV6 sequences used for the phylogenetic tree construction in Fig. 1.
FIG 3.
Diagnostic lineage/sublineage-specific SNPs identified from comparisons of the 48 most divergent HPV6 genomes. SNP positions relative to the genome of the prototype HPV6b isolate (PaVE ID HPV6REF), linearized at the first ATG of the E6 ORF, are displayed across lineages (A and B), sublineages (B1, B2, B3, B4, and B5), and intersublineages (B1/2/4, B2/4, and B3/5). A representative region (960 bp) for whole-genome-based phylogenetic clustering is marked with vertical lines.
Lineage- and sublineage-specific SNPs.
A total of 202 SNPs across 48 complete HPV6 genomes (the most variable genomes out of 190 complete genomes) were determined (Fig. 3). Analysis of diagnostic lineage-specific SNPs showed that lineages A and B had SNPs dispersed throughout all genomic regions. More detailed examination of SNP positions suggested that any genomic fragment of >500 bp could be used to discriminate between lineages A and B. SNPs specific for all five B sublineages were not dispersed across the whole genome. Instead, they clustered in specific genomic regions. For example, sublineage B3 had no diagnostic SNPs in the first 4,000 bp of the genome (Fig. 3). All identified HPV6 lineage- and sublineage-specific SNPs allowed correct (sub)lineage identification of all of the remaining 142/190 complete genome sequences.
Ethnogeographical and clinical associations for HPV6 variant lineages and sublineages.
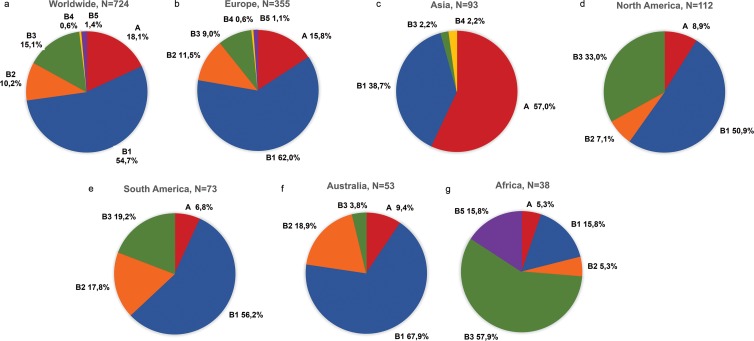
Figure 4 illustrates the global and continental distributions of HPV6 variant (sub)lineages among 724 HPV6 sequences. Lineage B was the most prevalent HPV6 lineage worldwide (593/724 sequences [81.9%]) (Fig. 4a), apart from in Asia, where genomic variants from lineage A predominated (53/93 sequences [57.0%]) (Fig. 4c). The most common sublineage was B1, which was found in 54.7% (396/724 sequences) of samples (Fig. 4a), ranging from 15.8% in Africa (Fig. 4g) to 67.9% in Australia (Fig. 4f). Sublineage B3 was the second most frequently detected sublineage globally (109/724 sequences [15.1%]) (Fig. 4a), with a higher prevalence in Africa (22/38 sequences [57.9%]) (Fig. 4g) than in other continents. Sublineage B2 was identified in all continents except Asia; sublineage B4 was found only in Europe and Asia, and sublineage B5 was found only in Europe and Africa.
FIG 4.
Worldwide and continental distributions of HPV6 variant lineage A and sublineages B1, B2, B3, B4, and B5. (a) Worldwide; (b) Europe; (c) Asia; (d) North America; (e) South America; (f) Australia; (g) Africa.
Bivariate analysis based on all continents demonstrated a statistically significant difference in the distribution of lineages A and B, indicating that HPV6 genetic variants from Asia had lower odds of belonging to lineage B than those of variants from Europe (P < 0.001), whereas variants from Africa and North and South America showed higher odds of belonging to sublineage B3 (P < 0.05 for each comparison). Multivariate logistic regression analysis confirmed statistical significance in the distribution of lineages A and B, showing lower odds for lineage B in variants from Asia than in variants from Europe (OR, 0.09; 95% confidence interval [95% CI], 0.05 to 0.17; P < 0.001); additionally, there were higher odds for sublineage B3 than for lineage A in Africa (OR, 29.84; 95% CI, 4.73 to 188.36; P < 0.001), North America (OR, 4.39; 95% CI, 1.28 to 15.04; P = 0.018), and South America (OR, 5.76; 95% CI, 1.77 to 18.76; P = 0.004) compared to the case in Europe (Table 4).
TABLE 4.
Multivariate logistic regression associations among geographic location (all continents), anatomical location of infection, and gender
| Characteristic (reference category) | Binomial logit model |
Multinomial logit model |
||||
|---|---|---|---|---|---|---|
| B vs A |
B1 vs A |
B3 vs A |
||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Continent (Europe) | ||||||
| Africa | 2.18 (0.45–10.47) | 0.331 | 0.19 (0.02–2.25) | 0.189 | 29.84 (4.73–188.36) | <0.001 |
| Asia | 0.09 (0.05–0.17) | <0.001 | 0.12 (0.06–0.22) | <0.001 | 0.04 (0.01–0.19) | <0.001 |
| Australia | 5.5 (0.72–42.28) | 0.101 | 6.23 (0.81–48.21) | 0.080 | 2.81 (0.23–33.88) | 0.416 |
| North America | 1.34 (0.43–4.12) | 0.614 | 0.9 (0.28–2.89) | 0.856 | 4.39 (1.28–15.04) | 0.018 |
| South America | 2.29 (0.86–6.11) | 0.098 | 1.99 (0.73–5.47) | 0.180 | 5.76 (1.77–18.76) | 0.004 |
| Gender (male) | ||||||
| Female | 1.57 (0.95–2.60) | 0.080 | 1.49 (0.88–2.50) | 0.134 | 2.89 (1.30–6.41) | 0.009 |
| Anatomical location (head and neck) | ||||||
| Anogenital region | 2.51 (1.30–4.83) | 0.006 | 2.12 (1.09–4.14) | 0.027 | 4.77 (1.61–14.18) | 0.005 |
Bivariate analysis of lineages A and B for associations with anatomical locations of infection (anogenital versus head and neck region) indicated higher odds for lineage B than for lineage A in anogenital infections, although associations were not statistically significant (P > 0.05). However, the results of multivariate logistic regression confirmed that lineage B was associated with infections in the anogenital region (OR, 2.51; 95% CI, 1.30 to 4.83; P = 0.006). Additionally, multivariate analysis showed an association of both sublineages B1 (OR, 2.12; 95% CI, 1.09 to 4.14; P = 0.027) and B3 (OR, 4.77; 95% CI, 1.61 to 14.18; P = 0.005) with anogenital infections in comparison to lineage A (Table 4).
Bivariate analysis revealed that lineage B was more common in females than in males (P = 0.002); in particular, sublineage B3 showed a higher association with females (P < 0.001). In multivariate logistic regression analysis, females showed marginally higher odds for lineage B than males in comparison to lineage A (P = 0.08) and higher odds for sublineage B3 than males in comparison to lineage A (OR, 2.89; 95% CI, 1.30 to 6.41; P = 0.009) (Table 4).
DISCUSSION
In this study, the following four genomic regions were initially sequenced from 530 HPV6 isolates (collected for the purpose of this study from 15 countries on six continents): the L1 ORF, which encodes the major capsid protein and is typically used in identification of HPV types; the E5a and E5b ORFs, which have previously shown the highest diversification trend among all HPV6 coding regions (17, 18); and LCR, which is considered the most variable HPV genomic region, harboring control sites for viral replication and gene expression. Following identification of the most divergent HPV6 E5a-E5b-L1-LCR genomic variants, a total of 130 HPV6 isolates were selected, completely sequenced, and phylogenetically evaluated together with 60 complete HPV6 genome sequences available in GenBank and PaVE.
HPV6 exhibited a maximum genomic difference of 1.6%, with only 48/190 (25.3%) complete genomes exhibiting >0.05% pairwise dissimilarity to other isolates. Overall, the HPV6 genomic diversity was lower than those of the two most important high-risk HPV types: HPV16 (2.3%) and HPV18 (2.1%) (29, 30). On the other hand, HPV6 shared similar genomic heterogeneity with some of the closest relatives of HPV16 and HPV18: HPV45 (1.5%), HPV70 (1.6%), HPV31 (1.4%), and HPV58 (1.7%) (30, 31). The global diversity of low-risk HPV types based on complete genomes is still a subject of investigation; nevertheless, previously published data suggest that HPV6 exhibits more genomic diversity than its closest relative, HPV11, with a maximum pairwise difference of 0.4% (19).
The direct inspection of complete genome sequences revealed the presence of several wobble nucleotides, suggesting that some individuals can potentially be coinfected with two different HPV6 genomic variants, which warrants further investigation.
The phylogenetic tree obtained from 190 complete genome sequences indicated the presence of two variant lineages, A and B, and five B sublineages (B1, B2, B3, B4, and B5, with B4 and B5 determined for the first time). Additionally, we showed that concatenated HPV6 E5a-E5b-L1-LCR sequences of approximately 2,800 bp displayed a tree topology that was completely congruent with that of the whole-genome sequences and that this region can therefore be considered a suitable representative region for whole-genome-based phylogenetic clustering of HPV6 isolates. In contrast, individual HPV6 ORFs, including the E6, E7, E5a, E5b, E4, and L1 ORFs, and the LCR noncoding region generated dichotomic tree topologies, identifying lineages A and B but not being reliable for identification of the B sublineages. The E1, E2, and L2 ORFs generated tree topologies that resembled the global phylogenetic tree topology, with the L2 ORF (1,380 bp) being the most informative single genomic region for phylogenetic identification of all currently known HPV6 variant lineages and sublineages. On identification of lineage/sublineage-specific SNPs, a shorter genomic segment in the L2 ORF (960 bp) was identified that totally reproduced the whole-genome phylogenetic tree topology. This representative genomic region or lineage/sublineage-specific SNPs can be used when complete HPV6 genome sequences are not available or when one is interested in accurate lineage/sublineage assignment only, rather than in studying the genetic variability of other genomic regions.
In order to investigate associations among HPV6 (sub)lineages, geographical location, anatomical location of infection, and gender, 724 HPV6 samples were evaluated. The analysis showed that HPV6 genomic variants from lineage B (“nonprototypic” HPV6 genomic variants) prevailed globally (14, 18), consisting mainly of variants from sublineage B1. Due to its global presence, it is possible that B1 is the oldest HPV6 sublineage, which disseminated during early human evolution and migrated to different parts of the world before other sublineages emerged.
Sublineage B2 was present in all continents except Asia, while sublineage B3 was detected significantly more frequently in Africa and North and South America. Sublineage B4 was identified only in Europe and Asia, and sublineage B5 was identified only in Africa and Europe, indicating a possible recent emergence and exchange of these specific sublineages between Europe and Asia and between Africa and Europe, respectively. Lineage A was identified in all continents but significantly predominated in Asia. Since our data set included an unbalanced number of samples per continent and country, further analyses on larger sets of HPV6 isolates from Asian and African countries are required to confirm these observations.
The reflection of coevolution of genomic variants with Homo sapiens, which was firmly illustrated with phylogenetic tree topologies of high-risk types HPV16 and HPV18 (32, 33), was not observed among HPV6 variants. The absence of strong geographical clustering of HPV6 genomic variants could be due to the sampling bias (African and Asian continents were underrepresented), strong intermixing of HPV6 variants in ethnic populations, or, more probably, a relatively lower replication rate of HPV6 than those of high-risk HPV types. A lower replication rate would result in slower molecular evolution and, consequently, in delayed SNP fixation throughout the HPV6 genome (16, 19, 31, 34, 35). The major HPV6 lineages we see today have therefore most likely existed relatively unchanged since the migration of Homo sapiens out of Africa. Many other HPV types have revealed unique European, African, and Asian variants, but the phylogenetic relationships of these HPV variants to the ethnogeographic origins of individuals were seldom as strong as those found for HPV16 and HPV18 (36), probably reflecting a different viral evolution.
Clinical associations for HPV6 variant lineages A and B and the most frequent sublineages, B1 and B3, were also determined. Multivariate logistic regression analysis revealed an association of lineage B, notably sublineages B1 and B3, with anogenital infections in comparison to lineage A. An association between sublineage B1 and anogenital infections was reported previously by Danielewski et al. (14), who analyzed the same set of Australian samples as that in our study, and was confirmed here with non-Australian samples and further expanded to sublineage B3. Furthermore, multivariate analysis showed that in comparison to males, females had higher odds for infection with genetic variants from sublineage B3 than with variants from lineage A. According to the unbalanced structure of the initial data set, presented in Table 2, all our statistical observations need to be interpreted carefully, and further investigations are warranted to establish associations between HPV6 (sub)lineages and specific HPV6-related disease outcomes.
In summary, we performed the largest global HPV6 genomic diversity study to date, contributing a total of 130 new, complete HPV6 genome sequences to available sequence repositories. Global phylogenetic analysis revealed the existence of two variant lineages and five variant sublineages, which showed some degree of ethnogeographic, gender, and/or disease predilection in their distribution. Lineage B prevailed globally and showed an association with anogenital infections, specifically for sublineages B1 and B3. Lineage A prevailed in Asia, and sublineage B3 prevailed in Africa and North and South America. Our analysis showed that females had higher odds for infection with genetic variants from sublineage B3 than males. HPV6 (sub)lineage-specific SNPs were identified to facilitate the rapid and reliable identification of HPV6 genomic variants in cases where complete genome sequences are unavailable, and a 960-bp representative region for whole-genome-based phylogenetic clustering within the L2 ORF was identified. Our study significantly expands the current knowledge of HPV6 genomic diversity and provides a valuable resource for future epidemiological, evolutionary, functional, pathogenicity, vaccination, and molecular assay development studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Research Grant 2014 from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to M.M.J. This work was also sponsored by the Slovenian Research Agency and the Ministry of Health, which funded M.M.J. through the Young Researcher Training Program (contract no. 1000-10-310163), and in part by the National Cancer Institute (grant CA78527) (R.D.B.), the Einstein-Montefiore Center for AIDS, funded by the NIH (grant AI-51519), and the Einstein Cancer Research Center (grant P30CA013330), funded by the National Cancer Institute.
We thank Ines Kmet and Anja Zagožen for technical laboratory support and Maja M. Lunar for technical advice and critical discussions. We thank Vanja Erčulj from RHO Sigma, Ljubljana, Slovenia, for support and help with statistical analysis. Specimens from Canada were obtained with the support of the Reseau FRSQ SIDA-Maladies Infectieuses.
Footnotes
Published ahead of print 16 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00621-14.
REFERENCES
- 1.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubin F, Prétet JL, Jacquard AC, Saunier M, Carcopino X, Jaroud F, Pradat P, Soubeyrand B, Leocmach Y, Mougin C, Riethmuller D. 2008. Human papillomavirus genotype distribution in external acuminata condylomata: a large French national study (EDiTH IV). Clin. Infect. Dis. 47:610–615. 10.1086/590560 [DOI] [PubMed] [Google Scholar]
- 3.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. 2009. A review of human carcinogens. Part B: biological agents. Lancet Oncol. 10:321–322. 10.1016/S1470-2045(09)70096-8 [DOI] [PubMed] [Google Scholar]
- 4.Garland SM, Steben M, Sings HL, James M, Lu S, Railkar R, Barr E, Haupt RM, Joura EA. 2009. Natural history of genital warts: analysis of the placebo arm of 2 randomized phase III trials of a quadrivalent human papillomavirus (types 6, 11, 16, and 18) vaccine. J. Infect. Dis. 199:805–814. 10.1086/597071 [DOI] [PubMed] [Google Scholar]
- 5.Komloš KF, Kocjan BJ, Košorok P, Luzar B, Meglič L, Potočnik M, Hočevar-Boltežar I, Gale N, Seme K, Poljak M. 2012. Tumor-specific and gender-specific pre-vaccination distribution of human papillomavirus types 6 and 11 in anogenital warts and laryngeal papillomas: a study on 574 tissue specimens. J. Med. Virol. 84:1233–1241. 10.1002/jmv.23318 [DOI] [PubMed] [Google Scholar]
- 6.Cubie HA. 2013. Diseases associated with human papillomavirus infection. Virology 445:21–34. 10.1016/j.virol.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 7.Cornall AM, Roberts JM, Garland SM, Hillman RJ, Grulich AE, Tabrizi SN. 2013. Anal and perianal squamous carcinomas and high-grade intraepithelial lesions exclusively associated with “low-risk” HPV genotypes 6 and 11. Int. J. Cancer 133:2253–2258. 10.1002/ijc.28228 [DOI] [PubMed] [Google Scholar]
- 8.Insinga RP, Liaw KL, Johnson LG, Madeleine MM. 2008. A systematic review of the prevalence and attribution of human papillomavirus types among cervical, vaginal, and vulvar precancers and cancers in the United States. Cancer Epidemiol. Biomarkers Prev. 17:1611–1622. 10.1158/1055-9965.EPI-07-2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miralles-Guri C, Bruni L, Cubilla AL, Castellsague X, Bosch FX, de Sanjosé S. 2009. Human papillomavirus prevalence and type distribution in penile carcinoma. J. Clin. Pathol. 62:870–878. 10.1136/jcp.2008.063149 [DOI] [PubMed] [Google Scholar]
- 10.Huebbers CU, Preuss SF, Kolligs J, Vent J, Stenner M, Wieland U, Silling S, Drebber U, Speel EJ, Klussmann JP. 2013. Integration of HPV6 and downregulation of AKR1C3 expression mark malignant transformation in a patient with juvenile-onset laryngeal papillomatosis. PLoS One 8:e57207. 10.1371/journal.pone.0057207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G. 2013. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 445:224–231. 10.1016/j.virol.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 12.Kiatpongsan S, Campos NG, Kim JJ. 2012. Potential benefits of second-generation human papillomavirus vaccines. PLoS One 7:e48426. 10.1371/journal.pone.0048426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combrinck CE, Seedat RY, Randall C, Roodt Y, Burt FJ. 2012. Novel HPV-6 variants of human papillomavirus causing recurrent respiratory papillomatosis in southern Africa. Epidemiol. Infect. 140:1095–1101. 10.1017/S0950268811001580 [DOI] [PubMed] [Google Scholar]
- 14.Danielewski JA, Garland SM, McCloskey J, Hillman RJ, Tabrizi SN. 2013. Human papillomavirus type 6 and 11 genetic variants found in 71 oral and anogenital epithelial samples from Australia. PLoS One 8:e63892. 10.1371/journal.pone.0063892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Matos RP, Sichero L, Mansur IM, do Bonfim CM, Bittar C, Nogueira RL, Küpper DS, Valera FC, Nogueira ML, Villa LL, Calmon MF, Rahal P. 2013. Nucleotide and phylogenetic analysis of human papillomavirus types 6 and 11 isolated from recurrent respiratory papillomatosis in Brazil. Infect. Genet. Evol. 16:282–289. 10.1016/j.meegid.2012.12.033 [DOI] [PubMed] [Google Scholar]
- 16.Heinzel PA, Chan SY, Ho L, O'Connor M, Balaram P, Campo MS, Fujinaga K, Kiviat N, Kuypers J, Pfister H. 1995. Variation of human papillomavirus type 6 (HPV-6) and HPV-11 genomes sampled throughout the world. J. Clin. Microbiol. 33:1746–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocjan BJ, Jelen MM, Maver PJ, Seme K, Poljak M. 2011. Pre-vaccination genomic diversity of human papillomavirus genotype 6 (HPV 6): a comparative analysis of 21 full-length genome sequences. Infect. Genet. Evol. 11:1805–1810. 10.1016/j.meegid.2011.06.022 [DOI] [PubMed] [Google Scholar]
- 18.Kocjan BJ, Poljak M, Cimerman M, Gale N, Potočnik M, Bogovac Ž, Seme K. 2009. Prevaccination genomic diversity of human papillomavirus genotype 6 (HPV 6). Virology 391:274–283. 10.1016/j.virol.2009.06.030 [DOI] [PubMed] [Google Scholar]
- 19.Burk RD, Chen Z, Harari A, Smith BC, Kocjan BJ, Maver PJ, Poljak M. 2011. Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages. Acta Dermatovenerol. Alp. Panonica Adriat. 20:113–123 http://www.zsd.si/ACTA/PUBLIC_HTML/acta-apa-11-3/2.pdf [PMC free article] [PubMed] [Google Scholar]
- 20.Platt AR, Woodhall RW, George AL., Jr 2007. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques 43:58, 60, 62. 10.2144/000112499 [DOI] [PubMed] [Google Scholar]
- 21.Kocjan BJ, Gale N, Hočevar Boltežar I, Seme K, Fujs Komloš K, Hošnjak L, Maver PJ, Jelen MM, Zupanič Pajnič I, Balažic J, Poljak M. 2013. Identical human papillomavirus (HPV) genomic variants persist in recurrent respiratory papillomatosis for up to 22 years. J. Infect. Dis. 207:583–587. 10.1093/infdis/jis733 [DOI] [PubMed] [Google Scholar]
- 22.Ure AE, Forslund O. 2012. Lack of methylation in the upstream region of human papillomavirus type 6 from aerodigestive tract papillomas. J. Virol. 86:13790–13794. 10.1128/JVI.01938-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 25.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution, version 4.08a. http://macclade.org/index.html [DOI] [PubMed]
- 29.Smith B, Chen Z, Reimers L, van Doorslaer K, Schiffman M, Desalle R, Herrero R, Yu K, Wacholder S, Wang T, Burk RD. 2011. Sequence imputation of HPV16 genomes for genetic association studies. PLoS One 6:e21375. 10.1371/journal.pone.0021375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Schiffman M, Herrero R, DeSalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2013. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One 8:e72565. 10.1371/journal.pone.0072565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Schiffman M, Herrero R, Desalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2011. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One 6:e20183. 10.1371/journal.pone.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho L, Chan SY, Burk RD, Das BC, Fujinaga K, Icenogle JP, Kahn T, Kiviat N, Lancaster W, Mavromara-Nazos P. 1993. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 67:6413–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong CK, Chan SY, Campo MS, Fujinaga K, Mavromara-Nazos P, Labropoulou V, Pfister H, Tay SK, ter Meulen J, Villa LL. 1993. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J. Virol. 67:6424–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Terai M, Fu L, Herrero R, DeSalle R, Burk RD. 2005. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. J. Virol. 79:7014–7023. 10.1128/JVI.79.11.7014-7023.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sichero L, Ferreira S, Trottier H, Duarte-Franco E, Ferenczy A, Franco EL, Villa LL. 2007. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer 120:1763–1768. 10.1002/ijc.22481 [DOI] [PubMed] [Google Scholar]
- 36.Bernard HU, Calleja-Macias IE, Dunn ST. 2006. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int. J. Cancer 118:1071–1076. 10.1002/ijc.21655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.