ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) has a significant contributory role in the development of three major human neoplastic or lymphoproliferative diseases: Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD). These diseases are associated with chromosomal instability, a hallmark of human cancer. The latency-associated nuclear antigen (LANA) encoded by KSHV plays a key role in regulating a number of cellular pathways critical for oncogenesis. KSHV LANA alone can induce the development of B-cell hyperplasia and lymphoma in mice expressing LANA. LANA also induces chromosomal instability, thus promoting oncogenesis. However, the precise mechanism underlying LANA-mediated chromosomal instability remains uncharted. Here we report that LANA promoted the induction of chromosomal instability and the formation of micronuclei and multinucleation through its interaction with one of the critical spindle checkpoint proteins, Bub1, and the resulting degradation of Bub1. This interaction occurs through the Knl and kinase domains of Bub1, identified as important for stability and degradation. These results suggest that LANA can dysregulate Bub1 activity, which leads to aberrant chromosome replication and aneuploidy, thus contributing to KSHV-mediated oncogenesis.
IMPORTANCE This work represents the first set of results identifying a novel mechanism by which LANA, a latency-associated antigen encoded by KSHV, can induce the degradation of Bub1, a spindle checkpoint protein that is important for spindle checkpoint signaling and chromosome segregation. The downregulation of Bub1 mediated by LANA resulted in chromosomal instability, a hallmark of cancer. We further investigated the specific domains of Bub1 that are required for the interaction between LANA and Bub1. The results demonstrated that the Knl and kinase domains of Bub1 are required for the interaction between LANA and Bub1. In addition, we also investigated the mechanism by which LANA promoted Bub1 degradation. Our results showed that LANA interacted physically with the anaphase-promoting complex (APC/C), thus promoting the degradation of Bub1 in a ubiquitin-dependent process.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), formally referred to as human herpesvirus 8 (HHV-8), is an enveloped double-stranded DNA tumor virus that was first discovered by representational differential analysis in 1994 (1). KSHV contributes not only to the development of KS but also to that of other lymphoproliferative disorders, including primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD) (2, 3). Like other human herpesviruses, KSHV exists in two replicative phases: a lytic and a latent phase. During the lytic phase, the majority of the KSHV genes are expressed, host cells are broken down, and KSHV infectious progeny virus particles are produced (4, 5). KSHV can establish latent infection after primary infection. During this latent phase, in order to evade the host immune surveillance, only a limited number of genes are expressed, such as the v-FLIP (ORF71), v-cyclin (ORF72), and latency-associated nuclear antigen (LANA) (ORF73) genes, as well as some microRNAs (miRNAs) (5, 6). The virus genome is maintained as a double-stranded circular DNA termed an episome, which is tethered to the host chromosomes through the interaction of LANA with a number of cellular proteins, including Bub1, centromere protein F (CENPF), and nuclear mitotic apparatus protein (NuMA), during cell division, ensuring that the viral genome is partitioned into new daughter cells (5–7).
KSHV-encoded ORF73, or LANA, is one of the predominant viral antigens highly expressed in latently infected cells (5, 8). LANA functions in activating as well as repressing cellular and viral gene transcription (9–16). In addition to modulating gene transcription, LANA plays a crucial role in KSHV episome replication and persistence in cell lines latently infected with KSHV (17–19). As an oncogenic protein encoded by KSHV, LANA has been shown to interact physically with, and inhibit the tumor suppressor functions of, the retinoblastoma protein pRb, as well as p53 and von Hippel-Lindau (VHL) protein, resulting in the inactivation of p53-dependent promoters and the induction of E2F-dependent genes (20–22). LANA also contributes to the immortalization of endothelial cells (23). Furthermore, LANA can deregulate and stabilize the expression of β-catenin by sequestering its inhibitor, glycogen synthase kinase 3β (GSK-3β) (24). Interestingly, the negative regulation of GSK-3β by LANA is essential for the promotion of S-phase entry in cells latently infected with KSHV or transiently transfected with LANA, which may be associated with KSHV-associated neoplasia. LANA can also activate or stabilize many oncoproteins, including c-Myc and c-Jun (25, 26).
Chromosome missegregation during cell division results in a loss or gain of chromosomes in the next generation of cells, which leads to aneuploidy and so contributes to the oncogenic process (27). A cellular surveillance system named the spindle assembly checkpoint (SAC) ensures that the chromosomes segregate correctly during each cell division by arresting cells in metaphase until every kinetochore of all the sister chromatids is correctly bound to the microtubules and all the chromosomes are aligned in the metaphase plate (28). The key protein components of the spindle checkpoint include Mad1, Mad2, Bub1, and BubR1, and these proteins are localized to unattached kinetochores during early mitosis (29, 30). Bub1, the first spindle checkpoint protein to dock at the kinetochore (31–33), contains an amino-terminal Knl binding domain, through which Bub1 interacts with the kinetochore protein Knl1 (34); a Bub3 binding domain, through which Bub1 binds to another spindle checkpoint protein, Bub3 (35); a carboxy-terminal kinase domain, which phosphorylates CDC20 (36); and two conserved motifs that contribute to spindle checkpoint signaling and chromosome segregation (37).
Bub1 has two crucial functions during the process of cell division. First, as a scaffold protein, Bub1 plays a critical role in spindle checkpoint signaling. Bub1 also recruits other spindle checkpoint proteins to the kinetochore, forming the mitotic checkpoint complex (MCC) (38). This can arrest cells at metaphase by inhibiting the functions of the anaphase-promoting complex (APC/C) (39). Second, Bub1 is a serine/threonine protein kinase, whose kinase activity is required for correct chromosome alignment and congression (28, 40, 41). In some eukaryotic cells, inhibition or inactivation of Bub1 results in severe chromosome segregation defects, including chromosome congression failure and chromosome lagging (41–44). These reports strongly suggest that the functions of Bub1 are highly conserved. Downregulation of Bub1 expression seems to play an important role in the development of human cancers. It has been reported that in some human cancers, including lung, colon, and pancreatic tumors, the protein levels of Bub1 are downregulated (45, 46). Mouse models mimicking these kinds of downregulation have led to the development of cancer in mice (47–49). These reports strongly suggested that downregulation of Bub1 can increase cancer risk. In the present study, we show that LANA can strongly induce chromosomal instability (CIN) and increase levels of multinucleation and the formation of micronuclei in PEL cells by inducing Bub1 degradation.
MATERIALS AND METHODS
Plasmids and antibodies.
pA3M-LANA, pA3F-LANA, pA3F-LANAΔSOCS, pA3M-Bub1, GFP-Bub1, HA-cul2, HA-cul5, His-Uba1, His-Ubc5a, and GST-ubiquitin have been described previously (22, 50). The pCS2-Cdc20 and pCS2-Cdh1 constructs were provided by Hongtao Yu (University of Texas Southwestern, Dallas, TX). Constructs expressing Bub1 with different domains deleted in pA3M or pGEX-2TK were prepared by PCR mutagenesis. Antibodies against MyC (9E10), hemagglutinin (HA) (12CA5), and LANA-1 were generated from hybridomas. A mouse anti-Flag monoclonal antibody (M2) was purchased from Sigma-Aldrich Corp. (St. Louis, MO). Rabbit anti-Bub1 and mouse anti-Cdh1 antibodies were purchased from Abcam (Cambridge, MA). The BJAB, DG75, BC-3, BCBL-1, JSC-1, BJAB-KSHV, BC-3 Shct, BC-3 ShBub1, BC-3-ShLANA, JSC-1 Shct, JSC-1 ShLANA, BJAB-RFP, and BJAB-RFP-LANA cell lines have been described previously (22, 50–52). BC-3-GFP, BC-3-Bub1, BJAB Shct, and BJAB ShBub1 cells were generated as described previously (52).
GST pulldown assay.
Glutathione S-transferase (GST) and GST fusion proteins were purified from BL21(DE3) as described previously (53). Beads coated with GST or GST fusion proteins were incubated with cell lysates for 6 h at 4°C. The beads were washed three times and were boiled in SDS sample buffer. The samples were then fractionated by SDS-PAGE for Western blot analysis.
RNA interference (RNAi).
The short hairpin oligonucleotides for Shct, ShBub1, and ShLANA have been described previously (50). The short hairpin RNA (shRNA) against Cdh1 (target sequence, GTGAACTTCCACAGGATTAAC) was constructed as described previously (51).
IP and Western blotting.
Immunoprecipitation (IP) and Western blotting were performed as described previously (22). Briefly, cells were collected and were lysed in lysis buffer (10 mM Tris, 1% NP-40, 2 mM EDTA, 150 mM NaCl [pH 7.5]) with protease inhibitor. For IP, lysates were incubated with the antibodies indicated in the figures and 30 μl of a 1:1 mixture of protein A/G Sepharose beads at 4°C overnight. After three stringent washes with radioimmunoprecipitation assay (RIPA) buffer, the beads were boiled and were subjected to SDS-PAGE for Western blotting. For all the co-IP experiments to demonstrate the interaction between LANA and Bub1, cells were treated with MG132 (20 μM) for 12 h before being harvested.
Ubiquitination assays.
To generate the His fusion proteins Ubca1, Ubc5a, and ubiquitin (Ub), BL21 bacterial cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at log phase (optical density at 600 nm [OD600], 0.6), and cells were incubated with shaking at 30°C overnight. Bacterial lysis and purification were performed according to the manual for Ni-nitrilotriacetic acid (NTA) agarose from Qiagen Inc. (Valencia, CA). In vivo and in vitro ubiquitination assays were performed as described previously (22).
Detection of chromosomal instability.
For the detection of chromosomal instability, cells were harvested, suspended in phosphate-buffered saline (PBS), spread on slides, and half dried. The cells were fixed with a fixative (3% paraformaldehyde [PFA] containing 0.1% Triton-100) for 30 min at room temperature (RT) and were then stained with 4′,6-diamidino-2-phenylindole (DAPI) for 1 h at RT. Then the cells were visualized and were examined for multinucleation and the presence of micronuclei. For quantitation of the percentage of multinucleation, 200 cells were counted.
Metaphase chromosome spread.
Nocodazole (0.1 μg/ml) was added to the cell culture medium, and cells were allowed to incubate for 12 h before harvesting. The cells were then treated with a hypotonic buffer (0.075 M KCl) for 30 min at RT and were fixed three times, for 10 min each time, in a fixative (3 parts of methanol and 1 part of acetic acid). Metaphase preparations were carried out as described previously (54).
Cell synchronization.
Cells were synchronized in different phases as described previously (55). Briefly, cells were synchronized in S phase by using a double thymidine block or in M phase by using a thymidine-nocodazole block. Cells were synchronized in G1 phase by growth in RPMI 1640 medium with 0.1% bovine growth serum (BGS) for 72 h.
Statistical analysis.
Each experiment was repeated at least three times. The mean scores were examined by using Student's t test. All statistical tests were performed using Microsoft Office Excel. A P value of <0.05 was considered to indicate a statistically significant difference. A P value of <0.01 was considered to indicate high statistical significance.
RESULTS
Bub1 levels are downregulated in LANA-expressing and KSHV-positive cell lines.
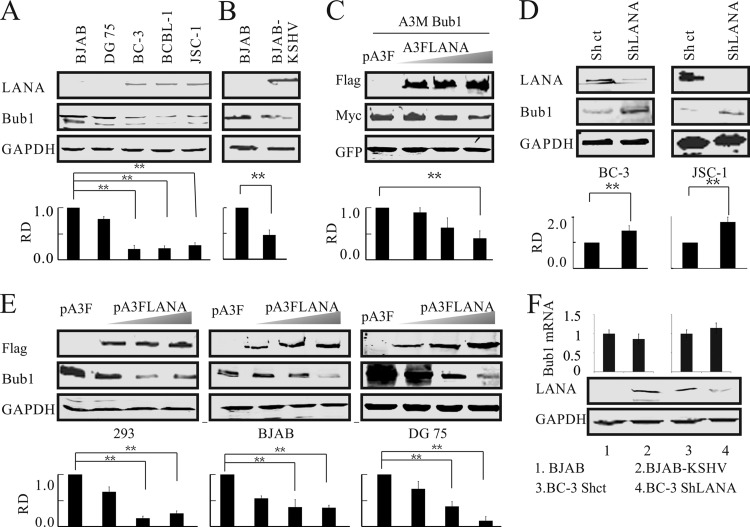
Previously, we demonstrated strong colocalization of LANA and Bub1 in KSHV-associated B-cell lymphoma (50). To further investigate whether LANA could regulate Bub1, a serine/threonine protein kinase with a critical role in mitotic spindle checkpoint establishment and chromosome congression, we examined the levels of Bub1 protein in KSHV-positive (BCBL-1, BC-3, and JSC-1) and KSHV-negative (BJAB and DG75) B-cell lines (Fig. 1A). Our data showed that Bub1 levels were significantly lower in KSHV-positive than in KSHV-negative cell lines. In fact, the levels in KSHV-positive cells were approximately 4-fold less than those in BJAB and DG75 cells (Fig. 1A). Interestingly, the intensity of a second band migrating just below Bub1 was also reduced in KSHV-positive or LANA-expressing cell lines, suggesting that Bub1 was modified posttranslationally. To rule out an inconsistency in genotypes among the different cell lines, the protein levels of Bub1 were also compared in cells of the same genotypic background: uninfected and KSHV-infected BJAB cells (Fig. 1B). Again, KSHV-infected BJAB cells showed >50% decreased levels of Bub1 protein, higher than the levels seen in the PEL cell lines but still significantly downregulated.
FIG 1.
Bub1 levels are downregulated in cells latently infected with KSHV and expressing LANA. (A) Western blotting was used to detect the endogenous protein levels of Bub1 in KSHV-negative (BJAB and DG75) and KSHV-positive (BC-3, BCBL-1, JSC-1) cell lines. (B) Level of Bub1 protein in BJAB cells infected with KSHV. (C) LANA decreases the level of exogenous Bub1 protein in HEK-293 cells. HEK-293 cells were electroporated with increasing amounts of Flag-tagged LANA, Myc-tagged Bub1, and a GFP plasmid. Forty-eight hours later, the cells were collected for Western blot analysis, which was performed using the indicated antibodies. GFP served as a control for protein loading. (D) LANA knockdown increases Bub1 accumulation. Cell lysates from KSHV-positive B cells (BC-3 and JSC-1) in which LANA or a luciferase control had been stably knocked down (ShLANA or Shct, respectively) were subjected to Western blot analysis. (E) LANA decreases the level of endogenous Bub1 protein in HEK-293 cells and KSHV-negative B cells. HEK-293 and KSHV-negative B cells (BJAB, DG75) were electroporated with increasing amounts of Flag-tagged LANA. Forty-eight hours after electroporation, the cells were collected for Western blot analysis. (F) KSHV infection or LANA overexpression does not affect the mRNA level of Bub1. Total RNAs were isolated from KSHV-infected B cells (BJAB-KSHV, BJAB) and from control knockdown and LANA knockdown BC-3 cells (BC-3 Shct and BC-3 ShLANA, respectively), and the mRNA levels of Bub1 were analyzed by real-time PCR. The relative densities (RD) of Bub1 were quantified and plotted against the signal obtained from the control after normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (for endogenous Bub1) or ectopically expressed GFP (for exogenous Bub1). Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**).
To determine whether Bub1 suppression was associated with LANA, Flag-tagged LANA was cotransfected with Myc-tagged Bub1 into HEK-293 cells. The Western blot results showed that ectopic Bub1 was suppressed in the presence of LANA in a dose-dependent manner (Fig. 1C). Further analysis using Western blot analysis showed that the Bub1 expression levels were rescued when LANA transcripts were knocked down by shRNA (Fig. 1D). To further investigate the role of LANA in the reduction of Bub1 levels, LANA was transfected into three KSHV-negative cell lines. In all cases, with increasing amounts of LANA, the levels of Bub1 were substantially reduced (Fig. 1E). These data suggested that Bub1 levels were significantly inhibited in the presence of LANA. Interestingly, no significant change in Bub1 transcript levels was observed in KSHV-infected BJAB cells or in the BC-3 ShLANA cell line (Fig. 1F). These results strongly suggested that the protein levels of Bub1 are regulated by LANA at the posttranslational level.
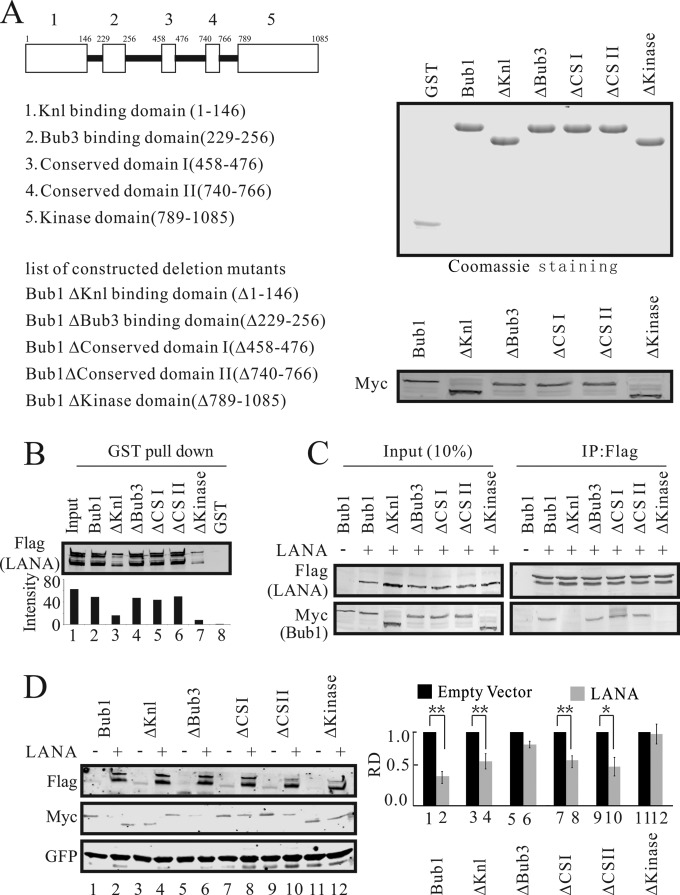
The Knl and kinase domains of Bub1 are important for its interaction with LANA.
Previously, we showed that Bub1 colocalizes with LANA in KSHV-positive cells, suggesting that LANA can interact directly with Bub1 (50). Here we investigated further the specific domain of Bub1 involved in its interaction with LANA (Fig. 2A). Bacterially expressed GST-fused Bub1 specific mutants and wild-type (WT) Bub1 (Fig. 2A) were incubated with lysates from HEK-293 cells transfected with Flag-tagged LANA (Fig. 2B). The bound Flag-tagged LANA was fractionated and was detected by Western blot analysis using the specific antibody M2. The results showed that the interaction between LANA and Bub1 was dramatically decreased when the Knl domain or the kinase domain was deleted (Fig. 2B). The interactions between LANA and Bub1 deletion mutants were further validated by coimmunoprecipitation (Fig. 2C). Similarly, we found that deletion of the Knl domain within Bub1 dramatically decreased the association between LANA and Bub1. Not only were the Knl and kinase domains necessary for LANA and Bub1 association, but the kinase domain was also required for LANA-mediated Bub1 degradation (Fig. 2D). Interestingly, when cotransfected with LANA, Bub1 and its mutants showed different levels of reduction. When the kinase domain was missing, LANA had little or no effect on Bub1 levels. Some effect was seen with Knl deletion, which was similar to that seen with the ΔCSI and ΔCSII domain mutants. Further, deletion of the Bub3 binding domain had no major effect in terms of Bub1 levels in the presence of LANA (Fig. 2D). Therefore, the Knl and kinase domains are important for the interaction of Bub1 with LANA, but interestingly, the kinase domain and the Bub3 domain both played roles in the stability of Bub1, as evidenced by the fact that deletion of these domains resulted in the inability of LANA to contribute to a reduction in Bub1 levels (Fig. 2D).
FIG 2.
The Knl and kinase domains of Bub1 are necessary for its interaction with LANA. (A) (Left) Schematic diagram of Bub1 mutants. (Right) (Top) GST-tagged proteins purified from Escherichia coli. (Bottom) Eukaryotic expression of these deletion mutants (Myc tagged). ΔCSI and ΔCSII, Bub1 mutants with deletions of conserved domain I and conserved domain II, respectively. (B) Beads coated with GST or with fusion proteins consisting of GST and Bub1 domain deletion mutant proteins were incubated with lysates from HEK-293 cells electroporated with pA3F-LANA for GST pulldown assays. The pulldown of LANA was detected by Western blotting using antibody M2. (C) Bub1 interacts with LANA through its kinase and Knl binding domains. HEK-293 cells were electroporated with pA3F-LANA and pA3M-Bub1 or its related domain deletion mutants. Forty-eight hours after electroporation, the cell lysates were prepared for IP with antibody M2 and Western blotting with antibodies 9E10 and M2. (D) HEK-293 cells were electroporated with pA3F-LANA, pA3M-Bub1 or its related domain deletion mutants, and GFP. Forty-eight hours later, cells were collected for Western blot analysis. GFP served as a control for protein loading. The relative densities (RD) of Bub1 and its related deletion mutants were quantified and plotted against the signal obtained from the control after normalization to GFP. Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**).
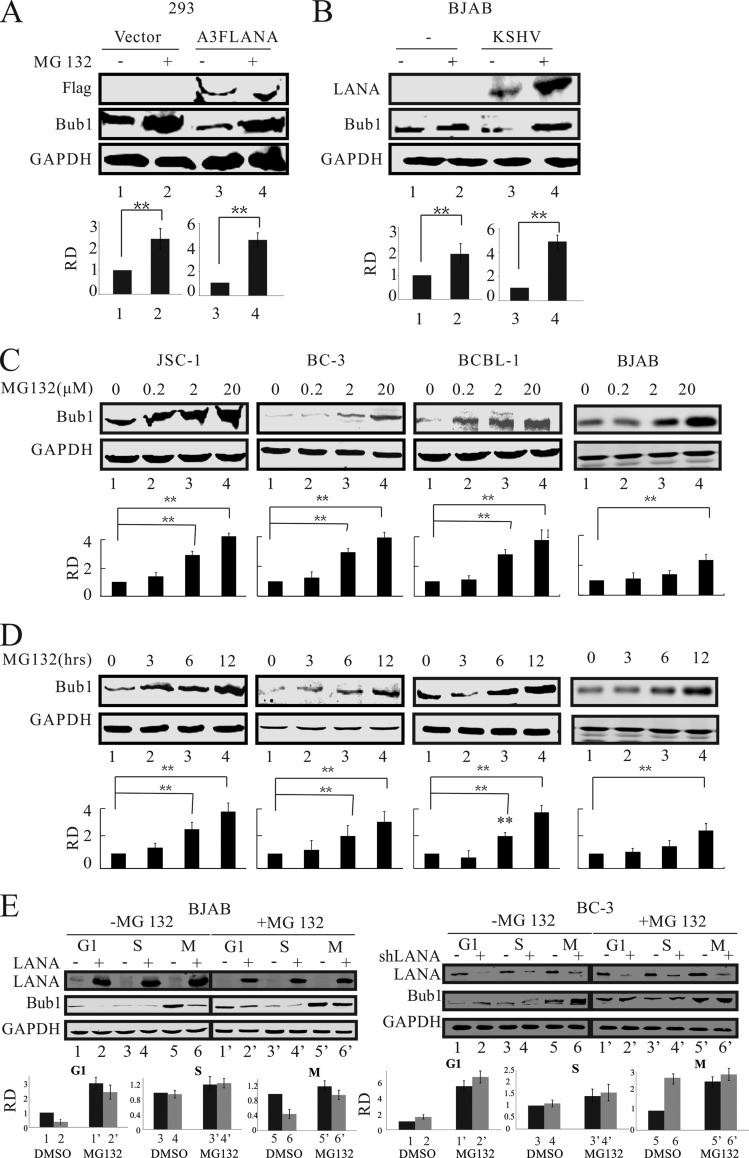
LANA promotes Bub1 degradation through the ubiquitin-mediated proteasome degradation pathway.
Our data presented above showed that Bub1 protein levels were dramatically reduced in the presence of LANA. Further, our previous studies showed that LANA can recruit an E3 ligase to degrade its target proteins (22). To determine whether Bub1 was targeted for ubiquitin-mediated proteasome degradation, we transfected HEK-293 cells with LANA and a control vector in the presence of the proteasome inhibitor MG132 (55). Analysis of Bub1 protein levels showed, as expected, a dramatic decrease in the presence of LANA (Fig. 3A, lane 3). This was rescued when cells were treated with MG132 in the presence of LANA (Fig. 3A, compare lanes 3 and 4). Bub1 levels were dramatically rescued relative to levels for the control in the presence of MG132 (Fig. 3A, lane 2). Furthermore, we also investigated the differences in Bub1 protein levels in the KSHV-negative B-cell line BJAB. As in HEK-293 cells, Bub1 protein levels in BJAB cells were dramatically reduced in the presence of LANA but were rescued when cells were treated with the proteasome inhibitor MG132 (Fig. 3B, lanes 2, 3, and 4). To determine Bub1 levels under physiological conditions in KSHV-positive cell lines (BC-3, BCBL-1, and JSC-1), we treated these cells with increasing doses of MG132 (0 to 20 μM) for 12 h. The results showed that the protein levels of Bub1 increased with increasing concentrations of MG132 (Fig. 3C). Further, we treated these cell lines with MG132 at 3-h intervals up to 12 h. The results demonstrated that the protein levels of Bub1 increased with increasing time associated with MG132 treatment (Fig. 3D). These studies showed that MG132 treatment can dramatically increase Bub1 protein levels in a dose-dependent and time-dependent manner in these KSHV-positive lymphoma cells. Similar experiments were performed with the KSHV-negative cell line BJAB (Fig. 3C and D), in which Bub1 protein levels showed a 2-fold increase in the presence of MG132, compared with an approximately 4-fold increase in KSHV-positive cell lines. This suggests that LANA can promote Bub1 degradation through the ubiquitin-proteasome pathway. To further determine in which phase LANA can promote Bub1 degradation, we synchronized the BC-3 Shct, BC-3-ShLANA, BJAB-RFP, and BJAB-RFP-LANA cell lines in the G, S, or M phase as described previously (56). We found that LANA can mediate Bub1 degradation in the G1 and M phases through a ubiquitin-dependent pathway (Fig. 3E). Interestingly, we found that the pattern of Bub1 degradation mediated by LANA is similar to what is found in cells without LANA or in cell lines in which LANA is knocked down (Fig. 3E, compare lanes 1, 2, 5, and 6 with lanes 1′, 2′, 5′, and 6′). This strongly suggested that LANA may target the Bub1 degradation pathway to enhance the degradation of Bub1.
FIG 3.
LANA-mediated Bub1 degradation is ubiquitin dependent. (A and B) LANA-transfected HEK-293 cells (A) and KSHV-infected BJAB cells (B) were either mock treated or treated with 20 μM MG132 for 12 h. The cells were then collected for Western blot analysis. (C) Cells were treated with MG132 for 12 h at 0, 0.2, 2, or 20 μM in a dose-dependent assay. The cells were then collected for Western blot analysis. (D) Cells were treated with MG132 at 20 μM for 0, 3, 6, or 12 h in a time course assay. The cells were then collected for Western blot analysis. (E) Cells were synchronized in the G1, S, or M phase as described in Materials and Methods. The cells were either mock treated or treated with MG132 (20 μM) for 12 h before they were harvested for Western blot analysis. The relative densities (RD) of Bub1 were quantified and plotted against the signal obtained from the control after normalization to GAPDH. Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**).
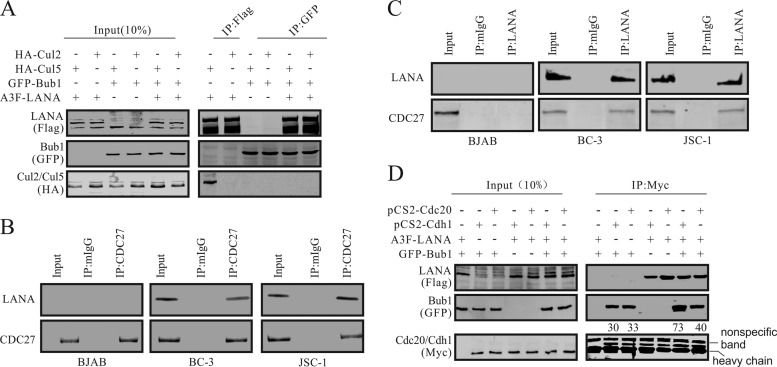
LANA is associated with the APC/C in KSHV-positive cell lines.
The data presented above showed that LANA can regulate Bub1 degradation via the ubiquitin-proteasome pathway. There are two E3 ligase complexes that may possibly be involved in LANA-mediated Bub1 degradation: the EC5S complex (22) or the APC/C. We performed a co-IP assay to further explore the possibility of interaction between Bub1 and the cullin component of the EC5S E3 ligase complex in HEK-293 cells. The results showed that the EC5S E3 ligase complex was not involved in LANA-mediated Bub1 degradation, although, as expected, LANA did associate with cullin 5 (Fig. 4A). Also, the co-IP showed that Bub1 did not interact with cullin 5, a scaffold protein of the EC5S complex, in the presence or absence of LANA, as revealed by comparison to the negative control, cullin 2 (Fig. 4A). To determine whether LANA can form a complex with a component of the APC/C, we used a Cdc27-specific antibody in two KSHV-positive cell lines, BC-3 and JSC-1, as well as in a KSHV-negative control B-cell line. We clearly observed that a complex formed with LANA, since Western blot analysis showed a clear signal for LANA in both the BC-3 and JSC-1 cell lines (Fig. 4B). As expected, the KSHV-negative cell line showed no signal for LANA after immunoprecipitation of Cdc27 (Fig. 4B). In the reverse co-IP assay, the result also showed that LANA can coprecipitate with Cdc27 in KSHV-positive BC-3 and JSC-1 cells (Fig. 4C). This shows that these two proteins can form a complex in KSHV-infected PEL cells and that LANA may be capable of functionally regulating Bub1 activities through its association in this complex with the APC/C. To investigate more closely the complex of LANA with the APC/C, we wanted to determine whether LANA can interact with the APC/C activators Cdh1 and Cdc20. Coimmunoprecipitation assays using the anti-Myc monoclonal antibody 9E10 demonstrated that Flag-tagged LANA can be clearly precipitated in a complex with Bub1 (as shown by the green fluorescent protein [GFP] antibody signal) along with Myc-tagged Cdc20 and Cdh1 (Fig. 4D). Interestingly, Bub1 associated with both Cdh1 and Cdc20. However, when LANA was present, the proportion of Bub1 protein in complex with Cdh1 was approximately 2-fold greater than that in complex with Cdc20 (Fig. 4D). This study indicated that LANA interacted with the APC/C and that this interaction accounts for the recruitment of a greater fraction of the Bub1 protein in KSHV-infected cell lines than in KSHV-negative cell lines.
FIG 4.
LANA associates with the APC/C. (A) Bub1 does not interact with the EC5S complex. HEK-293 cells were electroporated with expression vectors as shown. Forty-eight hours postelectroporation, cells were harvested, and the cell lysates underwent IP with antibody M2 or an anti-GFP antibody. IP pellets and portions (10%) of lysates (input) were boiled, fractionated by SDS-PAGE, and subjected to Western blot analysis with the indicated antibodies. (B and C) LANA interacts with the APC/C. Cell lysates were used for IP with LANA- or CDC27-specific antibodies. IP pellets and portions (10%) of lysates were loaded onto SDS-PAGE gels, followed by Western blot analysis with the indicated antibodies. (D) LANA interacts with the activator protein of the APC/C. HEK-293 cells were electroporated with expression vectors as shown. Thirty-six hours postelectroporation, cells were treated with MG132 (20 μm) for 12 h before harvesting. The cell lysates were used for IP with an anti-Myc antibody. IP pellets and portions (10%) of lysates were boiled, fractionated by SDS-PAGE, and subjected to Western blot analysis with specific antibodies.
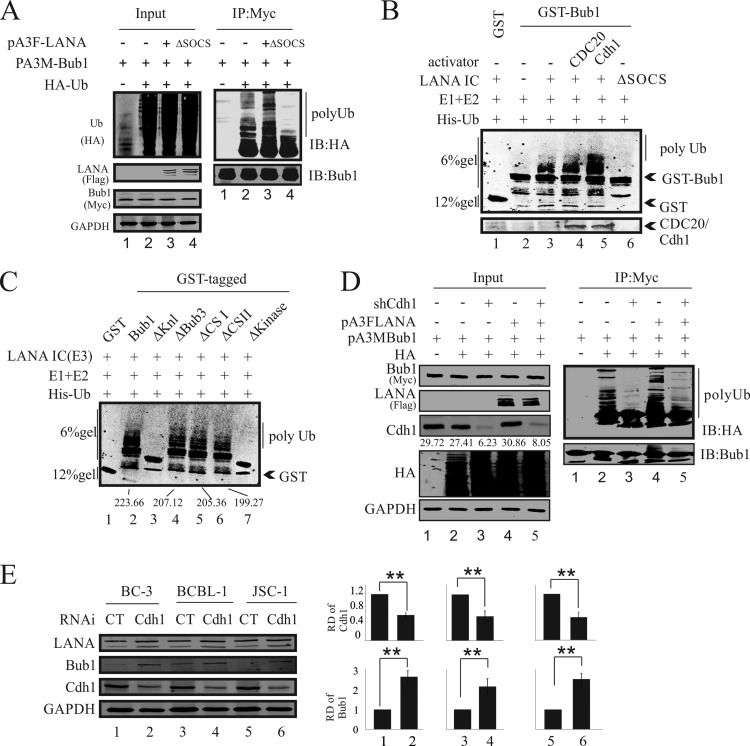
LANA promotes Bub1 ubiquitination.
To further explore the LANA-promoted degradation of Bub1 via the ubiquitin-mediated proteasome pathway, we investigated whether LANA-induced Bub1 ubiquitination led to Bub1 degradation. Here we performed both in vivo and in vitro ubiquitination assays in HEK-293 cells. HEK-293 cells were cotransfected with Flag-tagged LANA or LANAΔSOCS, a LANA dominant negative mutant, together with HA-tagged ubiquitin and Myc-tagged Bub1 (Fig. 5A). Immunoprecipitation with an anti-Myc antibody and Western blotting with an anti-HA antibody showed enhanced ubiquitination of Bub1, as evidenced by the increased intensity of the ladder in the presence of WT LANA (Fig. 5A, lane 3). However, a dramatic reduction in the intensity of the Ub ladder was found in the presence of the LANAΔSOCS polypeptide. These data strongly indicated that LANA can specifically promote Bub1 ubiquitination in cells. Furthermore, we performed an in vitro ubiquitination assay with bacterially expressed GST-Bub1 or its mutants and in vivo-overexpressed LANA. The LANA protein was immune affinity purified and was incubated with GST-Bub1, His-Uba1 (E1), His-Ubc5a (E2), and His-ubiquitin in the presence of ATP. The results revealed that the LANA immune complex (IC) can mediate the ubiquitination of Bub1 in vitro (Fig. 5B). It was clear that in the presence of the LANA IC in addition to the E1 and E2 enzymes, Bub1-GST was strongly ubiquitinated (Fig. 5B, compare lanes 2 and 3). Furthermore, when the APC coactivator Cdh1 was present, ubiquitination activity was significantly enhanced (Fig. 5B, compare lanes 3 and 5). Notably, the LANAΔSOCS polypeptide did not result in enhanced ubiquitination and showed a pattern similar to that seen with the control (Fig. 5B, compare lane 2 with lanes 3 and 4).
FIG 5.
LANA promotes Bub1 ubiquitination. (A) HEK-293 cells were electroporated with expression vectors as shown. Forty-eight hours after electroporation, the cells were treated with MG132 for another 6 h. Cell lysates were then prepared for IP with 9E10, followed by Western blot analysis with the indicated antibodies. IB, immunoblot. (B) LANA ubiquitinates Bub1 in vitro. Bacterially expressed GST-Bub1, His-Ubca1 (E1), and His-Ubc5a (E2) were incubated with a purified LANA immune complex (E3) in kinase buffer for 1 h at 30°C. The sample was then resolved on an SDS-PAGE gel for Western blot analysis. (C) The Knl and kinase domains are required for LANA-mediated Bub1 ubiquitination in vitro. Bacterially expressed GST-Bub1 or its related deletion mutants, His-Ubca1 (E1), and His-Ubc5a (E2) were incubated with a purified LANA immune complex (E3) in kinase buffer for 1 h at 30°C. The samples were then resolved on an SDS-PAGE gel for Western blot analysis. (D) HEK-293 cells were electroporated with expression vectors as shown. Seventy-two hours after electroporation, the cells were treated with MG132 for another 12 h. Cell lysates were then prepared for IP with 9E10, followed by Western blot analysis with the indicated antibodies. (E) The endogenous Cdh1 of KSHV-positive cell lines (BC-3, BCBL-1, and JSC-1) was knocked down, and the protein levels of Bub1 were monitored. The relative densities (RD) of Bub1 and Cdh1 were quantified and plotted against the signals obtained from the control after normalization to GAPDH. Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**).
To determine the involvement of the Bub1 domains in contributing to ubiquitination, the Bub1-GST mutants were compared to the WT. The ΔKnl and ΔKinase mutants showed an almost complete loss of ubiquitination activity. This strongly indicated that these two domains are critical for the stability of Bub1 in the context of LANA-mediated degradation (Fig. 5C, compare lane 2 with lanes 3 and 7).
To further support our hypothesis that LANA mediated Bub1 degradation by manipulating the APC/C, we knocked down the APC activator Cdh1, a protein that is necessary for the APC/C to recognize and degrade Bub1, in HEK-293 cells and KSHV-positive cell lines (BC-3, BCBl-1, JSC-1). In vivo ubiquitination assays were performed in Cdh1-depleted HEK-293 cells (Fig. 5D). Immunoprecipitation with anti-Myc and Western blotting with anti-HA showed dramatically decreased ubiquitination of Bub1 in Cdh1-depleted cell lines, as evidenced by the decreased intensity of the ladder (Fig. 5D, compare lanes 2 and 4 with lanes 3 and 5). Furthermore, we investigated the protein levels of Bub1 in Cdh1-depleted KSHV-positive cell lines (Fig. 5E). Significant increases in Bub1 protein levels were found in Cdh1 knockdown cell lines (Fig. 5E, compare lanes 1, 3, and 5 with lanes 2, 4, and 6).
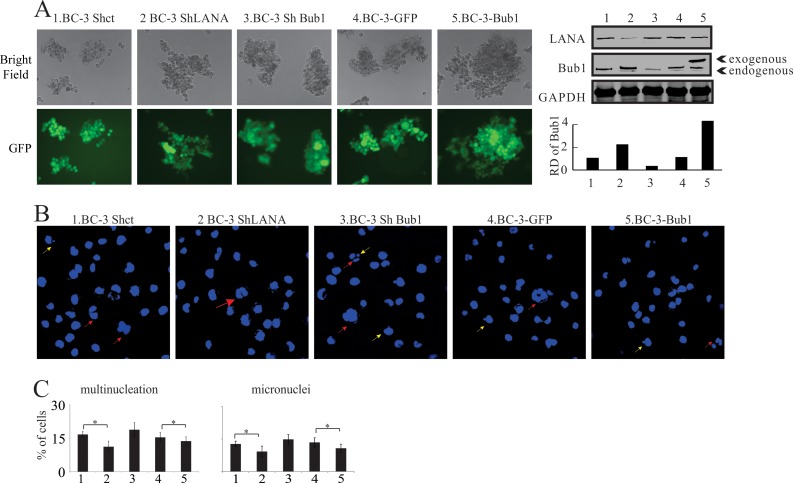
LANA-mediated Bub1 degradation leads to increases in the levels of micronucleus formation and multinucleation.
Bub1 is an important component of the SAC (46), and disruption of the SAC often leads to the formation of micronuclei and multinucleation (57). We had demonstrated previously that LANA can induce the formation of micronuclei and multinucleation (52), but the mechanism remained largely unknown. Here we evaluated the role of LANA-mediated Bub1 degradation in the formation of micronuclei and multinucleation. Cell lines in which Bub1 or LANA was knocked down or in which Bub1 was stably expressed were generated (Fig. 6A). Subsequently, these cells were stained with DAPI. Both Bub1-expressing cell lines and cell lines with stable knockdown of LANA showed significant decreases in the proportions of cells with micronuclei and multinucleation from their respective controls (Fig. 6B and C). The cell line with LANA stably knocked down exhibited a significant decrease in the percentage of cells with aneuploidy, having more than two nuclei, from approximately 17% in the control to less than 11%. Similar results were seen in the cell line stably expressing Bub1, where the proportion of cells with more than two nuclei exhibited a significant decrease from approximately 16% to 12%. In the BC-3 control cell lines, BC-3 Shct and BC-3-GFP, approximately 13% of the cells showed the micronucleus phenotype. In comparison, the BC-3 ShLANA and BC-3-Bub1 cell lines both showed decreased proportions of cell with micronuclei. The formation of micronuclei decreased in the LANA knockdown cell line from approximately 13% to 9%. In the BC-3-Bub1 cell line, the formation of micronuclei decreased from 13% to 10%. Interestingly, a higher number of micronuclei and a higher level of multinucleation were observed in Bub1 knockdown cell lines than in LANA knockdown or Bub1-expressing cell lines (Fig. 6B and C).
FIG 6.
Induction of micronuclei and multinucleation in various BC-3-derived cell clones. (A) (Left) BC-3-derived cell lines with stable expression of Bub1 or stable knockdown of Bub1 or LANA. The GFP signal appears in green in the bottom panels. (Right) (Top) Expression of LANA and Bub1 in the BC-3-derived cell lines. Lane numbers correspond to the numbering of the cell lines on the left. (Bottom) The relative densities (RD) of Bub1 were quantified and plotted against the signal obtained from the control after normalization to GAPDH. Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**). (B) Examples of micronuclei and multinucleation induced in BC-3 cells in which either LANA or Bub1 was knocked down or in which Bub1 was stably expressed. Yellow arrows indicate micronuclei, and red arrows indicate multinucleation. (C) Quantitation of cells with multinucleation (left) or micronuclei (right) among BC-3-derived cell clones. All assays were carried out in triplicate.
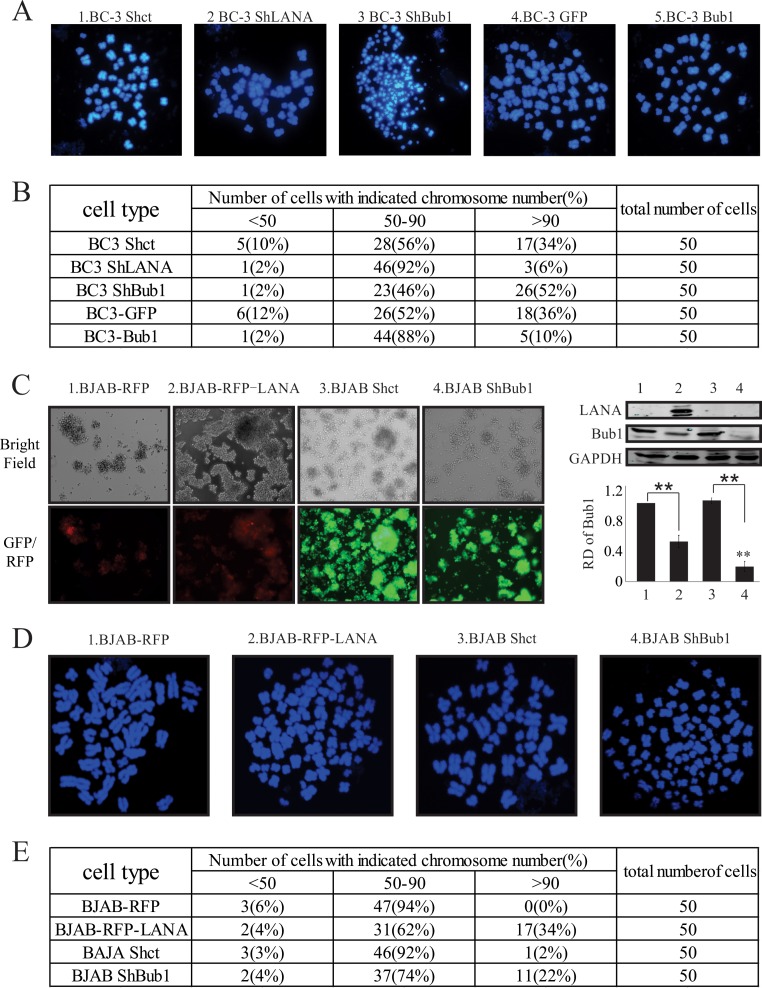
LANA-mediated Bub1 degradation increases chromosomal instability.
Disruption of the SAC leads to chromosomal missegregation and, as a consequence, causes aneuploidy (58–60). Therefore, we investigated the role of LANA-mediated Bub1 degradation in the process of aneuploidy. Here we performed a metaphase chromosome spread assay. A total of 50 metaphase cells were analyzed for each of the cell lines BC-3 Shct, BC-3 ShLANA, BC-3 ShBub1, BC-3-GFP, and BC-3-Bub1. The results are presented in Fig. 7. The chromosome number distribution, which is a marker of chromosomal instability (CIN), is shown for these cell lines (Fig. 7B). Forty-six of 50 (92%) BC-3 ShLANA cells had 50 to 90 chromosomes. Only 1 (2%) of the cells had fewer than 50 chromosomes, and 3 (6%) of the cells had more than 90 chromosomes. A similar distribution of chromosomes was found in the BC-3-Bub1 cell line, where 44 (88%) of the cells had 50 to 90 chromosomes. One (2%) of the cells had fewer than 50 chromosomes, and 5 (10%) of the cells had more than 90 chromosomes. However, in the BC-3 ShBub1 cell line, only 23 (46%) of the cells had 50 to 90 chromosomes. One (2%) of the cells had fewer than 50 chromosomes, and 26 (52%) of the cells had more than 90 chromosomes (Fig. 7B). In comparison, 28 (56%) BC-3 Shct cells and 26 (52%) BC-3-GFP cells had 50 to 90 chromosomes, 5 (10%) BC-3 Shct cells and 6 (12%) BC-3-GFP cells had fewer than 50 chromosomes, and 17 (34%) BC-3 Shct cells and 18 (36%) BC-3-GFP cells had more than 90 chromosomes (Fig. 7B). These data indicated that BC-3 ShBub1 cells showed greater heterogeneity in terms of chromosome number distribution than other cell lines. The BC-3 ShLANA and BC-3-Bub1 cell lines showed less heterogeneity in terms of chromosome number distribution (Fig. 7B). To further confirm the roles of LANA and Bub1 in CIN, BJAB-derived cell lines in which LANA was stably expressed or Bub1 was stably knocked down were generated (Fig. 7C). Metaphase spread experiments were performed on these cell lines (Fig. 7D). The results were similar to those we obtained with BC-3-derived cell lines: in the presence of LANA or upon depletion of Bub1, smaller numbers of cells had 50 to 90 chromosomes, and more cells had more than 90 chromosomes (Fig. 7E). These data clearly demonstrated that CIN was remarkably increased in a Bub1 knockdown cell line, indicating that LANA-mediated Bub1 degradation led to a substantial increase in chromosomal instability in KSHV-infected B-cell lymphoma cell lines.
FIG 7.
Metaphase chromosome spreads of BC-3- and BJAB-derived cell clones. (A) Representative metaphase spreads of the indicated BC-3-derived cell lines. (B) Distribution of chromosome numbers in the indicated BC-3-derived cell lines. Chromosomes were counted from metaphase spreads. (C) (Left) BJAB-derived cell lines with stable expression of LANA or stable knockdown of Bub1. (Right) (Top) Expression of LANA and Bub1 in the BJAB-derived cell lines. Lane numbers correspond to the numbering of the cell lines on the left. (Bottom) The relative densities (RD) of Bub1 were quantified and plotted against the signal obtained from the control after normalization to GAPDH. Statistical significance was evaluated by using P values of <0.05 (*) and <0.01 (**). (D) Representative metaphase spreads of the indicated BJAB-derived cell lines. (E) Distribution of chromosome numbers in the indicated BJAB-derived cell lines. Chromosomes were counted from metaphase spreads.
DISCUSSION
Chromosomal instability (CIN), a high rate of loss or gain of chromosomes, is a hallmark of most human cancers and causes cell aneuploidy (61). The molecular mechanism for CIN is still being explored. However, evidence to date has suggested that defects in the spindle checkpoint, which plays a critical role in the accuracy of chromosomal segregation during mitosis, will promote cell aneuploidy and lead to oncogenesis (62, 63). During cell division, the spindle checkpoint maintains genome stability by arresting cell division until all chromosomes are properly aligned on the metaphase equatorial plate. They are accurately attached to the microtubule spindle apparatus via their kinetochores (40). Unattached or incorrectly attached chromosomes activate the spindle checkpoint, resulting in blocked cell cycle progression (40). When all kinetochores are correctly attached to the spindles, the checkpoint is inactivated, and the cell cycle occurs normally (28).
Bub1 is the first checkpoint component docking at the kinetochore in early prophase and is recruited to the kinetochore through the direct interaction between its N-terminal Knl domain and blinkin, a member of the conserved KMN (KNL1/Mis12 complex/Ndc80 complex) family of kinetochore proteins (64). Studies show that Bub1 may act as a scaffold protein, which determines the kinetochore recruitment of a number of target proteins, including Cenp-E and Cenp-F, Bub3, Mad1, Mad2, Mad3, and other spindle checkpoint proteins (10, 11, 25, 29, 30). In humans, it has been reported that Bub1 is mutated or downregulated in colorectal cancers associated with chromosomal instability and in other aneuploid tumor types (45, 46).
The Knl domain of Bub1 is necessary for the kinetochore localization of Bub1 and is important for the regulation of the spindle checkpoint (65–67). The kinase domain of Bub1 not only can regulate the spindle checkpoint by phosphorylating the APC/C coactivator Cdc20 (36, 68) but also has a key role in chromosome alignment and segregation (69). Our data now show that the Knl domain and kinase domain of Bub1 are required not only for the interaction of the KSHV oncoprotein LANA and Bub1 but also for LANA-mediated Bub1 ubiquitination and for Bub1 degradation. This strongly suggests a role for LANA in suppressing Bub1 function through its Knl and kinase domains, and this suppression can result in chromosome instability and missegregation.
The activity of the APC/C is tightly controlled during the cell cycle (41). Its dysfunction can cause deregulation of mitosis (70). Recently, a few viral factors have been reported to interact physically with the APC/C, playing a modulatory role in its ubiquitin ligase activity (71, 72). Here we found that LANA encoded by KSHV was associated with the APC/C in KSHV-positive cell lines. Our results from co-IP assays of LANA and Cdc27 showed that LANA was associated with the APC/C. Further investigation suggested that Bub1 was recruited to the APC/C by LANA. These data strongly suggested that LANA is capable of manipulating the intrinsic Bub1 degradation pathway. This degradation is likely to compromise or weaken the formation of the spindle checkpoint and ultimately lead to uncontrolled activity of the APC/C, which will cause chromosome segregation even though the chromosomes are not correctly attached.
Our data showed that the protein levels of Bub1 were dramatically suppressed in KSHV-positive B-cell lymphoma cells or in cells stably expressing LANA. However, the Bub1 protein levels were increased in LANA-depleted KSHV-positive cell lines. This indicated that LANA can play a critical role in downregulating Bub1 in KSHV-positive B-cell lymphoma cell lines. Furthermore, depletion of Bub1 by lentivirus-delivered shRNA led to an increase in CIN and to multinucleation and the formation of micronuclei in KSHV-positive B-cell lymphoma cell lines. The depletion of LANA or the overexpression of Bub1 in KSHV-positive B-cell lymphoma cells showed decreased rates of CIN, multinucleation, and micronucleus formation. These data strongly suggested that LANA-mediated Bub1 degradation plays critical contributory roles in CIN and in the observed increases in levels of multinucleation and micronucleus formation in KSHV-positive B-cell lymphoma cells.
We have now provided new evidence that LANA can degrade Bub1 through manipulation of the APC/C, thus increasing CIN in KSHV-positive B-cell lymphoma cells. CIN is frequently observed in KSHV-associated cancer and plays an important role in the pathogenesis of many human cancers. Therefore, our results now provide an additional novel mechanism, elucidating a role for LANA in chromosome instability and oncogenesis through targeted degradation of Bub1 by recruitment and ubiquitination via the APC/C E3-Ub ligase and the proteasome degradation pathway. This mechanism is likely to be finely tuned in terms of Bub1 regulation at specific phases during mitosis when Bub1 can be associated with LANA; this association is decreased as the cell enters metaphase and is regained in telophase (50). These studies may also have therapeutic applications: small molecules or inhibitors of the microtubule formation of the kinetochore may be utilized to block infections by KSHV and other gammaherpesviruses and their persistence in associated cancer.
ACKNOWLEDGMENTS
We thank Hongtao Yu (University of Texas Southwestern, Dallas, TX) for kindly providing the pCS2-Cdc20 and pCS2-Cdh1 plasmids.
This project was supported by Public Health Service grants R01-CA-137894, R01-CA-171979, R01-CA-177423, P30-DK-050306, and P01-CA-174439 (to Erle S. Robertson). Erle S. Robertson is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print 16 April 2014
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- 2.Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O'Leary JJ. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274–1278. 10.1038/nm1295-1274 [DOI] [PubMed] [Google Scholar]
- 3.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648–2654 [PubMed] [Google Scholar]
- 4.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342–346. 10.1038/nm0396-342 [DOI] [PubMed] [Google Scholar]
- 5.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renne R, Lagunoff M, Zhong W, Ganem D. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An FQ, Compitello N, Horwitz E, Sramkoski M, Knudsen ES, Renne R. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862–3874. 10.1074/jbc.M407435200 [DOI] [PubMed] [Google Scholar]
- 10.Di Bartolo DL, Cannon M, Liu YF, Renne R, Chadburn A, Boshoff C, Cesarman E. 2008. KSHV LANA inhibits TGF-β signaling through epigenetic silencing of the TGF-β type II receptor. Blood 111:4731–4740. 10.1182/blood-2007-09-110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882–7892. 10.1128/JVI.75.17.7882-7892.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shamay M, Krithivas A, Zhang J, Hayward SD. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554–14559. 10.1073/pnas.0604469103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JS, Cotter MA, II, Robertson ES. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971–22978. 10.1074/jbc.M101890200 [DOI] [PubMed] [Google Scholar]
- 14.Krithivas A, Young DB, Liao G, Greene D, Hayward SD. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637–9645. 10.1128/JVI.74.20.9637-9645.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma SC, Borah S, Robertson ES. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348–10359. 10.1128/JVI.78.19.10348-10359.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, Howley PM. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919. 10.1128/JVI.00502-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker LL, Shankar P, Khan G, Freeman RB, Dezube BJ, Lieberman J, Thorley-Lawson DA. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283–288. 10.1084/jem.184.1.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verma SC, Lan K, Robertson E. 2007. Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312:101–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radkov SA, Kellam P, Boshoff C. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121–1127. 10.1038/80459 [DOI] [PubMed] [Google Scholar]
- 21.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894 [DOI] [PubMed] [Google Scholar]
- 22.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. 10.1371/journal.ppat.0020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Sugaya M, Atkins AM, Aquilino EA, Yang A, Borris DL, Brady J, Blauvelt A. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J. Virol. 77:6188–6196. 10.1128/JVI.77.11.6188-6196.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimuro M, Hayward SD. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3β. J. Virol. 77:8019–8030. 10.1128/JVI.77.14.8019-8030.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Martin HJ, Liao G, Hayward SD. 2007. The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol. 81:10451–10459. 10.1128/JVI.00804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An J, Sun Y, Rettig MB. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222–228. 10.1182/blood-2003-05-1538 [DOI] [PubMed] [Google Scholar]
- 27.Jallepalli PV, Lengauer C. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer 1:109–117. 10.1038/35101065 [DOI] [PubMed] [Google Scholar]
- 28.Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- 29.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millband DN, Campbell L, Hardwick KG. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 12:205–209. 10.1016/S0962-8924(02)02276-6 [DOI] [PubMed] [Google Scholar]
- 31.Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107:386–396. 10.1007/s004120050322 [DOI] [PubMed] [Google Scholar]
- 32.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 33.Cheeseman IM, Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 34.Kiyomitsu T, Obuse C, Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13:663–676. 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- 35.Taylor SS, Ha E, McKeon F. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1–11. 10.1083/jcb.142.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z, Shu H, Oncel D, Chen S, Yu H. 2004. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16:387–397. 10.1016/j.molcel.2004.09.031 [DOI] [PubMed] [Google Scholar]
- 37.Klebig C, Korinth D, Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858. 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams GL, Roberts TM, Gjoerup OV. 2007. Bub1: escapades in a cellular world. Cell Cycle 6:1699–1704. 10.4161/cc.6.14.4493 [DOI] [PubMed] [Google Scholar]
- 39.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323:1477–1481. 10.1126/science.1163300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillett ES, Espelin CW, Sorger PK. 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164:535–546. 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meraldi P, Sorger PK. 2005. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 24:1621–1633. 10.1038/sj.emboj.7600641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernard P, Hardwick K, Javerzat JP. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775–1787. 10.1083/jcb.143.7.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13:3029–3041. 10.1091/mbc.E02-04-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, Taylor SS. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell 13:566–579. 10.1016/j.devcel.2007.08.008 [DOI] [PubMed] [Google Scholar]
- 45.Shichiri M, Yoshinaga K, Hisatomi H, Sugihara K, Hirata Y. 2002. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res. 62:13–17 http://cancerres.aacrjournals.org/content/62/1/13.long [PubMed] [Google Scholar]
- 46.Hempen PM, Kurpad H, Calhoun ES, Abraham S, Kern SE. 2003. A double missense variation of the BUB1 gene and a defective mitotic spindle checkpoint in the pancreatic cancer cell line Hs766T. Hum. Mutat. 21:445. 10.1002/humu.9120 [DOI] [PubMed] [Google Scholar]
- 47.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267. 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16:475–486. 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schliekelman M, Cowley DO, O'Quinn R, Oliver TG, Lu L, Salmon ED, Van Dyke T. 2009. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 69:45–54. 10.1158/0008-5472.CAN-07-6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao B, Verma SC, Cai Q, Kaul R, Lu J, Saha A, Robertson ES. 2010. Bub1 and CENP-F can contribute to Kaposi's sarcoma-associated herpesvirus genome persistence by targeting LANA to kinetochores. J. Virol. 84:9718–9732. 10.1128/JVI.00713-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J, Jha HC, Verma SC, Sun Z, Banerjee S, Dzeng R, Robertson ES. 2014. Kasposi's sarcoma-associated herpesvirus-encoded LANA contributes to viral latent replication by activating phosphorylation of survivin. J. Virol. 88:4204–4217. 10.1128/JVI.03855-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Si H, Robertson ES. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697–709. 10.1128/JVI.80.2.697-709.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaul R, Verma SC, Robertson ES. 2007. Protein complexes associated with the Kaposi's sarcoma-associated herpesvirus-encoded LANA. Virology 364:317–329. 10.1016/j.virol.2007.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tien HF, Su IJ, Tang JL, Liu MC, Lee FY, Chen YC, Chuang SM. 1997. Clonal chromosomal abnormalities as direct evidence for clonality in nasal T/natural killer cell lymphomas. Br. J. Haematol. 97:621–625. 10.1046/j.1365-2141.1997.752711.x [DOI] [PubMed] [Google Scholar]
- 55.Cai Q, Robertson ES. 2010. Ubiquitin/SUMO modification regulates VHL protein stability and nucleocytoplasmic localization. PLoS One 5:e12636. 10.1371/journal.pone.0012636 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13:1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhivotovsky B, Kroemer G. 2004. Apoptosis and genomic instability. Nat. Rev. Mol. Cell Biol. 5:752–762. 10.1038/nrm1443 [DOI] [PubMed] [Google Scholar]
- 58.Scully R. 2010. The spindle-assembly checkpoint, aneuploidy, and gastrointestinal cancer. N. Engl. J. Med. 363:2665–2666. 10.1056/NEJMe1008017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanks S, Rahman N. 2005. Aneuploidy-cancer predisposition syndromes: a new link between the mitotic spindle checkpoint and cancer. Cell Cycle 4:225–227. 10.4161/cc.4.2.1419 [DOI] [PubMed] [Google Scholar]
- 60.Bharadwaj R, Yu H. 2004. The spindle checkpoint, aneuploidy, and cancer. Oncogene 23:2016–2027. 10.1038/sj.onc.1207374 [DOI] [PubMed] [Google Scholar]
- 61.Lengauer C, Kinzler KW, Vogelstein B. 1997. Genetic instability in colorectal cancers. Nature 386:623–627. 10.1038/386623a0 [DOI] [PubMed] [Google Scholar]
- 62.Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. 2009. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum. Mol. Genet. 18:2656–2669. 10.1093/hmg/ddp207 [DOI] [PubMed] [Google Scholar]
- 63.Shen KC, Heng H, Wang Y, Lu S, Liu G, Deng CX, Brooks SC, Wang YA. 2005. ATM and p21 cooperate to suppress aneuploidy and subsequent tumor development. Cancer Res. 65:8747–8753. 10.1158/0008-5472.CAN-05-1471 [DOI] [PubMed] [Google Scholar]
- 64.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharp-Baker H, Chen RH. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250. 10.1083/jcb.153.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589. 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- 67.Meraldi P, Draviam VM, Sorger PK. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7:45–60. 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 68.Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell 32:394–405. 10.1016/j.molcel.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vanoosthuyse V, Valsdottir R, Javerzat JP, Hardwick KG. 2004. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol. Cell. Biol. 24:9786–9801. 10.1128/MCB.24.22.9786-9801.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakayama KI, Nakayama K. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6:369–381. 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- 71.Heilman DW, Green MR, Teodoro JG. 2005. The anaphase promoting complex: a critical target for viral proteins and anti-cancer drugs. Cell Cycle 4:560–563. 10.4161/cc.4.4.1606 [DOI] [PubMed] [Google Scholar]
- 72.Wiebusch L, Bach M, Uecker R, Hagemeier C. 2005. Human cytomegalovirus inactivates the G0/G1-APC/C ubiquitin ligase by Cdh1 dissociation. Cell Cycle 4:1435–1439. 10.4161/cc.4.10.2077 [DOI] [PubMed] [Google Scholar]