ABSTRACT
Influenza is the cause of significant morbidity and mortality in pediatric populations. The contribution of pulmonary host defense mechanisms to viral respiratory infection susceptibility in very young children is poorly understood. As a surrogate to compare mucosal immune responses of infant and adult lungs, rhesus monkey primary airway epithelial cell cultures were infected with pandemic influenza A/H1N1 virus in vitro. Virus replication, cytokine secretion, cell viability, and type I interferon (IFN) pathway PCR array profiles were evaluated for both infant and adult cultures. In comparison with adult cultures, infant cultures showed significantly increased levels of H1N1 replication, reduced alpha interferon (IFN-α) protein synthesis, and no difference in cell death following infection. Age-dependent differences in expression levels of multiple genes associated with the type I IFN pathway were observed in H1N1-infected cultures. To investigate the pulmonary and systemic responses to H1N1 infection in early life, infant monkeys were inoculated with H1N1 by upper airway administration. Animals were monitored for virus and parameters of inflammation over a 14-day period. High H1N1 titers were recovered from airways at day 1, with viral RNA remaining detectable until day 9 postinfection. Despite viral clearance, bronchiolitis and alveolitis persisted at day 14 postinfection; histopathological analysis revealed alveolar septal thickening and intermittent type II pneumocyte hyperplasia. Our overall findings are consistent with the known susceptibility of pediatric populations to respiratory virus infection and suggest that intrinsic developmental differences in airway epithelial cell immune function may contribute to the limited efficacy of host defense during early childhood.
IMPORTANCE To the best of our knowledge, this study represents the first report of intrinsic developmental differences in infant airway epithelial cells that may contribute to the increased susceptibility of the host to respiratory virus infections. Despite the global burden of influenza, there are currently no vaccine formulations approved for children <6 months of age. Given the challenges of conducting experimental studies involving pediatric patients, rhesus monkeys are an ideal laboratory animal model to investigate the maturation of pulmonary mucosal immune mechanisms during early life because they are most similar to those of humans with regard to postnatal maturation of the lung structure and the immune system. Thus, our findings are highly relevant to translational medicine, and these data may ultimately lead to novel approaches that enhance airway immunity in very young children.
INTRODUCTION
Influenza remains a significant health threat for pediatric populations. The Centers for Disease Control and Prevention estimates that 20,000 children under the age of 5 years are hospitalized each year due to influenza-related illnesses (1, 2). During the 2009 H1N1 pandemic, children showed an increased risk of severe outcomes and a higher death rate than that of previous influenza seasons (3–7). We currently lack licensed vaccines and antiviral drugs for children <6 months of age, and traditional inactivated vaccines for school-aged children show limited efficacy (8, 9). A better understanding of the mucosal immune response to influenza virus infection in young children is necessary to identify specific deficiencies in young hosts and elucidate adjunctive methods for improving pediatric immunity against all respiratory pathogens.
Although the infant immune system provides some protection against infectious disease, its ability to mount a robust response to respiratory pathogens is limited (10–13). The reduced functional capacity of the infant immune system has been attributed to deficiencies in several immune cells, including T lymphocytes, B lymphocytes, and antigen-presenting cells (14–18). Circulating T cells and antigen-presenting cells in infants are impaired in a range of effector functions from cellular activation to cytokine production (11, 13, 19–23).
Airway epithelial cells play a central role in host immunity to respiratory pathogens, providing the first line of defense and regulating adaptive immune responses (24). A unique feature of primate species is the presence of a prolonged period of postnatal lung development, during which airway epithelial cells continue to mature and differentiate (25–27). In humans, levels of antimicrobial receptors and defensins in the airway epithelium have been shown to increase from birth through preschool age (18, 28, 29), and dramatic differences in endotoxin responsiveness have been demonstrated for infant compared to adult intestinal epithelial cells (30). We have recently reported that innate immune responses to endotoxin in the nonhuman primate airway epithelium are also variable with age (31). Collectively, these data suggest that chronological age can impact the immune response elicited by lung mucosa in infant versus adult hosts.
The mechanisms underlying the development of immune competency in human infant lungs are not well understood due to challenges and ethical concerns involved in studying pediatric patients. Thus, our understanding of mucosal immunity in early life has been acquired primarily through studies of human cord blood and neonatal rodents. Because the interconnected maturation of pulmonary and immune systems in humans is distinct from that of rodents, it is critical to address translational questions associated with the development of lung immunity in a primate species (32, 33). In this study, we hypothesized that intrinsic functional differences in infant respiratory mucosa contribute to enhanced susceptibility to influenza virus infection. This hypothesis was tested by conducting infection studies with the pandemic influenza virus A/H1N1 strain in primary airway epithelial cell cultures derived from infant and adult rhesus monkeys. To compare the in vitro infant airway epithelial cell responses to infection with in vivo airway responses, we assessed pulmonary and systemic responses following inoculation of the pandemic influenza virus A/H1N1 strain in infant rhesus monkeys.
MATERIALS AND METHODS
Virus.
The A/California/04/2009 H1N1 strain was obtained from the Centers for Disease Control and Prevention (Atlanta, GA) through the Lovelace Respiratory Research Institute. The stock was passaged twice in embryonated eggs and twice in Madin-Darby canine kidney (MDCK) cells. Viral stocks were stored in liquid nitrogen and thawed immediately before use.
In vitro H1N1 infection.
Infant (<12 months of age) and adult (>3 years of age) rhesus monkey tracheobronchial tissues were obtained from the California National Primate Research Center Pathology Unit. Primary airway epithelial cells were cultured on transwell inserts (Corning, Corning, NY) under air-liquid interface (ALI) conditions, as previously described (31). The apical surface of individual ALI culture wells was inoculated with 150 μl H1N1 influenza virus in ALI medium with 1 μg/ml trypsin at a multiplicity of infection (MOI) of 0, 0.1, 1, or 10. In addition, an inoculum at an MOI of 1 was UV inactivated as a control. The inoculum was removed from ALI culture wells after 1 h, and the apical epithelial cell surface was washed with sterile phosphate-buffered saline (PBS) (Sigma-Aldrich Corp.). ALI cultures were subsequently maintained for 6 to 96 h.
For poly(I·C) stimulations, the Toll-like receptor 3 ligand was diluted in ALI medium to a concentration of 10 μg/ml, and 100 μl was then applied to the apical surface. The stimulation medium was removed after 1 h, and cells were maintained in culture for 24 h.
Metabolic activity was measured with alamarBlue (Invitrogen/Life Technologies, Carlsbad, CA), which was diluted directly into the basolateral ALI medium of cultures with ∼16-h incubations at 37°C in 5% CO2. Fluorescence was read by using an excitation wavelength of 540 to 570 nm and an emission wavelength of 580 to 610 nm, according to the manufacturer's recommendations. Nonviable cells generate a lower fluorescence signal than healthy cells, and results were reported relative to those for age-matched, mock-infected controls for each time point.
In vivo H1N1 infection.
Infant rhesus macaque monkeys between 6 and11 months of age were inoculated with 1 × 108 50% tissue culture infective doses (TCID50) of H1N1 influenza A virus (n = 5) or control medium (n = 3) by intranasal and intratracheal instillation in a 1-ml total volume at day 0. While animals were under ketamine anesthesia, the nasal cavity was lavaged with 2 ml of PBS (Sigma-Aldrich, St. Louis, MO), and trachea samples were obtained with a Mini-Tip BD Culturette swab (BD Biosciences, San Jose, CA) that was inserted with the aid of a laryngoscope. Nasal lavage fluid, pharyngeal swab samples, trachea swab samples, and peripheral blood were collected on days −4, 1, 2, 3, 7, 9, 11, and 14 of infection. Rectal temperature was measured at each collection time point. All animals were seronegative for hemagglutinin-inhibiting antibodies to H1N1 influenza virus prior to the initiation of the study. Bronchoalveolar lavage fluid specimens were obtained at the time of necropsy by cannulation of the right caudal lung lobe and inflation with 20 ml sterile PBS. Care and housing of animals complied with the provisions of the Institute of Laboratory Animal Resources and conformed to practices established by the American Association for Accreditation of Laboratory Animal Care.
H1N1 virus quantitation.
Viral titers were determined by TCID50 assays on MDCK cells according to the method of Reed and Muench (34). Samples were incubated for 24 h, followed by visualization of influenza virus-infected cells by immunocytochemistry using an anti-influenza A virus nucleoprotein clone A1 and A3 blend (Millipore, Billerica, MA) and using 3′,3′-diaminobenzidine (Vector Laboratories, Burlingame, CA). Influenza virus matrix (M) gene RNA was measured by reverse transcription-PCR (RT-PCR) using SYBR green (Applied Biosystems, Carlsbad, CA). A standard curve of the full-length matrix gene of influenza virus A/California/04/2009 was amplified by RT-PCR with forward primer TTC ACA GCA TCG GTC TCA CAG ACA and reverse primer TCC AGC CAT CTG TTC CAT AGC CTT. The PCR products were cloned into pTarget (Promega, Madison, WI) for in vitro transcription with a MEGAshortscript kit (Applied Biosystems).
RNA isolation and analysis.
Cells were harvested in TRIzol reagent (Invitrogen), and reverse transcription was performed on total RNA extracted with random hexamer primers and MultiScribe reverse transcriptase (Applied Biosystems). mRNA levels of interleukin-8 (IL-8), alpha 6 interferon (IFN-α6), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were measured by TaqMan real-time PCR using commercially available primer-probe sets (Applied Biosystems) and analyzed on an Applied Biosystems Prism 7900 sequence detection system. Progressive dilution of purified human cDNA plasmid constructs (Origene, Rockville, MD) for each gene target was used to generate standard curves for the absolute quantitation of mRNA copy numbers. All reagents were tested to confirm comparable detection of both rhesus and human targets.
For the IFN-α/β PCR array (SABiosciences, Valencia, CA), airway epithelial cells were harvested in RLT buffer (Qiagen, Valencia, CA), and RNA was extracted according to the RNeasy minikit instructions. The RT2 first-strand kit was used to synthesize cDNA from 1,000 ng of RNA for use in a 96-well format, which was analyzed on a ViiA 7 real-time PCR system (Applied Biosystems). Analysis of the PCR array was conducted as previously reported, using RT2 PCR array online data analysis software, version 3.5 (31).
Cytokine ELISA.
IL-8 (eBiosciences, San Diego, CA) and IFN-α (panspecific; MABtech, Sweden) proteins were measured by enzyme-linked immunosorbent assays (ELISAs), according to the manufacturer's instructions. The limits of detection for ELISAs were 4 pg/ml (IL-8) and 7 pg/ml (IFN-α).
Immunocytochemistry staining of airway epithelial cell cultures.
For H1N1 immunostaining, epithelial cells on transwells were fixed with 10% phosphate-buffered formalin (Sigma-Aldrich Corp.) for 30 min. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 1 h, followed by a nonspecific block with Western blot-grade nonfat dried milk for 30 min (Lab Scientific, Highlands, NJ). The primary anti-influenza A virus nucleoprotein clone A1 and A3 blend antibody (Millipore, Billerica, MA) was incubated overnight at 4°C. Incubation with horseradish peroxidase-conjugated secondary antibody (Life Technologies, Grand Island, NY) was followed by 3′,3′-diaminobenzidine substrate development. The immunostained transwell membrane was mounted onto a microscope slide and imaged on an Olympus BX61 microscope with an Olympus DP72 color camera. For cleaved caspase-3 immunofluorescent staining, the following modifications were made: transwells were blocked in immunograde bovine serum albumin (Sigma-Aldrich Corp.) and incubated with Image-It (Invitrogen/Life Technologies) prior to overnight incubation with the caspase-3 primary antibody (cleaved ASP175; Cell Signaling Technologies, Danvers, MA). A goat anti-rabbit Alexa 488 secondary antibody (Invitrogen/Life Technologies) was applied for 45 min.
Leukocyte differentials.
Aliquots of lavage fluid and swab samples were cytocentrifuged, air dried, and stained with a modified Wright stain (Diff-Quik; Polysciences, Warrington, PA). The proportion of macrophages, monocytes, neutrophils, eosinophils, lymphocytes, and epithelial cells was determined by counting 300 cells per sample by light microscopy.
Histopathology.
The left caudal and right cranial lung lobes of mock- and influenza virus-infected infants were perfused via bronchial cannula for 1 h at 30-cm fluid pressure with 10% phosphate-buffered formalin (Sigma-Aldrich Corp.). The lung lobes were then processed and embedded in paraffin. Five-micrometer-thick paraffin sections were cut and stained with hematoxylin and eosin (H&E). Histopathology slides were read and scored by a board-certified veterinary pathologist. Images were obtained with an Olympus BX53 microscope mounted with an Olympus DP72 camera and Olympus CellSens software.
Pulmonary lesions were scored on a scale of 0 to 4, with a score of 0 indicating no lesions; 1 indicating minimal lesions, with inflammation centered on terminal bronchioles with little or no extension into the dependent alveoli; 2 indicating mild lesions, with inflammation centered on terminal bronchioles with some extension into the dependent alveoli; 3 indicating moderate lesions, with inflammation centered on terminal bronchioles with extension into the dependent alveoli accompanied by moderate alveolar epithelial injury and repair (large confluent lesions may be formed); and 4 indicating severe lesions, with extensive inflammation centered on terminal bronchioles and alveolar parenchyma with severe alveolar epithelial injury and repair resulting in extensive confluent lesions.
Statistics.
Data are reported as means ± standard errors of the means (SEM), unless otherwise noted. Time postinfection and age-dependent differences were evaluated for each infection condition separately by two-way analysis of variance (ANOVA) with multiple comparisons and a Holm-Sidak posttest. Unpaired two-tailed, parametric, or nonparametric t tests were also used where appropriate. GraphPad Prism 6.03 (GraphPad, La Jolla, CA) was used for statistical analysis. A P value of 0.05 or less was considered statistically significant.
RESULTS
Increased H1N1 replication in infant airway epithelial cell cultures.
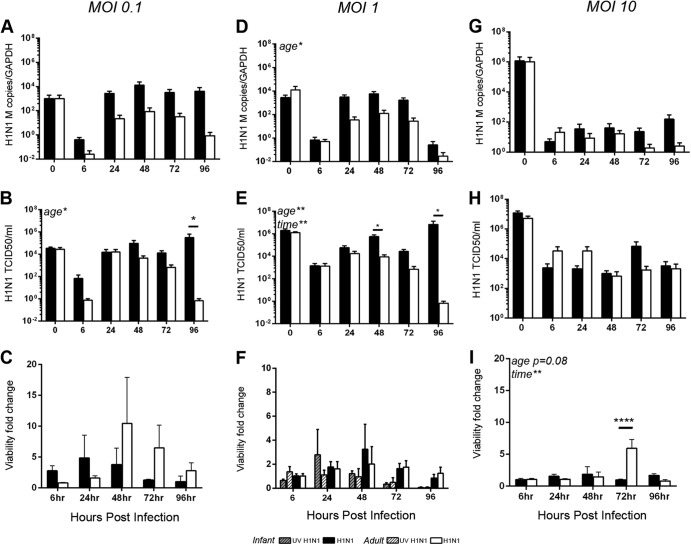
Little is known about the immunological factors that contribute to heightened infection susceptibility in very young children. As an approach to compare the mucosal innate immune responses to viral infection in infant and adult monkeys, we conducted comparative H1N1 influenza virus A/CA/04/09 infections with live or UV-inactivated virus in primary airway epithelial cell ALI cultures derived from infant (<1 year of age) and adult (>3 years of age) rhesus monkeys. At an MOI of 1, levels of viral RNA and replicating virus measured in wash specimens from the apical surface were increased in infant ALI cultures relative to those of adults, with significantly higher H1N1 titers being detected at 48 and 96 h postinfection (by two-way ANOVA for age and time postinfection) (Fig. 1D and E). Levels of replication-competent virus fluctuated at between 24 and 96 h postinfection in infant cultures, whereas levels of virus recovered from adult cultures appeared to progressively decline after 24 h. No replicating virus was detected in cultures infected with UV-inactivated virus. Age-specific differences in replicating H1N1 levels but not viral mRNA levels were also observed during infection with a lower dose (MOI 0.1), resulting in significantly higher virus titers in infant than in adult cultures at 96 h postinfection (by two-way ANOVA for age and time postinfection) (Fig. 1A and B). Similar to infant cultures infected at an MOI of 1, infant cultures infected at an MOI of 0.1 varied in the levels of virus recovered throughout the infection period, whereas adult virus titers tended to decline from 24 to 96 h. H1N1 infection of infant and adult cultures at an MOI of 10 resulted in similar viral replication and mRNA levels between age groups, with little to no increase over the infection period (Fig. 1G and H). At an MOI of 1, we also observed more H1N1 nucleoprotein immunostaining in infant ALI cultures than in cultures from adults (Fig. 2).
FIG 1.
H1N1 replication and cell viability in infant and adult airway epithelial cell cultures. Cultures from infant and adult monkeys were inoculated with live or UV-inactivated (UV) H1N1 virus at MOIs of 0.1, 1, and 10, followed by evaluation of viral replication and cell viability at 6, 24, 48, 72, and 96 h postinfection (n = 3 to 5 for each age group). The zero time point refers to the viral inoculum. (A, D, and G) Viral RNA and GAPDH levels in the apical wash fluid were measured by RT-PCR. Absolute numbers of H1N1 matrix (M) gene copies were calculated based on a standard curve and are reported relative to the GAPDH copy number. (B, E, and H) Replicating virus in apical wash samples at each time point was evaluated by TCID50 assays. As no viral RNA or replicating virus was detected in any culture infected with UV-inactivated virus, these were excluded from the graphs. (C, F, and I) Cell viability was assessed by alamarBlue assays. The fold change in fluorescence over that for mock-infected controls is reported for both UV-inactivated and live viruses at each time point. Bar graphs represent means and SEM. Statistically significant results of a two-way ANOVA comparing age and time postinfection are indicated at the top left of the graphs, while horizontal bars show results of Holm-Sidak posttests. *, P < 0.05; **, P < 0.005; ****, P < 0.0001.
FIG 2.

Influenza A virus nucleoprotein staining in infant and adult epithelial cell cultures. Representative mock- and H1N1-infected (MOI of 1) epithelial cell cultures at 24 h postinfection were immunostained with anti-influenza virus nucleoprotein immunoglobulin and visualized with 3′,3′-diaminobenzidine. (A and C) Infant cultures; (B and D) adult cultures. Images were collected at a ×20 magnification. Bar = 100 mm. Arrowheads indicate positive staining.
To determine if cell death had an impact on viral replication levels, viability assays were conducted using ALI cultures. Infant and adult airway epithelial cell cultures showed no significant difference in cell viability when infected with live or UV-inactivated virus (MOI of 1) at any time point from 6 to 96 h postinfection (Fig. 1F). Immunostaining for the apoptosis marker cleaved caspase-3 showed more fluorescence-positive cells in H1N1-infected (MOI of 1) infant and adult epithelial cell cultures than in mock-infected controls, peaking at 6 h postinfection; overall, the cleaved-caspase-3 staining patterns were similar in H1N1-infected infant and adult cultures (Fig. 3). There was also no clear trend for age-specific differences in cell viability for infection at an MOI of 0.1 (Fig. 1C). However, in cultures infected at an MOI of 10, time was a significant source of variation, and age was near significance (P = 0.08), for metabolic activity, which peaked at 72 h in adult cells (by two-way ANOVA for age and time postinfection) (Fig. 1I).
FIG 3.
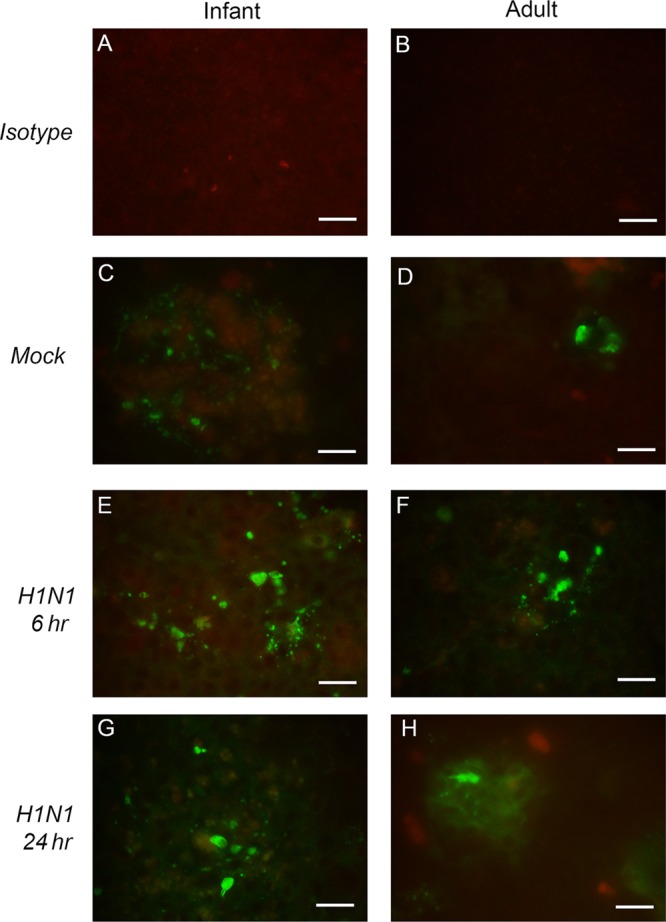
Cleaved-caspase-3 immunofluorescent staining in infant and adult epithelial cell cultures. Representative mock- and H1N1-infected (MOI of 1) epithelial cell cultures were stained with mouse monoclonal antibody to cleaved caspase-3 and visualized with an Alexa 488 fluorescent secondary antibody to detect apoptotic cells. (A and B) Isotype controls are included for comparison. (C to H) Mock-infected cultures were evaluated at 24 h postinfection (C and D), and H1N1-infected cultures were evaluated at 6 h (E and F) and 24 h (G and H) postinfection. Images were collected at a ×20 magnification. Bar = 100 mm.
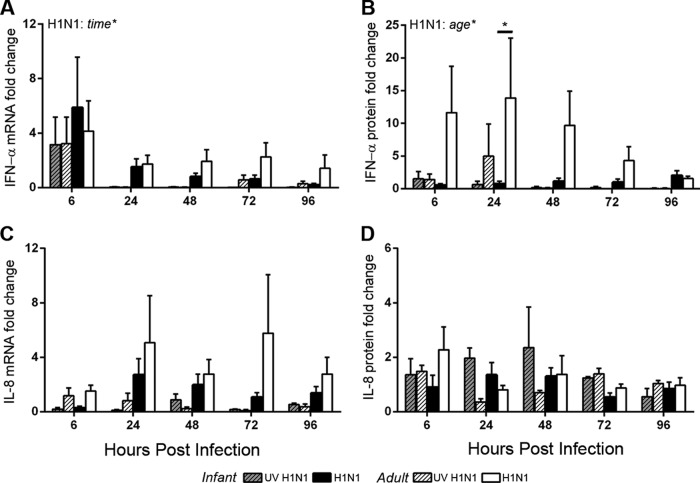
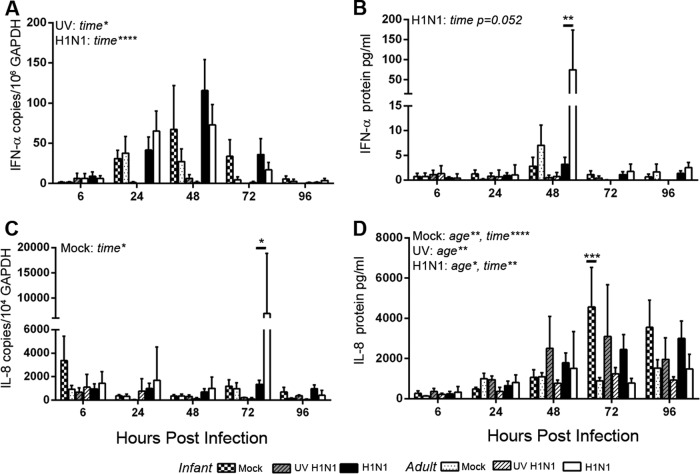
Evaluation of the antiviral cytokine IFN-α revealed an increase in mRNA levels over those of mock-infected controls at 6 h postinfection in both infant and adult airway epithelial cell cultures infected with live or UV-inactivated virus at an MOI of 1 (Fig. 4A). Infection-induced changes in IFN-α mRNA expression levels were lower at subsequent time points for both age groups, with no age-dependent differences being detected (by two-way ANOVA for age and time postinfection; live and UV-inactivated virus statistical tests were done separately). In contrast, age significantly impacted the secretion of IFN-α protein in basolateral media from H1N1-infected ALI cultures (by two-way ANOVA for time and age) (Fig. 4B). Little to no induction of IFN-α protein was detected in H1N1-infected infant cultures relative to mock-infected cultures, whereas adult cultures showed increased levels above those of mock-infected controls by 6 h postinfection. The total IFN-α protein level in the basolateral medium peaked at 48 h postinfection in H1N1-infected cultures, with significantly lower levels in infants than in adults (by two-way ANOVA for age and time postinfection) (Fig. 5B). No age-dependent difference in IFN-α mRNA or protein induction was detected in cultures infected with UV-inactivated virus. We also assessed the impact of age on IL-8 expression in ALI cultures; levels of induction of IL-8 mRNA for both live and UV-inactivated H1N1 infections were generally lower in infant than in adult cultures at most infection time points, but these differences did not reach statistical significance (by two-way ANOVA for age and time postinfection; live and UV-inactivated virus statistical tests were done separately) (Fig. 4C). Similarly, the induction of IL-8 protein with H1N1 infection relative to mock infection did not vary significantly with age (by two-way ANOVA for age and time postinfection) (Fig. 4D). Independent of infection status, IL-8 protein concentrations in the basolateral medium of infant cultures increased over the 96-h time course, whereas adult cultures showed minimal changes (by two-way ANOVA for age and time postinfection) (Fig. 5D). We also compared the cytokine responses to Toll-like receptor 3 ligand binding after 24 h following treatment with poly(I·C) in infant and adult airway epithelial cell ALI cultures (Fig. 6). As with H1N1 infection, we detected transcription of IFN-α mRNA but no IFN-α protein induction with poly(I·C) treatment in infant cultures. This was in contrast to the robust IFN-α protein response to poly(I·C) observed in adult cultures (P < 0.05 by Student's t test for infants versus adults). IL-8 mRNA and protein were induced by poly(I·C) treatment in both infant and adult airway epithelial cell cultures, with no age-dependent differences being noted at 24 h posttreatment.
FIG 4.
Infant and adult airway epithelial cell cytokine responses to H1N1 infection. IFN-α and IL-8 cytokine responses were measured at 6, 24, 48, 72, and 96 h after mock infection and after UV-inactivated and live H1N1 virus infection at an MOI of 1. (A and C) Transcripts of cytokine genes and GAPDH were measured in epithelial cells by RT-PCR, and absolute gene copy numbers relative to the GAPDH copy number were calculated based on standard curves for IFN-α (A) and IL-8 (C). (B and D) Cytokine protein secretion into the basolateral medium was measured by ELISAs for pan-IFN-α (B) and IL-8 (D). Values are reported as the fold change over values for average age-matched mock-infected controls for each time point. Absolute mRNA and protein levels are reported in Fig. 5. Bar graphs represent means and SEM. Statistically significant results from two-way ANOVA comparing age and time postinfection for live and UV-inactivated viruses (analyzed separately) are indicated in at the top left of the graphs. The horizontal bars show results of Holm-Sidak posttests. *, P < 0.05.
FIG 5.
Absolute values for infant and adult airway epithelial cell cytokine responses to H1N1 infection. IFN-α and IL-8 cytokine responses were measured at 6, 24, 48, 72, and 96 h after mock infection and UV-inactivated and live H1N1 virus infections at an MOI of 1 (n = 3 to 6). Cytokine and GAPDH mRNA levels were measured in airway epithelial cells by RT-PCR. (A and C) Gene copy numbers relative to GAPDH copy numbers were calculated based on standard curves for IFN-α (A) and IL-8 (C). (B and D) Cytokine protein secretion into the basolateral medium was measured by ELISAs for pan-IFN-α (B) and IL-8 (D). Bar graphs represent means and SEM. Statistically significant results from two-way ANOVA comparing age and time postinfection were determined for each condition separately and are indicated at the top left of the graphs (mock, UV-inactivated virus, or live virus). Horizontal bars show results of Holm-Sidak posttests. *, P < 0.05; **, P < 0.05; ****, P < 0.0001.
FIG 6.

Infant and adult airway epithelial cell cytokine responses to poly(I·C) treatment. IFN-α and IL-8 cytokine responses were measured at 24 h after treatment with 10 mg/ml poly(I·C) in infant and adult airway epithelial cell cultures (n = 4). Transcripts of cytokine genes and GAPDH were measured in epithelial cells by RT-PCR, and absolute gene copy numbers relative to the GAPDH copy number were calculated based on standard curves. Cytokine protein secretion into the basolateral medium was measured by an ELISA. Values are reported as the fold change over the average values for age-matched unstimulated controls. Bar graphs represent means and SEM. *, P < 0.05 by Student's t test comparing infant and adult values.
Targeted RT-PCR arrays for IFN-α and IFN-β pathways were used to further characterize the differential innate immune response to H1N1 infection in infant and adult primary airway epithelial cell ALI cultures. In mock-infected cultures, we observed multiple genes that were expressed at a >2-fold difference in adult versus infant cultures, although only the expression of MAL (myelin and lymphocyte protein) was found to be statistically significant (Table 1). MAL encodes an epithelial cell membrane lipoprotein known to be involved in apical transport and was expressed at >5-fold-lower levels in infant than in adult airway epithelial cells. After 24 h of H1N1 infection, infant cultures showed significantly increased mRNA levels of CXCL10, OAS1, OAS2, ISG15, and the apoptotic genes IFI27 and PTTG1 relative to mock-infected controls; none of these genes showed significantly increased levels in H1N1-infected adult cultures (by Student's t tests) (Fig. 7). For direct comparison of H1N1 infection-induced gene expression levels for IFN-α and IFN-β pathways in infant cultures relative to adult cultures, the levels of several genes, including CIITA, CASP1, and RPL13A, were significantly lower in infant cultures (by Student's t test) (Table 1). PTTG1 was the only gene that showed a significantly higher level in H1N1-infected infant cultures than in adult cultures. Many genes in the array showed no age-dependent differences in mock- or H1N1-infected epithelial cell cultures (see Tables S1 and S2 in the supplemental material).
TABLE 1.
Comparison of alpha and beta interferon pathway gene expression levels in infant and adult airway epithelial cell culturesa
| Gene | Infection | Avg CT (SD) |
Fold regulation | P value | |
|---|---|---|---|---|---|
| Infants | Adults | ||||
| MAL | − | 28.05 (1.1) | 25.44 (1.1) | −5.94 | 0.03 |
| IFNAR2 | − | 27.44 (0.6) | 28.14 (0.4) | 1.67 | 0.03 |
| CD70 | − | 27.07 (0.4) | 27.64 (0.5) | 1.53 | 0.05 |
| CIITA | + | 32 (0.44) | 30.13 (1.0) | −3.57 | 0.05 |
| CASP1 | + | 25.47 (0.67) | 24.26 (0.52) | −2.27 | 0.02 |
| RPL13A | + | 29.19 (0.42) | 28.07 (1.1) | −2.17 | 0.05 |
| PTTG1 | + | 21.96 (0.53) | 23.17 (0.90) | 2.34 | 0.02 |
Shown are significantly regulated genes from an RT2 profiler PCR array for alpha and beta interferons comparing infant and adult cultures. − indicates mock-infected cultures, and + indicates H1N1 at an MOI of 1 at 24 h postinfection. Also shown are average cycle threshold (CT) values, with standard deviations in parentheses (n = 4 per group). Fold regulation represents fold change values, with values of <1 indicating a negative number. Fold changes in infant gene expression levels over adult levels were calculated as 2−[(delta CT infant) − (delta CT adult)], where delta CT is the CT value of the gene of interest minus the mean CT value of housekeeping genes. The P value was determined based on a two-tailed t test of replicate 2(−delta CT) values.
FIG 7.
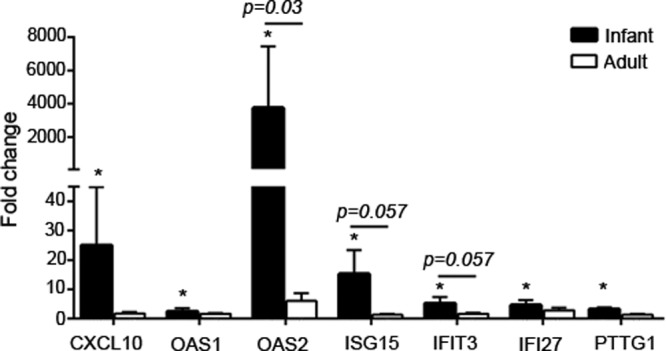
IFN-α and -β pathway gene expression in H1N1-infected airway epithelial cell cultures. IFN-α and -β array data are graphed for all mRNAs that were significantly modulated at 24 h post-H1N1 infection in infant and/or adult cultures. The average fold changes in gene expression levels and SEM in H1N1-infected animals compared to age-matched, mock-infected controls are graphed for infant (n = 4) and adult (n = 4) epithelial cell cultures. *, P < 0.05 by Student's t test for infection-induced fold changes. The horizontal bar and indicated P values are for comparisons between infant and adult fold changes in gene expression values determined by Student's t tests.
H1N1 viral loads and clinical outcomes for infant monkeys.
To assess infant susceptibility to H1N1 influenza virus infection in vivo, 6- to 11-month-old rhesus monkeys were inoculated with H1N1 A/California/04/09 (n = 5) or control medium (n = 3) by administration into upper airways. Animals were monitored longitudinally for viral replication, clinical symptoms, and inflammation over a 14-day infection time course. Replicating virus was recovered from nasal lavage fluid samples of all H1N1-infected animals at day 1 postinfection, with mean viral titers exceeding 4 × 104 TCID50/ml (Fig. 8A). Replicating virus remained detectable in 2 of the 5 animals to day 7 postinfection in nasal lavage fluid and pharyngeal swab samples. Trachea swab samples resulted in a low level of recovery of replicating virus, with only 2 of the 5 animals testing positive by TCID50 assays (data not shown), but viral RNA was detected by RT-PCR in all trachea samples from H1N1-infected animals through day 3 postinfection (Fig. 8B). Replicating virus was cleared from the nose and pharynx by day 9 postinfection, with low to undetectable levels of viral RNA being observed in lavage fluid and swab samples at this time point.
FIG 8.

H1N1 replication and elevated body temperature in infant monkeys. Infant monkeys were inoculated at day 0 with H1N1 virus or control medium. (A) The nasal cavity was serially lavaged postinoculation, and H1N1 virus replication was measured by TCID50 assays. Unique symbols are used to represent each individual animal, and values are plotted as the TCID50 per ml of nasal lavage fluid on a log scale, with means ± SEM indicated. (B) Viral RNA in nasal lavage fluid cell pellets as well as trachea and pharyngeal swab samples for each animal throughout the infection time course was assessed by RT-PCR. Absolute copy numbers of the H1N1 matrix (M) gene per copy of GAPDH were calculated based on a standard curve, and the averages (and SEM) were graphed for each sample type. Values are plotted on a log scale. (C) Rectal temperature was measured in degrees Fahrenheit prior to inoculation and at days 1, 2, 3, 7, 9, 11, and 14 postinoculation in H1N1- and mock-infected animals. The average temperature (±SEM) is graphed for mock-infected and H1N1-infected animals. *, P < 0.05 for infection status by two-way ANOVA for infection and time, with a significant difference at day 3; **, P < 0.005 by the Holm-Sidak posttest.
Body temperature was significantly different in H1N1-infected compared to mock-infected animals, with a mean of 103.1°F for H1N1-infected infants at day 3 postinfection (by two-way ANOVA for infection status and time) (Fig. 8C). Increased body temperature was maintained in 2 H1N1-infected animals for 2 days despite treatment with acetaminophen. Clinical evaluation of animals by California National Primate Research Center staff veterinarians detected wheezing in 3 of 5 H1N1-infected animals at between days 2 and 7 postinfection.
Airway inflammation in H1N1-infected infant monkeys.
There was a modest increase in upper airway inflammation in H1N1-infected animals, with a trend toward increased numbers of total leukocytes per ml recovered from nasal lavage fluids compared to mock-infected animals (P = 0.056 for infection status by two-way ANOVA) (Fig. 9A). Infection status accounted for 14.7% of the total variance in the nasal lavage fluid leukocyte counts. The phenotype of nasal lavage fluid leukocytes changed slightly during the evaluation period, with a significant influx of neutrophils (P = <0.05 for time by two-way ANOVA) (Fig. 9B). Time accounted for 35.8% of the total variance; however, there was no significant difference in neutrophil frequency between mock- and H1N1-infected animals. The frequency of lavage fluid monocytes was significantly increased over that for mock-infected animals (P = <0.05 for infection status by two-way ANOVA) (Fig. 9C). Lymphocyte frequencies in nasal lavage fluid were more variable and did not reach statistical significance at any infection time point (Fig. 9D). Greater differences were observed in the compositions of inflammatory cells recovered from trachea swab samples of H1N1- and mock-infected animals, with significantly increased frequencies of macrophages, monocytes, and lymphocytes as a result of infection (P = <0.05 for infection status by two-way ANOVA) (Fig. 9E, G, and H). In trachea swab samples, the neutrophil frequency did not change as a result of infection status or time (Fig. 9F). In the bronchoalveolar lavage fluid at day 14 postinfection, lymphocyte numbers appeared slightly elevated in H1N1-infected infants; however, only the numbers of monocytes were significantly increased over those in mock-infected animals (P < 0.05 by Student's t test) (Fig. 9I). H1N1 infection did not significantly alter total white blood cell counts or change the proportion of neutrophils, lymphocytes, monocytes, or eosinophils in the peripheral blood (Table 2).
FIG 9.
Upper airway inflammation following H1N1 influenza virus infection. (A) Leukocytes were quantitated in nasal cavity lavage fluid prior to and after inoculation of H1N1- and mock-infected infants. Total leukocyte numbers were calculated for each animal at days 1, 2, 3, 7, 9, 11, and 14 postinfection, and the averages (±SEM) were graphed for both groups. (A to H) Leukocyte differentials were conducted on cytospins of nasal lavage fluid (A to D) and trachea swab samples (E to H), and the frequencies of neutrophils (B and F), monocytes (C and G), lymphocytes (D and H), and macrophages (E) were graphed for influenza virus- and mock-infected infants across the infection time course. *, P < 0.05 for either time or infection status by two-way ANOVA. (I)The leukocyte composition of the bronchoalveolar (lung) lavage fluid at day 14 postinfection was also assessed in cytospins from mock- and H1N1-infected animals, and the percent frequencies of total lavage cells for each subtype observed are graphed. PMNs, polymorphonuclear leukocytes. **, P = 0.008 by Student's t test comparing mock- and H1N1-infected animals.
TABLE 2.
White blood cell counts and differentials in influenza virus- and mock-infected animalsa
| Day | Infection type | Mean WBC count/μl (103) ± SEM | Mean % of WBC count ± SEM |
|||
|---|---|---|---|---|---|---|
| Neutrophils | Monocytes | Lymphocytes | Eosinophils | |||
| 0 | H1N1 | 8.8 ± 1.8 | 47.2 ± 6.5 | 1.2 ± 0.5 | 51.4 ± 6.5 | 0.2 ± 0.2 |
| Mock | 11.9 ± 1.1 | 62.7 ± 3.8 | 3.0 ± 1.0 | 31.3 ± 4.1 | 3.0 ± 1.1 | |
| 1 | H1N1 | 7.7 ± 0.9 | 66.7 ± 7.6 | 4.8 ± 0.6 | 28.2 ± 7.6 | 0.4 ± 0.2 |
| Mock | 9.7 ± 1.1 | 49.3 ± 0.8 | 3.7 ± 0.9 | 40.3 ± 7.6 | 0 | |
| 2 | H1N1 | 5.6 ± 0.4 | 39.8 ± 5.2 | 2.8 ± 0.8 | 56.4 ± 5.8 | 1.0 ± 0.5 |
| Mock | 11.1 ± 2.5 | 53.3 ± 2.9 | 2.0 ± 0.0 | 44.7 ± 2.9 | 0 | |
| 3 | H1N1 | 8.6 ± 1.9 | 52.6 ± 11.6 | 1.8 ± 0.9 | 44.6 ± 11.7 | 1.0 ± 0.4 |
| Mock | 12.3 ± 2.4 | 60.0 ± 3.0 | 0.5 ± 0.5 | 35.0 ± 0.0 | 4.5 ± 2.5 | |
| 7 | H1N1 | 8.3 ± 1.1 | 41.6 ± 8.1 | 5.4 ± 0.7 | 52.0 ± 7.8 | 0.9 ± 0.7 |
| Mock | 10.5 ± 1.9 | 57.7 ± 7.2 | 4.0 ± 1.5 | 38.3 ± 8.6 | 0 | |
| 14 | H1N1 | 3.5 ± 0.5 | 37.2 ± 7.5 | 1.2 ± 0.4 | 56.8 ± 8.9 | 0.8 ± 0.2 |
| Mock | 5.1 + 0.7 | 43.3 ± 8.7 | 2.3 ± 0.9 | 53.7 ± 8.6 | 0.6 ± 0.6 | |
Complete white blood cell (WBC) counts and cell differentials were assessed by the CNPRC Clinical Laboratory Staff. The averages and standard errors are reported for mock-infected (n = 3) and influenza virus-infected (n = 5) infant rhesus macaques.
The airway and systemic proinflammatory cytokine responses following H1N1 infection were evaluated by ELISA and RT-PCR. There was a modest but not statistically significant increase in IFN-α protein levels in the nasal lavage fluid of H1N1-infected compared to mock-infected animals during the first few days of sampling, but this trend was reversed at later time points (Fig. 10A). In serum, the induction of IFN-α protein in H1N1-infected compared to mock-infected infants did not reach statistical significance (Fig. 10B). IFN-α mRNA levels were highly variable in peripheral blood and not consistently detected in nasal lavage fluid cell pellets regardless of infection status (data not shown). In contrast to the IFN-α response, IL-8 protein (Fig. 10C and D) and mRNA (data not shown) levels were reduced in both nasal lavage fluid and serum samples of H1N1-infected animals compared to mock-infected animals.
FIG 10.
IFN-α and IL-8 responses following H1N1 infection. (A and B) IFN-α protein levels were measured longitudinally in the nasal lavage fluid (A) and serum (B) by ELISAs and graphed on a log scale. (C and D) IL-8 protein levels at multiple time points postinfection in nasal lavage fluid (C) and serum (D) samples were determined by ELISAs. Graphs represent average values (±SEM) graphed on a log scale. *, P < 0.05 for infection status by two-way ANOVA for infection and time.
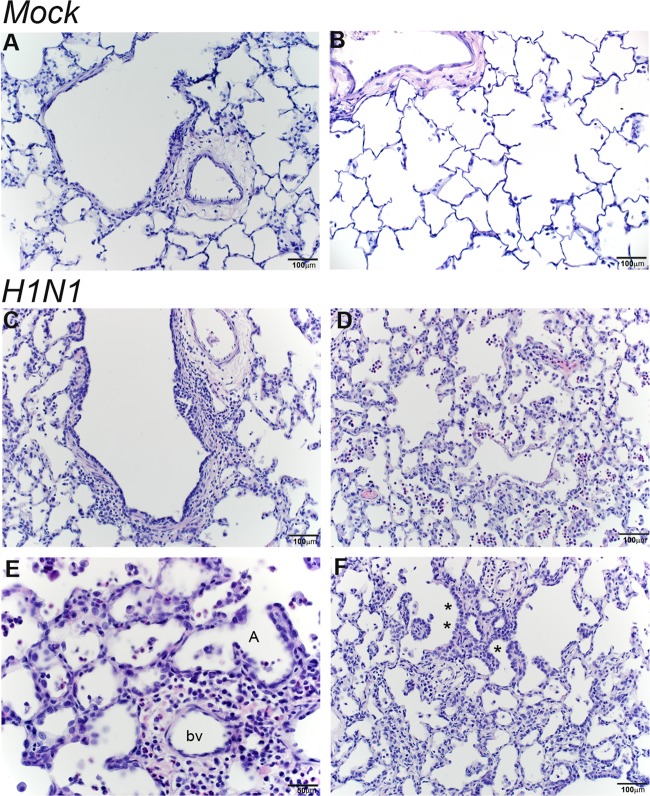
Recovery of the airways following clearance of infection was evaluated by histopathology of H&E-stained lung sections from mock- and H1N1-infected animals at 14 days postinfection. Lesions were scored on a scale from 1 to 4, with 1 being mild and 4 being severe (Table 3). Four of the five H1N1-infected animals had a pulmonary lesion score of 2 or higher, with influenza virus-induced lung lesions centered on the terminal bronchioles. The mildest lesions had some expansion of the bronchiolar wall and the adjacent perivascular zone by an inflammatory cell infiltrate composed predominantly of macrophages and small lymphocytes, with fewer plasma cells and neutrophils (Fig. 11C). In more severe lesions, inflammation extended out into the dependent alveoli. Alveolar septa were thickened by a mixed population of inflammatory cells, and alveolar spaces also contained inflammatory infiltrates, predominantly alveolar macrophages and a small number of neutrophils (Fig. 11D and E). In some areas, alveolar inflammation was accompanied by type II cell hyperplasia (Fig. 11F). In regions with resolved inflammation, the alveolar septae remained prominent and thickened, and the alveoli were intermittently lined by type II pneumocytes or plump type I pneumocytes with rounded nuclei protruding into the alveolus. The mock-challenged animals did not have any significant pulmonary lesions, although there were scattered clusters of alveolar macrophages in some histology sections of the lungs (Fig. 11A and B).
TABLE 3.
Pulmonary histopathology of influenza virus-infected infant monkeys
| Animal | Infection status | Pulmonary lesion scorea |
|---|---|---|
| 1 | H1N1 | 2 |
| 2 | H1N1 | 1.5 |
| 3 | H1N1 | 2.5 |
| 4 | H1N1 | 3 |
| 5 | H1N1 | 3 |
| 6 | Mock | 0 |
| 7 | Mock | 0 |
| 8 | Mock | 0 |
Lesion score scale where 0 indicates no lesions, 1 indicates minimal lesions, 2 indicates mild lesions, 3 indicates moderate lesions, and 4 indicates severe lesions.
FIG 11.
Persistent inflammation in respiratory bronchioles and alveoli at day 14 post-H1N1 infection. Histological sections from the lungs of mock- and H1N1-infected infant monkeys at 14 days postinoculation were stained with H&E. (A and B) Normal terminal bronchiole (A) and normal alveolar parenchyma (B) from mock-infected animals. (C to E) H1N1-infected animals show bronchiolar walls expanded by inflammation (macrophages and small lymphocytes, with fewer neutrophils) (C) and alveolar inflammation (predominantly neutrophils and macrophages) with thickened alveolar septae (D and E). (F) Alveolar septal thickening with some type II pneumocyte hyperplasia, marked with an asterisk, was also observed for H1N1-infected animals. bv, blood vessel; A, alveoli.
DISCUSSION
The immunological basis for the increased frequency and enhanced severity of respiratory virus infection observed in early life is poorly defined. To begin to investigate the immune mechanisms of influenza virus infection susceptibility during early childhood, we conducted comparative in vitro infections of infant and adult rhesus macaque airway epithelial cell cultures with pandemic H1N1. We also assessed whether infant rhesus monkeys could be infected in vivo by inoculation with pandemic H1N1.
We focused on the contribution of airway epithelial cells as essential mediators of host defense. Our studies showed that infant airway epithelial cells supported higher levels of H1N1 replication for an extended period of time than their adult counterparts during infection at an MOI of 1 or 0.1 (Fig. 1B and E). Increased viral replication in infant cultures did not appear to be due to age-dependent differences in cell death, as markers of cellular viability and apoptosis were similar in H1N1-infected epithelial cells from both age groups (Fig. 1C and F and 3). Variability in cell viability assays was noted for both infant and adult airway epithelial cells, which may reflect differences in plating efficiency during the initiation of primary cultures. Infant airway epithelial cells appeared to be capable of initiating an antiviral response at the transcriptional level, as evidenced by the observed increase in IFN-α and several type I IFN pathway gene mRNA levels at between 6 and 24 h in H1N1-infected cultures (Fig. 4A and 7). However, there was a lack of IFN-α protein induction with H1N1 infection as well as with poly(I·C) treatment in infant cultures (Fig. 4B and 6). It is important to consider that our current study was limited to IFN-α, and with recent evidence suggesting a critical role for IFN-λ in the control of influenza A virus infections in respiratory epithelium (35, 36), it will be important in the future to assess type III interferon in an age-dependent manner. Analysis of IL-8 in parallel did not reflect the age-dependent response of IFN-α (Fig. 4C and D), indicating that the expression of all cytokines in response to virus is not universally regulated by chronological age.
Our results suggest the existence of an intrinsic developmental mechanism in infant airway epithelial cells that is responsible for attenuated IFN-α protein production following viral infection. Infant airway epithelial cells were capable of sensing virus via Toll-like receptor 3, as demonstrated by the induction of IL-8 mRNA and protein following poly(I·C) treatment (Fig. 6). The observed attenuated IFN-α protein response in infant airway epithelial cells may be related to their sensitivity to the influenza virus-specific immune evasion and subversion properties reported previously for the influenza virus protein NS1 (37). To further explore the type I IFN molecular pathways that distinguish infant airway epithelial cells from adult airway epithelial cells, we measured mRNA expression levels of 84 genes associated with IFN-α and -β signaling pathways in ALI cultures. In uninfected cells, the MAL level was significantly reduced by almost 6-fold in infant cultures relative to adult cultures. MAL encodes a proteolipid that has been shown to function in signaling, epithelial cell differentiation, and apical transport of proteins, including influenza virus hemagglutinin (38–41). More recent studies described MAL as a putative tumor suppressor gene with a role in promoting apoptosis (42). The contribution of MAL to infant airway epithelial cell influenza virus susceptibility is unclear, and although its mRNA level was constitutively reduced in infant cultures, expression levels of MAL and several other type I IFN pathway genes were found to be increased at 24 h post-H1N1 infection (Fig. 7). Comparatively, the majority of genes evaluated in this pathway were either unchanged or downregulated in adult cultures at this infection time point (Table 1; see also Tables S1 and S2 in the supplemental material).
Age-dependent differences in the host response to pandemic H1N1 in rhesus macaques and in ferrets have recently been reported. Josset et al. examined late-life immunity to influenza in aged rhesus monkeys, which is highly relevant to our pediatric studies, as a mirror-image pattern has been used by other investigators to describe immune function at the beginning and near the end of life (43). Consistent with our observations of infant airway epithelium, aged rhesus monkeys showed increased viral loads and distinct innate immune responses to H1N1 compared to young adults (44). In contrast, ferrets showed no significant age-associated difference in H1N1 titers, although newly weaned ferrets had consistently higher viral loads than adult ferrets (45). The viral replication discrepancy among these studies is potentially related to species-specific differences between ferrets and primates or may be due to the different pandemic H1N1 strains used. Nevertheless, newly weaned ferrets did show differential interferon responses to pandemic H1N1 compared to adult ferrets, which is consistent with our findings for airway epithelial cells derived from infant and adult monkeys.
Our study is the first to demonstrate pandemic H1N1 infection in infant rhesus monkeys; however, an important limitation is the lack of parallel in vivo studies in adult rhesus monkeys for direct comparison of susceptibility. Based upon previously reported findings for adult monkeys infected with either seasonal or pandemic H1N1 strains, peak viral titers in the nasal lavage fluid of H1N1-infected infants in this study appear to be 1 to 2 logs higher (46–50). However, it should be emphasized that it is difficult to directly correlate parameters of infection and inflammation observed in this study with those of previous reports of adult rhesus monkeys based on differences in dose, inoculation methods, and sampling procedures. Surprisingly, unlike most influenza virus infection models, H1N1-infected infant monkeys showed only modest airway neutrophil infiltration associated with infection from days 1 to 3 (Fig. 9B and F). Furthermore, the level of the neutrophil chemoattractant IL-8 remained low in nasal lavage fluid during infection (Fig. 10C). Neutrophils were observed in nasal lavage fluid and trachea swab samples, where there was a statistically significant change over time in lavage fluid only, suggesting that the presence of this leukocyte phenotype may be due to repeated sampling, as mock-infected animals also showed signs of neutrophil influx in this compartment. A recent ferret study indicated that the lack of neutrophil lung recruitment in pandemic H1N1 infection may be age related given the significantly reduced neutrophil-induced inflammatory response in newly weaned compared to adult ferrets (45). An additional unexpected finding was the influx of monocytes into the upper airways early in infection and the marked increase in this leukocyte population in bronchoalveolar lavage fluid, which remained until day 14 postinfection (Fig. 9C, G, and I). The timing of monocyte appearance in the upper airways coincides with the decline of viral replication, suggesting that this population may contribute to the eradication of H1N1 and recovery of infant airways. Although we did not immunophenotype the monocytes in our model, these cells may be similar to a monocyte population recently identified in the nasal lavage fluid of influenza-infected pediatric patients, which was thought to suppress mucosal cytokine inflammation (51).
Recent reports suggest a higher pathogenic potential for pandemic than for seasonal H1N1. Most experimental infection models have evaluated lung histopathology at early time points, before viral clearance, and described pulmonary lesions as being more severe and prolonged than those associated with seasonal H1N1 strains (47, 50, 52). Indeed, pulmonary inflammation was still present in our H1N1-infected infants 7 days after viral clearance. At day 14 postinfection, lesions were focused predominantly around terminal bronchioles, with extension into dependent alveoli (Fig. 11). In addition, there were histological lesions indicative of significant alveolar injury and repair in H1N1-infected animals. It is generally accepted that a subset of type II pneumocytes is able to proliferate in response to type I pneumocyte injury and then differentiate into type I cells to affect alveolar repair (53). In addition, a population of airway stem cells was recently shown to contribute to alveolar regeneration following sublethal influenza virus infection in mice (54). The alveolar injury in our model may be a direct effect of virus infection, as pandemic H1N1 has been shown to infect both type I and type II cells (47, 55, 56). Alternatively, the observed epithelial cell injury may be caused by inflammation, as the cells in the terminal bronchioles and alveoli are specifically vulnerable to inflammation-induced injury. Regardless of the etiology, alveolar damage such as that detected in the H1N1-infected infant monkeys in this study may lead to secondary bacterial infection. These findings are consistent with epidemiologic data suggesting that influenza infection is more likely to progress to bronchiolitis and pneumonia in infant hosts (1, 2, 57). However, comparative in vivo infections of adult rhesus monkeys with pandemic H1N1 will be necessary to fully understand the contribution of age to influenza pathogenicity.
Taken together, our data indicate that H1N1 viral replication is prolonged in infant airway epithelium and that secretion of IFN-α is attenuated relative to that of adults. Our experimental results support future mechanistic studies to identify adjunctive strategies capable of overcoming the immune deficits of the immature airway mucosa; this may ultimately translate into novel approaches to enhance the efficacy of pediatric vaccines for respiratory pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sarah Davis, Paul-Michael Sosa, Linda Hirst, Miles Christensen, Wilhelm Von Morgenland, Irma Cazares-Shaw, and Jody Burton for excellent technical support during this project. The California National Primate Research Center Pathology Unit also provided technical assistance. We thank Candace Burke and Carolyn Black for their helpful discussions during the preparation of the manuscript.
C.C.C., K.S.H., and L.A.M. conceived of and designed the experiments. C.C.C., J.R.R., J.E.G., and T.T.W. acquired and analyzed the data.
This work was supported NIH grant RR000169, TRDRP grant 20FT-0075, NIH grant HL097087, and EPA STAR grant 832947.
Footnotes
Published ahead of print 16 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00188-14.
REFERENCES
- 1.Laraya-Cuasay LR, DeForest A, Huff D, Lischner H, Huang NN. 1977. Chronic pulmonary complications of early influenza virus infection in children. Am. Rev. Respir. Dis. 116:617–625 [DOI] [PubMed] [Google Scholar]
- 2.Ajayi-Obe EK, Coen PG, Handa R, Hawrami K, Aitken C, McIntosh ED, Booy R. 2008. Influenza A and respiratory syncytial virus hospital burden in young children in East London. Epidemiol. Infect. 136:1046–1058. 10.1017/S0950268807009557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myles PR, Semple MG, Lim WS, Openshaw PJ, Gadd EM, Read RC, Taylor BL, Brett SJ, McMenamin J, Enstone JE, Armstrong C, Bannister B, Nicholson KG, Nguyen-Van-Tam JS. 2012. Predictors of clinical outcome in a national hospitalised cohort across both waves of the influenza A/H1N1 pandemic 2009-2010 in the UK. Thorax 67:709–717. 10.1136/thoraxjnl-2011-200266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libster R, Bugna J, Coviello S, Hijano DR, Dunaiewsky M, Reynoso N, Cavalieri ML, Guglielmo MC, Areso MS, Gilligan T, Santucho F, Cabral G, Gregorio GL, Moreno R, Lutz MI, Panigasi AL, Saligari L, Caballero MT, Egues Almeida RM, Gutierrez Meyer ME, Neder MD, Davenport MC, Del Valle MP, Santidrian VS, Mosca G, Garcia Dominguez M, Alvarez L, Landa P, Pota A, Bolonati N, Dalamon R, Sanchez Mercol VI, Espinoza M, Peuchot JC, Karolinski A, Bruno M, Borsa A, Ferrero F, Bonina A, Ramonet M, Albano LC, Luedicke N, Alterman E, Savy V, Baumeister E, Chappell JD, Edwards KM, Melendi GA, Polack FP. 2010. Pediatric hospitalizations associated with 2009 pandemic influenza A (H1N1) in Argentina. N. Engl. J. Med. 362:45–55. 10.1056/NEJMoa0907673 [DOI] [PubMed] [Google Scholar]
- 5.O'Riordan S, Barton M, Yau Y, Read SE, Allen U, Tran D. 2010. Risk factors and outcomes among children admitted to hospital with pandemic H1N1 influenza. CMAJ 182:39–44. 10.1503/cmaj.091724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon S, Louie J, Siniscalchi A, Rico E, Richter D, Hernandez R, Lynfield R, Stoute L, Landers K, Brady U, Pascoe N, Vernon V, Haupt T, Moore C, Schieve L, Peacock G, Boyle C, Finelli L, Uyeki T, Dhara R, Fowlkes A, Christensen D, Jarquin V, CDC 2009. Surveillance for pediatric deaths associated with 2009 pandemic influenza A (H1N1) virus infection—United States April-August 2009. MMWR Morb. Mortal. Wkly. Rep. 58:941–947 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5834a1.htm [PubMed] [Google Scholar]
- 7.Martic J, Savic N, Minic P, Pasic S, Nedeljkovic J, Jankovic B. 2011. Novel H1N1 influenza in neonates: from mild to fatal disease. J. Perinatol. 31:446–448. 10.1038/jp.2010.194 [DOI] [PubMed] [Google Scholar]
- 8.Greenberg ME, Lai MH, Hartel GF, Wichems CH, Gittleson C, Bennet J, Dawson G, Hu W, Leggio C, Washington D, Basser RL. 2009. Response to a monovalent 2009 influenza A (H1N1) vaccine. N. Engl. J. Med. 361:2405–2413. 10.1056/NEJMoa0907413 [DOI] [PubMed] [Google Scholar]
- 9.Zhu FC, Wang H, Fang HH, Yang JG, Lin XJ, Liang XF, Zhang XF, Pan HX, Meng FY, Hu YM, Liu WD, Li CG, Li W, Zhang X, Hu JM, Peng WB, Yang BP, Xi P, Wang HQ, Zheng JS. 2009. A novel influenza A (H1N1) vaccine in various age groups. N. Engl. J. Med. 361:2414–2423. 10.1056/NEJMoa0908535 [DOI] [PubMed] [Google Scholar]
- 10.Holt PG, Jones CA. 2000. The development of the immune system during pregnancy and early life. Allergy 55:688–697. 10.1034/j.1398-9995.2000.00118.x [DOI] [PubMed] [Google Scholar]
- 11.Wilson CB, Kollmann TR. 2008. Induction of antigen-specific immunity in human neonates and infants. Nestle Nutr. Workshop Ser. Pediatr. Program 61:183–195 [DOI] [PubMed] [Google Scholar]
- 12.PrabhuDas M, Adkins B, Gans H, King C, Levy O, Ramilo O, Siegrist CA. 2011. Challenges in infant immunity: implications for responses to infection and vaccines. Nat. Immunol. 12:189–194. 10.1038/ni0311-189 [DOI] [PubMed] [Google Scholar]
- 13.Marchant A, Goldman M. 2005. T cell-mediated immune responses in human newborns: ready to learn? Clin. Exp. Immunol. 141:10–18. 10.1111/j.1365-2249.2005.02799.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velilla PA, Rugeles MT, Chougnet CA. 2006. Defective antigen-presenting cell function in human neonates. Clin. Immunol. 121:251–259. 10.1016/j.clim.2006.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granberg C, Hirvonen T, Toivanen P. 1979. Cell-mediated lympholysis by human maternal and neonatal lymphocytes: mother's reactivity against neonatal cells and vice versa. J. Immunol. 123:2563–2567 [PubMed] [Google Scholar]
- 16.Wegmann TG, Lin H, Guilbert L, Mosmann TR. 1993. Bidirectional cytokine interactions in the maternal-fetal relationship: is successful pregnancy a TH2 phenomenon? Immunol. Today 14:353–356. 10.1016/0167-5699(93)90235-D [DOI] [PubMed] [Google Scholar]
- 17.Firth MA, Shewen PE, Hodgins DC. 2005. Passive and active components of neonatal innate immune defenses. Anim. Health Res. Rev. 6:143–158. 10.1079/AHR2005107 [DOI] [PubMed] [Google Scholar]
- 18.Marodi L. 2006. Innate cellular immune responses in newborns. Clin. Immunol. 118:137–144. 10.1016/j.clim.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 19.Holt PG, Upham JW, Sly PD. 2005. Contemporaneous maturation of immunologic and respiratory functions during early childhood: implications for development of asthma prevention strategies. J. Allergy Clin. Immunol. 116:16–24; quiz 25. 10.1016/j.jaci.2005.04.017 [DOI] [PubMed] [Google Scholar]
- 20.Haines CJ, Giffon TD, Lu LS, Lu X, Tessier-Lavigne M, Ross DT, Lewis DB. 2009. Human CD4+ T cell recent thymic emigrants are identified by protein tyrosine kinase 7 and have reduced immune function. J. Exp. Med. 206:275–285. 10.1084/jem.20080996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan J, Reen DJ. 2001. Human recent thymic emigrants—identification, expansion, and survival characteristics. J. Immunol. 167:1970–1976 http://www.jimmunol.org/content/167/4/1970.long [DOI] [PubMed] [Google Scholar]
- 22.Bertotto A, Gerli R, Lanfrancone L, Crupi S, Arcangeli C, Cernetti C, Spinozzi F, Rambotti P. 1990. Activation of cord T lymphocytes. II. Cellular and molecular analysis of the defective response induced by anti-CD3 monoclonal antibody. Cell. Immunol. 127:247–259 [DOI] [PubMed] [Google Scholar]
- 23.Levy O. 2007. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat. Rev. Immunol. 7:379–390. 10.1038/nri2075 [DOI] [PubMed] [Google Scholar]
- 24.Vareille M, Kieninger E, Edwards MR, Regamey N. 2011. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 24:210–229. 10.1128/CMR.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burri PH. 1984. Fetal and postnatal development of the lung. Annu. Rev. Physiol. 46:617–628. 10.1146/annurev.ph.46.030184.003153 [DOI] [PubMed] [Google Scholar]
- 26.Plopper CG, Weir AJ, Nishio SJ, Cranz DL, St George JA. 1986. Tracheal submucosal gland development in the rhesus monkey, Macaca mulatta: ultrastructure and histochemistry. Anat. Embryol. (Berl.) 174:167–178. 10.1007/BF00824332 [DOI] [PubMed] [Google Scholar]
- 27.Van Winkle LS, Fanucchi MV, Miller LA, Baker GL, Gershwin LJ, Schelegle ES, Hyde DM, Evans MJ, Plopper CG. 2004. Epithelial cell distribution and abundance in rhesus monkey airways during postnatal lung growth and development. J. Appl. Physiol. 97:2355–2363; discussion 2354. 10.1152/japplphysiol.00470.2004 [DOI] [PubMed] [Google Scholar]
- 28.Yerkovich ST, Wikstrom ME, Suriyaarachchi D, Prescott SL, Upham JW, Holt PG. 2007. Postnatal development of monocyte cytokine responses to bacterial lipopolysaccharide. Pediatr. Res. 62:547–552. 10.1203/PDR.0b013e3181568105 [DOI] [PubMed] [Google Scholar]
- 29.Renz H, Brandtzaeg P, Hornef M. 2011. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 12:9–23. 10.1038/nri3112 [DOI] [PubMed] [Google Scholar]
- 30.Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. 2006. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203:973–984. 10.1084/jem.20050625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniar-Hew K, Clay CC, Postlethwait EM, Evans MJ, Fontaine JH, Miller LA. 2013. Innate immune response to LPS in airway epithelium is dependent on chronological age and antecedent exposures. Am. J. Respir. Cell Mol. Biol. 49:710–720. 10.1165/rcmb.2012-0321OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plopper C, St George J, Cardoso W, Wu R, Pinkerton K, Buckpitt A. 1992. Development of airway epithelium. Patterns of expression for markers of differentiation. Chest 101:2S–5S [PubMed] [Google Scholar]
- 33.DeMaria MA, Casto M, O'Connell M, Johnson RP, Rosenzweig M. 2000. Characterization of lymphocyte subsets in rhesus macaques during the first year of life. Eur. J. Haematol. 65:245–257. 10.1034/j.1600-0609.2000.065004245.x [DOI] [PubMed] [Google Scholar]
- 34.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. Lond. 27:493–497 [Google Scholar]
- 35.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84:5670–5677. 10.1128/JVI.00272-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khaitov MR, Laza-Stanca V, Edwards MR, Walton RP, Rohde G, Contoli M, Papi A, Stanciu LA, Kotenko SV, Johnston SL. 2009. Respiratory virus induction of alpha-, beta- and lambda-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy 64:375–386. 10.1111/j.1398-9995.2008.01826.x [DOI] [PubMed] [Google Scholar]
- 37.Haye K, Burmakina S, Moran T, Garcia-Sastre A, Fernandez-Sesma A. 2009. The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J. Virol. 83:6849–6862. 10.1128/JVI.02323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Belmonte F, Puertollano R, Millan J, Alonso MA. 2000. The MAL proteolipid is necessary for the overall apical delivery of membrane proteins in the polarized epithelial Madin-Darby canine kidney and Fischer rat thyroid cell lines. Mol. Biol. Cell 11:2033–2045. 10.1091/mbc.11.6.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank M. 2000. MAL, a proteolipid in glycosphingolipid enriched domains: functional implications in myelin and beyond. Prog. Neurobiol. 60:531–544. 10.1016/S0301-0082(99)00039-8 [DOI] [PubMed] [Google Scholar]
- 40.Cheong KH, Zacchetti D, Schneeberger EE, Simons K. 1999. VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc. Natl. Acad. Sci. U. S. A. 96:6241–6248. 10.1073/pnas.96.11.6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marazuela M, Acevedo A, Adrados M, Garcia-Lopez MA, Alonso MA. 2003. Expression of MAL, an integral protein component of the machinery for raft-mediated apical transport, in human epithelia. J. Histochem. Cytochem. 51:665–674. 10.1177/002215540305100512 [DOI] [PubMed] [Google Scholar]
- 42.Cao W, Zhang ZY, Xu Q, Sun Q, Yan M, Zhang J, Zhang P, Han ZG, Chen WT. 2010. Epigenetic silencing of MAL, a putative tumor suppressor gene, can contribute to human epithelium cell carcinoma. Mol. Cancer 9:296. 10.1186/1476-4598-9-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kollmann TR, Levy O, Montgomery RR, Goriely S. 2012. Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity 37:771–783. 10.1016/j.immuni.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Josset L, Engelmann F, Haberthur K, Kelly S, Park B, Kawoaka Y, Garcia-Sastre A, Katze MG, Messaoudi I. 2012. Increased viral loads and exacerbated innate host responses in aged macaques infected with the 2009 pandemic H1N1 influenza A virus. J. Virol. 86:11115–11127. 10.1128/JVI.01571-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang SS, Banner D, Degousee N, Leon AJ, Xu L, Paquette SG, Kanagasabai T, Fang Y, Rubino S, Rubin B, Kelvin DJ, Kelvin AA. 2012. Differential pathological and immune responses in newly weaned ferrets are associated with a mild clinical outcome of pandemic 2009 H1N1 infection. J. Virol. 86:13187–13201. 10.1128/JVI.01456-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll TD, Matzinger SR, Genesca M, Fritts L, Colon R, McChesney MB, Miller CJ. 2008. Interferon-induced expression of MxA in the respiratory tract of rhesus macaques is suppressed by influenza virus replication. J. Immunol. 180:2385–2395. 10.4049/jimmunol.180.4.2385 [DOI] [PubMed] [Google Scholar]
- 47.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025. 10.1038/nature08260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinfurter JT, Brunner K, Capuano SV, III, Li C, Broman KW, Kawaoka Y, Friedrich TC. 2011. Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates. PLoS Pathog. 7:e1002381. 10.1371/journal.ppat.1002381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boonnak K, Paskel M, Matsuoka Y, Vogel L, Subbarao K. 2012. Evaluation of replication, immunogenicity and protective efficacy of a live attenuated cold-adapted pandemic H1N1 influenza virus vaccine in non-human primates. Vaccine 30:5603–5610. 10.1016/j.vaccine.2012.06.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herfst S, van den Brand JM, Schrauwen EJ, de Wit E, Munster VJ, van Amerongen G, Linster M, Zaaraoui F, van Ijcken WF, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Andeweg AC, Kuiken T. 2010. Pandemic 2009 H1N1 influenza virus causes diffuse alveolar damage in cynomolgus macaques. Vet. Pathol. 47:1040–1047. 10.1177/0300985810374836 [DOI] [PubMed] [Google Scholar]
- 51.Oshansky CM, Gartland AJ, Wong SS, Jeevan T, Wang D, Roddam PL, Caniza MA, Hertz T, Devincenzo JP, Webby RJ, Thomas PG. 2014. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am. J. Respir. Crit. Care Med. 189:449–462. 10.1164/rccm.201309-1616OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Safronetz D, Rockx B, Feldmann F, Belisle SE, Palermo RE, Brining D, Gardner D, Proll SC, Marzi A, Tsuda Y, Lacasse RA, Kercher L, York A, Korth MJ, Long D, Rosenke R, Shupert WL, Aranda CA, Mattoon JS, Kobasa D, Kobinger G, Li Y, Taubenberger JK, Richt JA, Parnell M, Ebihara H, Kawaoka Y, Katze MG, Feldmann H. 2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J. Virol. 85:1214–1223. 10.1128/JVI.01848-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fehrenbach H. 2001. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2:33–46. 10.1186/rr36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, Wang DY, Lim B, Chow VT, Crum CP, Xian W, McKeon F. 2011. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147:525–538. 10.1016/j.cell.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nin N, Sanchez-Rodriguez C, Ver LS, Cardinal P, Ferruelo A, Soto L, Deicas A, Campos N, Rocha O, Ceraso DH, El-Assar M, Ortin J, Fernandez-Segoviano P, Esteban A, Lorente JA. 2012. Lung histopathological findings in fatal pandemic influenza A (H1N1). Med. Intensiva 36:24–31. 10.1016/j.medin.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 56.Weinheimer VK, Becher A, Tonnies M, Holland G, Knepper J, Bauer TT, Schneider P, Neudecker J, Ruckert JC, Szymanski K, Temmesfeld-Wollbrueck B, Gruber AD, Bannert N, Suttorp N, Hippenstiel S, Wolff T, Hocke AC. 2012. Influenza A viruses target type II pneumocytes in the human lung. J. Infect. Dis. 206:1685–1694. 10.1093/infdis/jis455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glezen WP, Taber LH, Frank AL, Gruber WC, Piedra PA. 1997. Influenza virus infections in infants. Pediatr. Infect. Dis. J. 16:1065–1068. 10.1097/00006454-199711000-00012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.