ABSTRACT
We investigated the migration patterns of hepatitis C virus (HCV) in China. Partial E1 and/or NS5B sequences from 411 volunteer blood donors sampled in 17 provinces and municipalities located in five large regions, the north-northeast, northwest, southwest, central south, and southeast, were characterized. The sequences were classified into eight subtypes (1a, n = 3; 1b, n = 183; 2a, n = 83; 3a, n = 30; 3b, n = 44; 6a, n = 55; 6n, n = 10; 6v, n = 1) and a new subtype candidate. Bayesian evolutionary analysis by sampling trees of the E1 sequences of the five major subtypes revealed distinct migration patterns. Subtype 1b showed four groups: one is prevalent nationwide with possible origins in the north-northeast; two are locally epidemic in the central south and northwest, respectively, and have spread sporadically to other regions; and the fourth one is likely linked to the long-distance dispersion among intravenous drug users from the northwest. Subtype 2a showed two groups: the larger one was mainly restricted to the northwest and seemed to show a trend toward migration via the Silk Road; the smaller one was geographically mixed and may represent descendants of those that spread widely during the contaminated plasma campaign in the 1990s. Subtype 3a exhibited three well-separated geographic groups that may be epidemically unrelated: one showed origins in the northwest, one showed origins in the southwest, and the other showed origins in the central south. In contrast, subtype 3b had a mixture of geographic origins, suggesting migrations from the southwest to the northwest and sporadically to other regions. Structurally resembling the tree for subtype 3a, the tree for subtype 6a showed four groups that may indicate migrations from the central south to southeast, southwest, and northwest. Strikingly, no subtype 6a strain was identified in the north-northeast.
IMPORTANCE With a population of greater than 1.3 billion and a territory of >9.6 million square kilometers, China has a total of 34 provinces and municipalities. In such a vast country, the epidemic history and migration trends of HCV are thought to be unique and complex but variable among regions and are unlikely to be represented by those observed in only one or at best a few provinces and municipalities. However, due to the difficulties in recruiting patients, all previous studies for this purpose have been based only on data from limited regions, and therefore, geographical biases were unavoidable. In this study, such biases were greatly reduced because we utilized samples collected from volunteer blood donors in 17 provinces and municipalities. To our knowledge, this is the first study in which the HCV isolates represented such a large portion of the country, and thus, the results should shed light on the current understanding of HCV molecular epidemiology.
INTRODUCTION
Hepatitis C virus (HCV) is a single-stranded positive RNA virus that has been categorized into the Hepacivirus genus of the Flaviviridae family. Taxonomically, the virus is classified into six confirmed genotypes and one provisional genotype, while each genotype, except for genotypes 5 and 7, is further divided into a number of subtypes (1). Different genotypes have shown distinct geographic distribution patterns. In general, genotypes 1, 2, and 3 are prevalent worldwide, while genotypes 4 and 5 are primarily restricted to Africa (2) and genotype 6 is endemic to Southeast Asia (3–6). However, such patterns are constantly evolving as a result of modern transmission and global travel.
HCV has caused infections in an estimated 170 million people worldwide, or 3% of the global population (7). In approximately 70% to 85% of the infected individuals, the infections are characterized by the establishment of chronic hepatitis, which produces a major risk of developing liver cirrhosis and hepatocellular carcinoma (8). Among populations and geographic regions, the frequency of HCV infection varies considerably, with Asia displaying significantly higher levels than the global average (9). China is a major Asian country with over 1.3 billion people where the frequency of HCV infection has been reported to be 3.2% overall and 3.1% in rural areas (10, 11). Namely, over 40 million people in China are infected with HCV, but the historical reasons for this high HCV prevalence and the migrations that have affected the current HCV genotype distribution patterns are not fully understood.
Although six genotypes (genotypes 1 to 6), 18 subtypes (subtypes 1a, 1b, 1c, 2a, 2b, 2f, 3a, 3b, 4d, 5a, 6a, 6e, 6g, 6k, 6n, 6u, 6v, and 6w), and a number of unassigned variants have been detected in China, over 95% of these isolates belong to five major subtypes: 1b, 2a, 3a, 3b, and 6a (12–19). Among them, subtype 1b is predominant nationwide, accounting for approximately 75% of all HCV infections, and this is followed by 2a. However, in Guangdong Province, in the south, 6a is increasingly prevalent (51.5% among HCV-infected intravenous drug users [IDUs], 49.7% among HCV-infected volunteer blood donors, and 17.1% among HCV-infected patients with chronic liver disease) and has replaced 2a as the second predominant subtype (12–14). On the other hand, in Yunnan Province, in the southwest, genotype 3 is thriving and cocirculating with multiple different HCV lineages (17). Because the 6a sequences detected in Vietnam are more diverse than those characterized in China, we performed a Bayesian evolutionary analysis by sampling trees (BEAST) of the 6a sequences from both countries and found that the ancestral origin of 6a was in Vietnam and that it was subsequently introduced into China. With the sequences sampled from different provinces, we further demonstrated that 6a has recently disseminated from Guangdong Province to other provinces. However, such a migration pattern appears to have been restricted to the southern half of China (12). Based on a collection of HCV sequences representing subtypes 1b, 2a, 3a, 3b, and 6a obtained in our previous studies, we performed BEAST to correlate these HCV strains with significant historical events. The explosive population growth of these five major subtypes of HCV in China was consistently shown to have occurred from 1993 to 2000. This corresponds to a period during which contaminated blood transfusions were common, largely due to a procedural error in an officially encouraged plasma campaign. Using a parametric model, we estimated the HCV population growth rates during this period and suggested that certain barriers to efficient viral transmission were removed, allowing nationwide dissemination (20). Although these findings support the belief that the plasma campaign founded the high HCV prevalence in China, the current HCV genotype distribution patterns could also have been affected by subsequent human migrations. For example, there has been a recent tide of migration toward the coastal regions, where fast economic development has led the economic growth in the country for decades and therefore attracted millions of migrants and immigrants. We hypothesized that such changes to the HCV genotype distribution pattern can be vitally reflected in phylogeographic trees reconstructed using the sequences of the five major HCV subtypes sampled from various provinces. By applying the Markov chain Monte Carlo (MCMC) algorithm implemented in BEAST software, we can retrospectively estimate these changes.
As the third largest country in the world by land area, China has a total of 34 province-level administrative units, which include four municipalities, 22 provinces, five autonomous regions, two special administrative districts (Hong Kong and Macao), and the island of Taiwan. Over such a vast territory, the origins and epidemic histories of various HCV strains are thought to be unique and complex but variable among regions and are unlikely to be represented by those observed in only one or at best a few administrative units. However, due to the difficulties involved with the collection of samples from most of these regions, all previous studies for this purpose have been based on data from only limited areas (12–15, 17, 21). For a better representation, here we utilized samples collected from volunteer blood donors in 17 such province-level regions of China. To our knowledge, this is the first study in which the HCV isolates studied represented such a large portion of the territory of China, and thus, this study should shed light on the current understanding of the HCV molecular epidemiology in the country.
MATERIALS AND METHODS
Subjects and specimens.
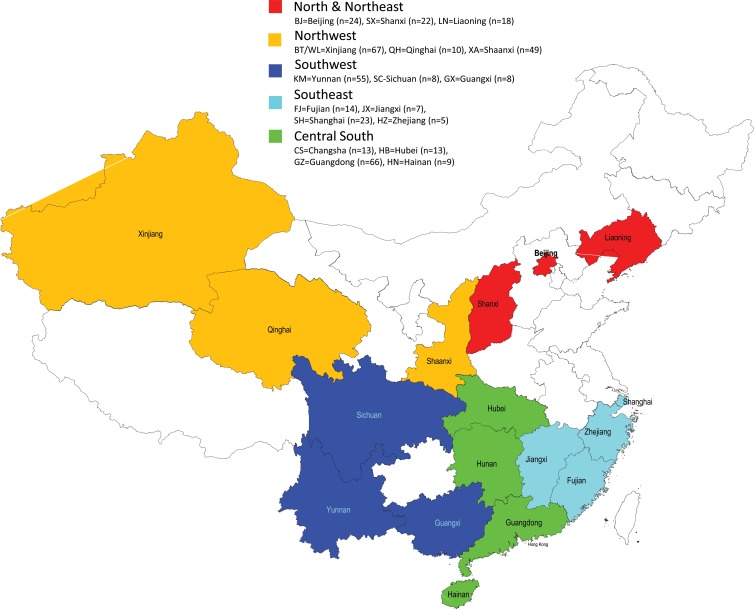
Serum samples were kindly provided by blood centers in 17 provinces and municipalities located in five larger regions: the north-northeast, northwest, southwest, central south, and southeast (Fig. 1). All samples were collected from HCV-infected volunteer blood donors identified during mandatory screening for hepatitis B virus, HCV, and HIV-1 prior to blood donation. The collection process was completed during blood donation campaigns implemented from January 2007 to April 2010. Only those samples that tested positive for HCV RNA were retained, while all others were discarded. This resulted in samples from a total of 411 donors being included in this study. The study was approved by the ethical review committee of the Guangzhou Blood Center. The guidelines set by this committee were strictly followed.
FIG 1.
A map highlighting the 17 provinces and municipalities where the HCV samples were collected from volunteer blood donors. For easier distinction, the 17 provinces and municipalities were divided into five larger regions: (i) the north-northeast (Beijing, Shanxi, and Liaoning), (ii) the northwest (Xinjiang, Qinghai, and Shaanxi), (iii) the southwest (Sichuan, Yunnan, and Guangxi), (iv) the central south (Hunan, Hubei, Guangdong, and Hainan), and (v) the southeast (Fujian, Jiangxi, Zhejiang, and Shanghai). Accordingly, five colors, red, yellow, indigo, green, and blue, were used to mark these five regions on the map, and this color scheme is indicated above the map.
Sequence amplification and phylogenetic analysis.
Partial HCV sequences in two routinely amplified regions, E1 and NS5B, were characterized as previously described (12, 13, 15). They correspond to nucleotides 739 to 1310 and 8267 to 8630, respectively, in the H77 genome. After the sequences were determined, they were aligned using BioEdit software (http://www.mbio.ncsu.edu/bioedit/). Prior to phylogenetic tree reconstruction, the best-fitting substitution model was selected using the jModeltest program on the basis of the Akaike information criterion (22). Consistent with our recent results (12), GTR+I+Γ was found to be the best model for all of the sequence data sets. Under this model, the maximum likelihood (ML) trees were heuristically searched using the subtree pruning and regrafting (SPR) algorithm and the nearest-neighbor interchange (NNI) perturbation algorithm implemented in PhyML software, with which bootstrap analyses were performed in 500 replicates (23). After NEXUS tree files were generated, the ML tree topology was displayed using the FigTree program (3).
Phylogeographic tree analysis.
The E1 sequences obtained were assembled into five data sets, representing five major subtypes, subtypes 1b, 2a, 3a, 3b, and 6a. On the basis of these five data sets, Bayesian coalescent analyses were performed individually using the MCMC algorithm implemented in BEAST software (version 1.6.1). Briefly, the best combination of the GTR+I+Γ substitution model, the Bayesian skyline coalescent model, and the uncorrelated exponential clock model was selected because this combination always outperformed other combinations (3, 12). However, an evolutionary rate of 1.02 × 10−3 ± 2.03 × 10−5 substitution per site per year was used as the prior rate. This rate was determined in one of our recent studies using a group of subtype 1b sequences from the same genomic region analyzed in this study (24). After these parameters were set in the BEAUTi program, XML files were generated and applied to BEAST software for analysis. The latter ran MCMC processes for each of 300 million states and generated a tree for every 10,000 states. To assess the sampling convergence of the MCMC procedures, the estimated effective sampling sizes (ESSs) were evaluated. In this study, when all of the ESSs were ≥200, sufficient sampling was considered to have been achieved. The program Tracer (version 1.5) was used to interpret the MCMC chains and output the posterior trees. To generate phylogeographic trees in a decreasing node order, all the branches were positioned so that the shorter the branches were, the higher they were positioned in the tree and vice versa. The resulting posterior tree files were deciphered using the FigTree program.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study were deposited in GenBank with the following accession numbers: KF585503 to KF586331.
RESULTS
HCV sequencing.
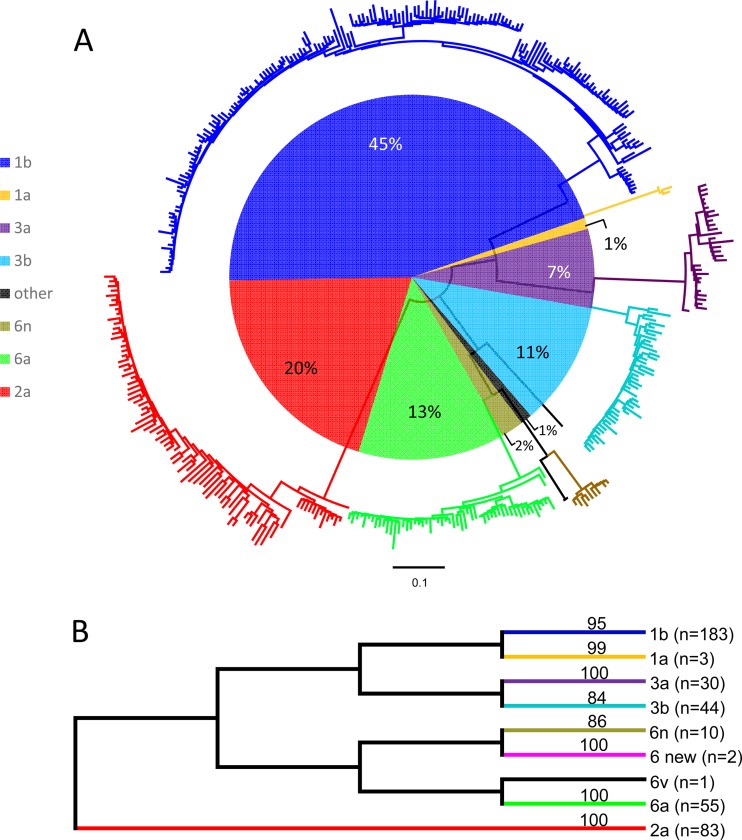
In this study, partial sequences of the E1 and NS5B regions were successfully amplified from 411 and 405 donors, respectively, and sequenced. The resulting sequences were classified into eight HCV subtypes, in addition to a new subtype candidate. Among them, subtype 1b was predominant (183 E1 sequences and 181 NS5B sequences were of this subtype), followed by subtypes 2a (83 E1 sequences and 82 NS5B sequences), 6a (55 E1 sequences and 53 NS5B sequences), 3b (44 sequences for each region), 3a (30 sequences for each region), 6n (10 sequences for each region), 1a (3 E1 sequences and 2 NS5B sequences), and 6v (1 sequence for each region). The number of donors recruited in each province where the HCV subtypes were characterized is shown in Table 1 and Fig. 1.
TABLE 1.
HCV genotype distribution in different provinces
| Subtype | No. of donors |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | Fujian | Guangdong | Guangxi | Hainan | Zhejiang | Hubei | Hunan | Jiangxi | Yunnan | Liaoning | Qinghai | Shanghai | Shanxi | Sichuan | Shaanxi | Xinjiang | Total | |
| 1a | 1 | 2a | 3 | |||||||||||||||
| 1b | 13 | 5 | 25 | 2 | 4 | 1 | 9 | 7 | 5 | 5a | 14 | 6 | 11 | 19 | 8a | 17 | 32 | 183 |
| 2a | 10a | 1 | 3 | 1 | 1 | 3 | 1 | 7 | 3 | 1 | 6 | 2 | 23 | 21 | 83 | |||
| 3a | 1 | 2 | 5 | 1 | 9 | 3 | 9 | 30 | ||||||||||
| 3b | 3 | 1 | 1 | 1 | 2 | 23 | 3 | 1 | 1 | 4 | 4 | 44 | ||||||
| 6a | 5 | 29 | 3 | 3 | 2 | 1 | 2 | 1 | 4 | 2b | 2 | 1 | 55 | |||||
| 6n | 1 | 1 | 4 | 1 | 3 | 10 | ||||||||||||
| 6v | 1 | 1 | ||||||||||||||||
| 6c | 2 | 2 | ||||||||||||||||
| Total | 24 | 14 | 66 | 8 | 9 | 5 | 13 | 13 | 7 | 55 | 18 | 10 | 23 | 22 | 8 | 49 | 67 | 411 |
NS5B sequence was lacking from one donor.
NS5B sequences were lacking from two donors.
6, a possible new subtype of genotype 6.
Two isolates might have been a new subtype of genotype 6. This was determined by coanalyses with reference sequences from all assigned subtypes and unclassified variants of genotype 6. Because the sequences of the two variants showed substantial genetic differences from all of the reference sequences, the variants may have qualified as a new subtype. However, three closely related isolates were not identified and a full-length genome was not characterized; therefore, a new subtype designation was not assigned (data not shown) (21).
Phylogenetic analysis.
A circular phylogenetic tree and a topology tree are shown in Fig. 2. Both trees were reshaped from an ML tree (not shown) reconstructed using the 411 E1 sequences determined in this study. They revealed a considerable diversity of HCV isolates representing eight subtypes and a new subtype candidate. In the circular tree, sequences of the same subtypes were grouped closely, and subtypes differed from each other considerably. When sequences formed clusters at the subtype level, significant bootstrap support of ≥84% was shown in the topology tree.
FIG 2.
(A) Circular form of a phylogenetic tree based on the partial E1 sequences amplified from the 411 blood donors. Different subtypes are shown in different colors, as indicated on the left of the tree. A horizontal ruler with a length of 0.1 nucleotide per site is shown at the bottom of the tree as a guide to measure the genetic distances. The pie chart inside the tree indicates the percentages of the different HCV subtypes (indicated in the same color coding used for the circular tree) into which the 411 isolates were classified. (B) Topology tree converted from the circular form of the tree shown in panel A. Each branch represents a single subtype or equivalent and is labeled to the right with the number of isolates in parentheses and at the top with the level of bootstrap support; otherwise, a branch represents a single isolate.
Figure S1 in the supplemental material shows two ML trees. They were reconstructed using 183 sequences of the E1 region and 181 sequences of the NS5B region that were all classified as subtype 1b. These two trees were consistent, as sequences of identical isolates were classified at similar positions, verifying the reliability of the sequencing results. In both trees, four major groups, groups A, B, C, and D, were indicated. In the E1 tree, the four groups showed bootstrap support values of 67%, 72%, 94%, and 76%, respectively, while in the NS5B tree, the bootstrap support values were 72%, 66%, 54%, and 80%, respectively. Among these four groups, groups A and B have been described previously (15), while groups C and D were newly designated in this study. Consistent with our previous reports (13), group A contained subtype 1b isolates from all provinces but Qinghai. In contrast, group B contained sequences mostly from the central south and was interspersed with a few sequences from other regions. The same was observed for group C, but with the majority of the sequences in group C being from the northwest.
Figure S2A in the supplemental material presents two ML trees reconstructed for the 83 subtype 2a isolates detected: one with the E1 sequences and the other with the NS5B sequences. As shown in both trees, a large fraction of these isolates were from the northwest. The E1 tree was divided into two groups, groups L and S, but only group S showed a significant bootstrap support of 92%. However, in the NS5B tree, the two groups appeared to be inseparable. A finding that was consistent between the two trees was the inclusion of sequences mostly from the northwest in group L. In contrast, the sequences in the rest of the tree appeared to show a mixture of geographic origins. Regardless, these findings were not supported by significant bootstrap support.
For a better comparison, sequences of both subtype 3a and subtype 3b were presented in combined trees (see Fig. S2B in the supplemental material). Although three groups were observed for subtype 3a, they showed no significant bootstrap support. Among the three groups, one included sequences mostly from the northwest, one included sequences mostly from the southwest, and the third one included sequences mostly from the central south. In contrast, the subtype 3b sequences appeared to be more monophyletic. A common feature of both the 3a and 3b subtypes is that they more frequently originated in the northwest, southwest, and central south than in the north-northeast and southeast.
Compared to the geographic sources of the above-mentioned subtypes, the majority of subtype 6a sequences were from the central south, a few were from the southwest and southeast, and fewer still were from the northwest. Strikingly, no subtype 6a isolates were from the north-northeast. Three previously denoted groups, groups I, II, and III (12, 13), were observed in the E1 tree but had bootstrap support values of only 27%, 57%, and 58%, respectively. However, in the NS5B tree, these three groups were inseparable. A small number of subtype 6n isolates were identified, and they appeared to be very closely related genetically (see Fig. S2C in the supplemental material).
Phylogeographic analysis.
To show the differences in the time of divergence among the HCV subtypes characterized, time-scaled phylogenetic trees were reconstructed after sampling the 411 E1 sequences using BEAST software (25). Five trees representing subtypes 1b, 2a, 3a, 3b, and 6a, respectively, were generated. Because information about their geographic origins was also provided, these time-scaled trees represent phylogeographic trees.
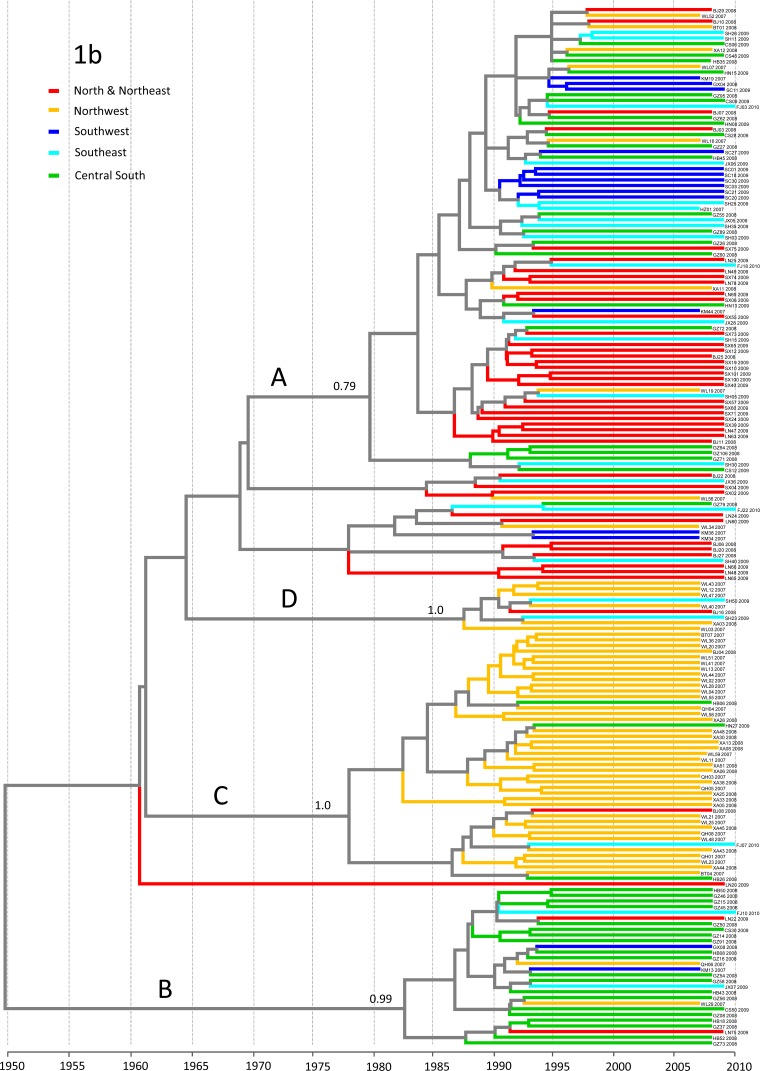
Figure 3 presents a phylogeographic tree reconstructed on the basis of the 183 E1 sequences of subtype 1b. Four groups, groups A, B, C, and D, were indicated, showing significant posterior probabilities of 0.79, 0.99, 1.00, and 1.00, respectively. Compared with the phylogenetic tree shown in Fig. S1 in the supplemental material, here, the time-scaled tree displayed the geographic distribution patterns and migration trends more robustly, with the Bayesian analysis providing additional support to validate the tree's topology. The sequences in group A presented a substantial mixture of geographic origins, and group A included sequences from all provinces but Qinghai. Within this group, subsets characteristic of origins in single geographic regions were observed, but they showed no significant posterior values. As a whole, group A seemed to indicate a trend of subtype 1b migration from the northern half to the southern half of China, but it more likely suggests a simultaneous dissemination nationwide. In contrast, group B featured sequences mainly from the central south, in particular, Guangdong Province, indicating that 1b strains are locally epidemic in this region and occasionally spread to other regions. Compared with groups A and B, group D and, especially, group C were characterized by sequences mostly from the northwest. Both group C and group D are geographic lineages that were newly identified in this study. Two additional groups appeared between groups A and D, but the posterior probabilities were not significant; they included sequences mostly from the north-northeast.
FIG 3.
Phylogeographic tree estimated with the E1 sequences of subtype 1b isolates. Branches are colored according to their geographic origins, indicated in the upper left. When an internal branch led to two sequences that were from different geographic regions, this internal branch and all of its upper internal branches were colored in gray; otherwise, the internal branches were colored the same as the ending branches. This rule was also applied to the tree in Fig. 4. Posterior probabilities of >0.70 are shown at the respective nodes. Below the tree is a time scale from 1950 to 2010, which indicates the time of HCV origin and evolution.
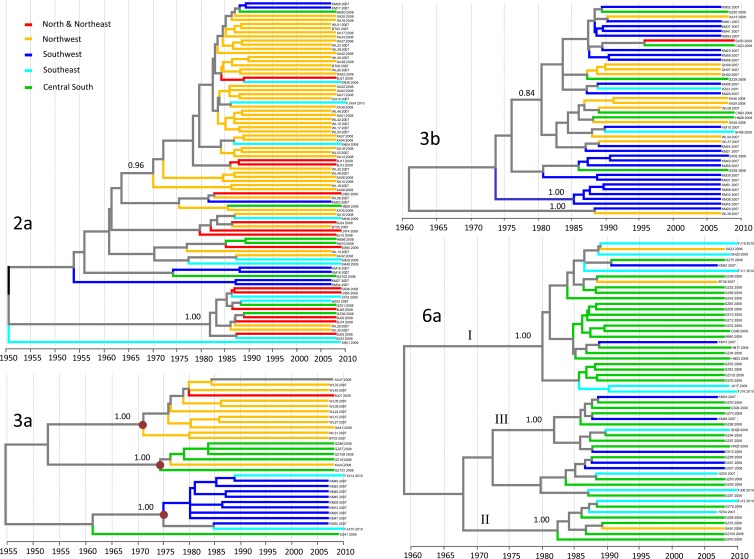
Excluding subtype 1b, phylogeographic trees for the other four HCV subtypes are all shown in Fig. 4. As mentioned above, in the phylogenetic tree for subtype 2a (see Fig. S2 in the supplemental material), two groups were visible, but they had no significant bootstrap support. However, in the phylogeographic tree presented here, these two groups showed significant posterior probabilities of 0.96 and 1.00. The larger group had the majority of its sequences originating in the northwest, while the smaller one displayed branches with sequences with a mixture of geographic origins. Between these two groups, dozens of sequences formed five other loose groups, but they were not geographically related and showed no significant posterior probabilities. This may indicate randomly sampled isolates.
FIG 4.
Phylogeographic trees estimated with the E1 sequences of subtype 2a, 3a, 3b, and 6a isolates. Branches are colored according to their geographic origins, indicated in the upper left. Posterior probabilities of >0.70 are shown at the respective nodes. Below each tree is shown a time scale from 1950 to 2010 (subtype 2a), 1955 to 2010 (subtype 3a), and 1960 to 2010 (subtypes 3b and 6a) indicating the time of origin and evolution of the respective HCV subtypes.
Compared to the two trees described above for subtypes 1b and 2a, the tree reconstructed for the subtype 3a sequences appeared to be very clean and structured in an orderly manner, showing three clearly separated geographic groups, in addition to a single branch. The three groups each contained sequences originating almost exclusively in a single geographic region, the northwest, southwest, or central south, and each had a full posterior probability of 1.00. Among these sequences, only one sequence was from the north-northeast and two sequences were from the southeast.
In contrast to subtype 3a, the phylogeographic tree for subtype 3b showed sequences with a greater mixture of geographic origins. The tree could roughly be divided into two subsets. The smaller subset was located at the tree base and contained only two branches but showed a full posterior probability of 1.00. With a less significant posterior probability of 0.62, the larger subset appeared to show a migration trend from the southwest to the northwest and sporadically to the central south, with a few migrations to the north-northeast and the southeast. Within this subset, three groups could be further classified and showed posterior probabilities of 0.66 to 1.00. Among them, the lower two groups were characterized by almost exclusive origins in the southwest, while the upper group showed substantial geographic interspersion.
The phylogeographic tree reconstructed for subtype 6a appeared to largely resemble that for subtype 3a; however, it included more sequences. Most of the sequences were from the central south, and all were segregated into four groups. Of these groups, three groups, corresponding to groups I, II, and III, which we recently numbered (13), showed full posterior probabilities of 1.00. On the basis of this tree only, the migration trend appeared to have been from the central south to the southeast, followed in frequency by that to the southwest and to the northwest. Strikingly, no subtype 6a isolates were identified in the north-northeast.
DISCUSSION
In this study, phylogeographic trees were reconstructed for HCV subtypes 1b, 2a, 3a, 3b, and 6a. The tree for 1b showed four major groups, groups A, B, C, and D, characteristic of different geographic distribution patterns and migration trends. Except for group A, which contained sequences with a substantial mixture of geographic origins, indicating its nationwide prevalence, the other three groups each contained sequences mostly from a single region. Group B was more prevalent in the central south and frequently spread to other regions, and both group C and group D were more common in the northwest and occasionally appeared outside that region. In one of our earlier studies, we included the worldwide subtype 1b sequences in a coanalysis (15) in which we first designated groups A and B, which are both unique to China. In this study, we found that groups C and D are also unique to China. We further revealed that eight sequences from patients (sequences with GenBank accession numbers AY835104, AY835105, AY835106, AY835107, JX676856, JX677041, JX676903, and JX677124) and two from blood donors (sequences with GenBank accession numbers GQ205717 and GQ205748) in our other studies (4, 6, 12) were classified into group C, while 13 IDU sequences detected in Xinjiang (from isolates XJB2, XJB8, XJB9, XJB12, XJB13, XJB16, XJB17, XJB21, XJB27, XJB31, XJB34, XJB38, and XJXU2) (26) and four sequences detected from IDUs in Hubei (sequences with GenBank accession numbers EF185933, EF185970, EF185990, and EF185989) (27) were classified into group D (see Fig. S3 in the supplemental material). These results seem to indicate that groups A, B, and C may include sequences from the general population of China and likely represent naturally transmitted subtype 1b strains. However, group D may preferentially include isolates from the IDU network.
Likely due to independent evolution, varied selective pressures, and differences in living and environmental conditions and transmission routes, different subtype 1b strains could have been selected in different geographic regions or different population subsets. Some strains, such as group A isolates, may have been highly selected for and widely spread in a dense manner. Some strains, such as group B and C isolates, may have predominantly circulated in certain regions due to a better geographic fitness for those regions. Other strains, such as the group D isolates, may have acquired a propensity for being transmitted via the IDU network.
The phylogeographic tree for subtype 2a showed two statistically well supported groups. The larger one featured sequences almost exclusively sampled in the northwest, while the smaller one showed sequences with a mixture of geographic origins. The former may suggest a common source of 2a strains that have been selected and highly disseminated in the northwest for some historical reasons. However, likely due to temporal or spatial restrictions, those strains have not been transmitted nationwide. In contrast, the smaller group may indicate descendants from a single lineage that is evolutionarily younger but has spread more widely at a lower density. Inclusion of the 2a sequences from a previous study indicated that the smaller group is closely related to a collection of 2a isolates sampled from the former commercial plasma donors who had played active roles in a contaminated plasma campaign in China during the 1990s (see Fig. S3 in the supplemental material) (28). Those 2a isolates could represent descendants from that epidemic event.
The first report about the HCV genotype distribution in China revealed that subtype 2a represented the second major HCV subtype (21). This situation persisted to 2002, when we showed that 2a was the second most predominant subtype in cities in the northern part of the country. However, this was not the case in cities in the southwest and south, where the percentages of other HCV subtypes appeared to be high (15, 17). In the present study, we also found that 2a represented the first major subtype among the volunteer blood donors in Shaanxi Province, accounting for 46.94% (23/49 donors), and the second major subtype among the donors in Xinjiang Province, accounting for 31.34% (21/67 donors). Although the percentages of subtype 2a were also very high in Beijing (10/24 donors, 41.67%) and Shanghai (6/23 donors, 26.08%), these are both centralized municipalities in China in which considerable portions of their inhabitants come from around the country. Why some 2a strains are predominant yet restricted to the northwest may be ascribed to sampling bias; i.e., only convenience samples other than the population-based samples were analyzed in this study. Nevertheless, Xinjiang Province is an autonomous administrative region in China that features a variety of ethnic minorities. These minorities are diverse and have unique cultures, traditions, and living behaviors which are markedly different from those of people living in other regions. More importantly, Xinjiang Province borders Central Asian countries, in particular, Afghanistan, which has been known to be a global drug manufacturing center in recent decades. Overland drug trafficking routes have been indicated to run from Afghanistan and across China (29), overlapping the ancient Silk Road. Through Xinjiang Province, this Silk Road used to link Central and Western Asia to Xi'an City, the capital of ancient China, which is now the capital city of Shaanxi Province, where we obtained 49 samples for this study. Reports have shown the dissemination of HIV-1 and HCV strains from Western and Central Asian countries to China via these drug trafficking routes (26, 30). This may alternatively explain why in the present study many subtype 2a strains isolated from volunteer blood donors in the two provinces of Xinjiang and Shaanxi were closely related, even though the provinces are geographically separated.
The phylogeographic tree for subtype 3a showed three clear separate geographic groups: one had origins almost exclusively in the northwest, one had origins almost exclusively in the southwest, and the third one had origins almost exclusively in the central south. In contrast, only one isolate had its origin in the north-northeast, and two had their origins in the southeast. This pattern supports the hypothesis that subtype 3a could have been introduced into China from different neighboring countries. In addition, the three groups showed common ancestors of similar ages and were led by internal branches that were well separated. This may indicate that these three groups represent three unrelated lineages that diverged before their introduction into China.
In contrast to the tree for subtype 3a, the phylogeographic tree for subtype 3b showed sequences with a mixture of geographic origins, to a certain extent. This indicates a trend for migration from the southwest to the northwest and sporadically to the central south, with a few spills into the southeast and the north-northeast. In relation to a previous finding that 3b was detected more often among IDUs than among other people (31), we may speculate that its migration from the southwest to other regions was primarily by transmission via the IDU network. This is consistent with the known drug trafficking routes in Yunnan Province that link the Golden Triangle in Southeast Asian countries to China (17, 27, 31). This is also consistent with the finding that IDUs have played important roles in mediating the migration of HIV-1 across China (32).
The phylogeographic tree for subtype 6a appears to show a trend for migration from the central south to the southeast, followed in frequency by that to the southwest and to the northwest. Strikingly, no 6a isolates were identified in the north-northeast. It has been estimated in one of our recent studies that the origin of 6a in China was Vietnam (12). From Vietnam, 6a was first introduced to the southwest provinces of China and then disseminated to Guangdong Province in the south, where 6a became locally epidemic and then spread to other provinces (12). However, the tree in this study appears to show that 6a had its origin in the central south, particularly in Guangdong Province, whereupon further spread to other regions, such as the southwest, later occurred. Actually, this tree showed only the recent trend of 6a migration, because we did not include the sequences from earlier studies (12, 31). Transmission via the IDU network could have played a critical role in the earlier introduction of 6a from Vietnam to the southwest provinces of China and then to Guangdong Province (12). However, the recent dissemination of 6a from the central south, particularly from Guangdong Province, to other regions may not be sufficiently explained by this transmission via the IDU network because these 6a sequences were isolated from volunteer blood donors who were not IDUs. Currently, Guangdong Province is playing a critical role in the economic development of China. The fast economic development has attracted millions of migrant laborers and visitors from across the country. In turn, these people have served as carriers for disseminating the 6a viruses from Guangdong Province back to their hometowns across the country (4, 5).
This report represents the largest of its kind conducted in China. Compared to previous studies, the current one showed strength in three aspects. First, the sequences analyzed represent those of HCV strains that are currently prevalent in most parts of China. As we know, with a population of greater than 1.3 billion and a territory of over 9.6 million square kilometers, an extensive survey of the molecular epidemiology of HCV across the country is hard to achieve. For this reason, almost all previous studies were based on data from single or at best a few provinces, and therefore, statistical biases were unavoidable. However, in this study, such bias was greatly reduced because half of the provinces of China were represented. Second, in this study, all the samples were collected from volunteer blood donors. Compared to previous studies that used samples from either IDUs, paid blood donors, or patients with chronic liver disease, the results from this study will better represent the otherwise healthy general population and natural HCV transmission. Third, with the exception of four samples, all of the HCV isolates in this study were characterized using sequences from two separate genomic regions, E1 and NS5B. With this strategy, the genotyping and sequencing results were reliably validated, as the criteria proposed in the recent consensus paper on HCV classification and nomenclature were strictly followed (2).
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by a grant from NIAID/NIH (5 R01 AI080734-03A) to Ling Lu, a grant from the National Natural Science Foundation of China (no. 81273145), and a grant from the 12th Five-Year National Science & Technology Major Project (no. 2012ZX10004702) to Yongshui Fu.
We have no potential conflicts of interest to report.
Footnotes
Published ahead of print 9 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00414-14.
REFERENCES
- 1.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188. 10.1099/vir.0.80401-0 [DOI] [PubMed] [Google Scholar]
- 2.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- 3.Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS, Lu L, Newton PN, Klenerman P. 2009. Genetic history of hepatitis C virus in East Asia. J. Virol. 83:1071–1082. 10.1128/JVI.01501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pybus OG, Markov PV, Wu A, Tatem AJ. 2007. Investigating the endemic transmission of the hepatitis C virus. Int. J. Parasitol. 37:839–849. 10.1016/j.ijpara.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 5.Pham VH, Nguyen HD, Ho PT, Banh DV, Pham HL, Pham PH, Lu L, Abe K. 2011. Very high prevalence of hepatitis C virus genotype 6 variants in southern Vietnam: large-scale survey based on sequence determination. Jpn. J. Infect. Dis. 64:537–539 [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Chan PK, Tam JS, Tang JW. 2011. A possible geographic origin of endemic hepatitis C virus 6a in Hong Kong: evidences for the association with Vietnamese immigration. PLoS One 6:e24889. 10.1371/journal.pone.0024889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705–714. 10.7326/0003-4819-144-10-200605160-00004 [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. 1997. Hepatitis C. Wkly. Epidemiol. Rec. 72(10):65–69 [PubMed] [Google Scholar]
- 9.Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, Amarapurkar D, Chen CH, Dou X, El Khayat H, Elshazly M, Esmat G, Guan R, Han KH, Koike K, Largen A, McCaughan G, Mogawer S, Monis A, Nawaz A, Piratvisuth T, Sanai FM, Sharara AI, Sibbel S, Sood A, Suh DJ, Wallace C, Young K, Negro F. 2011. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 31(Suppl 2):61–80. 10.1111/j.1478-3231.2011.02540.x [DOI] [PubMed] [Google Scholar]
- 10.Lei X, Shigeko N, Deng X, Wang S, Qin S, Liu L, Tang H, Zhao L, Lei B, Yoshihiro A. 1999. Prevalence of hepatitis C virus infection in the general population and patients with liver disease in China. Hepatol. Res. 14:135–143. 10.1016/S1386-6346(98)00119-3 [DOI] [Google Scholar]
- 11.Xia GL, Liu CB, Cao HL, Bi SL, Zhan MY, Su CA, Nan JH, Qi XQ. 1996. Prevalence of hepatitis B and C virus infections in the general Chinese population: results from a nationwide cross-sectional seroepidemiologic study of hepatitis A, B, C, D, and E virus infections in China, 1992. Int. Hepatol. Commun. 5:62–73. 10.1016/S0928-4346(96)82012-3 [DOI] [Google Scholar]
- 12.Fu Y, Qin W, Cao H, Xu R, Tan Y, Lu T, Wang H, Tong W, Rong X, Li G, Yuan M, Li C, Abe K, Lu L, Chen G. 2012. HCV 6a prevalence in Guangdong Province had the origin from Vietnam and recent dissemination to other regions of China: phylogeographic analyses. PLoS One 7:e28006. 10.1371/journal.pone.0028006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Wang Y, Xia W, Pybus OG, Qin W, Lu L, Nelson K. 2011. New trends of HCV infection in China revealed by genetic analysis of viral sequences determined from first-time volunteer blood donors. J. Viral Hepat. 18:42–52. 10.1111/j.1365-2893.2010.01280.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L, Tong W, Yuan M, Lu T, Li C, Lu L. 2013. An increased diversity of HCV isolates were characterized among 393 patients with liver disease in China representing six genotypes, 12 subtypes, and two novel genotype 6 variants. J. Clin. Virol. 57:311–317. 10.1016/j.jcv.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L, Nakano T, He Y, Fu Y, Hagedorn CH, Robertson BH. 2005. Hepatitis C virus genotype distribution in China: predominance of closely related subtype 1b isolates and existence of new genotype 6 variants. J. Med. Virol. 75:538–549. 10.1002/jmv.20307 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Xia X, Li C, Maneekarn N, Xia W, Zhao W, Feng Y, Kung HF, Fu Y, Lu L. 2009. A new HCV genotype 6 subtype designated 6v was confirmed with three complete genome sequences. J. Clin. Virol. 44:195–199. 10.1016/j.jcv.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 17.Xia X, Lu L, Tee KK, Zhao W, Wu J, Yu J, Li X, Lin Y, Mukhtar MM, Hagedorn CH, Takebe Y. 2008. The unique HCV genotype distribution and the discovery of a novel subtype 6u among IDUs co-infected with HIV-1 in Yunnan, China. J. Med. Virol. 80:1142–1152. 10.1002/jmv.21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia X, Zhao W, Tee KK, Feng Y, Takebe Y, Li Q, Pybus OG, Lu L. 2008. Complete genome sequencing and phylogenetic analysis of HCV isolates from China reveals a new subtype, designated 6u. J. Med. Virol. 80:1740–1746. 10.1002/jmv.21287 [DOI] [PubMed] [Google Scholar]
- 19.Xu R, Tong W, Gu L, Li C, Fu Y, Lu L. 2013. A panel of 16 full-length HCV genomes was characterized in China belonging to genotypes 1-6 including subtype 2f and two novel genotype 6 variants. Infect. Genet. Evol. 20:225–229. 10.1016/j.meegid.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Tong W, Gu L, Li C, Lu T, Tee KK, Chen G. 2013. The current hepatitis C virus prevalence in China may have resulted mainly from an officially encouraged plasma campaign in the 1990s: a coalescence inference with genetic sequences. J. Virol. 87:12041–12050. 10.1128/JVI.01773-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Okamoto H, Tsuda F, Nagayama R, Tao QM, Mishiro S. 1993. Prevalence, genotypes, and an isolate (HC-C2) of hepatitis C virus in Chinese patients with liver disease. J. Med. Virol. 40:254–260. 10.1002/jmv.1890400316 [DOI] [PubMed] [Google Scholar]
- 22.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 23.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 24.Yuan M, Lu T, Li C, Lu L. 2013. The evolutionary rates of HCV estimated with subtype 1a and 1b sequences over the ORF length and in different genomic regions. PLoS One 8:e64698. 10.1371/journal.pone.0064698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Zhang C. 2011. Phylogeographic analyses reveal a crucial role of Xinjiang in HIV-1 CRF07_BC and HCV 3a transmissions in Asia. PLoS One 6:e23347. 10.1371/journal.pone.0023347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng JS, Wang X, Liu MQ, Zhou DJ, Gong J, Xu HM, Chen JP, Zhu HH, Zhou W, Ho WZ. 2008. Genetic variation of hepatitis C virus in a cohort of injection heroin users in Wuhan, China. Virus Res. 135:191–196. 10.1016/j.virusres.2008.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CH, Zhou JK, Liu L, Jiang RM, Cao YQ, Mu ZY, Zhang Y. 2009. Investigating genotype of HCV distribution among residents in a “blood donation” village in Hebei Province. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 23:8–10 (In Chinese.) [PubMed] [Google Scholar]
- 29.United Nations Office on Drugs and Crime. 2011. The global Afghan opium trade: a threat assessment. United Nations Office on Drugs and Crime, Vienna, Austria: http://www.unodc.org/documents/data-and-analysis/Studies/Global_Afghan_Opium_Trade_2011-web.pdf [Google Scholar]
- 30.Zhang L, Chen Z, Cao Y, Yu J, Li G, Yu W, Yin N, Mei S, Li L, Balfe P, He T, Ba L, Zhang F, Lin HH, Yuen MF, Lai CL, Ho DD. 2004. Molecular characterization of human immunodeficiency virus type 1 and hepatitis C virus in paid blood donors and injection drug users in China. J. Virol. 78:13591–13599. 10.1128/JVI.78.24.13591-13599.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garten RJ, Zhang J, Lai S, Liu W, Chen J, Yu XF. 2005. Coinfection with HIV and hepatitis C virus among injection drug users in southern China. Clin. Infect. Dis. 41(Suppl 1):S18–S24. 10.1086/429491 [DOI] [PubMed] [Google Scholar]
- 32.Bao Y, Liu Z. 2009. Current situation and trends of drug abuse and HIV/AIDS in China. HIV Ther. 3:237–240. 10.2217/hiv.09.4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.