ABSTRACT
The influenza A virus nuclear export protein (NEP) plays crucial roles in the nuclear export of the viral ribonucleoprotein complex through the chromosome region maintenance 1 (CRM1)-mediated cellular protein transport system. However, the detailed mechanism of NEP nucleocytoplasmic trafficking remains incompletely understood. Here, we investigated the subcellular localization of NEP from two strains of H1N1 influenza A virus and found that 2009 swine-origin H1N1 influenza A virus A/California/04/2009 (CA04) NEP displayed a distinct cellular distribution pattern, forming unique nuclear aggregates, compared to A/WSN/33 (H1N1) (WSN) NEP. Characterization of the nucleocytoplasmic transport pathways of these two NEPs showed that they both enter the nucleus by passive diffusion but are exported through the nuclear export receptor CRM1-mediated pathway with different efficiencies. The two identified nuclear export signals (NESs) on the two NEPs functioned similarly despite differences in their amino acid sequences. Using a two-hybrid assay, we confirmed that the CA04 NEP interacts less efficiently with CRM1 and that a threonine residue at position 48 is responsible for the nuclear aggregation. The present study revealed the dissimilarity in subcellular NEP transport processes between the 2009 pandemic (H1N1) influenza A virus CA04 and the laboratory-adapted H1N1 virus WSN and uncovered the mechanism responsible for this difference.
IMPORTANCE Because the efficiency of the nucleocytoplasmic transport of viral components is often correlated with the viral RNA polymerase activity, propagation, and host range of influenza viruses, the present study investigated the subcellular localization of NEP from two strains of H1N1 influenza virus. We found that the NEPs of both A/California/04/2009 (H1N1) (CA04) and A/WSN/33 (H1N1) (WSN) enter the nucleus by passive diffusion but are exported with different efficiencies, which were caused by weaker binding activity between the CA04 NEP and CRM1. The results of the present study revealed characteristics of the nuclear import and export pathways of NEP and the mechanism responsible for the differences in the cellular distribution of NEP between two H1N1 strains.
INTRODUCTION
Influenza A viruses are major human and animal pathogens that cause seasonal epidemics and occasional pandemic infections, posing a severe public health threat. Although wild aquatic fowl are their nature host, influenza A viruses occasionally cross the species barrier to infect domestic birds and certain mammalian species, including humans (1). The genome of type A influenza virus consists of eight segments of single-stranded, negative-sense RNA that are bound to viral RNA polymerases (PB2, PB1, and PA) and the nucleoprotein (NP) to form viral ribonucleoprotein (vRNP) complexes and encodes 14 viral proteins (2–5). Influenza A virus is unusual among RNA viruses for performing its RNA synthesis in the nucleus of infected cells (6). After invading a host cell, influenza virus delivers its vRNPs into the nucleus, where the viral RNAs are transcribed and replicated. After synthesis in the cytoplasm, the viral RNA polymerases and NP are transported into the nucleus to form new vRNPs (7). The assembly of viral components and the process of budding, however, occur at the plasma membrane. Therefore, progeny vRNPs must be exported from the nucleus to the cytoplasm at the late stage of infection to complete the viral life cycle.
Active transport of macromolecules across the nuclear membrane is often accomplished with the involvement of certain transport receptors (8). Studies have shown that the nuclear export of influenza vRNPs is mediated by the cellular protein CRM1 (chromosome region maintenance 1) (9), a member of the importin β superfamily of nuclear transport receptors, and can be blocked by the specific and irreversible inhibitor leptomycin B (LMB) (10–13). The matrix protein (M1) is considered necessary in this process, along with the viral nuclear export protein (NEP), which was formerly called nonstructural protein 2 (NS2) (14, 15). NEP is encoded by the spliced-form mRNA derived from the eighth vRNA segment of the influenza A virus genome (16–18). NEP interacts with certain components of the nuclear pore complex (NPC) and the nuclear export receptor CRM1 (19, 20). An anti-NEP antibody can block the export of vRNPs when microinjected into the nucleus of an infected cell, and recombinant viruses lacking NEP expression are deficient for the nuclear export of vRNPs (15). Evidence from several research groups confirms that the CRM1-interacting region of NEP is located in its N-terminal domain, though mutation of a putative nuclear export signal (NES) between residues 12 and 21 (NES1) does not abolish this interaction, suggesting that another portion of NEP may participate in the export process (20, 21). Recently, a second CRM1-dependent NES in the NEP of an H5N1 avian influenza virus was discovered between residues 31 and 40 (NES2) that is crucial for viral propagation and the nuclear export of vRNPs (22). Evidence from that study demonstrates that both NES1 and NES2 contribute to the subcellular localization of NEP.
Because the efficiency of the nucleocytoplasmic transport of viral components is often correlated with the viral RNA polymerase activity, propagation, and host range of influenza viruses, the present study investigated the subcellular localization of NEP from two strains of H1N1 influenza virus. We found that the NEPs of both A/California/04/2009 (H1N1) (CA04) and A/WSN/33 (H1N1) (WSN) enter the nucleus by passive diffusion but are exported with different efficiencies caused by weaker binding activity between the CA04 NEP and CRM1. The results of the present study revealed characteristics of the nuclear import and export pathways of NEP and the mechanism responsible for the difference in the levels of cellular distribution of NEP between two H1N1 strains.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Madin-Darby canine kidney (MDCK) cells, human embryonic kidney (293T) cells, and human adenocarcinomic alveolar basal epithelial (A549) cells were grown in Dulbecco modified Eagle medium (DMEM) (Gibco-BRL, Inc., Gaithersburg, MD) containing 10% fetal bovine serum (FBS) (Gibco-BRL, Inc., Gaithersburg, MD) and antibiotics at 37°C with 5% CO2. Influenza viruses A/WSN/33 (H1N1) (WSN) and A/California/04/2009 (H1N1) (CA04) were propagated in MDCK cells.
Anti-NP rabbit polyclonal antibody and anti-M1 mouse monoclonal antibody were obtained by immunizing animals with hexahistidine-tagged NP and M1 as previously described (23, 24). Anti-c-myc (9E10) and anti-FLAG (M2) mouse monoclonal antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) and Sigma-Aldrich (St. Louis, MO), respectively. The anti-NEP rabbit polyclonal antibody was purchased from GenScript USA Inc. (Piscataway, NJ). The goat anti-mouse tetramethyl rhodamine isocyanate (TRITC)-conjugated and goat anti-rabbit fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were purchased from Baihuizhongyuan Biotechnology Inc. (Beijing, China).
Plasmid construction.
The cDNAs encoding the WSN and CA04 NEP genes were amplified by reverse transcription-PCR (RT-PCR) of viral RNA exacts, and the products were digested and inserted between the EcoRI and XhoI sites of the eukaryotic protein expression plasmid pCMV-myc or the SalI and EcoRV sites of the pBIND vector for mammalian two-hybrid assays. The pACT-hCRM1 plasmid for mammalian two-hybrid assays was kindly provided by Ze Chen. To construct the simian virus 40 large T-antigen (SVLT)-NEP chimera protein, DNA fragments encoding SVLT and NEP were amplified with a Thr-Thr-Thr-Thr-Gly-Ser linker downstream of the SVLT coding region and subcloned into the KpnI and XhoI sites of a modified pCDNA3.0 plasmid which encodes an N-terminal FLAG tag.
Transfection, energy depletion, and LMB treatment.
293T cells grown on glass coverslips in 24-well tissue culture plates to 70% to 90% confluence were transfected with plasmids (0.8 μg of plasmid DNA per well) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were cultured for 24 h to allow gene expression.
For energy depletion assays, the cells were incubated in an energy depletion medium composed of glucose-free DMEM (Invitrogen), 6 mM 2-deoxy-d-glucose (Sigma), and 10 mM sodium azide (Sigma) and supplemented with 10% FBS for 3 h at 37°C prior to fixation.
For LMB treatment, cells were incubated in DMEM containing 11 ng/ml LMB (Sigma) and 10% FBS for 3 h before fixation. Cells treated with solvent were used as negative controls.
In vitro transport assay.
In vitro transport assays were performed as described by Adam et al. (25). 293T cells grown on coverslips were permeabilized by immersion in ice-cold transport buffer containing 40 mg/ml digitonin for 5 min. Then, cells were washed and stored in cold transport buffer. The coverslips were then inverted over a drop of complete transport mixture on a sheet of parafilm in a humidified box. The complete transport mixture contained 50% to 75% fluorescent protein diluted with transport buffer to give the following final conditions: approximately 25 to 35 mg/ml fluorescence-tagged fusion protein in transport buffer (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 5 mM sodium acetate, 2 mM dithiothreitol [DTT], 1.0 mM EGTA, 1 mM ATP, 5 mM creatine phosphate [Sigma], 20 U/ml creatine phosphokinase [Sigma], and protease inhibitor cocktail [Roche]). The entire box was then floated in a water bath at 30°C. At the end of the assay, each coverslip was rinsed and mounted in a small amount of transport buffer with DAPI (4′,6-diamidino-2-phenylindole) and observed using a confocal microscope.
Fluorescence microscopy.
At the indicated time points, cells were rinsed in phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min at room temperature, and permeabilized with PBS-T (PBS containing 0.5% Triton X-100) for 15 min. To detect the indicated proteins, the coverslips were blotted within PBS-T and incubated with the corresponding antibodies for 1 h at 37°C. After five washes with PBS-T, the cells were incubated for 1 h at 37°C with appropriate secondary antibodies, washed with PBS-T, and counterstained with DAPI. Then, the coverslips were rinsed with PBS and mounted on a glass slide. Cell samples were imaged using a Olympus FV500 confocal laser scanning microscope (LSCM).
Mammalian two-hybrid assay.
293T cells were cotransfected with 300 ng pBIND fusion construct, 300 ng pACT fusion construct, and 300 ng reporter gene construct pG5luc (Promega) per well in 24-well culture plates using Lipofectamine 2000 to analyze the interaction of various NEP proteins with CRM1 in mammalian cells. Twenty-four hours after transfection, the cells were harvested, washed twice with cold PBS, and lysed with passive lysis buffer (Promega). Luciferase assays were performed using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Renilla luciferase activity was used to correct for the variation in transfection efficiency. After sequential quantification of firefly and Renilla luciferase activities in cell lysates, the binding activity between the two proteins was calculated as the ratio of firefly activity to Renilla activity. Experiments were performed in triplicate, and luciferase activity was determined using a T20/20 luminometer (Turner Designs, Sunnyvale, CA).
RESULTS
The subcellular localization of CA04 NEP is distinct from that of WSN.
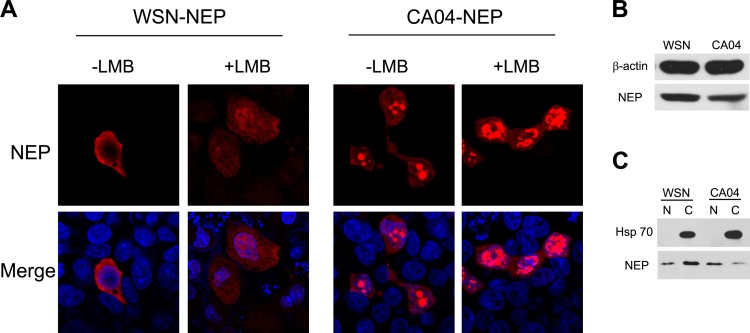
In April 2009, a swine-origin A (H1N1) influenza virus containing a previously undescribed combination of gene segments was isolated from humans (26). The mechanism for human adaption of this influenza virus is still not fully understood. Because NEP plays a compensating role for human adaption of the H5N1 avian influenza virus by regulating the activity of viral RNA polymerase (27), we attempted to characterize the NEPs from one of the first isolated 2009 (H1N1) influenza A viruses, A/California/04/2009 (CA04). First, its subcellular localization was compared to that of the laboratory-adapted H1N1 virus WSN. 293T cells were transfected with plasmids encoding WSN-NEP and CA04-NEP, respectively. The cells were treated with cycloheximide to block new protein synthesis and with LMB or vehicle, as indicated, at 24 h posttransfection and processed for immunofluorescence microscopy. In contrast to the cytoplasm distribution of WSN-NEP, the CA04 NEP displayed distinct nuclear aggregation in transfected cells. This nuclear accumulation was increased by LMB treatment for both proteins, indicating that the CRM1-mediated nuclear export pathway is commonly used by NEP from different viral strains (Fig. 1A). Western blotting revealed that the two proteins had similar expression levels in the whole cells (Fig. 1B). The nuclear and cytoplasmic fractionation also confirmed that the CA04-NEP had a preference for nuclear localization (Fig. 1C).
FIG 1.
The cellular localization pattern of CA04-NEP is different from that of WSN-NEP when overexpressed in cells. (A) N-terminally c-Myc-tagged NEP from WSN or CA04 was transiently expressed in 293T cells. At 24 h posttransfection, the cells were treated with 100 ng/μl of cycloheximide for 3 h to block protein synthesis. LMB (11 nM) was added to the medium along with the cycloheximide for LMB treatment. (B and C) Western blotting confirmed the different cellular localizations of the two NEPs. 293T cells transfected with WSN-NEP or CA04-NEP were lysed with whole-cell lysis buffer (B) or nucleus (“N”)-cytoplasm (“C”) fractionation buffer (C) at 24 h posttransfection and prepared for Western blotting. An anti-c-Myc monoclonal antibody was used for protein detection. DAPI (4′-6-diamidino-2-phenylindole) was used for nuclear staining. Results shown are representative images.
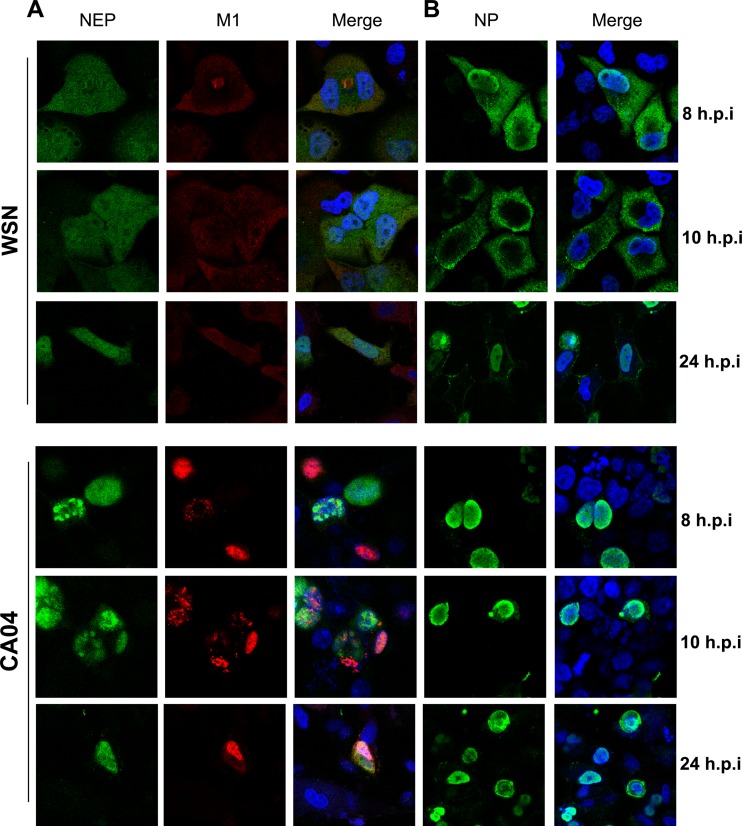
We next examined this phenomenon in cells infected with influenza viruses. A549 cells were infected with WSN or CA04 at a multiplicity of infection (MOI) of 1, and the cells were fixed at 8 or 10 h postinfection, respectively. Localization of M1 and NEP within the same cells was detected with corresponding antibodies. Due to the detection limit of fluorescently tagged secondary antibodies, the distribution of NP was separately stained with an anti-NP polyclonal antibody at the same time points. As shown in Fig. 2A, at 8 and 10 h postinfection, NEP was evenly distributed through the nucleus and cytoplasm at the late phase of the viral life cycle within cells infected with WSN virus. WSN-M1 was mainly located in the cytoplasm and colocalized with NEP. As in transiently transfected 293T cells, CA04-NEP was aggregated inside the nucleus of infected A549 cells at the indicated time points, and the nuclear export of CA04–M1 was appreciably restricted in NEP-expressing cells compared to that of WSN. As an indication of levels of progeny vRNPs, the localization of NP within infected cells was also analyzed (Fig. 2B). At 8 h postinfection in WSN-infected A549 cells, NP was located at the perinuclear domain and cytoplasm but was predominantly cytoplasmic at 10 h postinfection, indicating that the nuclear export process of newly assembled vRNPs was achieved. In contrast, the NP of CA04 was mainly located in the nucleus of infected cells at the same time points, which likely resulted from the nuclear aggregation of NEP and M1 because these two proteins are mediators for the nuclear export of progeny vRNPs. These results demonstrated that the CA04 virus has a NEP subcellular localization pattern distinct from that of the WSN virus, as well as impaired nuclear transport of vRNPs.
FIG 2.
Different subcellular distributions of viral proteins in CA04- and WSN-infected cells. A549 cells were infected with WSN or CA04 at an MOI of 1 and fixed at 8 h, 10 h, and 24 h postinfection (h.p.i). The subcellular localization of NEP, M1 (A), and NP (B) was examined with corresponding antibodies. DAPI was used for nuclear staining.
The NEPs of WSN and CA04 enter the nucleus by passive diffusion.
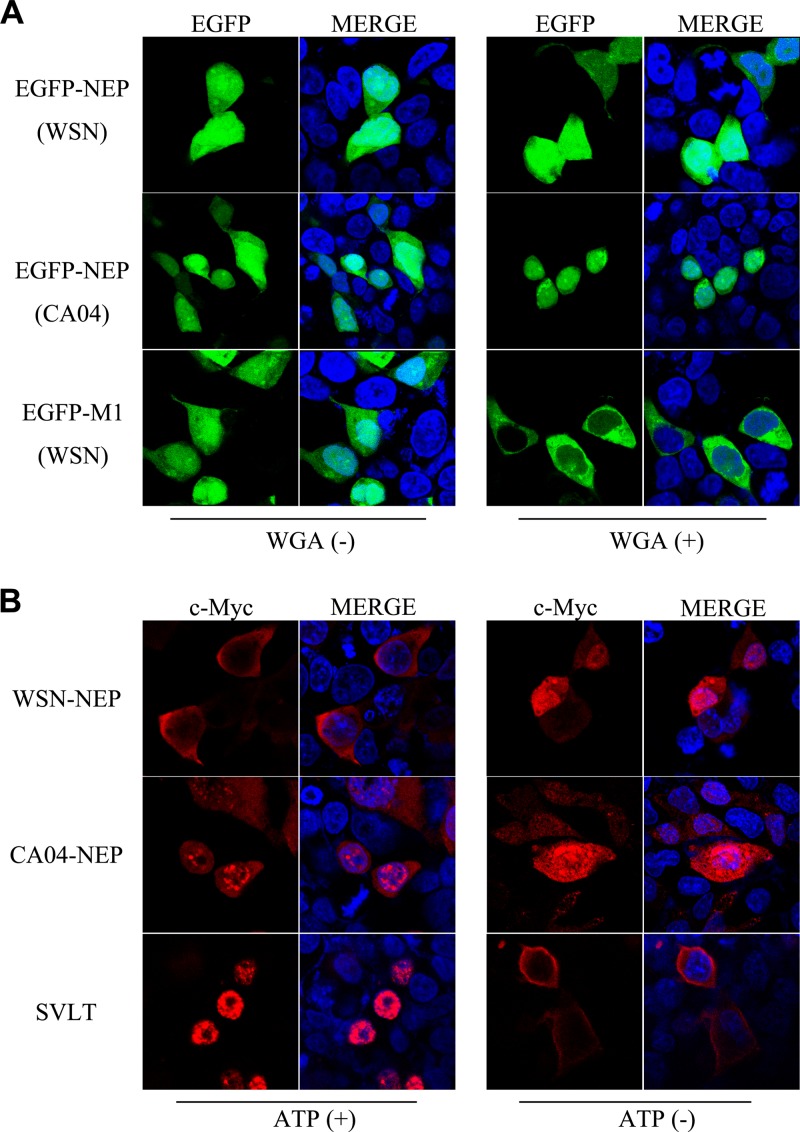
To determine the mechanism for the localization differences between the NEPs of the two viral strains, we first examined the NEP nuclear import pathway. Indeed, despite substantial evidence supporting the crucial role NEP plays during the nuclear export process of influenza vRNPs, the nuclear import mechanism of NEP remains unclear. Analysis of the amino acid sequences of the WSN and CA04 NEPs predicted the absence of nuclear localization signals (NLSs), suggesting that their nuclear import is passive. An in vitro NEP transport assay was performed to characterize their transport mode. An enhanced green fluorescent protein-NEP (EGFP-NEP) fusion protein was initially expressed in 293T cells. The cells were then lysed, and the extracts were added to 293T cells permeabilized by digitonin treatment with all of the components needed for the in vitro transport assay. Wheat germ agglutinin (WGA), a lectin that can inhibit NLS-dependent active nuclear transport, was used to investigate whether the NEPs entered the nucleus by active transport (28). Fluorescence microscopy imaging revealed that both EGFP-NEP fusion proteins were able to enter the nucleus of permeabilized cells. The EGFP and WSN-M1 fusion protein was used as a positive control in the assay. In agreement with previous studies suggesting that M1 undergoes NLS-dependent, importin-mediated nuclear import (29), WGA strongly blocked the nuclear accumulation of EGFP-M1 in treated cells, whereas the nuclear localization of EGFP-NEP remained unaffected (Fig. 3A). To further confirm the energy requirement of NEP transport, the subcellular localization of NEP was observed in an energy depletion assay. Under these conditions, Ran-dependent active nucleocytoplasmic transport is blocked, whereas passive diffusion is unaffected (30). The large T-antigen of simian virus 40 (SVLT) possesses a classic NLS mediating its nuclear import through a Ran-dependent pathway and thus was used as a positive control in the assay (31); its nuclear import was completely inhibited under energy depletion conditions. N-terminally myc-tagged WSN-NEP or CA04-NEP was expressed in 293T cells for use in the same assay. Energy depletion of the transfected cells resulted in enhanced accumulation inside the nucleus of both proteins, demonstrating that they are able to passively diffuse from the cytoplasm into the nucleus of transfected cells.
FIG 3.
NEP enters the nucleus by passive diffusion. (A) In vitro NEP transport assay. EGFP-NEP (upper panel) and EGFP-M1 (lower panel) fusion proteins were separately expressed in 293T cells, and cell lysates in transport buffer were added to digitonin-treated 293T cells. For WGA treatment (WGA+), the permeabilized cells were incubated in transport buffer containing 50 μg/ml WGA before addition of cell lysates. Images revealed fluorescence signals of EGFP from fusion proteins. (B) Energy depletion assay of the nucleocytoplasmic transport of NEP. N-terminally myc-tagged NEP was expressed in 293T cells. N-terminally FLAG-tagged SVLT was expressed as a positive control. At 24 h posttransfection, the cells were incubated with energy depletion medium (ATP−) or normal culture medium (ATP+) for 3 h before fixation and visualization. Anti-myc and anti-FLAG monoclonal antibodies were used for the detection of the indicated proteins.
The nuclear export activity of CA04-NEP is less efficient than that of WSN-NEP.
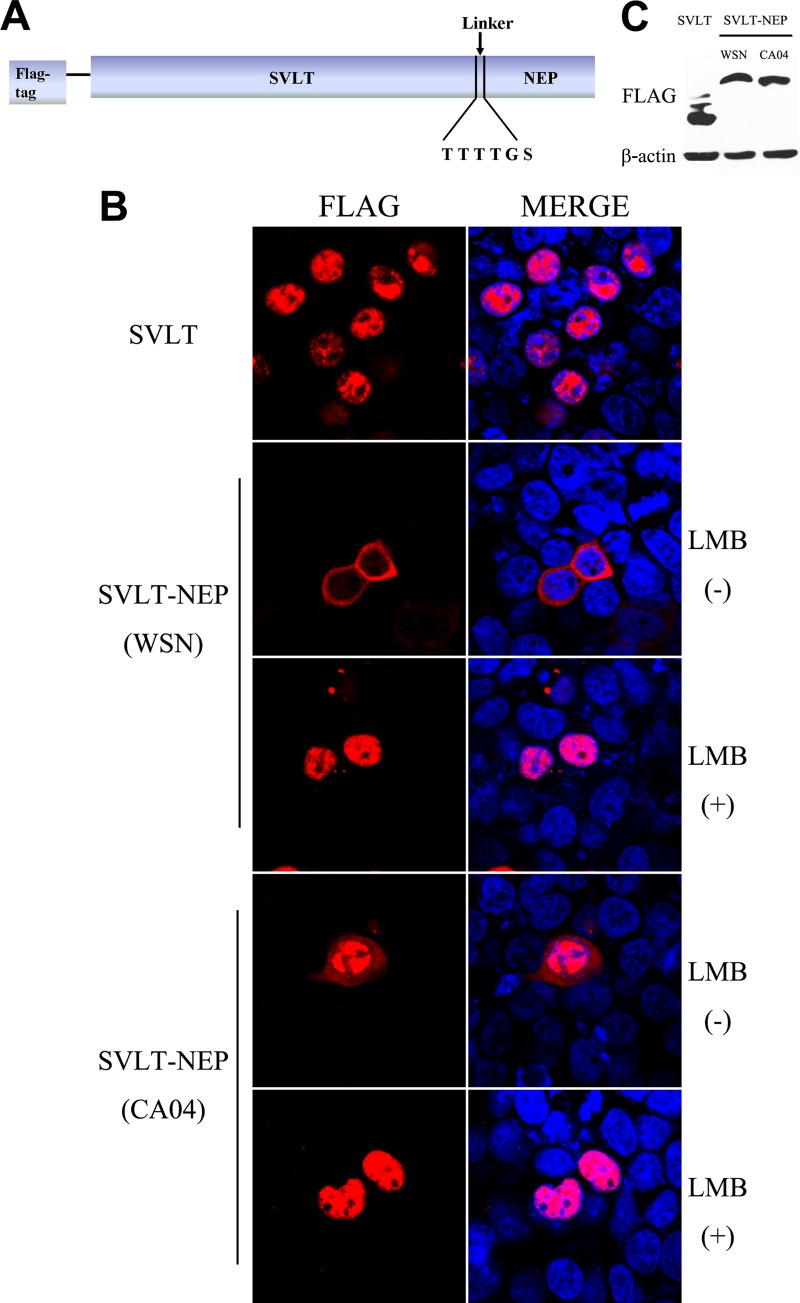
Because both CA04-NEP and WSN-NEP passively enter the nucleus, the difference in subcellular localization between the two proteins might be caused during nuclear export. To address this issue, a vector expressing a chimeric SVLT-linker-NEP protein was constructed (Fig. 4A). This fusion protein is imported into the nucleus immediately after synthesis in the cytoplasm due to the SVLT NLS and then exported through the NES on the NEP. The chimeric protein between SVLT and WSN-NEP displayed a predominantly cytoplasmic distribution in transfected 293T cells, whereas only a very small amount of the chimeric protein of SVLT-CA04-NEP was transported out of the nucleus (Fig. 4B). LMB treatment completely restrained both types of fusion protein inside the nucleus, suggesting that the observed cytoplasmic localization occurred due to nuclear export mediated by CRM1 and, thus, that CA04-NEP export is less efficient. Western blot analysis was carried out to confirm the intactness of fusion proteins, and the result showed that all three constructs were expressed at similar levels and in accordance with the expected molecular size (Fig. 4C).
FIG 4.
Evaluation of the nuclear export activity of SVLT-NEP fusion proteins. (A) Schematic representation of the SVLT-NEP chimeric protein. (B) 293T cells were transfected with plasmids encoding SVLT, SVLT-NEP (WSN), or SVLT-NEP (CA04). At 24 h posttransfection, the cells were treated with 100 ng/μl cycloheximide for 3 h to block protein synthesis. LMB (11 nM) was added to the medium along with the cycloheximide for LMB treatment. (C) A Western blot assay of 293T cells transfected with FLAG-SVLT or SVLT-NEP fusion protein constructs was carried out at 24 h posttransfection. An anti-FLAG monoclonal antibody was used to detect the target proteins.
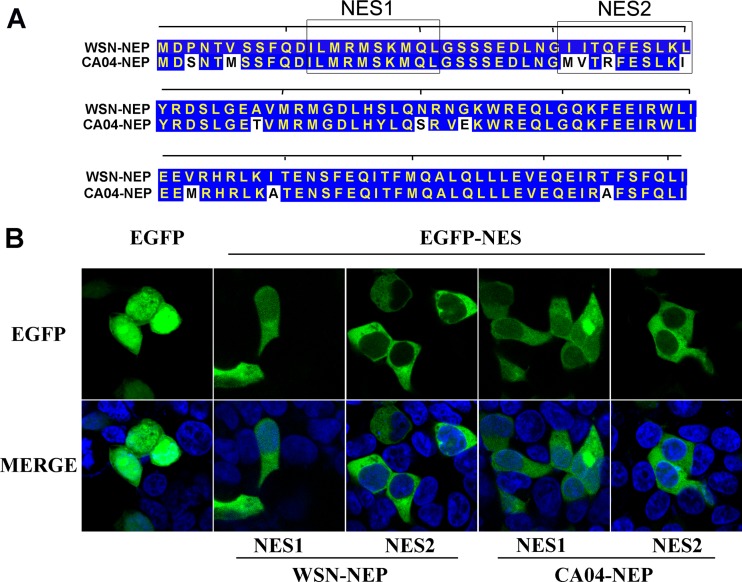
NES1 and NES2 on the two NEPs have similar functions.
To further clarify the mechanism responsible for the difference in nuclear export efficiency between WSN-NEP and CA04-NEP, we compared the functions of their NESs. Data from previous studies reveal that two leucine-rich, CRM1-dependent NESs are present in the N-terminal domain of NEP (20, 22). Alignment of the two NES motifs within WSN-NEP and CA04-NEP showed that four residues in NES2 differ between the NEPs, including replacement of a highly conserved leucine at position 40 of WSN-NEP with an isoleucine in CA04-NEP (Fig. 5A). To characterize their functions, oligonucleotides encoding these NES motifs were synthesized and cloned into the pEGFP-C1 vector to express EGFP-NES fusion proteins. When expressed in cells, functional NESs bind to CRM1 and direct the fusion protein from the nucleus to the cytoplasm. As shown in Fig. 5B, EGFP alone was evenly distributed in the nucleus and cytoplasm due to passive diffusion. Despite the discrepancy in the amino acid sequences, the EGFP-NES1 and EGFP-NES2 fusions of the two NEPs exhibited similar intracellular localization patterns in transfected cells. These results demonstrate that NES1 and NES2 on the NEPs from WSN and CA04 have comparable activities with respect to exporting the fusion proteins from nucleus into the cytoplasm as an independent functional unit.
FIG 5.
The NESs on WSN-NEP and CA04-NEP have comparable functions. (A) Alignment of amino acid sequences of WSN-NEP and CA04-NEP. (B) 293T cells were transfected with plasmids expressing fusion proteins of EGFP and NESs. At 24 h posttransfection, the cells were treated with 100 ng/μl cycloheximide for 3 h to block protein synthesis prior to fixation. The cells were then permeabilized with PBS-T, mounted with DAPI-containing medium, and imaged by fluorescence microscopy.
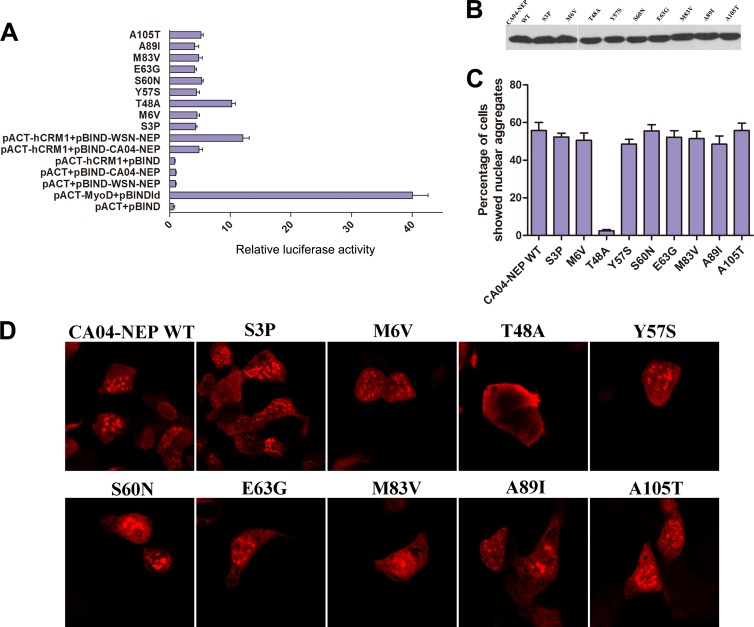
Nuclear aggregation of CA04-NEP is caused by inefficient interaction with CRM1.
The nuclear export of molecules through the CRM1-mediated pathway normally requires direct interaction between the receptor and the cargo. To evaluate the interaction between WSN-NEP or CA04-NEP and CRM1, a two-hybrid assay in mammalian cells was performed. 293T cells were cotransfected with pBIND-WSN-NEP or CA04-NEP, pACT-CRM1, and the reporter plasmid pG5luc, encoding firefly luciferase, as indicated. We found that the binding activity of CA04-NEP with CRM1 was ∼50% of that between WSN-NEP and CRM1, indicating that the efficiency of CRM1-mediated nuclear export of CA04-NEP was lower than that of WSN-NEP, which may have caused the nuclear aggregation of CA04-NEP (Fig. 6A). The expression levels of CA04-NEP and WSN-NEP were also assessed by Western blotting to rule out the possibility that the nuclear aggregation of CA04-NEP was caused by overexpression in the transfected cells (Fig. 6B).
FIG 6.
The nuclear aggregation of CA04-NEP is caused by inefficient interaction with Crm1. (A) Mammalian cell two-hybrid assays were used to detect the binding of NEPs (and their mutants) with Crm1. 293T cells were cotransfected with the same amounts of pBIND-NEP encoding point mutants, pACT-Crm1, and pG5luc. At 24 h posttransfection, the cells were lysed, and luciferase activity assays were performed. The length of each bar represents the relative level of luciferase activity, calculated as the ratio of firefly activity to Renilla activity. Results shown are the means ± standard deviations (SD) from three independent experiments. (B) Western blotting with an anti-NEP monoclonal antibody confirmed that all constructs expressed similar levels of NEP. (C and D) 293T cells were transfected with a plasmid expressing myc-tagged CA04-NEP or point mutants. At 24 h posttransfection, the cells were treated with 100 ng/μl cycloheximide for 3 h to block protein synthesis and processed for immunofluorescence microscopy. (C) The numbers of cells transfected with myc-CA04-NEP or point mutants exhibiting nuclear aggregation were counted, and ratios to total examined cells were calculated. Approximately 90 to 100 cells were counted for each replicate. Results shown are the averages from three independent experiments. (D) Cellular localizations of wild-type and point mutant proteins were detected with anti-myc monoclonal antibodies, and images representing the whole population are shown.
Next, a series of single-amino-acid mutants of CA04-NEP was constructed by introducing substitutions with the corresponding residues in WSN-NEP. The cellular localization of the CA04-NEP mutants and their binding to CRM1 were examined. For each mutant, over 100 cells were examined, and the ratios of cells exhibiting nuclear NEP aggregation were calculated. Among all tested mutants, the T48A substitution in CA04-NEP decreased nuclear aggregation (Fig. 6D). Furthermore, the mammalian two-hybrid assay confirmed that the T48A mutation increased the binding of CA04-NEP to CRM1 to a level similar to that seen with WSN-NEP (Fig. 6A). These results indicated that CA04-NEP is restricted inside the nucleus, resulting in aggregate formation, due to an inefficient interaction with CRM1, and that the T48 residue in CA04-NEP was crucial for the reduced CRM1 binding activity and the cellular localization distinct from that of WSN-NEP.
DISCUSSION
As influenza A viruses are among the very few known RNA viruses that replicate in the nucleus, regulation of the nuclear transport of viral proteins and RNP in both directions is crucial. Though investigations into this topic have been reported, many questions remain to be answered. In the present study, we compared the pathways taken by the NEP of two strains of H1N1 influenza A virus to achieve nucleocytoplasmic translocation. For the first time, we demonstrated that CA04-NEP prefers to form aggregates inside the nucleus of transfected or infected cells due to inefficient binding to the nuclear export receptor CRM1.
Generally, proteins with molecular masses of <40 kDa can passively shuttle through nuclear pore complexes (32). Though the molecular mass of NEP is much lower than 40 kDa, there is no experimental evidence confirming its nuclear localization pathway. By using in vitro transport and energy depletion assays, we found that the nuclear import of WSN-NEP and CA04-NEP is NLS and energy independent. Structural research indicating that NEP exists as a compact monomer in solution also supports our deduction that it passively diffuses into the nucleus (33).
The crystal structure of the protease-resistant C-terminal domain (amino acids 54 to 121) of NEP has been solved, but there is no structural information available for the protease-sensitive N-terminal domain (amino acids 1 to 53) (21). Though locating outside the NES region, the threonine at position 48 in CA04-NEP may reduce the steric accessibility of NES2 for binding to CRM1. Because the two NESs on NEP may work coordinately for the interaction with CRM1, this could result in the restricted nuclear export activity of CA04-NEP.
It is important for us to investigate the relationship between the cellular localization of CA04-NEP and its impact on the viral replication and pathogenicity. Unfortunately, due to the lack of a reverse genetic system and a vRNP reconstitution system of CA04 virus, we are unable to determine the effect of this phenotype on viral infectivity directly at this time. Under these conditions, we replaced the NS gene in the WSN reverse genetic system with the CA04-NS gene and performed a virus rescue assay to investigate if the aggregation of NEP had an effect on the replication of WSN virus. The titers and the growth rates of the rescued recombinant viruses were comparable, whereas the patterns of localization of NEP were quite different (data not shown). Although these results suggest that the nuclear aggregation of NEP has no significant impact on the replication of WSN, the possibility cannot be ruled out that this phenotype is correlated with the infectivity and pathogenicity of CA04 virus, because, according to a study by Manz et al., the regulation of viral polymerase activity by NEP involves direct association with the PB2 subunit (27). NEP also interacts with several nuclear proteins, including nucleolin, ribosomal proteins, and histones, and participates in the regulation of the transcription and replication of the viral genome (34–37). The presence of the nucleus-aggregated form of CA04-NEP implies a unique interactome with host cellular factors or a regulatory function for viral polymerase activity that is worthy of further study.
In summary, we investigated and compared the characteristics of nucleocytoplasmic transport of NEP from different strains of H1N1 influenza A virus and provided new perspectives for research into the biological functions of NEP during the influenza virus life cycle.
ACKNOWLEDGMENTS
This study was funded by grants from National Key Technologies Research and Development Program of China (2013ZX10004-610), Ministry of Science and Technology of China Program 973 (grant no. 2012CB955501, 2011CB504705, and 2012CB518903), the National Natural Science Foundation of China (NSFC) (grant no. 81101254, 81101253, and 31370200), and the Key Research Program of the Chinese Academy of Sciences (grant no. KSZD-EW-Z-005). Wenjun Liu is a principal investigator of the National Natural Science Foundation of China Innovative Research Group (grant no. 81021003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 16 April 2014
REFERENCES
- 1.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samji T. 2009. Influenza A: understanding the viral life cycle. Yale J. Biol. Med. 82:153–159 [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439. 10.1016/j.chom.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. 10.1126/science.1222213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998. 10.1371/journal.ppat.1002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chase GP, Rameix-Welti MA, Zvirbliene A, Zvirblis G, Gotz V, Wolff T, Naffakh N, Schwemmle M. 2011. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 7:e1002187. 10.1371/journal.ppat.1002187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulo S, Akarsu H, Ruigrok RW, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 124:12–21. 10.1016/j.virusres.2006.09.013 [DOI] [PubMed] [Google Scholar]
- 8.Görlich D, Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607–660. 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- 9.Watanabe K, Takizawa N, Katoh M, Hoshida K, Kobayashi N, Nagata K. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77:31–42. 10.1016/S0168-1702(01)00263-5 [DOI] [PubMed] [Google Scholar]
- 10.Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308–311. 10.1038/36894 [DOI] [PubMed] [Google Scholar]
- 12.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540–547. 10.1006/excr.1998.4136 [DOI] [PubMed] [Google Scholar]
- 13.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U. S. A. 96:9112–9117. 10.1073/pnas.96.16.9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130. 10.1016/0092-8674(91)90576-K [DOI] [PubMed] [Google Scholar]
- 15.O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296. 10.1093/emboj/17.1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inglis SC, Barrett T, Brown CM, Almond JW. 1979. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc. Natl. Acad. Sci. U. S. A. 76:3790–3794. 10.1073/pnas.76.8.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb RA, Choppin PW. 1979. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. U. S. A. 76:4908–4912. 10.1073/pnas.76.10.4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb RA, Choppin PW, Chanock RM, Lai CJ. 1980. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc. Natl. Acad. Sci. U. S. A. 77:1857–1861. 10.1073/pnas.77.4.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Huang S, Chen Z. 2010. Human cellular protein nucleoporin hNup98 interacts with influenza A virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J. Gen. Virol. 91:2474–2484. 10.1099/vir.0.022681-0 [DOI] [PubMed] [Google Scholar]
- 20.Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758. 10.1093/emboj/19.24.6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akarsu H, Burmeister WP, Petosa C, Petit I, Muller CW, Ruigrok RW, Baudin F. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 22:4646–4655. 10.1093/emboj/cdg449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, Chen J, Chen Q, Wang H, Yao Y, Chen J, Chen Z. 2013. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 87:767–778. 10.1128/JVI.06519-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Liu X, Zhao Z, Xu C, Zhang K, Chen C, Sun L, Gao GF, Ye X, Liu W. 2011. Cyclophilin E functions as a negative regulator to influenza virus replication by impairing the formation of the viral ribonucleoprotein complex. PLoS One 6:e22625. 10.1371/journal.pone.0022625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koestler TP, Rieman D, Muirhead K, Greig RG, Poste G. 1984. Identification and characterization of a monoclonal antibody to an antigen expressed on activated macrophages. Proc. Natl. Acad. Sci. U. S. A. 81:4505–4509. 10.1073/pnas.81.14.4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam SA, Marr RS, Gerace L. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. 10.1083/jcb.111.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WMX, Skepner XE, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke D, Fouchier R, Pappas AC, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair P, Yu C, Keene KM, Dotson PDJ, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mänz B, Brunotte L, Reuther P, Schwemmle M. 2012. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 3:802. 10.1038/ncomms1804 [DOI] [PubMed] [Google Scholar]
- 28.Yoneda Y, Imamoto-Sonobe N, Yamaizumi M, Uchida T. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586–595. 10.1016/0014-4827(87)90297-7 [DOI] [PubMed] [Google Scholar]
- 29.Ye Z, Robinson D, Wagner RR. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69:1964–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwoebel ED, Ho TH, Moore MS. 2002. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 157:963–974. 10.1083/jcb.200111077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt MK, Mann K. 1987. Glycosylation of simian virus 40 T antigen and localization of glycosylated T antigen in the nuclear matrix. Virology 156:268–281. 10.1016/0042-6822(87)90407-7 [DOI] [PubMed] [Google Scholar]
- 32.Peters R. 2005. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic 6:421–427. 10.1111/j.1600-0854.2005.00287.x [DOI] [PubMed] [Google Scholar]
- 33.Lommer BS, Luo M. 2002. Structural plasticity in influenza virus protein NS2 (NEP). J. Biol. Chem. 277:7108–7117. 10.1074/jbc.M109045200 [DOI] [PubMed] [Google Scholar]
- 34.de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot PE, Ciancia C, Perrin-Cocon L, Bartenschlager R, Andre P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 9:e1003440. 10.1371/journalppat.1003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorai T, Goto H, Noda T, Watanabe T, Kozuka-Hata H, Oyama M, Takano R, Neumann G, Watanabe S, Kawaoka Y. 2012. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc. Natl. Acad. Sci. U. S. A. 109:4615–4620. 10.1073/pnas.1114728109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullido R, Gomez-Puertas P, Saiz MJ, Portela A. 2001. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J. Virol. 75:4912–4917. 10.1128/JVI.75.10.4912-4917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robb NC, Smith M, Vreede FT, Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398–1407. 10.1099/vir.0.009639-0 [DOI] [PubMed] [Google Scholar]