Abstract
APOBEC3 proteins are restriction factors that induce G→A hypermutation in retroviruses during replication as a result of cytidine deamination of minus-strand DNA transcripts. However, the mechanism of APOBEC inhibition of murine leukemia viruses (MuLVs) does not appear to be G→A hypermutation and is unclear. In this report, the incorporation of mA3 in virions resulted in a loss in virion reverse transcriptase (RT) activity and RT fidelity that correlated with the loss of virion-specific infectivity.
TEXT
APOBEC3G (hA3G) in humans and APOBEC3 (mA3) in mice are cytidine deaminases that act on single-stranded DNA during reverse transcription, resulting in G→A hypermutation of newly synthesized proviral DNA (1, 2). Although exogenous murine leukemia viruses (MuLVs) are relatively insensitive to the actions of mA3 (2–7), several studies have reported partial inhibition of exogenous MuLVs after incorporation of mA3 (2, 3, 6, 8–11). Furthermore, the finding that Rfv3, a resistance gene for Friend erythroleukemia, encodes mA3 and is responsible for a decreased infectious titer of the Friend MuLV (Fr-MuLV) (11–13) strongly suggests that mA3 inhibits the replication of exogenous MuLVs in vivo. Exogenous ecotropic MuLVs, such as the Fr-MuLV and Moloney MuLV (Mo-MuLV), are inhibited through mechanisms that do not appear to involve cytidine deamination (2, 9, 10). In this study, the effects of mA3 on the infectivity, reverse transcriptase (RT) activity, and frequency of mutations of the ecotropic MuLV CasFrKP were examined.
Virion-associated mA3 suppresses CasFrKP MuLV infectivity.
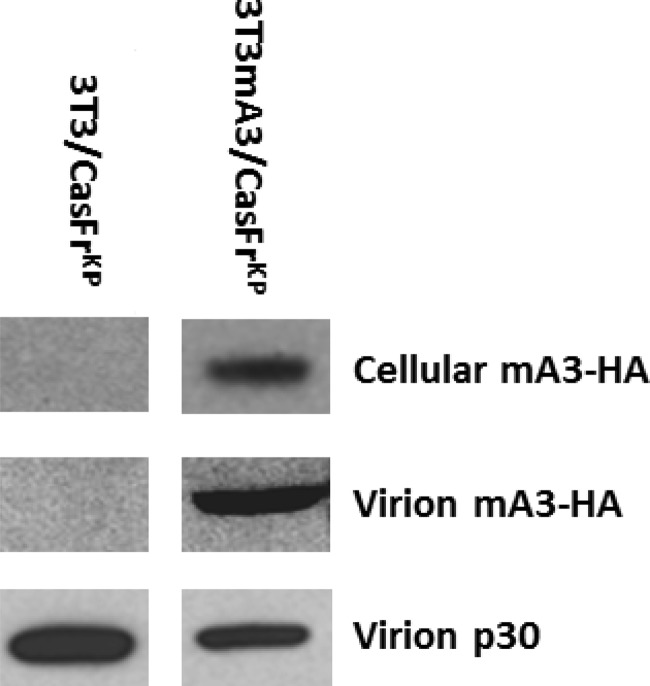
In order to examine the effects of mA3 incorporated into MuLV virions, we derived clonal cell lines infected with CasFrKP (14). The 3T3mA3 cells were derived by transfection of a plasmid encoding the full-length mA3 derived from the BALB/c mouse strain and was tagged at the C termini with hemagglutinin (HA) (5). Infected clonal cell lines were obtained from 3T3 cells as well as from 3T3 cells expressing mA3 (3T3mA3) and were designated 3T3/CasFrKP and 3T3mA3/CasFrKP, respectively. In agreement with earlier reports (2, 7, 9, 13, 15), the clonal cell line expressing mA3 (3T3mA3/CasFrKP) released virions that had incorporated an easily detectable level of mA3 (Fig. 1).
FIG 1.
Incorporation of mA3 into virions released from clonal cell lines. The expression of mA3-HA in the clonal cell line 3T3mA3/CasFrKP was determined from an immunoblot of a cellular extract containing 15 μg protein and developed using an anti-HA monoclonal antibody. Virion mA3-HA released from 3T3mA3/CasFrKP cells was determined from an immunoblot of a gel of virion proteins contained in 1.5 ml of culture supernatant using the anti-HA antibody, while virion p30 was determined from immunoblots of gels of virion proteins contained in 0.075 ml of culture supernatant developed with a monoclonal antibody to p30. Exposure times for the immunoblots of virion p30 were 1 h, while immunoblots of virion mA3-HA were exposed for 5 min. Parallel analyses of 3T3/CasFrKP cells and virions devoid of mA3-HA were included as controls.
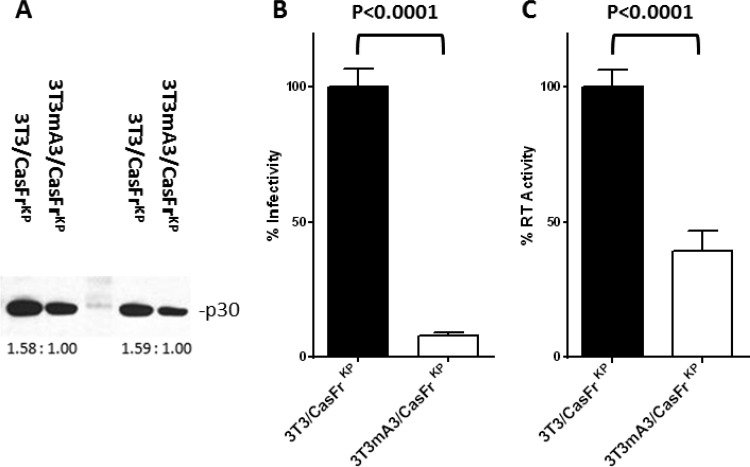
The infectivity of CasFrKP containing mA3 was compared to that of CasFrKP devoid of mA3 by a focal immunofluorescence assay (FIA) (16) and normalized for virion number using the level of p30 CA protein (Fig. 2A). The specific infectivity of virions released from cells expressing mA3 exhibited over a 90% reduction in infectivity (Fig. 2B), corroborating earlier studies of inhibition by mA3 (2, 3, 6, 8–11).
FIG 2.
Influence of virion-incorporated mA3 on specific infectivity and virion RT activity. (A) Virion p30 was determined from immunoblots of gels of virion proteins contained culture supernatant from 3T3/CasFrKP and 3T3mA3/CasFrKP cells. Ratios of the intensities of the bands determined by ImageJ densitometry are shown below the sets of bands. Two different levels of each sample were loaded, resulting in sets of bands with nearly identical ratios of intensity. The average of the ratios was used to normalize the infectivity and RT activity determinations. (B) The infectivity of virions released from cells during an 8-h interval was determined by the FIA and normalized to the levels of p30 present in the samples. The determinations are expressed as a percentage of the mean specific infectivity exhibited by virions released from cells not expressing mA3 (3T3/CasFrKP). Error bars represent the standard errors of 3 to 6 determinations. The P value determined by Student's unpaired t test is indicated. P values of <0.05 were considered significant. (C). The RT activities of virions released from cells during an 8-h interval were determined and normalized to the levels of p30 present in the samples. RT activity was determined for virions contained in 2.5 ml of supernatant media of the samples assayed for infectivity and p30. The determinations are expressed as a percentage of the mean specific infectivity exhibited by virions released from cells not expressing mA3 (3T3/CasFrKP). Error bars represent the standard errors of 3 to 6 determinations. The P value determined by Student's unpaired t test is indicated. P values of <0.05 were considered significant.
Decrease in RT activity in virions containing mA3.
A recent study examined the efficiency of virion reverse transcription by monitoring the appearance of strong-stop DNA during the course of the RT reaction using virions isolated from C57BL/6 and BALB/c mice as well as those from mA3 knockout (KO) mice (17). Both mouse strains exhibited a similar decrease in RT activity compared to KO mice, indicating an effect of endogenous mA3. C57BL/6 and BALB/c mice express different allelic forms of mA3, suggesting that both forms inhibit RT to a similar extent. These results may reflect a direct effect of mA3 on the enzymatic activity of RT or an indirect effect, such as interference of primer binding (18). To assess the effect of virion-incorporated mA3 on the enzymatic activity of RT, we compared the RT activity using a colorimetric assay (Roche Applied Science; no. 11468120910) with exogenous poly(A)/oligo(dT) as the substrate/primer. As was the case with infectivity, the RT activity was significantly lower in virions containing mA3 released from 3T3mA3/CasFrKP cells than that in virions released from 3T3/CasFrKP cells (Fig. 2C). These results suggest an inhibition of the specific activity of the enzyme, although a decrease in the level of fully active RT in the virion cannot be excluded.
Incorporation of mA3 into virions results in a decrease of RT fidelity.
The clonal cell lines utilized were obtained at a low multiplicity of infection, such that only a small percentage of the clonal lines were infected. This procedure served to minimize virion genomic heterogeneity as well as yield infected clones with and without mA3 expression. Viruses released from the clonal cell lines (3T3/CasFrKP and 3T3mA3/CasFrKP) were used to infect new 3T3 cells, and 8 h after infection, cellular DNA was isolated and amplified by PCR using Pfx50 polymerase, which exhibits a very low error rate (∼2 × 10−6) (Invitrogen). Amplicons from the env gene region were cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen) and subsequently sequenced using the amplification primers CasFrKP6805FOR (TTGAGAGAGTACACTAGTC) and CasFrKP7886RC (TCTGTTCCTGACCTTGATC).
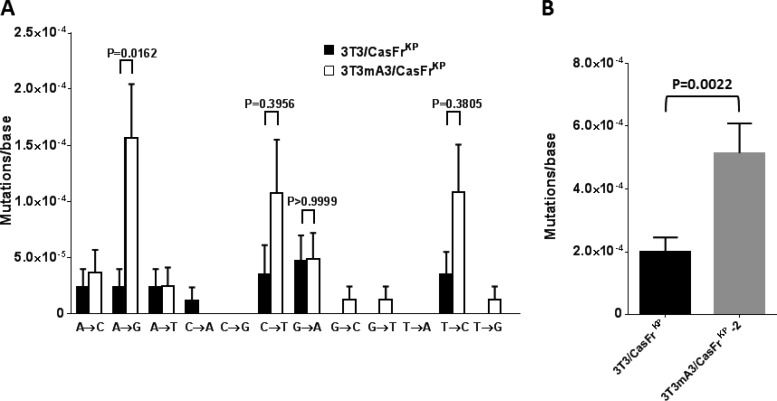
In agreement with other reports (2, 3, 6, 8–11), there was no significant increase in G→A mutations attributed to mA3 (Fig. 3A). Transversion mutations were infrequent; however, increases in transition mutations other than G→A mutations were observed. Notably A→G mutations were significantly higher in infections by the mA3-containing virus. C→T and T→C transition mutations also appeared elevated but did not reach significance (P < 0.05). The inclusion of all of the mutations to calculate overall mutation rates yielded a very significant difference attributable to mA3 (Fig. 3B).
FIG 3.
Influence of mA3 on RT fidelity. 3T3 cells were infected with viruses released from each of the clonal cell lines, and the nucleotide sequences of env gene transcripts synthesized during the first 8 h were determined. (A) The rates of all transition and transversion mutations are shown. Error bars represent the standard errors of detected mutations on approximately 90 transcripts from each of the infections, totaling about 85,000 bases analyzed for each virus. P values determined by one-way ANOVA using Sidak's multiple comparisons test are indicated for the transition mutations. P values of <0.05 were considered significant. (B) The rates of all transition and transversion mutations shown in panel A were combined to calculate the overall mutation rates. Error bars represent the standard errors of detected mutations. The P value determined by Student's unpaired t test is indicated. P values of <0.05 were considered significant.
A bias in the mutation rate in retroviruses has been reported in which the frequency of incurred G→A mutations differs in different regions of the viral genome (19–21). We compared the overall mutation rates in transcripts of regions of the gag, pol, and env genes generated after infection of 3T3 cells with mA3-containing viruses. No significant differential effect of mA3 was observed, suggesting the absence of G→A hypermutation in other regions.
Sequence context of mutations effected by mA3 in CasFrKP.
Flanking sequences of mutations effected by mA3 were examined to determine if any sequence consensus was evident. The analyses revealed only four instances of flanking sequences that were significantly different from the expected frequency (Fig. 4). It is unclear if these deviations reflect preferred targets of mA3. The context of the G→A mutations of CasFrKP (GGN) was distinct from that observed with mA3-induced G→A hypermutation (GAA) (2, 4, 22, 23) and is unlikely to involve cytidine deaminase activity.
FIG 4.
Consensus 3′ and 5′ sequences of mA3-induced transition mutations. The incidence of flanking bases for each of the transition mutations observed in transcripts obtained 8 h after infection by mA3-containing viruses is shown. Analyses include mutations detected in approximately 90 transcripts totaling about 85,000 bases. Significant elevation of the frequency of bases at each position compared to the expected frequency was calculated by the two-tailed binomial test with probability values of <0.05% considered significant (*, P = 0.01 to 0.05; **, P = 0.001 to 0.01).
The incidence of G→A transition mutations was unchanged, while other transition mutations were elevated in the presence of mA3. A paucity of target sequences in CasFrKP could reflect the low G→A mutation rate. In contrast to other MuLVs, AKV undergoes mA3-mediated G→A hypermutation (2). A comparison of putative mA3 target sequences from the AKV genome to those of the CasFrKP sequence did not reveal substantial differences in the number of preferred sites. It remains unclear why the incidence of G→A mutations was unchanged, while the incidence of other transition mutations was increased in transcripts from the mA3-containing virions.
The results of this study suggest an effect of virion-incorporated mA3 on the RT of the virus that affects the activity of the transcription process as well as the fidelity of the enzyme. A loss of fidelity of RT as a result of mA3 incorporation has not been previously reported and may represent another cytidine deaminase-independent mechanism by which APOBEC proteins act to inhibit retroviral replication. CasFrKP, like other exogenous MuLVs, encodes a glycosylated Gag protein (gGag) that partially counteracts the action(s) of mA3. Stavrou et al. (17) have recently observed an inhibition of RT activity by mA3 that is counteracted by gGag. It would be of interest to determine if the gGag protein influences RT fidelity.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, NIAID, and the Department of Biomedical Sciences, University of Cagliari, Monserrato (Cagliari), Italy.
We thank Sue Priola, Kim Hasenkrug, Karin Peterson, and Clayton Winkler for helpful discussions.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Chiu YL, Greene WC. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353. 10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- 2.Langlois MA, Kemmerich K, Rada C, Neuberger MS. 2009. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J. Virol. 83:11550–11559. 10.1128/JVI.01430-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abudu A, Takaori-Kondo A, Izumi T, Shirakawa K, Kobayashi M, Sasada A, Fukunaga K, Uchiyama T. 2006. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 16:1565–1570. 10.1016/j.cub.2006.06.055 [DOI] [PubMed] [Google Scholar]
- 4.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396. 10.1016/j.cub.2004.06.057 [DOI] [PubMed] [Google Scholar]
- 5.Browne EP, Littman DR. 2008. Species-specific restriction of Apobec3-mediated hypermutation. J. Virol. 82:1305–1313. 10.1128/JVI.01371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201–8207. 10.1128/JVI.79.13.8201-8207.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. 2010. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J. Virol. 84:10933–10936. 10.1128/JVI.01023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groom HC, Yap MW, Galao RP, Neil SJ, Bishop KN. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166–5171. 10.1073/pnas.0913650107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rulli SJ, Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J. Virol. 82:6566–6575. 10.1128/JVI.01357-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Martinez S, Aloia AL, Harvin D, Mirro J, Gorelick RJ, Jern P, Coffin JM, Rein A. 2012. Studies on the restriction of murine leukemia viruses by mouse APOBEC3. PLoS One 7:e38190. 10.1371/journal.pone.0038190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 321:1343–1346. 10.1126/science.1161121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith DS, Guo K, Barrett BS, Heilman KJ, Evans LH, Hasenkrug KJ, Greene WC, Santiago ML. 2011. Noninfectious retrovirus particles drive the APOBEC3/Rfv3 dependent neutralizing antibody response. PLoS Pathog. 7:e1002284. 10.1371/journal.ppat.1002284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. 2008. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J. Virol. 82:10998–11008. 10.1128/JVI.01311-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monk RJ, Malik FG, Stokesberry D, Evans LH. 1992. Direct determination of the point mutation rate of a murine retrovirus. J. Virol. 66:3683–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455–463. 10.1016/j.virol.2008.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sitbon M, Nishio J, Wehrly K, Lodmell D, Chesebro B. 1985. Use of a focal immunofluorescence assay on live cells for quantitation of retroviruses: distinction of host range classes in virus mixtures and biological cloning of dual-tropic murine leukemia viruses. Virology 141:110–118. 10.1016/0042-6822(85)90187-4 [DOI] [PubMed] [Google Scholar]
- 17.Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. 2013. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. U. S. A. 110:9078–9083. 10.1073/pnas.1217399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 81:11322–11331. 10.1128/JVI.00162-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chelico L, Pham P, Calabrese P, Goodman MF. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13:392–399. 10.1038/nsmb1086 [DOI] [PubMed] [Google Scholar]
- 20.Poss M, Ross HA, Painter SL, Holley DC, Terwee JA, Vandewoude S, Rodrigo A. 2006. Feline lentivirus evolution in cross-species infection reveals extensive G-to-A mutation and selection on key residues in the viral polymerase. J. Virol. 80:2728–2737. 10.1128/JVI.80.6.2728-2737.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442. 10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]
- 22.Jern P, Stoye JP, Coffin JM. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3:2014–2022. 10.1371/journal.pgen.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit V, Guetard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian JP. 2009. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J. Mol. Biol. 385:65–78. 10.1016/j.jmb.2008.10.043 [DOI] [PubMed] [Google Scholar]