ABSTRACT
Respiratory syncytial virus (RSV), a member of the Paramyxoviridae family of nonsegmented, negative-sense, single-stranded RNA genome viruses, is a leading cause of lower respiratory tract infections in infants, young children, and the elderly or immunocompromised. There are many open questions regarding the processes that regulate human RSV (hRSV) assembly and budding. Here, using cryo-electron tomography, we identified virus particles that were spherical, filamentous, and asymmetric in structure, all within the same virus preparation. The three particle morphologies maintained a similar organization of the surface glycoproteins, matrix protein (M), M2-1, and the ribonucleoprotein (RNP). RNP filaments were traced in three dimensions (3D), and their total length was calculated. The measurements revealed the inclusion of multiple full-length genome copies per particle. RNP was associated with the membrane whenever the M layer was present. The amount of M coverage ranged from 24% to 86% in the different morphologies. Using fluorescence light microscopy (fLM), direct stochastic optical reconstruction microscopy (dSTORM), and a proximity ligation assay (PLA), we provide evidence illustrating that M2-1 is located between RNP and M in isolated viral particles. In addition, regular spacing of the M2-1 densities was resolved when hRSV viruses were imaged using Zernike phase contrast (ZPC) cryo-electron tomography. Our studies provide a more complete characterization of the hRSV virion structure and substantiation that M and M2-1 regulate virus organization.
IMPORTANCE hRSV is a leading cause of lower respiratory tract infections in infants and young children as well as elderly or immunocompromised individuals. We used cryo-electron tomography and Zernike phase contrast cryo-electron tomography to visualize populations of purified hRSV in 3D. We observed the three distinct morphologies, spherical, filamentous, and asymmetric, which maintained comparable organizational profiles. Depending on the virus morphology examined, the amount of M ranged from 24% to 86%. We complemented the cryo-imaging studies with fluorescence microscopy, dSTORM, and a proximity ligation assay to provide additional evidence that M2-1 is incorporated into viral particles and is positioned between M and RNP. The results highlight the impact of M and M2-1 on the regulation of hRSV organization.
INTRODUCTION
Respiratory syncytial virus (RSV) is a member of the Paramyxoviridae family and contains a continuous, single-stranded negative-sense RNA genome (Fig. 1). Paramyxoviruses are of worldwide importance to human and animal populations. Several highly contagious viruses belong to this family, including the human pathogens measles, mumps, and RSV, as well as the zoonotic viruses Hendra and Nipah (1). Human RSV (hRSV) is the most common cause of bronchiolitis and pneumonia in children under 12 months of age. In the United States, there are between 75,000 and 125,000 children hospitalized each year due to complications associated with RSV infection (2). It has been estimated that, globally, there are 64 million cases of RSV infections annually that culminate in 253,500 deaths (3, 4). Although researchers are actively developing numerous treatments, no effective vaccine has been developed or licensed (5–9).
FIG 1.
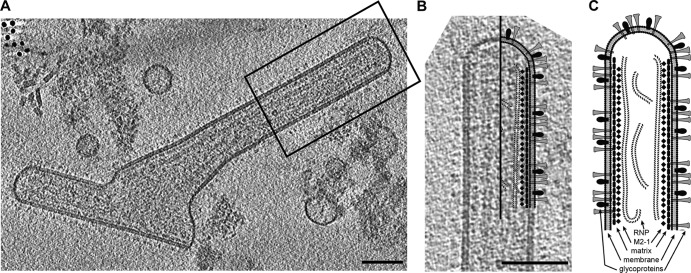
The architecture of an hRSV viral particle. (A) A 7.5-nm central slice from a tomographic reconstruction of a filamentous hRSV viral particle. The boxed region highlights the area of the virus used in panel B. (B) Schematic of hRSV superimposed over the selected area of the hRSV viral particle from panel A. (C) Schematic representation of the architecture of an hRSV virus; the main structural components, the glycoproteins, the viral membrane, the matrix protein layer, the linker (M2-1), and RNP are indicated. Scale bars, 100 nm.
RSV and human metapneumovirus (HMPV) are grouped together in the Paramyxoviridae subfamily Pneumovirinae based on their sequence homology, protein activity, and morphology. The RSV genome consists of 10 open reading frames (ORFs) that encode 11 structural and nonstructural proteins (10). The first seven genes found in all paramyxoviruses translate into seven structural proteins. The nucleoprotein (N), phosphoprotein (P), and RNA-dependent RNA polymerase (L) encapsulate the viral RNA to form a helical assembly termed the ribonucleoprotein complex (RNP). This structure protects the RNA and forms the minimal replication machinery. RSV possesses three integral membrane proteins: the receptor attachment glycoprotein (G), the fusion protein (F), and a short hydrophobic (SH) protein. The G protein is involved in viral attachment to the host cell, while the F protein is responsible for fusion. The SH protein forms a pentameric ion channel (11).
Much like other paramyxoviruses (12–16), the polymerization of the matrix protein (M) is regarded as the main force that drives RSV assembly and budding. M is a membrane-associated protein that consists of both positively charged and hydrophobic domains that are important for cytoplasmic membrane binding (17). M is essential for virus particle formation. Recent studies of an M-null mutant indicated that the absence of M impairs the formation of long viral filaments (18). This may illustrate that the presence of M or the polymerization of M at the budding sites promotes the elongation of filamentous viruses. M also interacts with the amino-terminal domain of G (19) and with the cytoplasmic tail of the F protein (20) and coordinates their recruitment to sites of assembly and budding. For some paramyxoviruses, matrix proteins have been shown to be sufficient for budding of viruslike particles (VLPs) in tissue culture, as is the case for PIV1 (21), Sendai (22), and RSV (20). However, other paramyxoviruses, such as Mumps (16) and PIV5 (23), require an accessory protein, e.g., F or N, for maximum budding process efficiency.
The three additional genes contained in the genomes of Pneumovirus genus members are the two nonstructural proteins (NS1 and NS2) and M2. NS1 and NS2 are type I interferon antagonists (10). The M2 gene encodes two distinct proteins, M2-1 and M2-2, which have been shown to function in genome transcription and replication (24–26). More specifically, the M2-1 protein serves two distinct roles. First, M2-1 is a transcriptional regulator and switches RNA synthesis from protein transcription to genome replication. Second, M2-1 has a complementary structural function, in which it has been shown to colocalize with cytoplasmic inclusions contained within infected cells, associating with the RNP complex through P (27) and with the N-terminal domain of M (28). In a recent study, it was suggested that M2-1 was contained in virus particles and proposed to be ∼13 nm underneath the membrane (29). However, no evidence has yet been presented regarding the relationship between M, M2-1, and RNP, within isolated or cell-associated viral particles.
To determine the structure and organization of hRSV, we have in this study combined several state-of-the-art imaging and analysis technologies. We used cryo-electron tomography (cryo-ET) and a newly developed approach, Zernike phase contrast (ZPC) cryo-ET, to visualize the architecture of hRSV in its native state. ZPC cryo-electron microscopy (cryo-EM) has been shown to produce higher-contrast images of biological specimens collected in near-to-focus imaging conditions (30, 31). Recently, ZPC cryo-ET has been applied to studies of several bacteriophage and small macromolecular complexes (32–34), thin edges of eukaryotic cells (35), and whole bacteria (36) and bacteriophage-infected bacteria (37). These reports illustrate the significant improvements that are achievable in contrast and resolution of three-dimensional (3D) reconstructions of unstained biological specimens in the native, hydrated state when imaged by ZPC cryo-ET.
To evaluate the presence and localization of M2-1 in filamentous viruses and to correlate with our cryo-ET analyses, we used confocal microscopy and direct stochastic optical reconstruction microscopy (dSTORM). The dSTORM technique utilizes two different laser wavelengths, one to convert organic fluorophores in a sample to a stable dark state, and another to periodically excite a small fraction of the fluorophores into a bright state. These bright spots are then localized (38). Through repeated cycles of excitation, localization, and deactivation, the sample can be imaged with a lateral resolution of ∼30 nm and an axial resolution of ∼75 nm. This method allowed for the specific localization of viral proteins at a resolution less than the width of typical RSV viral particles (∼200 nm). In addition, we utilized a proximity ligation assay (PLA) to measure the specific protein-protein and protein-RNA interactions in hRSV viruses. PLA utilizes rolling-circle amplification between two oligonucleotide-labeled secondary antibodies to generate discrete bright puncta at interaction sites with ∼30-nm resolution (39). It should be noted that PLA is not a colocalization technique per say. If a punctum is generated, then this indicates that an interaction (within ∼30 nm) has occurred; however, due to the size and shape of the DNA produced during rolling-circle amplification, it may not colocalize perfectly with surrounding molecules. By using peptide-tagged RNA probes against viral genomic RNA, we were also able to identify interactions between viral proteins, namely, M and M2-1, and RNA in hRSV viral particles (40).
MATERIALS AND METHODS
Virus preparation for cryo-ET.
The A2 strain of RSV was grown in HEp-2 cells under batch conditions and purified to a viral particle count of ∼6 ×109 viral particles/ml (50% tissue culture infective dose [TCID50] titer, 106 TCID50/ml). Aliquots of the purified virus suspended in a 10 mM Tris-150 mM NaCl-1 mM EDTA (pH 7.5) buffer were acquired from Advanced Biotechnologies Inc. (Columbia, MD).
Cryo-ET preparation and data collection.
The purified virus suspension was flash-frozen (plunge frozen) onto glow-discharged Quantifoil grids (Quantifoil Micro Tools GmbH, Jena, Germany) in liquid ethane by using either a Vitrobot Mark III or Mark IV system (FEI, Hillsboro, OR). Aliquoted 10 nm bovine serum albumin (BSA)-treated or protein A-treated colloidal gold was added to the sample and applied to the grid as a fiduciary marker.
Cryo-ET data collection was performed by using two microscopes: a JEOL JEM-2200FS 200-kV FEG-TEM (JEOL Ltd., Japan) with a Zernike-style phase plate system, an in-column Omega-style energy filter (slit width, 20 eV), and a 4k-by-4k Gatan Ultrascan charge-coupled-device (CCD) camera (Gatan, Pleasanton, CA) at the Emory University Robert P. Apkarian Integrated Electron Microscopy Core or an FEI Titan Krios 300-kV FEG-TEM (FEI, Hillsboro, OR) with a GIF-2002 energy filter (slit width, 20 eV) coupled to a 2k-by-2k MultiScan Gatan Ultrascan CCD camera at the Rocky Mountain Laboratories, DIR, NIAID. Images were acquired with pixel sizes of 0.764 nm and 0.45 nm on the specimen. A total electron dose of ∼120 e−/A2 was fractionated over individual tilt series ranging from −62° to +62°. Data were collected at a defocus ranging between 4 and 6 μm under focus. Tilt series images were taken automatically with 2 tilt increments by using either SerialEM (41) or Xplore3D, the FEI automated tomography data collection software.
Cryo-ET image processing and analysis.
Three-dimensional (3D) reconstructions (tomograms) were generated using IMOD (42), and the data were binned 2-fold during the process. The tomograms were further binned and denoised by either bilateral filtering or nonlinear anisotropic diffusion as implemented in the Bsoft suite (43). Final analyses of the tomograms were with a voxel size range of 1.5 nm to 0.9 nm. Direct volume rendering, z-slice progressions, and quantitative rendering were done with Amira 5.4.1 (Visualization Sciences Group). Averaged linear profiles were generated from multiple representative hRSV particles for the three classes of hRSV particle morphologies using the ImageJ software package (44). Graphical analyses were performed using Prism (GraphPad Software), while Prism, StatPlus, and Microsoft Excel were used for statistical analysis.
Matrix protein coverage.
To assess the amount of M on the different particle types, the viral membrane was segmented using an Amira total surface segmentation tool. Then regions with visible M and no M were segmented as separate materials. This process was repeated throughout each tomogram. The amount of M coverage was calculated as a percentage of the total surface area determined by the surface area measuring tool in Amira.
Calculation of membrane curvature.
To study the membrane curvature, radius1 (r1) and radius2 (r2) were measured locally on equally sized regions for spherical viruses without M, spherical viruses with M, and filamentous viruses using ImageJ (44). The two principle curvature values (c) were calculated (c1 = 1/r1 and c2 = 1/r2). The total curvature (J = c1 + c2) and the Gaussian curvature (K = c1 × c2) were also determined (45).
RNA imaging probes.
RNA imaging probe design is described in more detail elsewhere (46). Briefly, 2′-O-methyl RNA/DNA chimeric nucleic acid ligands containing a 5′-biotin modification (Biosearch Technologies) were labeled with either Cy3B NHS ester (GE Healthcare) or DyLight 650 NHS esters (Pierce). Unbound dye was removed via centrifugation in a 3-kDa filter (Millipore) and stored at −20°C until further use. To assemble probes, labeled oligonucleotides were mixed with neutravidin (Pierce) in a 5:1 molar ratio and allowed to react at room temperature for 1 h. Unbound oligonucleotides were removed via centrifugation in a 30-kDa filter (Millipore).
Cell culture.
HEp-2 human epithelial cells (American Tissue Culture Collection CCL-23) were maintained in Dulbecco's modified Eagle's medium (DMEM; Lonza) supplemented with 10% fetal bovine serum (HyClone) and 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen). Cells were plated on no. 1.5 coverslips (Electron Microscopy Sciences) 1 day prior to infection or imaging.
Probe delivery.
Cells were washed in Dulbecco's phosphate-buffered saline (DPBS) without Ca2+ and Mg2+ (Lonza) and subsequently incubated with Opti-MEM (Invitrogen) containing 30 nM RNA imaging probes and 0.2 U/ml− activated streptolysin O (Sigma) for 10 min at 37°C. Afterwards, the Opti-MEM was replaced with complete growth medium for 15 min at 37°C before fixation or isolation of the virus.
Virus propagation for fluorescence imaging.
RSV strain A2 (American Tissue Culture Collection VR-1544) was amplified in low-passage-number HEp-2 cells when the cells were >80% confluent. The medium was removed from cells, the cells were washed with DPBS (without Ca2+ and Mg2+), and virus was added at a multiplicity of infection (MOI) of 0.01 for 1 h before fresh medium was added to the inoculum. Cell-associated virus was harvested by scraping when the cells displayed a high degree of cytopathic effect (CPE) (approximately 90%, or around 96 h postinfection), vortexed briefly, aliquoted, and stored at −80°C. For RNA imaging probe-labeled virus, RNA imaging probes were delivered to cells according to the probe delivery method section before the cells were scraped. Viral titers were quantified by plaque assay.
Immunostaining.
Cells or virus on coverslips were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 10 min at room temperature before permeabilization with 0.2% Triton X-100 (Sigma) for 5 min at room temperature. This step was followed by blocking with 5% bovine serum albumin for 30 min at 37°C, washing with PBS, primary antibody incubation for 30 min at 37°C, washing with PBS, secondary antibody incubation for 30 min at 37°C, and a final washing with PBS. Multiple antibody labeling was done simultaneously after checking for cross-reactivity. Cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI), and all samples were mounted with Prolong Gold (Invitrogen). For virus on glass, before fixation, virus was centrifuged through 5-μm-pore-size and 0.45-μm-pore-size filters (Millipore) at 5,000 × g and 4°C for 4 min and 1 min, respectively, to isolate single filaments from infected cells. This size fraction was then spun down onto poly-l-lysine (Sigma)-coated coverslips via centrifugation at 3,007 × g and 4°C for 30 min. Virus was then fixed and immunostained using the above-described method. For simultaneous staining of M and M2-1 (both antibodies raised in a mouse), several steps were added. After permeabilization and blocking, cells were incubated with M2-1 primary, then rabbit anti-mouse Fab fragment (Jackson ImmunoResearch), then a donkey anti-rabbit secondary antibody, followed by a light fixation in 1% paraformaldehyde in PBS for 10 min at room temperature. All incubations were for 30 min at 37°C, and cells were washed twice with PBS between each incubation. Afterwards, cells were stained for viral M and F according to the standard protocol given above.
Antibodies.
Primary antibodies used were human anti-RSV F (Palivizumab; MedImmune), mouse anti-RSV M (B135; a gift from James E. Crowe Jr.'s lab), mouse anti-RSV M2-1 (5H5; Abcam). Secondary antibodies used were donkey anti-mouse Alexa Fluor 488 (Invitrogen), donkey anti-human Alexa Fluor 488 (Jackson ImmunoResearch), donkey anti-rabbit Cy3 (Jackson ImmunoResearch), donkey anti-mouse Alexa Fluor 647 (Invitrogen), and donkey anti-human Alexa Fluor 647 (Jackson ImmunoResearch).
Fluorescent imaging.
Images for fixed cell experiments were taken on an LSM 710 laser scanning confocal microscope (Zeiss) with a ×63-magnification, 1.4-numerical-aperture (NA) plan-Apochromat objective. Alexa Fluor 488 and Cy3 channels were taken on separate tracks to minimize bleedthrough; DAPI and Cy5 were taken on the same track. Lasers used were 405 nm, 488 nm, 555 nm, and 635 nm. Z stacks were taken in increments of 0.38 μm, with the pinhole set to 1 Airy disk. Linear contrast enhancement and rolling-ball fine-filter remove-noise tasks were made in Volocity (PerkinElmer).
dSTORM imaging.
Superresolution images were recorded with a Vutara SR 200 (Vutara, Inc., Salt Lake City, UT) commercial microscope based on the single molecule localization (SML) biplane FPALM technology (47, 48). Viral proteins and MTRIP probes were imaged using 647-nm and 561-nm excitation lasers, respectively, and a 405-nm activation laser in a photoswitching buffer comprised of 20 mM cysteamine and oxygen scavengers (glucose oxidase and catalase) in 50 mM Tris-10 mM buffer at pH 8.0. Images were recorded using a ×60-magnification, 1.2-NA water immersion objective (Olympus) and an Evolve 512 EM CCD camera (Photometrics) with gain set at 50, frame rate at 50 Hz, and maximal powers of 647-nm, 561-nm, or 405-nm lasers set at 8, 8, or 0.05 kW/cm2, respectively. Total number of frames acquired per channel ranged from 2,000 to 4,000 frames. Data were analyzed by the Vutara SRX software (version 4.07). Single molecules were identified by their brightness frame by frame after removing the background. Identified particles were then localized in three dimensions by fitting the raw data in a customizable region of interest (typically 16 by 16 pixels) centered on each particle in each plane with a 3D model function which was obtained from recorded bead data sets. Fit results were stored as data lists for further analysis. The image resolution capable of being achieved experimentally is 20 nm laterally (x and y) and 50 nm axially (z).
Proximity ligation assay.
Virus was filtered and spun down onto glass coverslips as mentioned above. Proximity ligation assay (PLA) was performed according to the Duolink II kit manufacturer's instructions (Olink Bioscience) with slight modifications to the diluents. Briefly, virus was fixed with 1% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 5 min, both at room temperature. Virus was blocked for 30 min at 37°C with a solution containing 0.5% Tween 20 (CalBioChem), 0.1% gelatin (Aurion), 2% donkey serum (VWR), and 1% bovine serum albumin in PBS. Virus was then incubated for 30 min at 37°C with primary antibodies (titrated to minimize nonspecific background) diluted in a solution containing 0.25% gelatin, 0.5% donkey serum, and 1% BSA in PBS followed by secondary PLA probe antibodies diluted in a 0.05% Tween 20 solution in PBS. Following a wash, the ligation, amplification, and subsequent washing steps were performed according to the manufacturer's protocol. Virus was then immunostained as described above with a polyclonal goat anti-RSV primary (Abcam) to label viral particles. For detecting PLA between M and M2-1 (both mouse antibodies), the same steps for immunostaining with a Fab fragment were performed. To measure PLA between either M or M2-1 and the RSV RNA genome, we isolated FLAG-tagged MTRIP-labeled virus from HEp-2 cells.
RESULTS
RSV morphology.
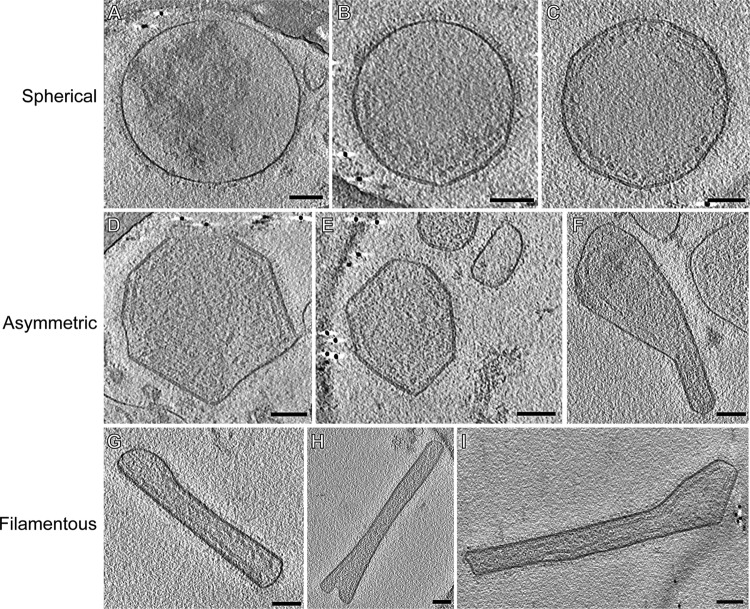
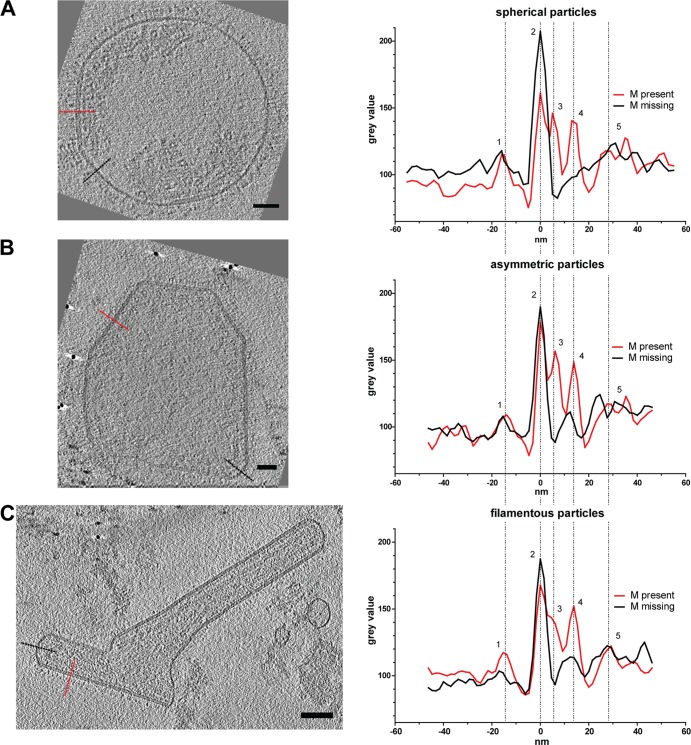
In order to assess the general morphology of hRSV purified from Hep-2 cell culture supernatant, 146 tomograms of viral particles was analyzed. In agreement with Liljeroos et al. (29), three morphological variants were observed in the purified viral samples: asymmetric, filamentous, and spherical (Fig. 1, 2, and 3; see also Movies S1 and S2 in the supplemental material). Despite the differences in shape, all particles shared similar ultrastructural arrangement. The organization of the virus particles was examined by the application of linear profile measurements across regions of the viruses (Fig. 4). All virus types contained areas with the glycoproteins protruding from the viral membrane, the matrix (M) protein associated with the membrane, and the ribonucleoprotein complex (RNP) within the viral particle. Where M was present, there was a visible layer underneath the viral membrane as previously described for measles virus (49), hRSV (29), and PIV1 (50). The center-to-center distance between the viral membrane and the M layer measured approximately 5 nm (Fig. 4), which is in close agreement with earlier studies (29). In all virus morphologies examined, we observed an additional density, which was previously attributed to being M2-1 (29), located ∼13 nm from the viral membrane and in between M and the RNP layers (Fig. 4). The mean RNP width, as determined by center-to-center density measurements, was 11 (±1.4) nm, correlating with the published crystal structure of nucleocapsid (N) of 10 nm (51, 52). In regions where M was absent, neither RNP nor M2-1 was present proximal to the viral membrane.
FIG 2.
Basic morphological characterization of the three hRSV morphologies. Representative particles of the three morphology categories found in the RSV sample: spherical (A to C), asymmetric (D to F), and filamentous (G to I). Common structural features present in the 3D reconstructions of all hRSV morphologies include the surface glycoproteins, the viral membrane, the matrix protein layer, and RNP. Images are 7.5-nm central slices from tomographic reconstructions. Scale bars, 100 nm.
FIG 3.
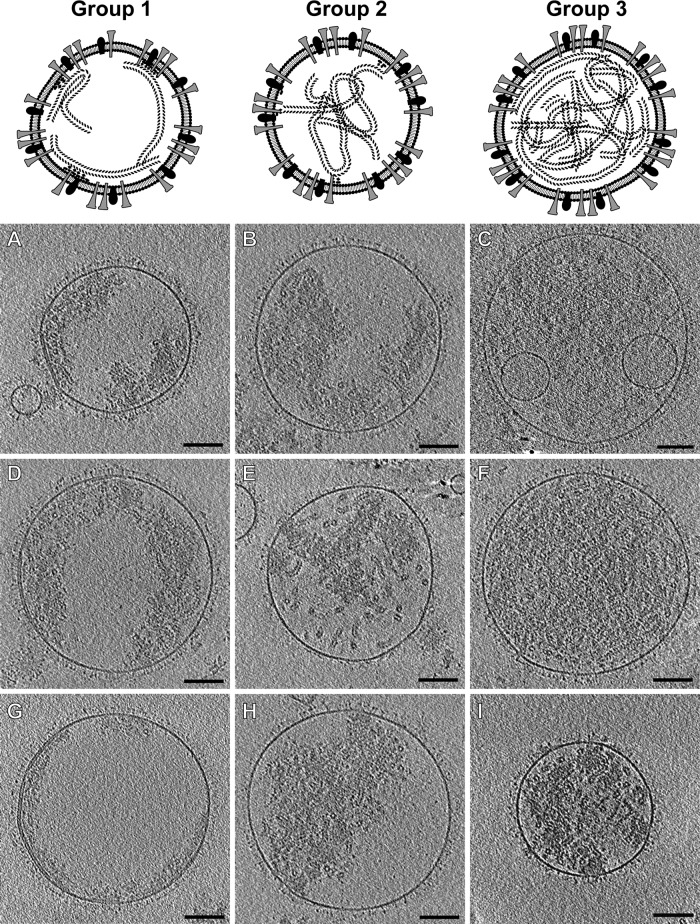
Subclassification of spherical hRSV viral particles. Top, models that represent the major structural features present in each group. Slices (4.5 nm) from tomographic reconstructions of spherical hRSV viral particles are presented for group 1 (A, D, and G), group 2 (B, E, and H), and group 3 (C, F, and I). Scale bars, 100 nm.
FIG 4.
Linear profiles through hRSV particles. Slices (4.5 nm) from tomographic reconstructions of hRSV viral particles indicate the regions used to generate the linear profiles from the three hRSV morphologies (spherical [A], asymmetric [B], and filamentous [C]). The red dotted line marks regions with M, and the black dotted line marks regions without M. Linear profile graphs show the density peaks in the presence (red) or absence (black) of M in the viral particles. Peaks of the different structural components are marked in the density profile: surface glycoproteins (1), viral membrane (2), matrix (3), M2-1 (4), and RNP (5). Scale bars, 50 nm.
The highest variability in terms of organization and content was observed among the spherical particles. The viruses were classified into three groups based on the presence and distribution of M and RNP within the particles (Fig. 3). Group 1 viruses (Fig. 3A, D, and G) contained extensive patches of M, creating the characteristic layer beneath the membrane. Groups 2 and 3, however, had no visible stretches of M yet contained RNP. Between groups 2 and 3, RNP orientation and quantity varied widely. The RNP of group 2 viruses (Fig. 3B, E, and H) was dispersed throughout the particles with few, usually only three to four, contact points with the membrane. The RNP of group 3 viruses (Fig. 3C, F, and I) was densely packed within the viral particles, which made it difficult to resolve other internal structural features native to hRSV. The arrangement of RNP was so dense that no interaction points between RNP and M could be detected. Interestingly, the organization of the asymmetric and filamentous viruses was better regulated than that of the spherical viruses (Fig. 2 and 4). The amount of RNP in the asymmetric and filamentous viruses was more consistent and there was a greater quantity of M associated with the viral membrane than in the spherical viruses. However, the asymmetric viruses had more irregularly shaped patches of M than the filamentous viruses, which were almost completely lined with M. A few viruses exhibited some novel structural components that included the presence of lipid vesicles inside the virus (Fig. 3C), which has been noted in Sendai virus (53), and empty blebs connected to the virus through glycoprotein-covered tubules (Fig. 3A), as has also been observed in Marburg virus (54).
Analysis of the hRSV RNP.
We quantified the number and distribution of genomic RNPs in all three virus morphologies because of the wide variation in RNP content noted upon the initial examination of the tomograms and previous accounts that have indicated that paramyxoviruses incorporate more than one genome copy into an assembled virus (53, 55). RNP was computationally tracked through the 3D volume of each viral particle examined. In a subset of the viral particles, RNP was partially traced in small regions because of the unpredictable trajectories of RNP through the 3D volume, the thickness and density of the viruses, and the effect of the missing wedge (56). However, in some particles, we were able to track the entire RNP. Using the previously determined genome length, pitch, and subunits per helix turn of the hRSV RNP helix (51, 52), an average complete genome length of 1.48 μm was calculated and used in our analysis.
The number of RNPs varied significantly among the spherical viral particles. RNP of the group 1 viruses was observed running in nearly parallel trajectories along the membrane; ∼11 (±1.4)-nm cross sections of RNP were resolved in individual slices through central regions of the viruses (Fig. 3A, D, and G). In group 2 viruses, RNP did not have a defined course, filled large regions of the central volume, and did not exhibit many contact points with the viral membrane (Fig. 3B, E, and H). Group 3 viruses contained large numbers of RNPs, which were flexible enough to fill all the available space within the virus (Fig. 3C, F, and I). Using 1.48 μm as the average genome length for hRSV, we assessed that, in spherical viruses, the genome content varied from one to nine copies (Fig. 5). When examining the tomograms of the asymmetric particles, RNP was noted to be associated with the viral membrane and the center of the virus was devoid of material. The asymmetric viruses had less variation in the quantity of RNP and contained a maximum of five genome copies per virus (Fig. 5). Most of the filamentous viruses contained two genomes (10 out of 18 viruses examined), with the maximum of three genome copies per particle observed (Fig. 5). We also determined that for the three virus morphologies, there was a significant positive correlation between the amount of RNP contained and the virus volume (for filamentous, R2 = 0.23, P = 0.04; for asymmetric, R2 = 0.31, P = 0.005; for spherical, R2 = 0.21, P = 0.012).
FIG 5.
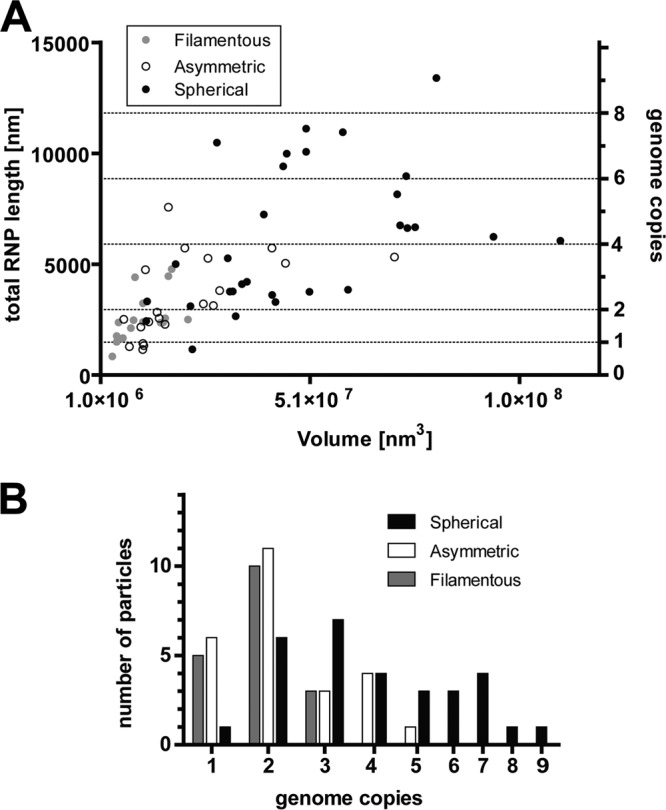
The RNP content of the hRSV viral particles. (A) Graph of the correlation between the traced RNP length and viral particle volume in the spherical, asymmetric, and filamentous virus morphologies. Based on previous studies, the average length of one full genome was calculated to be 1.48 μm. With this, the number of genomes per particle was determined. The left y axis notes the total RNP length in nm, the right y axis corresponds to the number of genome copies, and the x axis is the calculated volume of the viral particles. (B) Graph representing the frequency of the different genome numbers in the three RSV morphologies.
Arrangement of the matrix protein layer.
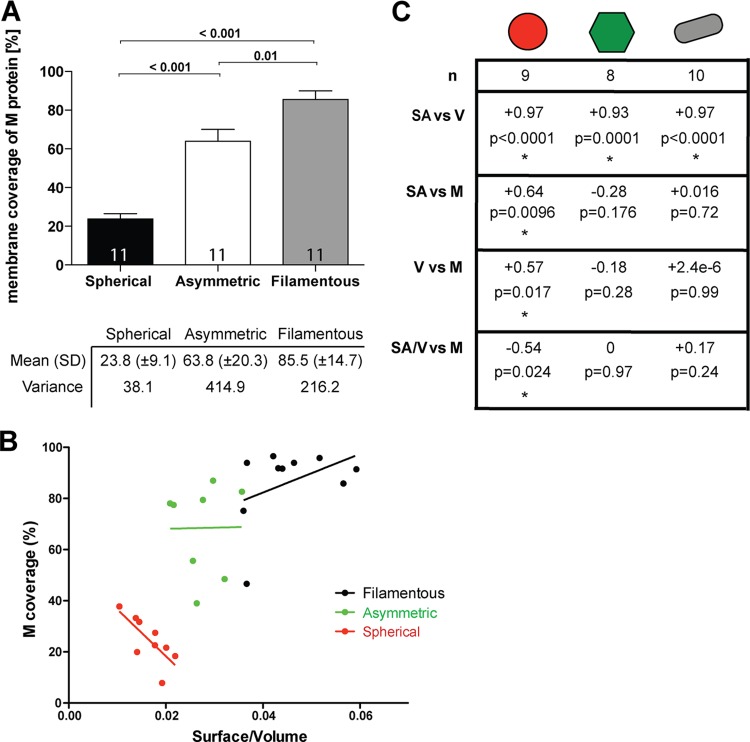
From the linear profiles of all three hRSV morphologies, we identified regions with and without M. In all viruses, M formed a distinct layer ∼5 nm away from the viral membrane (Fig. 4). We noted that M coverage varied significantly between the three virus morphologies. We quantified the amount of M as a percentage of total membrane surface area (Fig. 6A). The surface area of the membrane covered with M was calculated to be ∼24% for spherical viruses, ∼64% for asymmetric viruses, and ∼85.5% for the filamentous viruses (Fig. 6A). This suggests that the more M that is present in a virus, the more likely that it will be filamentous. This observation correlates with previous findings, where M was reported to be required for filament formation (18). To further assess the impact of M on virus structure, the surface area-to-volume (SA/V) ratio of the viruses was calculated and its relationship was compared to M coverage (Fig. 6B and C). It is well established that when spheres and cylinders have the same radius (r) and volume, the surface area ratio is 2:3. Likewise, upon examination of the SA/V ratio, for the sphere the ratio is 3/r and for the cylinder, which includes the additional height (h) measurement, the ratio is (2r + 2h)/rh. Therefore, when we consider filamentous (or cylindrical) and spherical viral particles, it was within reason to expect that, as supported by our calculations, filamentous hRSV particles had, on average, a 1.5× greater SA/V ratio than their spherical hRSV counterparts. M had a significantly negative impact on the SA/V ratio in the spherical particles, suggesting that M did not contribute to particle growth in spherical particles. In fact, the larger hRSV spheres had less M. Asymmetric particles had a higher proportion of M than the spherical particles, but this proportion was highly variable. We concluded that M did not control the SA/V ratio in asymmetric particles. Most of the filamentous particles had close to 100% M coverage, with only the ends lacking M. The slight positive correlation between the amount of M and the SA/V ratio suggests that M is required for the growth of the filament, which agrees with previous studies (18, 29, 57, 58).
FIG 6.
Effect of the matrix protein on hRSV particle morphology. (A) Graph of the amount of M coverage on the different particle morphologies. The number of particles included in the study and the P value of the t test are indicated. Mean and variance are marked in the table below. (B) Graph demonstrates the correlation between M coverage and surface area/volume ratio in the different particle types. (C) Relevant statistics for panel B are shown, displaying the number of particles in the study, R2, and the P value for the linear regression model. Asterisks mark statistically significant correlation.
In addition to the visible layer beneath the membrane, the presence of M was also associated with the presence of flattened membrane surfaces, even in spherical virus particles that had less than 20% M coverage. To study the effect of M on virus particle shape, the membrane curvature was determined (Table 1) (45). In all particle types, when M was present along the viral membrane, regions presented the cylindrical architecture, which was straight along one axis and positively curved in the orthogonal axis (Table 1). Spherical viral particles that did not contain M were positively curved in both axes and did not deviate from the character of a traditional spherical object (Table 1).
TABLE 1.
Effect of M on the membrane curvaturea
| Shape or type | J | K |
|---|---|---|
| Plane | 0 | 0 |
| Cylinder | + | 0 |
| Sphere | + | + |
| Saddle | + | − |
| Spherical without M | +0.014 | +0.00004 |
| Spherical with M | +0.008 | 0 |
| Filamentous | +0.02 | 0 |
The table displays the J and K values for shapes governed by curvature along multiple axes (45) (top four rows) and displays the calculated J (J = c1 + c2) values of the hRSV viral particle types that were examined and the K (K = c1 × c2) values for the three viral particle types (bottom three rows). A larger calculated value indicates a membrane with greater curvature. Curvatures, c1 and c2, were measured on local, equivalent segmented areas for each hRSV particle type. +, positive; −, negative.
M2-1 placement in hRSV.
In the raw tomograms of hRSV particles, we observed a regularly spaced density localized between the M protein layer and RNP; using linear profile analysis, we confirmed that the spacing of the density was in agreement with the recent work of Liljeroos et al. (29). In their analysis, the density was referred to as M2-1 based on several reports that have illustrated that there is an interaction between M and M2-1 and the observation that M does not bind RNP directly (59–61). However, the previous colocalization analyses were performed in cells and not with purified viral particles; therefore, the exact localization of M2-1 compared to other macromolecules within intact viruses had not been determined. While a peak was present in the linear profiles that could be attributed to M2-1, we were unable to resolve additional details regarding its structure or orientation using conventional defocus phase contrast cryo-ET. Therefore, we used ZPC cryo-ET to determine the placement and periodicity of the M2-1 protein in the viral particles. From the ZPC cryo-ET data, we observed small individual densities between the M layer and RNP (Fig. 7A; see also Movie S2 in the supplemental material). The densities were ∼12 nm from the membrane (Fig. 7A), which corresponded to the M2-1 density present in the linear profiles from the conventional cryo-ET data (Fig. 4). The discrete M2-1 masses maintained a regular spacing between each other of 12.6 (±2.1) nm (Fig. 7B).
FIG 7.
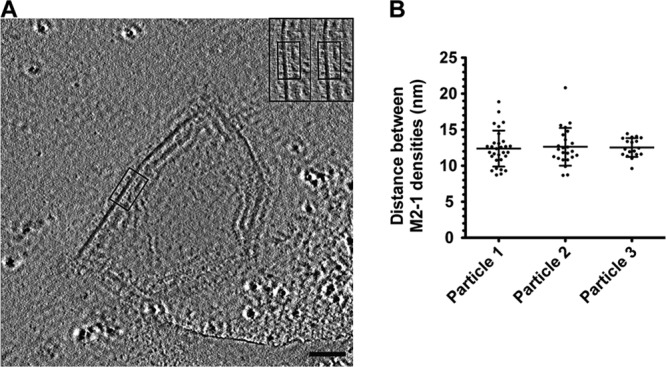
Spacing of M2-1 in hRSV viral particles collected using ZPC cryo-ET. (A) A 7.64-nm slice from a tomographic reconstruction of an hRSV viral particle used for measuring M2-1 spacing. Boxed region highlights the location of M2-1 densities in the slice. The inset is a magnified view of the boxed region; red dots mark the M2-1 densities. Scale bar, 100 nm. (B) Graph of the distances measured between M2-1 densities in three hRSV viral particles. The mean and standard deviation are marked.
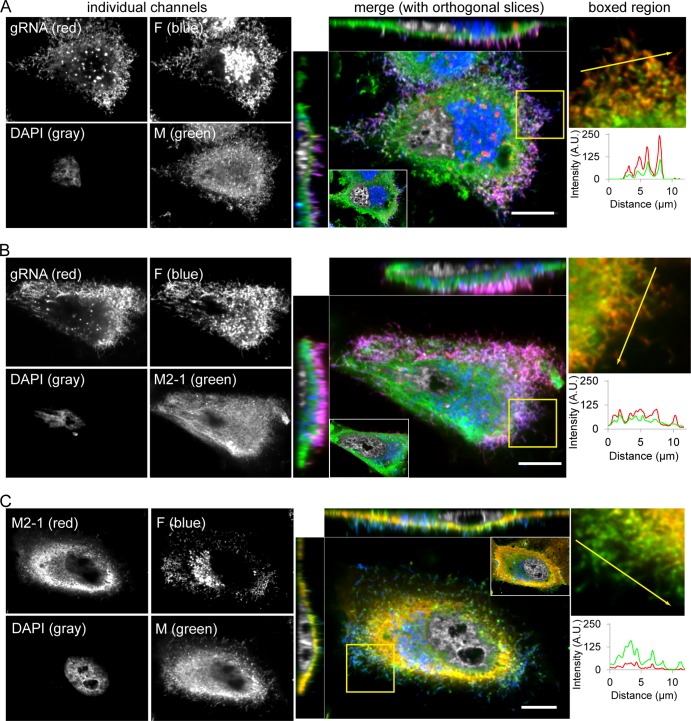
To confirm and correlate with the cryo-ET and ZPC cryo-ET findings regarding the localization patterns of M2-1, we imaged M, M2-1, F, and viral genomic RNA in hRSV-infected HEp-2 cells using laser scanning confocal microscopy (Fig. 8). From these images, we found that the M protein was distributed throughout the cell's cytoplasm, as well as within viral filaments (indicated by viral genomic RNA and F protein) extending from the cell surface (Fig. 8A and C). We also found that M2-1, in addition to being in the cytoplasm as shown by Li et al. (28), was located in viral filaments, albeit at lower quantities than M (Fig. 8B and C). This finding supports the hypothesis that M2-1 is part of the structure of a mature hRSV virion.
FIG 8.
M and M2-1 immunostaining and confocal fluorescence microscopy. (A) HEp2 cells infected with RSV A2 (MOI of 1) had RNA imaging probes targeting the RSV genomic RNA (red) delivered to them at 24 hpi and were fixed and immunostained for the viral F (blue) and M (green) proteins. Also included is a DAPI nuclear stain (gray). To the right, a merged image of an xy plane near the top of the cell is shown with orthogonal slices through the middle of the cell. Inset shows merged image of an xy plane near the bottom of the cell (to emphasize regularity of cell nuclei); boxed region shows magnified image of filamentous virions protruding from the cell membrane with the F and DAPI channels removed to emphasize the clarity of colocalization. Fluorescence intensity profile plot along arrow in the boxed region is also shown. (B) RSV-infected HEp-2 cells with RNA imaging probes (red) and immunostaining for viral F (blue) and M2-1 (green) proteins. (C) RSV-infected HEp-2 cells immunostained for viral F (blue), M (green), and M2-1 (red) proteins. Single optical plane, laser scanning confocal images shown. Scale bars, 10 μm.
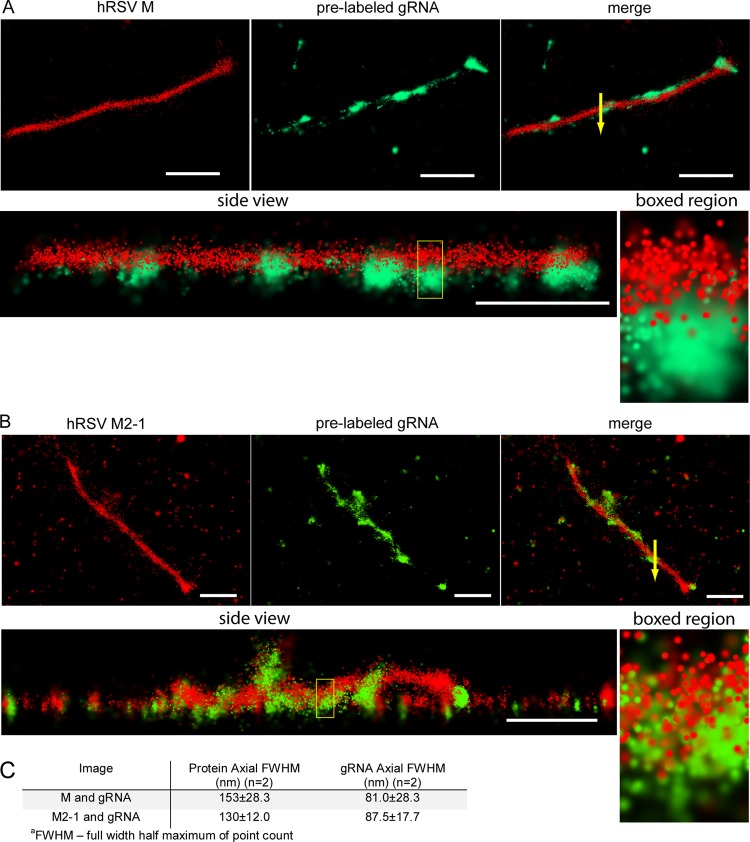
To further investigate the localization of M2-1, we imaged filamentous viral particles isolated from infected cell monolayers using dSTORM (Fig. 9). Both the M protein (Fig. 9A) and the M2-1 protein (Fig. 9B) were distributed throughout the length of the filament; the M was distributed continuously, and the M2-1 coincided more frequently with the genomic RNA. This was more evident in side views where there was both protein and genomic RNA (Fig. 9A and B, side views and boxed regions). The M protein appeared to surround the top of the genomic RNA, while M2-1 was in closer proximity to the genomic RNA than M and followed the genomic RNA distribution closely. This observation was quantified by measuring the full-width half-maximum (FWHM) of the point count of individual filaments (Fig. 9C), which showed the FWHM of M and M2-1 to be greater than that of the RNA, with the M2-1 FWHM being slightly smaller than the M FWHM. These results suggest that both M and M2-1 are nearer to the viral envelope and outside the genomic RNA layer, with M2-1 being closer to the genomic RNA than M.
FIG 9.
dSTORM imaging of RSV filamentous virions. (A) Single RSV filamentous virion on glass, with RNA imaging probes targeting the RSV genomic RNA (green) delivered to it prior to isolation from cells and immunostained for viral M protein (red). A merged image of M and RNA and a side view are also shown. Magnified images of the boxed region in the side view are shown to the right. Arrow in the merge represents an example 100-nm cross section used to measure the axial FWHM. (B) Single RSV filamentous virion on glass with RNA imaging probes that was immunostained for the viral M2-1 protein. Views similar to that in Fig. 10A are shown. (C) Table containing axial FWHM of the point count for the viral protein and RNA in the 100-nm boxed region of the filaments shown in panels A and B. This measurement was repeated for 2 filaments for each protein. Scale bars are 1 μm for whole filaments.
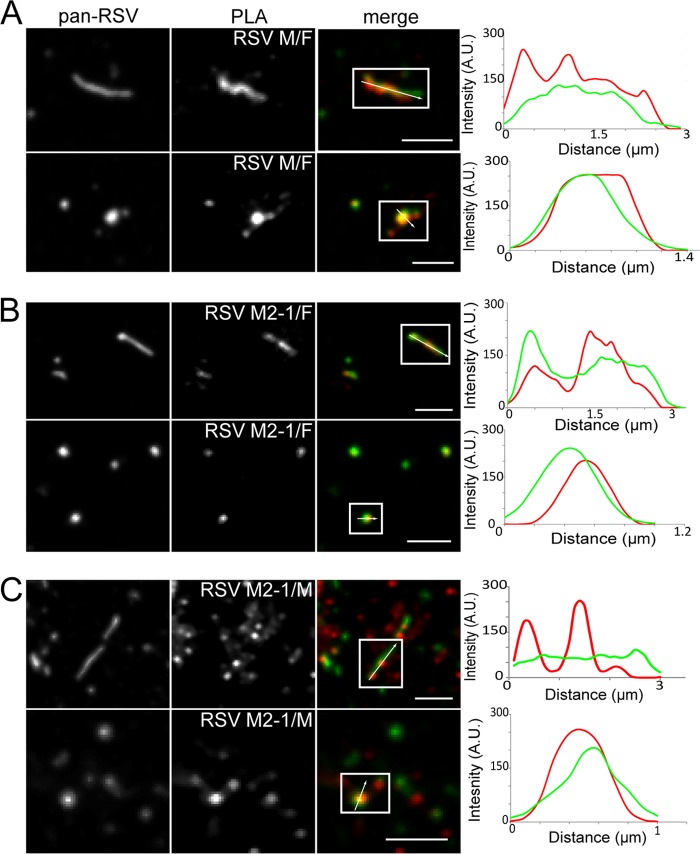
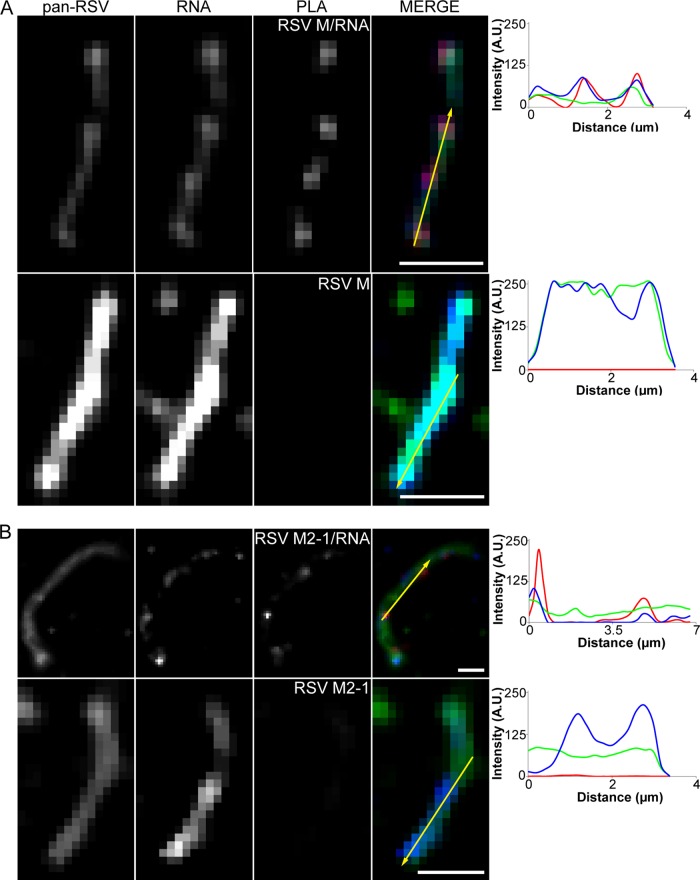
Additionally, we investigated the interactions between F and M, F and M2-1, and M and M2-1 in isolated viral particles using PLA (Fig. 10). All three proteins appeared to be within very close proximity of each other, as indicated by nearly continuous PLA signal even along more filamentous virion morphologies. No PLA signal was detected between either M or M2-1 and green fluorescent protein (GFP) or under a no-primary-antibody control for the M and M2-1 data (controls; data not shown). These data indicate that both M and M2-1 are in very close proximity to F and to each other in mature hRSV viral particles. We also measured the interaction between M and M2-1 and hRSV genomic RNA by utilizing FLAG-tagged RNA probes (Fig. 11). PLA signal, representing an interaction between both M and M2-1 and the RNA, corresponds extremely well with the fluorescent signal from the RNA probes (different wavelengths) compared to no-FLAG-tag controls where there is no PLA signal. This result suggests that both M and M2-1 are within very close proximity to the RNA, and it is not highly dependent on RNA localization within the particles, indicating the RNA is often very close to the membrane, corroborating the cryo-ET findings.
FIG 10.
Proximity ligation assay between F and M, F and M2-1, and M and M2-1. RSV virions on glass were assayed for interactions between F and M, F and M2-1, and M and M2-1 by PLA. The virions were also immunostained for pan-RSV for context. Note that PLA signal is an indication of two molecules in close proximity and may not colocalize with staining. From left to right, column 1 shows pan-RSV staining, column 2 shows PLA signal between F and M (A), F and M2-1 (B), and M and M2-1 (C), column 3 shows the merged image of columns 1 and 2 with pan-RSV signal in green and PLA signal in red, and column 4 shows the fluorescence intensity profile plot along the arrow. The first rows show filamentous virion morphologies, while second rows show spherical virion morphologies. Boxes indicate viral particles of interest; scale bars, 2.5 μm.
FIG 11.
Proximity ligation assay between RNA and either M or M2-1. RSV virions labeled with FLAG-tagged MTRIPs on glass were assayed for interactions between FLAG and either M (A) or M2-1 (B) by PLA. The virions were also immunostained for pan-RSV for context. Note that PLA signal is an indication of two molecules in close proximity and may not colocalize with staining. From left to right, column 1 shows pan-RSV staining, column 2 shows RNA probes targeting the RSV genome, column 3 shows PLA between M and either FLAG-tagged or control MTRIPs (A, rows 1 and 2, respectively) and between M2-1 and either FLAG-tagged or control MTRIPs (B, rows 1 and 2, respectively), column 4 shows the merge of columns 1, 2, and 3 with pan-RSV signal in green, RNA signal in blue, and PLA signal in red, and column 5 shows the fluorescence intensity profile plot along the arrow. Scale bars, 2 μm.
DISCUSSION
Impact of culture and purification conditions on hRSV morphology.
We examined purified preparations of the A2 strain of hRSV that contained a mixture of spherical, asymmetric, and filamentous particles. The spherical viruses were present in the greatest numbers, which confirmed the results of a recent study (29). However, it is still unclear which viral morphology is most prevalent in an active infection in humans. Early scanning electron microscopy (SEM) and transmission electron microscopy (TEM) studies of budding hRSV viral particles indicated that there might be a larger number of filamentous viruses versus other virus morphologies (18, 62–68). One explanation for the difference between budding and purified virus samples could be that viral filaments are transformed to spherical viruses during the purification process through disruption of the extended M layer. This hypothesis is partially supported by the studies of Liljeroos et al., which demonstrated that an hRSV sample lost infectivity and viral particles transitioned to spheres when the sample temperature was cycled (29). Studies of Sendai virus have also indicated that perhaps there may be less variation in virus particle size and the morphology of viruses purified at early time points postinfection (69). However, when early harvest viruses were incubated at 37°C or stored at 4°C for several days and then examined by EM, the virus morphology became more irregular and suggested that the viruses aged after budding. Additionally, the morphology of the virus could vary depending on when the virus was purified postinfection. Electron microscopy and electron tomography analyses of Marburg virus (70) illustrated that there was a larger population of spherical virus particles than filamentous virus particles present in cell culture supernatants at longer time points postinfection. The populations that contained the greatest quantity of spherical Marburg virus particles also displayed reduced infectivity. This could indicate that the timing of assembly and budding events became unsynchronized due to poorer regulation of cellular processes because of reduced cell fitness. If it is indeed the case for hRSV and other paramyxoviruses that assembly of the virus changes over time and is perhaps cell type and virus, as well as virus strain, specific, further investigations to correlate the morphology of purified hRSV, and other viruses, at specified time points combined with using a range of cell types will be required to establish the relationships that exist.
An alternative view could be that there is a difference in morphology between budded and released particles compared to those that remain cell associated. It was recently shown that influenza viruses might utilize multiple pathways for host cell infection (71). Long influenza virus particles utilize the macropinocytosis pathway for infection instead of infecting target cells via the common receptor-mediated endocytosis pathway used by budded viruses (71). This may be an alternative infection method used by many viruses that remain cell associated. Our study and the report of Liljeroos et al. (29) were examinations of hRSV virus purified from the supernatant of infected cell cultures, while the previous studies were of either budding hRSV or a purified sample that contained both supernatant and cell-associated viruses (18, 62–68). Additional studies of hRSV during an active infection of permissive cells will be required to determine whether the different virus morphologies may support a range of successful infection pathways.
The M protein and its regulation of virus assembly and morphology.
It is well known that matrix proteins are essential coordinators of assembly and direct morphological variation in many enveloped viruses (29, 57, 72–76). Despite the extreme pleomorphicity observed with hRSV particles, all morphologies had similarly structured regions that were governed by the presence of the M protein along the viral membrane. M formed extended layers approximately 5 nm underneath the viral membrane. The presence of M influenced the organization of M2-1 and RNP, as they were membrane proximate only when the M layer was visible. Their spacing from the membrane was also regulated, ∼13 nm for M2-1 and 27 nm for RNP. RNP ran parallel to the membrane in all three of the virus morphologies only when M was present, which left an empty volume in the center of many of the viruses examined. Likewise, the densities associated with M2-1 were apparent only when M and RNP were localized together along the membrane. Our findings support the previous observation that M influences particle shape, as the presence of M was associated with elongated and filamentous particles. Changes in the quantity of M present along membranes correlated with explicit differences in particle structure. This suggested that M actively shapes the membrane and influences particle assembly and budding. Similarly, we demonstrated that M actively formed the membrane into cylindrical (or filamentous) shapes, and the widths of the cylindrical regions of the filamentous particles varied between 71 nm and 166 nm.
Quantity and organization of RNP in hRSV.
We traced RNP in the viral particles and found that the number of RNPs was variable and dependent on virus morphology. In filamentous hRSV particles less than 2 μm in length, RNP incorporation was highly regulated. A majority of the filamentous virus particles contained enough measurable RNP that was equivalent to two genome copies. Viral filaments much longer than 2 μm were not examined in 3D by cryo-ET. By cryo-EM 2D imaging, we observed filamentous particles ranging from 2 μm to 10 μm in length. The extremely long viral filaments contained electron densities similar to the RNP density found in the shorter viral filaments (G. Kiss and E. R. Wright, unpublished data). Using dSTORM, we confirmed that long viral filaments over 2 μm in length were filled with RNP, which illustrates that long viral filaments may contain more copies of the hRSV genome. Further studies are needed to clarify the organization of the long filaments. Future research using nanogold versions of the fluorescent RNA imaging probes utilized in this work (46) should allow us to gain further insight into the orientation, sequence, and structure of the genomic RNAs incorporated into the hRSV virion.
Asymmetric and spherical particles supported a broader variation in the number of incorporated genome copies. The average genome copy number per particle was two for asymmetric viral particles and four for spherical viral particles. Our data are consistent with previous findings that paramyxoviruses consistently contain more than one genome copy (53, 55) and with results from quantitative PCR and viral particle counts which revealed that purified hRSV particles have, on average, 1.8 genome copies per particle (B. S. Graham, unpublished data). We also specifically addressed the likelihood that a percentage of the viral particles in the purified samples may include shorter RNP strands. It is known that several enveloped viruses may incorporate truncated genomes into assembling viral particles, which are classified as defective interfering (DI) particles (77–81). DI particles are unable to replicate by themselves and, when present, negatively compete with full-length genomes for the replication machinery. In a recent cryo-ET publication on Sendai virus, DI particles were observed along with viruses that contained complete genomes (53).
M2-1 forms a link between M and RNP.
Using conventional defocus phase contrast (DPC) cryo-ET, we detected the M2-1 signal in linear profiles through all three morphological classes of the virus particles. The application of ZPC cryo-ET allowed us to resolve the individual M2-1 densities between the M layer and RNP. We measured the distance between the M2-1 densities and found a regular, 12.6-nm spacing. The regularity of the M2-1 spacing could be attributed to either a linkage associated with a well-ordered M layer or dependence on the interaction of M2-1 with P from the RNP complex. Producing viruses with nanogold-labeled viral proteins and RNA and analyzing them by cryo-ET could test these hypotheses in future work. Using confocal and dSTORM microscopy, we confirmed that M2-1 is indeed present in viral filaments and that it exists in a layer in close proximity to M as well as the hRSV genome. Additionally, using PLA, we demonstrated that both M and M2-1 are within 30 nm of each other and the F protein in viral filaments.
In summary, the present results provide strong evidence that M, M2-1, and RNP are linked together in all hRSV particle morphologies. Our analyses also revealed that hRSV particles with higher percentages of M associated with the viral membrane were more likely to maintain the filamentous architecture, while lower levels of M corresponded to spherical or asymmetric particles. However, the reasons for such a high pleomorphicity are still not clear. A recent publication showed evidence of how the filamentous particles are more successful in transport and trafficking in the respiratory tract, which can explain the propensity for respiratory viruses to have a filamentous morphology (82) but leaves the purpose of the other observed morphologies unclear. Other studies speculate that the purpose of the extreme morphologies is to distract the immune system from the infectious particles, such as Archetti bodies associated with influenza (83). The reported difference in virion shape and organization in early- and late-harvest Sendai virus samples suggests that host cell factors can also contribute to the virus morphology (84). Additional cryo-ET, fluorescence light microscopy (fLM), and virological studies are needed to clarify these questions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeannette Taylor of the Emory University Robert P. Apkarian Integrated Electron Microscopy Core for assistance. We are grateful to Kirk Easley of the Emory CFAR biostatistics and biomedical informatics core for help with statistical analysis.
This work was supported in part by Emory University, Children's Healthcare of Atlanta, the Center for AIDS Research at Emory University (P30 AI050409), the Georgia Research Alliance, and the James B. Pendleton Charitable Trust to E.R.W., NSF grant 0923395 to E.R.W., and public health service grants R21AI101775 to E.R.W. and R01GM094198 to P.J.S.
Footnotes
Published ahead of print 23 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00256-14.
REFERENCES
- 1.Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DMH, Peter M. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Mullooly JP, Bridges CB, Thompson WW, Chen J, Weintraub E, Jackson LA, Black S, Shay DK. 2007. Influenza- and RSV-associated hospitalizations among adults. Vaccine 25:846–855. 10.1016/j.vaccine.2006.09.041 [DOI] [PubMed] [Google Scholar]
- 3.Nair H, Verma VR, Theodoratou E, Zgaga L, Huda T, Simoes EA, Wright PF, Rudan I, Campbell H. 2011. An evaluation of the emerging interventions against respiratory syncytial virus (RSV)-associated acute lower respiratory infections in children. BMC Public Health 11(Suppl 3):S30. 10.1186/1471-2458-11-S3-S30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krilov LR. 2011. Respiratory syncytial virus disease: update on treatment and prevention. Expert Rev. Anti Infect. Ther. 9:27–32. 10.1586/eri.10.140 [DOI] [PubMed] [Google Scholar]
- 6.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749–1759. 10.1056/NEJMoa043951 [DOI] [PubMed] [Google Scholar]
- 7.Dowell SF, Anderson LJ, Gary HE, Erdman DD, Plouffe JF, File TM, Marston BJ, Breiman RF. 1996. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174:456–462. 10.1093/infdis/174.3.456 [DOI] [PubMed] [Google Scholar]
- 8.Falsey AR, Walsh EE. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371–384. 10.1128/CMR.13.3.371-384.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. 1999. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA 282:1440–1446. 10.1001/jama.282.15.1440 [DOI] [PubMed] [Google Scholar]
- 10.Huang YT, Collins PL, Wertz GW. 1985. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope-associated protein. Virus Res. 2:157–173. 10.1016/0168-1702(85)90246-1 [DOI] [PubMed] [Google Scholar]
- 11.Gan SW, Tan E, Lin X, Yu D, Wang J, Tan GM, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K, Torres J. 2012. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J. Biol. Chem. 287:24671–24689. 10.1074/jbc.M111.332791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shnyrova AV, Ayllon J, Mikhalyov II, Villar E, Zimmerberg J, Frolov VA. 2007. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J. Cell Biol. 179:627–633. 10.1083/jcb.200705062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J. Virol. 80:12070–12078. 10.1128/JVI.01743-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J. Virol. 80:11062–11073. 10.1128/JVI.00726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 4:1. 10.1186/1743-422X-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83:7261–7272. 10.1128/JVI.00421-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Money Va McPhee HK, Mosely Ja. Sanderson JM, Yeo RP. 2009. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc. Natl. Acad. Sci. U. S. A. 106:4441–4446. 10.1073/pnas.0805740106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitra R, Baviskar P, Duncan-Decocq RR, Patel D, Oomens AG. 2012. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J. Virol. 86:4432–4443. 10.1128/JVI.06744-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghildyal R, Li D, Peroulis I, Shields B, Bardin PG, Teng MN, Collins PL, Meanger J, Mills J. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 86:1879–1884. 10.1099/vir.0.80829-0 [DOI] [PubMed] [Google Scholar]
- 20.Baviskar PS, Hotard AL, Moore ML, Oomens AG. 2013. The respiratory syncytial virus fusion protein targets to the perimeter of inclusion bodies and facilitates filament formation by a cytoplasmic tail-dependent mechanism. J. Virol. 87:10730–10741. 10.1128/JVI.03086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384–11391. 10.1128/JVI.75.23.11384-11391.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952–3964. 10.1128/JVI.76.8.3952-3964.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran T-L, Castagné N, Dubosclard V, Noinville S, Koch E, Moudjou M, Henry C, Bernard J, Yeo RP, Eléouët J-F. 2009. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner. J. Virol. 83:6363–6374. 10.1128/JVI.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowton VM, McGivern DR, Fearns R. 2006. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J. Gen. Virol. 87:1805–1821. 10.1099/vir.0.81786-0 [DOI] [PubMed] [Google Scholar]
- 26.Cheng X, Park H, Zhou H, Jin H. 2005. Overexpression of the M2–2 protein of respiratory syncytial virus inhibits viral replication. J. Virol. 79:13943–13952. 10.1128/JVI.79.22.13943-13952.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason SW, Aberg E, Lawetz C, DeLong R, Whitehead P, Liuzzi M. 2003. Interaction between human respiratory syncytial virus (RSV) M2-1 and P proteins is required for reconstitution of M2-1-dependent RSV minigenome activity. J. Virol. 77:10670–10676. 10.1128/JVI.77.19.10670-10676.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Jans Da Bardin PG, Meanger J, Mills J, Ghildyal R. 2008. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J. Virol. 82:8863–8870. 10.1128/JVI.00343-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. 2013. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc. Natl. Acad. Sci. U. S. A. 110:11133–11138. 10.1073/pnas.1309070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danev R, Nagayama K. 2001. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy 88:243–252. 10.1016/S0304-3991(01)00088-2 [DOI] [PubMed] [Google Scholar]
- 31.Nagayama K, Danev R. 2008. Phase contrast electron microscopy: development of thin-film phase plates and biological applications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:2153–2162. 10.1098/rstb.2008.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murata K, Liu X, Danev R, Jakana J, Schmid MF, King J, Nagayama K, Chiu W. 2010. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure 18:903–912. 10.1016/j.str.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danev R, Kanamaru S, Marko M, Nagayama K. 2010. Zernike phase contrast cryo-electron tomography. J. Struct. Biol. 171:174–181. 10.1016/j.jsb.2010.03.013 [DOI] [PubMed] [Google Scholar]
- 34.Hosogi N, Shigematsu H, Terashima H, Homma M, Nagayama K. 2010. Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J. Struct. Biol. 173:67–76. 10.1016/j.jsb.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 35.Fukuda Y, Nagayama K. 2011. Zernike phase contrast cryo-electron tomography of whole mounted frozen cells. J. Struct. Biol. 177:484–489. 10.1016/j.jsb.2011.11.018 [DOI] [PubMed] [Google Scholar]
- 36.Guerrero-Ferreira RC, Wright ER. 2013. Zernike phase contrast cryo-electron tomography of whole bacterial cells. J. Struct. Biol. 185:129–133. 10.1016/j.jsb.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, Nagayama K, Schmid MF, King JA, Chiu W. 2013. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 502:707–710. 10.1038/nature12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Linde S, Loschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6:991–1009. 10.1038/nprot.2011.336 [DOI] [PubMed] [Google Scholar]
- 39.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 40.Jung J, Lifland AW, Zurla C, Alonas EJ, Santangelo PJ. 2013. Quantifying RNA-protein interactions in situ using modified-MTRIPs and proximity ligation. Nucleic Acids Res. 41:e12. 10.1093/nar/gks837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 42.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- 43.Heymann JB. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169. 10.1006/jsbi.2001.4339 [DOI] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmerberg J, Kozlov MM. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell. Biol. 7:9–19. 10.1038/nrm1784 [DOI] [PubMed] [Google Scholar]
- 46.Santangelo PJ, Lifland AW, Curt P, Sasaki Y, Bassell GJ, Lindquist ME, Crowe JE., Jr 2009. Single molecule-sensitive probes for imaging RNA in live cells. Nat. Methods 6:347–349. 10.1038/nmeth.1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mlodzianoski MJ, Juette MF, Beane GL, Bewersdorf J. 2009. Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy. Opt. Express 17:8264–8277. 10.1364/OE.17.008264 [DOI] [PubMed] [Google Scholar]
- 48.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. 2008. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5:527–529. 10.1038/nmeth.1211 [DOI] [PubMed] [Google Scholar]
- 49.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. 2011. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc. Natl. Acad. Sci. U. S. A. 108:18085–18090. 10.1073/pnas.1105770108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coronel EC, Takimoto T, Murti KG, Varich N, Portner A. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117–1123. 10.1128/JVI.75.3.1117-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagné N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eléouët J-F, Rey Fa. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283. 10.1126/science.1177634 [DOI] [PubMed] [Google Scholar]
- 52.El Omari K, Dhaliwal B, Ren J, Abrescia NGA, Lockyer M, Powell KL, Hawkins AR, Stammers DK. 2011. Structures of respiratory syncytial virus nucleocapsid protein from two crystal forms: details of potential packing interactions in the native helical form. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67:1179–1183. 10.1107/S1744309111029228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loney C, Mottet-Osman G, Roux L, Bhella D. 2009. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of Sendai virus. J. Virol. 83:8191–8197. 10.1128/JVI.00693-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bharat TAM, Riches JD, Kolesnikova L, Welsch S, Krähling V, Davey N, Parsy M-L, Becker S, Briggs JAG. 2011. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 9:e1001196. 10.1371/journal.pbio.1001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. 2002. Polyploid measles virus with hexameric genome length. EMBO J. 21:2364–2372. 10.1093/emboj/21.10.2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimm R, Barmann M, Hackl W, Typke D, Sackmann E, Baumeister W. 1997. Energy filtered electron tomography of ice-embedded actin and vesicles. Biophys. J. 72:482–489. 10.1016/S0006-3495(97)78689-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liljeroos L, Butcher SJ. 2013. Matrix proteins as centralized organizers of negative-sense RNA virions. Front. Biosci. 18:696–715. 10.2741/4132 [DOI] [PubMed] [Google Scholar]
- 58.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42:1416–1429. 10.1016/j.biocel.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D, Jans DA, Bardin PG, Meanger J, Mills J, Ghildyal R. 2008. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J. Virol. 82:8863–8870. 10.1128/JVI.00343-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asenjo A, Calvo E, Villanueva N. 2006. Phosphorylation of human respiratory syncytial virus P protein at threonine 108 controls its interaction with the M2-1 protein in the viral RNA polymerase complex. J. Gen. Virol. 87:3637–3642. 10.1099/vir.0.82165-0 [DOI] [PubMed] [Google Scholar]
- 61.Cuesta I, Geng X, Asenjo A, Villanueva N. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J. Virol. 74:9858–9867. 10.1128/JVI.74.21.9858-9867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bächi T. 1988. Direct observation of the budding and fusion of an enveloped virus by video microscopy of viable cells. J. Cell Biol. 107:1689–1695. 10.1083/jcb.107.5.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bächi T, Howe C. 1973. Morphogenesis and ultrastructure of respiratory syncytial virus. J. Virol. 12:1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. 2002. Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J. Gen. Virol. 83:753–757 [DOI] [PubMed] [Google Scholar]
- 65.Gower TL, Pastey MK, Peeples ME, Collins PL, Mccurdy LH, Hart TK, Guth A, Johnson TR, Graham BS. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 79:5326–5336. 10.1128/JVI.79.9.5326-5336.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mccurdy LH, Graham BS. 2003. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 77:1747–1756. 10.1128/JVI.77.3.1747-1756.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeffree CE, Brown G, Aitken J, Su-Yin DY, Tan B-H, Sugrue RJ. 2007. Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology 369:309–323. 10.1016/j.virol.2007.08.007 [DOI] [PubMed] [Google Scholar]
- 68.Jeffree C. 2003. Distribution of the attachment (G) glycoprotein and GM1 within the envelope of mature respiratory syncytial virus filaments revealed using field emission scanning electron microscopy. Virology 306:254–267. 10.1016/S0042-6822(02)00016-8 [DOI] [PubMed] [Google Scholar]
- 69.Kim J, Hama K, Miyake Y, Okada Y. 1979. Transformation of intramembrane particles of HVJ (Sendai virus) envelopes from an invisible to visible form on aging of virions. Virology 95:523–535. 10.1016/0042-6822(79)90506-3 [DOI] [PubMed] [Google Scholar]
- 70.Welsch S, Kolesnikova L, Krahling V, Riches JD, Becker S, Briggs JA. 2010. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 6:e1000875. 10.1371/journal.ppat.1000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rossman JS, Leser GP, Lamb RA. 2012. Filamentous influenza virus enters cells via macropinocytosis. J. Virol. 86:10950–10960. 10.1128/JVI.05992-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoenen T, Biedenkopf N, Zielecki F, Jung S, Groseth A, Feldmann H, Becker S. 2010. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J. Virol. 84:7053–7063. 10.1128/JVI.00737-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dick RA, Kamynina E, Vogt VM. 2013. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J. Virol. 87:13598–13608. 10.1128/JVI.01659-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. 2011. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174:11–22. 10.1016/j.jsb.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez M, Greenwald DL, de la Torre JC. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443–11448. 10.1128/JVI.78.20.11443-11448.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wirblich C, Tan GS, Papaneri A, Godlewski PJ, Orenstein JM, Harty RN, Schnell MJ. 2008. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J. Virol. 82:9730–9738. 10.1128/JVI.00889-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lazzarini RA, Keene JD, Schubert M. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145–154. 10.1016/0092-8674(81)90298-1 [DOI] [PubMed] [Google Scholar]
- 78.Treuhaft MW, Beem MO. 1982. Defective interfering particles of respiratory syncytial virus. Infect. Immun. 37:439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valdovinos MR, Gomez B. 2003. Establishment of respiratory syncytial virus persistence in cell lines: association with defective interfering particles. Intervirology 46:190–198. 10.1159/000071461 [DOI] [PubMed] [Google Scholar]
- 80.Treuhaft MW. 1983. A colorimetric assay for quantification of defective interfering particles of respiratory syncytial virus. J. Gen. Virol. 64(Part 6):1301–1309 [DOI] [PubMed] [Google Scholar]
- 81.Hall WW, Martin SJ, Gould E. 1974. Defective interfering particles produced during the replication of measles virus. Med. Microbiol. Immunol. 160:155–164. 10.1007/BF02121722 [DOI] [PubMed] [Google Scholar]
- 82.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. 2007. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2:249–255. 10.1038/nnano.2007.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vijayakrishnan S, Loney C, Jackson D, Suphamungmee W, Rixon FJ, Bhella D. 2013. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS Pathog. 9:e1003413. 10.1371/journal.ppat.1003413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haywood AM. 2010. Membrane uncoating of intact enveloped viruses. J. Virol. 84:10946–10955. 10.1128/JVI.00229-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.