ABSTRACT
Resistance to various human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs) challenges the effectiveness of therapies in treating HIV-1-infected individuals and AIDS patients. The virus accumulates mutations within the protease (PR) that render the PIs less potent. Occasionally, Gag sequences also coevolve with mutations at PR cleavage sites contributing to drug resistance. In this study, we investigated the structural basis of coevolution of the p1-p6 cleavage site with the nelfinavir (NFV) resistance D30N/N88D protease mutations by determining crystal structures of wild-type and NFV-resistant HIV-1 protease in complex with p1-p6 substrate peptide variants with L449F and/or S451N. Alterations of residue 30's interaction with the substrate are compensated by the coevolving L449F and S451N cleavage site mutations. This interdependency in the PR–p1-p6 interactions enhances intermolecular contacts and reinforces the overall fit of the substrate within the substrate envelope, likely enabling coevolution to sustain substrate recognition and cleavage in the presence of PR resistance mutations.
IMPORTANCE Resistance to human immunodeficiency virus type 1 (HIV-1) protease inhibitors challenges the effectiveness of therapies in treating HIV-1-infected individuals and AIDS patients. Mutations in HIV-1 protease selected under the pressure of protease inhibitors render the inhibitors less potent. Occasionally, Gag sequences also mutate and coevolve with protease, contributing to maintenance of viral fitness and to drug resistance. In this study, we investigated the structural basis of coevolution at the Gag p1-p6 cleavage site with the nelfinavir (NFV) resistance D30N/N88D protease mutations. Our structural analysis reveals the interdependency of protease-substrate interactions and how coevolution may restore substrate recognition and cleavage in the presence of protease drug resistance mutations.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) protease (PR) plays an essential role in the viral life cycle by processing Gag and GagProPol viral polyproteins, resulting in a mature virus. Therefore, nine FDA-approved protease inhibitors (PIs) target PR. PIs are an integral component of highly active antiretroviral therapy (HAART) (1–3), which also includes reverse transcriptase inhibitors and, more recently, integrase inhibitors. HAART has significantly improved the prognosis for HIV-1-infected individuals. Nevertheless, the development of resistance is a major cause for antiretroviral therapy failure.
Evolution of PI resistance occurs due to a combination of factors: the lack of a proofreading mechanism of the viral reverse transcriptase, a high viral replication rate, and selective pressure of PIs. Mutations occur within the active/substrate binding site of the PR (4) and compromise inhibitor potency. These mutations lower the efficacy of the PIs by several orders of magnitude yet allow the PR to process its substrates, Gag and GagProPol. Resistance thus reflects a change in molecular recognition by the protease where a balance is maintained between (i) substrate recognition and cleavage and (ii) inhibition by PIs.
HIV-1 Gag processing is a highly regulated and ordered process that is essential for the production of mature virions, where HIV-1 PR recognizes and cleaves 10 specific sites within Gag (5–7). The sites recognized by the PR are nonhomologous and asymmetric, but they occupy a conserved volume within the active site of the PR (8). This volume forms the basis for PR-substrate recognition, and we previously defined this conserved volume as the “substrate envelope.” PIs that protrude beyond the substrate envelope are susceptible to resistance (9).
PI-resistant viruses accumulate mutations in both the viral PR and its substrates, Gag and GagProPol (10–12). While some of these mutations may compensate for loss of viral fitness and improve efficiency of Gag processing by the resistant PR (11, 13), we previously found that they may also directly modulate drug resistance (14). Frequently, these mutations arise within the Gag cleavage sites, particularly the NC-p1 and p1-p6 sites (15–18). Cleavage site mutations have been specifically associated with several major protease resistance mutations (14, 16, 17), suggesting that as the virus evolves resistance to protease inhibitors with prolonged protease inhibitor therapy, evolution of cleavage sites could be a fairly frequent mechanism for maintaining viral fitness.
Previously, we reported that the p1-p6 cleavage site coevolves with the D30N/N88D PR mutations (14, 15) in HIV-1 subtype B. The D30N mutation at the PR active site arises specifically in response to nelfinavir (NFV) inhibition, both in viral cultures and in patients being treated with NFV, and is often accompanied by the N88D secondary mutation, causing severe resistance to NFV (19). Our inhibitor-bound co-crystal structures revealed that the N88D secondary mutation interacts with residue 30 to orient the side chain away from the active site (20) and thereby disrupts the interaction between residue 30 and the inhibitor. This effect is even more pronounced in subtype AE, where N88S pulls D30 out of the active site (21). We also showed that D30 not only is key for inhibitor binding but also is essential for recognition of the p1-p6 cleavage site (8, 15, 22). Consequently, the D30N mutation, which lowers affinity for NFV (23), also likely compromises p1-p6 recognition, leading to coevolution of this cleavage site (15). Such p1-p6 cleavage site mutations commonly observed with D30N/N88D PR are L449F, S451N, and P453L (14), and they may help restoring the fit of the substrate in the consensus substrate envelope (24). Mutations are observed at either Gag 449 or 451, but simultaneous occurrence of both mutations is infrequent (15).
The current study focused on elucidating the structural rationale for the coevolution of the p1-p6 cleavage site with the NFV-resistant D30N/N88D HIV-1 protease. We determined crystal structures of wild-type (WT) and D30N/N88D HIV-1 proteases in complex with four p1-p6 substrate variants, L449F, S451N, L449F/S415N, and wild type. (The double mutant substrate, L449F/S415N, was included to elucidate its infrequent occurrence.) Structural analyses of PR and substrate complexes showed tighter packing around coevolved substrates and an interdependency of interactions between the PR and the p1-p6 substrate. The coevolved p1-p6 substrates fit better within the substrate envelope in the NFV-resistant PR complexes, as we have previously observed with the coevolved NC-p1 (13).
MATERIALS AND METHODS
Nomenclature.
HIV-1 protease (WT or D30N/N88D) and the p1-p6 substrate variants (WT, L449F, S451N, and L449F/S451N) in the PR-substrate complexes are distinguished as subscripts. For example, WT protease in complex with WT p1-p6 is denoted PRWT–p1-p6WT, and mutant protease in complex with the S451N mutation in p1-p6 is denoted PRD30N/N88D–p1-p6S451N. The two monomers of the protease are denoted a and b.
Substrate peptides.
Decameric peptides corresponding to the p1-p6 processing site within the Gag polyprotein (amino acids 444 to 453) and its variants were purchased from Quality Controlled Biochemicals, Inc., Hopkinton, MA. The following peptide variants were used in the X-ray crystallographic structural studies described in the subsequent sections (Fig. 1): p1-p6WT (RPGNFLQSRP), p1-p6L449F (RPGNFFQSRP), p1-p6S451N (RPGNFLQNRP), and p1-p6L449F/S451N (RPGNFFQNRP).
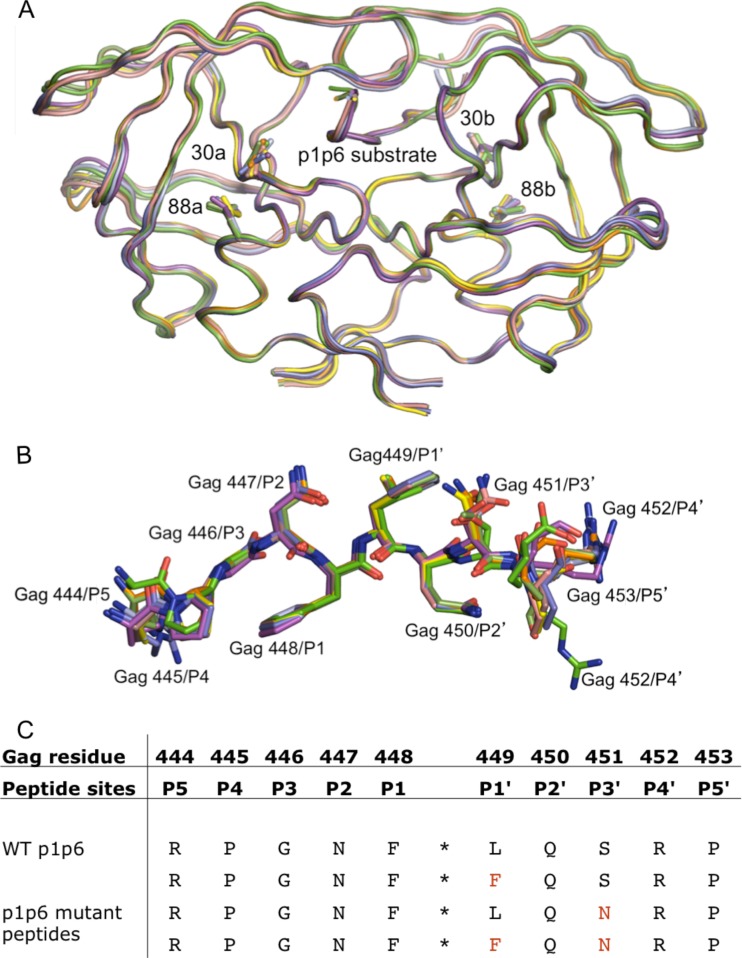
FIG 1.
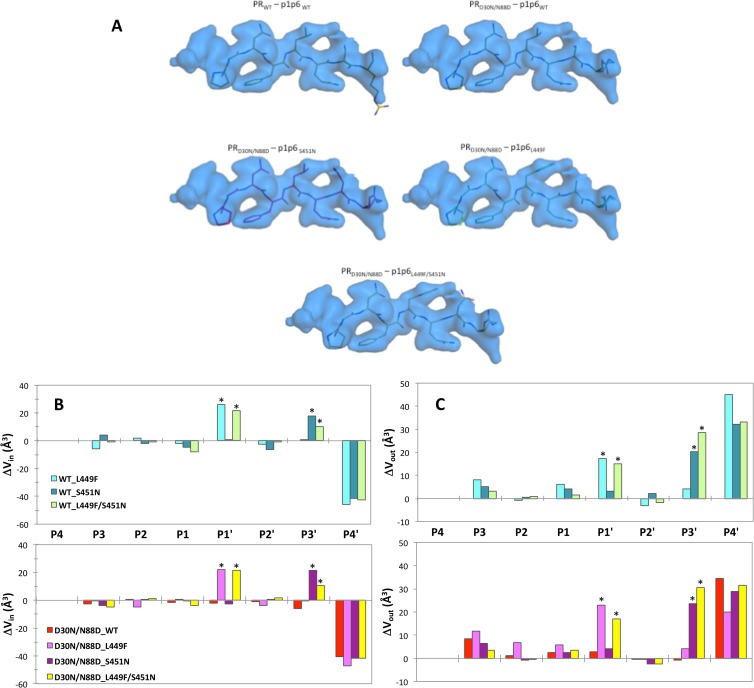
Overall backbone structures of the coevolved and mutant complexes are similar to that of PRWT–p1-p6WT. (A) Superposition of the WT and D30N/N88D PRs in complex with p1-p6 variants. (B) Superposition of the p1-p6 peptide in the active site of the PR variants. Note the different orientations of the Gag452 side chain. (C) The different p1-p6 peptides used in this study with their Gag numbering as well as the peptide site numbering.
D30N/N88D protease gene construction.
The clade B wild-type (WT) protease gene was constructed synthetically using codons optimized for protein expression in Escherichia coli as previously described, with the Q7K mutation introduced to prevent autoproteolysis (25). The D25N mutation was introduced to inactivate the protease and prevent substrate cleavage, which has previously been shown to have little impact on the structure of the protease (26); this construct hence will be referred to as WT throughout. The D30N and N88D protease mutations were then introduced sequentially into the gene. Mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by DNA sequencing.
Protein expression and purification.
The protease gene was subcloned into the heat-inducible pXC35 expression vector (American Type Culture Collection [ATCC], Manassas, VA) and transformed into E. coli TAP-106 cells. Expression and purification of the inactive WT and D30N/N88D protease variants were performed as previously described (27). The protease used in this study was further purified with a Pharmacia Superdex 75 fast-performance liquid chromatography (FPLC) column equilibrated with refolding buffer (50 mM sodium acetate [pH 5.5], 10% glycerol, 5% ethylene glycol, and 5 mM dithiothreitol).
Crystallization and data collection.
The crystals used in this study were obtained under multiple conditions. Protease solutions of between 1.0 and 2.2 mg/ml were equilibrated with a 10-fold molar excess of the p1-p6 peptide variants, individually, for 1 h on ice. Crystals were grown by the hanging-drop vapor diffusion method over a reservoir solution consisting of one of the following three buffer solutions: (i) 126 mM sodium phosphate buffer (pH 6.2), 63 mM sodium citrate, and 20% to 32% ammonium sulfate; (ii) 0.1 to 0.5 M morpholineethanesulfonic acid (MES) monohydrate buffer (pH 6.5), 0.1 to 0.4 M ammonium sulfate, and 20 to 31% polyethylene glycol monomethyl ether (PEG MME) 5000; or (iii) 0.1 M citrate (pH 5.5) and 0.5 to 3.0 M ammonium sulfate. X-ray diffraction data for PRD30N/N88D–p1-p6S451N were collected on the BioCARS beamline 14-BMC at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) at a wavelength of 0.9 Å with a Quantum 315 charge-coupled device (CCD) X-ray detector (Area Detector Systems Corporation, Poway, CA). Diffraction data for the other 5 complexes were collected on the BioCARS beamline 14-IDB at a wavelength of 1.033 Å with a Mar 165 CCD X-ray detector (Rayonix, LLC, Evanston, IL). All data were collected under cryo-cooled conditions.
Structure solution and crystallographic refinement.
The data were indexed and scaled using HKL-2000 software (HKL Research, Charlottesville, VA). Structure determination and refinement were carried out using programs within the CCP4 program suite (28) as previously described (29). The Matthews coefficient was calculated for all structures to determine the number of molecules in the asymmetric unit. For complexes with two molecules in the asymmetric unit, structure solution was carried out using the molecular replacement package Phaser (30), whereas for complexes with one molecule in the asymmetric unit, AMoRe was used (31). The structure of darunavir (inhibitor) in complex with active protease was used as the starting model (1T3R) (32). ARP/wARP was used to build solvent molecules into the electron density (33). Refinement was carried out using a combination of TLS (34) and restrained refinement using Refmac5 (35). Each cycle of refinement was followed by model building and real-space refinement performed using COOT, a molecular graphics software (36). For all complexes, the peptides were built into the Fo − Fc electron density after two rounds of refinement cycles. The working R (Rwork) and its cross validation (Rfree) were monitored throughout refinement. The quality of the structures was assessed using MolProbity (37).
Analysis of PR–p1-p6 complex structures.
Structures of the complexes were compared by superposition of Cα atoms of the active site (residues 24 to 26) and the terminal regions (residues 85 to 90) from both monomers onto a previously solved structure of WT protease in complex with WT p1-p6 (PRWT–p1-p6WT) (PDB 1KJF) (8). The structure of PRD30N/N88D–p1-p6L449F could not be obtained by crystallization and therefore was modeled in silico starting from the PRD30N/N88D–p1-p1WT structure as a template. On this template, the L449 side chain was removed and the position of a Phe side chain was predicted using Prime (38). After side chain replacement, the structural model was refined by energy minimization with the Impref program implemented in Maestro (39). Structures were visualized using PyMOL molecular graphics software (40). In crystals with two molecules in the asymmetric unit (Table 1), structural differences between the two molecules were visualized by distance difference plots, as described previously (29). In all such cases, these plots indicate minor differences between the two molecules (data not shown), and the calculated root mean square deviations (RMSDs) range between 0.14 and 0.19 Å (for the entire protease) and between 0.11 and 0.16 Å (for the p1-p6 peptide). Therefore, in the subsequent structural analyses, only one molecule from the asymmetric units was chosen based on (i) the unambiguous location of side chains for Gag residues 445 to 452 within the electron density and (ii) lower temperature factors for the p1-p6 substrate.
TABLE 1.
Crystallographic statistics for the PR–p1-p6 complexes
| Parameter | Value for: |
||||||
|---|---|---|---|---|---|---|---|
| PRWT–p1-p6WTa | PRD30N/N88D–p1-p6WT | PRWT–p1-p6L449F | PRWT–p1-p6S451N | PRD30N/N88D–p1-p6S451N | PRWT–p1-p6L449F/S451N | PRD30N/N88D–p1-p6L449F/S451N | |
| PDB code | 1KJF | 4OBG | 4OBH | 4OBJ | 4OBF | 4OBK | 4OBD |
| Data collection | |||||||
| Space group | P212121 | P21 | P21 | P212121 | P21 | P212121 | P21 |
| a (Å) | 51.29 | 51.67 | 51.14 | 51.19 | 51.55 | 50.94 | 51.47 |
| b (Å) | 59.07 | 60.11 | 62.79 | 58.3 | 60.07 | 58.41 | 59.54 |
| c (Å) | 61.81 | 60.2 | 61.46 | 61.36 | 60.23 | 61.46 | 59.36 |
| b angle | 90 | 99.09 | 98.03 | 90 | 99.25 | 90 | 98.6 |
| Z | 4 | 8 | 8 | 4 | 8 | 4 | 8 |
| Temp (°C) | −80 | −80 | −80 | −80 | −80 | −80 | −80 |
| Resolution (Å) | 2 | 1.78 | 1.85 | 1.75 | 1.68 | 1.65 | 1.9 |
| Total no. of reflections | 41,786 | 133,292 | 113,050 | 109,140 | 167,489 | 155,872 | 126,355 |
| No. of unique Reflections | 12,376 | 34,288 | 28,931 | 14,456 | 41,265 | 22,296 | 32,879 |
| Rmerge (%) | 6.7 | 6.8 | 7.3 | 4.5 | 6.6 | 8.7 | 6.4 |
| Completeness (%) | 93.4 | 99.2 | 99.7 | 96 | 99.8 | 98.1 | 98.8 |
| Crystallographic refinement | |||||||
| R (%) | 20.3 | 20.04 | 16.08 | 14.35 | 17.97 | 18.00 | 20.01 |
| Rfree (%) | 25.1 | 25.46 | 20.88 | 18.29 | 22.89 | 21.08 | 24.44 |
| RMSDb | |||||||
| Bond length (Å) | 0.006 | 0.009 | 0.007 | 0.009 | 0.009 | 0.009 | 0.009 |
| Bond angle | 1.3 | 1.404 | 1.554 | 1.551 | 1.46 | 1.589 | 1.441 |
From reference 8.
RMSD, root mean square deviation
Determination of hydrogen bonds.
Hydrogen bonds were determined using Maestro (39). A hydrogen bond is defined by a distance of less than 3.5 Å between donor and acceptor atoms and a donor-hydrogen-acceptor angle greater than 120° and less than 180°.
Estimation of vdW potential.
Interactions between protease and the p1-p6 peptide were computed as described previously (41) using a simplified Lennard-Jones potential, V(r), with the relation 4ε[(σ/r)12 − (σ/r)6], where r is the protease–p1-p6 interatomic distance and ε and σ are the depth of the potential well and the collision diameter, respectively, for each protease-substrate atom pair. V(r) is computed for all possible protease–p1-p6 atom pairs within 6.0 Å, and potentials for nonbonded pairs separated less than a distance at the minimum of the potential were equated to −ε. Using this simplified potential for each nonbonded protease–p1-p6 atom pair, the total van der Waals (vdW) contact energy, ΣV(r), was then computed for each protease and p1-p6 residue. Only residues 445 to 452 of the p1-p6 substrate variants were analyzed, as the substrate residues beyond these positions are not located within the binding site.
Calculation of Vin and Vout.
The substrate envelope was previously calculated using a three-dimensional grid-based approach as described earlier (41). The available crystal structures of the wild-type substrates (MA-CA, CA-p2, p2-NC, NC-p1, p1-p6, RT-RH, and RH-IN, with PDB accession codes 1KJ4, 1F7A, 1KJ7, 1TSU, 1KJF, 1KJG, and 1KJH, respectively) were aligned on the WT protease bound to CA-p2 (1F7A) (42) using Cα atoms of residues 24 to 26 and 85 to 90 with PyMOL. A grid was placed on the active site of the protease, and each grid cell was assigned an initial occupancy of zero. Whenever a grid cell was within the van der Waals volume of any substrate atom, the occupancy of the grid cell was increased by one. The grid occupancy was normalized by the total number of crystal structures after all the wild-type substrate volumes were mapped onto the grid. The fit of a substrate within this substrate envelope for a given PR complex structure was then evaluated by Vin and Vout (volumes of the substrate within and outside the substrate envelope, respectively), as described in detail elsewhere (41). Only the P4 to P4′ regions of the substrates were analyzed, as the substrate residues beyond these positions do not have a significant consensus volume. The Vin and Vout for the p1-p6 variants from all the complexes were compared to those for the WT complex to estimate their fit within the substrate envelope.
RESULTS
The four p1-p6 substrate variant peptides (WT, L449F, S451N, and L449F/S451N) were crystallized in complex with WT and NFV-resistant D30N/N88D HIV-1 protease (Fig. 1). Although the simultaneous occurrence of L449F and S451N mutations is infrequent (15), the double mutant was included in this study to investigate why the individually favored mutations do not cooccur and to better understand p1-p6 coevolution. Crystal structures were solved for all complexes (with the exception of PRD30N/N88D–p1-p6L449F, whose structure was modeled [see Materials and Methods]) to a resolution of between 1.6 and 1.9 Å (Table 1). The structures were analyzed to reveal the changes in protease-substrate interactions and fit within the substrate envelope as a result of protease resistance and substrate coevolution mutations.
Overall structure of the complexes.
The overall structures of all six complexes are similar to the previously solved PRWT–p1-p6WT structure, with RMSDs of 0.10 to 0.26 Å for Cα atoms (Fig. 1A). The protease complexes display the most pronounced backbone shifts in the flap-hinge region (residues 35 to 43) and at the 20s loop (RMSDs of 0.36 of 0.40 Å). These changes correlate with the known flexibility of these loop regions and changes in crystallographic space groups (Table 1). Despite these minor differences, the backbones of the complexes superpose very well with the WT structure (Fig. 1A).
For all structures used in the analyses, including the previously solved PRWT–p1-p6WT, at least 8 substrate residues that are important for binding, Gag 445 to 452, are unambiguously located within the electron density map. All the p1-p6 substrate peptides are bound to the protease in the same extended conformation as shown in previous studies (8, 42). The mutant p1-p6 peptides differ from the PRWT–p1-p6WT structure in that the side chain for Gag R452 is significantly reoriented, although the side chains of Gag residues 447 through 451 are relatively unchanged (Fig. 1B). Despite high overall similarity of the backbone, the structures display additional subtle rearrangements within the active site.
Hydrogen bonds between PR and p1-p6 are compensated for in mutant complexes.
The structures of the WT and D30N/N88D PR complexes with p1-p6 variants contain an extensive network of hydrogen bonds between PR and the p1-p6 peptide, both backbone to backbone and side chain to side chain, which plays a key role in substrate recognition and cleavage. Backbone-to-backbone PR–p1-p6 hydrogen bonds are conserved across all structures in this study as in WT PR complexes with other substrate peptides (Table 2) (8). Similarly, five water molecules, including three that bridge the peptide to the protease, are also conserved across all six structures.
TABLE 2.
Hydrogen bonds observed in the PR–p1-p6 complexes
| p1-p6 atom | PR atom | PR chain | Bond length (Å) in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PRWT–p1-p6WT | PRWT–p1-p6L449F | PRWT–p1-p6S451N | PRWT–p1-p6L449F/S451N | PRD30N/N88D–p1-p6WT | PRD30N/N88D–p1-p6L449F | PRD30N/N88D–p1-p6S451N | PRD30N/N88D–p1-p6L449F/S451N | |||
| P445, O | R8, NH2 | B | None | 2.7 | 2.9 | 2.8 | 2.7 | 2.9 | None | 2.9 |
| G446, N | G48, O | A | 3.0 | 2.8 | 3.0 | 3.1 | 2.9 | 3.1 | 2.9 | 2.9 |
| G446, O | D29, N | B | 2.8 | 2.9 | 2.8 | 2.8 | 3.0 | 2.9 | 2.8 | 2.8 |
| N447, N | G48, O | A | 3.0 | 2.7 | 2.9 | 2.9 | 2.8 | 2.9 | 2.9 | 2.9 |
| F448, N | G27, O | A | 2.9 | 2.9 | 2.8 | 2.8 | 3.0 | 3.0 | 2.8 | 2.7 |
| F448, O | N/D25, ND2 | B | 2.7 | 2.7 | 2.7 | 2.7 | 2.9 | 2.9 | 2.8 | 2.8 |
| Q450, N | G27, O | B | 2.9 | 3.1 | 3.0 | 3.0 | 2.9 | 3.1 | 2.9 | 3.0 |
| Q450, O | D29, N | B | 3.1 | 3.2 | 3.1 | 3.1 | 3.0 | 3.3 | 3.0 | 3.1 |
| Q450, OE1 | D/N30, N | B | 2.7 | 2.9 | 2.7 | 2.8 | 2.7 | 2.9 | 2.8 | 2.8 |
| Q450, NE2 | N30, OD2 | B | 3.0 | 3.0 | 2.8 | 2.8 | 3.0 | 3.3 | 3.0 | 2.9 |
| N/S451, N | G48, O | A | 3.0 | 2.9 | 2.8 | 2.9 | 2.8 | 3.0 | 2.8 | 3.0 |
| N/S451, O | G48, N | B | 2.8 | 3.3 | 3.0 | 3.0 | 3.3 | 3.4 | 3.3 | 3.4 |
| S451, OG | G48, O | B | None | None | None | None | 3.2 | 3.0 | None | None |
| R452, N | D29, OD2 | B | 3.8a | 3.0 | 3.0 | 3.0 | 2.8 | 3.0 | 2.8 | 2.8 |
| R452, NE | D30, OD2 | B | 3.0 | None | None | None | None | None | None | None |
| L/F451, N; N447, OD1 | None | None | 3.3 | 3.1 | 3.1 | 3.2 | 3.1 | 3.5 | 3.2 | 3.2 |
Length is given for comparison, even though it is longer than the 3.5-Å cutoff criterion.
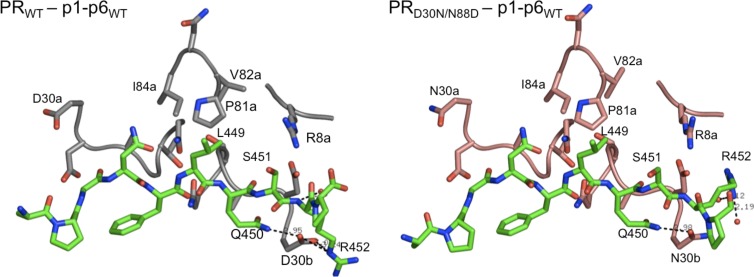
The most significant change in hydrogen bonds is observed around residue PR 30b due to a mutation either at residue 30 in the PR or at the p1-p6 substrate compared to the PRWT–p1-p6WT structure. In the PRWT–p1-p6WT structure, D30b hydrogen bonds with the side chains of Gag Q450 and R452 (Fig. 2). In all other complexes analyzed, the residue 30b hydrogen bond with Q450 is conserved but the one with Gag R452 is lost due to the altered orientation of the latter. Instead, Gag R452 makes a new hydrogen bond as the backbone N moves closer to PR D29b (Table 2). This change is asymmetric and does not involve the other monomer of the protease, which does not make hydrogen bonds at positions 29 and 30 with the p1-p6 peptide. Therefore, the hydrogen bond with D30b lost due to the mutations in either PR or the substrate appears to be compensated for by enhanced interactions with D29b.
FIG 2.
Altered orientation of p1-p6 R452 and loss of interactions with PR residue 30 in the active site of D30N/N88D protease. Wild-type PR is in gray, D30N/N88D PR is in pink, and the p1-p6 peptide is in green.
Packing around the mutant substrate is enhanced in all complex structures.
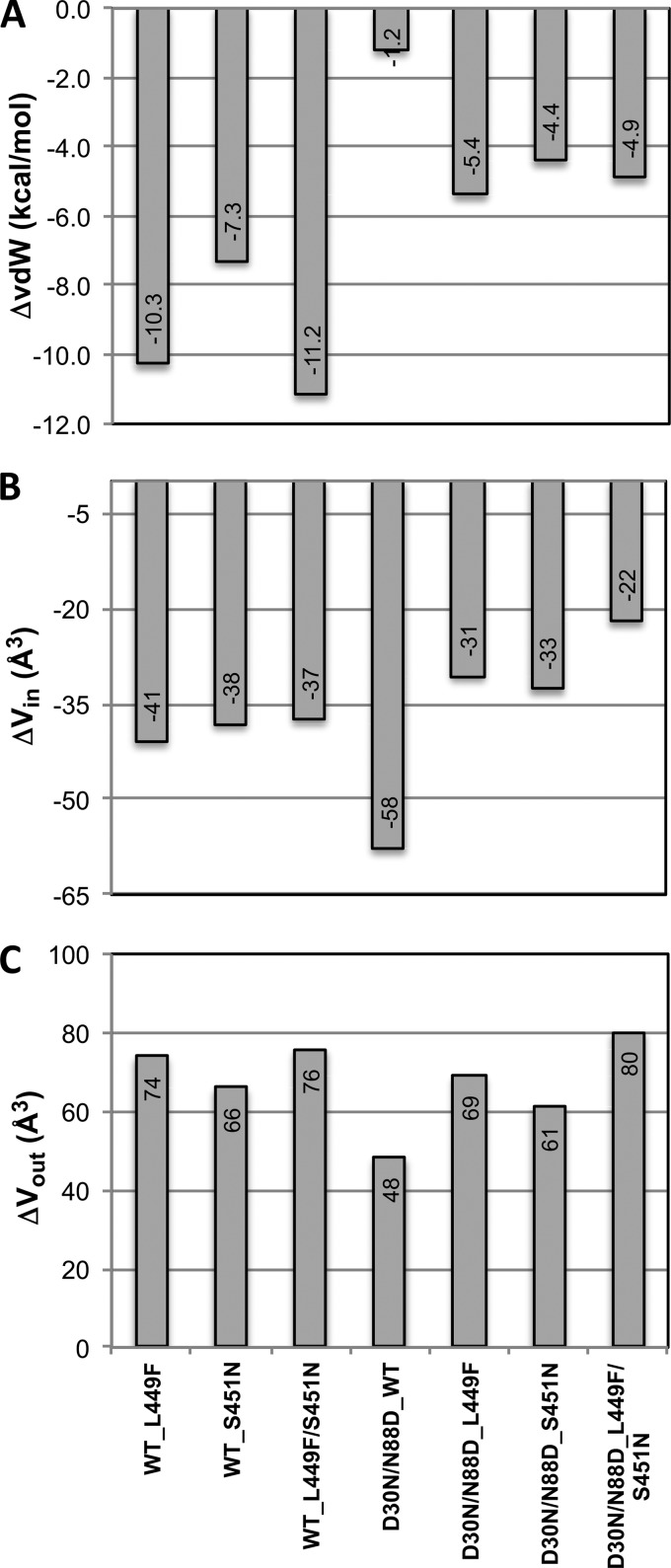
Packing within the active site was evaluated by analyzing the vdW interactions between the PR and the substrate, which are a key component for substrate recognition and cleavage by the protease. The total vdW interaction energies relative to those of the WT complex are all more favorable (more negative) in the mutant complexes (Fig. 3A). Notably, the PR resistance mutations D30N/N88D alone do not significantly change the total vdW interaction value, suggesting that the substrate mutations are responsible for enhanced overall packing. WT PR makes more favorable contacts with the mutant p1-p6 substrates than the D30N/N88D variant. The total gain in vdW interactions of substrates with the L449F mutation is almost 2-fold larger in D30N/N88D than in WT protease. In fact, p1-p6 with the L449F mutation is a better substrate for WT enzyme (11, 16).
FIG 3.
Change in mutant structures relative to the wild-type complex (PRWT–p1-p6WT). (A) Overall vdW interactions between p1-p6 and protease. (B and C) Vin (B) and Vout (C) for the fit of p1-p6 within the substrate envelope.
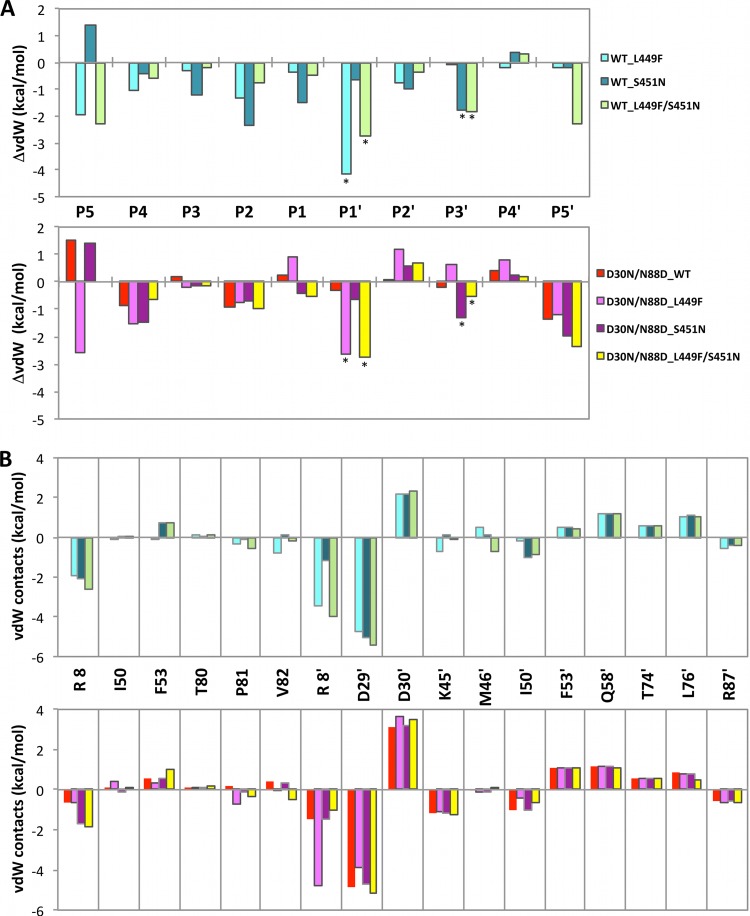
Although the overall packing around the substrate is more extensive in the mutant complexes, the vdW contacts are drastically reduced for protease residue 30b (Fig. 4B), mainly due to an altered orientation of p1-p6 R452 (Fig. 1B and 2; positive values at P4′ position in Fig. 4A). Despite conservation of the hydrogen bond between PR residue 30b and Gag Q450, the vdW interactions are reduced in the coevolved complexes (positive values at P2′). However, these losses are compensated for by the increased interactions of PR D29b (Fig. 4B). The packing around the substrate in this monomer is also rearranged, with interactions lost with residues 53, 58, 74, and 76 and enhanced with 8, 45, 50, and 87. There are few interactions between D30a and the p1-p6 peptide, and very little change is observed around the other monomer due to the D30N mutation. As a result of this asymmetric interaction between PR and the p1-p6 substrate, compensatory mutations in p1-p6 are clustered from the C terminus to the scissile bond.
FIG 4.
Changes in vdW contacts between the protease and p1-p6 substrate in mutant complex structures with respect to the WT complex (PRWT–p1-p6WT). WT and D30N/N88D protease complexes are in upper and lower panels in cool and warm colors, respectively. (A) Change in contacts of substrate p1-p6 residues. Asterisks below data bars indicate mutation sites. (B) Change in contacts of protease residues. ′, residues in monomer b. Significant enhancement in contacts of protease residue D29b compensates for reduced interactions at residue 30b in all mutant structures.
The changes in vdW interactions are also asymmetric for the substrate around the cleavage site (Fig. 4A). The effect of mutations in either the PR or substrate is not confined to the points of mutation but instead propagates over all residues within the p1-p6 peptide. WT protease packs tighter around the whole core of the peptide (P3–P3′) in mutant p1-p6 complexes relative to WT substrate, especially at the sites of mutations (Fig. 4A, upper panel, asterisks). This increase is reflected in the enhanced overall vdW contacts described above and correlates with more efficient processing of mutant p1-p6 by WT protease (11, 16). Although the general trends in repacking around the substrate are similar to those in the WT protease case, D30N/N88D protease has decreased contacts with Gag residue 450 (Fig. 4A, lower panel, P2′) and has less tightening of the substrate packing, as reflected in the smaller overall change in vdW energies (Fig. 3A). Interestingly, despite repacking around the substrate, the enhanced and reduced contacts almost completely compensate for each other to yield no considerable overall change in WT substrate vdW interactions with D30N/N88D protease. Thus, although the per-residue vdW contacts are altered irrespective of mutations in the PR or the p1-p6, the overall change in vdW contact potentials of the mutant substrates, and not the WT, are favorable, due to the correlated changes in interactions between the different substrate sites.
Fit within the substrate envelope is improved by coevolution.
Substrate fit within the substrate envelope for all the mutant complexes was investigated in terms of the volumes that lie within (Vin) and outside (Vout) the substrate envelope. The differences in Vin and Vout relative to those for the PRWT–p1-p6WT structure were computed for all mutant complexes (Fig. 3B and C). The D30N/N99D mutations cause a worse fit of the WT substrate within the envelope, with a decrease of 58 Å3 in Vin. The substrate mutations compensated for this loss by almost half in the coevolved structures (Fig. 3B).
In all complexes, including the coevolved structures, a mutation(s) either in the PR or p1-p6 reduced Vin (Fig. 3B) and increased Vout (Fig. 3C), suggesting worse fit within the substrate envelope. However, this apparent overall worse fit is dominated by changes at the P4′ position (Fig. 5B), where the Gag R452 has rearranged as discussed above (Fig. 1) and protrudes beyond the substrate envelope. In fact, D30N/N88D protease mutations do not significantly impact the fit of P3–P3′ moieties within the envelope, whereas substrate mutations better fill the substrate envelope at the P1′ and P3′ positions while at the same time causing protrusions beyond the envelope (positive values with asterisks in Fig. 5B and C, respectively). The double mutation in the substrate (L449F/S451N) does not improve the fit within the envelope (Vin) any further than the individual mutations. Rather, the presence of the bulky F449 side chain shifts N451 to protrude more outside the envelope (Vout) (Fig. 5A). This worse fit of the double mutant (L449F/S451N) within the substrate envelope may be the underlying reason why these two mutations do not simultaneously occur in patient sequences (15), as there is no selective advantage to substrate recognition to have both changes simultaneously.
FIG 5.
(A) Fit of p1-p6 substrates into the substrate envelope. (B and C) Change in Vin (B) and Vout (C) in mutant structures with respect to the WT complex (PRWT–p1-p6WT). WT and D30N/N88D protease complexes are in upper and lower panels in cool and warm colors, respectively. Asterisks above data bars indicate mutation sites.
DISCUSSION
Drug resistance to HIV-1 PIs reflects a change in molecular recognition, where a fine balance exists between loss of inhibitor binding to the PR and substrate recognition and processing of Gag/GagProPol. When this equilibrium is disturbed, the substrate sites coevolve and allow the virus to maintain adequate fitness and contribute to drug resistance (11, 12, 14, 15). Our analysis of the crystal structures reveals the molecular basis of p1-p6 substrate coevolution in response to the D30N/N88D protease mutations selected under the pressure of NFV inhibition.
The D30N/N88D resistance mutations in protease cause substantial loss of substrate interactions at residue 30b (Fig. 5 and Table 2). D30N is one of the few mutations that cause a change in the charge of an active-site side chain, which disrupts the electrostatic and H bond interactions with Gag R452. D30 is also involved in an H bond network involving N88 in WT protease (21). The secondary N88D mutation restores the net charge of the protease, as well as causing additional loss of substrate contacts by shifting residue 30 away from the active site. Coevolution of p1-p6 substrate to include bulkier side chains at Gag positions 449 and 451 enhances substrate packing at the active site, mainly at the mutation sites, but the effects propagate throughout the substrate as well. Hence, the compensation mechanism for the lost intermolecular interactions due to drug resistance protease mutations is not simply selecting substrate mutations to restore those interactions through coevolution but also gaining other types of interactions throughout the active site.
Coevolution mutations also help reinstate the substrate's fit within the substrate envelope. The consensus volume occupied by substrates bound to HIV-1 protease, or the substrate envelope, defines the substrate recognition motif. We have established that when inhibitors protrude out of this envelope to contact protease residues, those sites are selected for primary drug resistance mutations (13). Similarly, we propose that substrates deviating from the consensus are more susceptible to coevolution with drug resistance mutations (41). The p1-p6 substrate protrudes out the envelope with a Vout higher than expected based on the sequence's molecular volume (41), which renders this substrate susceptible to coevolution. The NFV resistance mutations also cause a decrease in Vin, likely reducing efficiency of p1-p6 recognition and cleavage and causing the virus to coevolve mutations at this site. In agreement with our previous molecular modeling and dynamics predictions (24), the crystal structures we present here confirm that the substrate coevolution compensates for the decrease in Vin due to protease resistance mutations and optimizes the fit of p1-p6 within the substrate envelope.
The changes in interactions between the different p1-p6 substrate residues and the protease due to mutations are interdependent and are not simply confined to sites of mutations. The total vdW interaction potentials of the WT p1-p6 cleavage site with NFV-resistant D30N/N88D protease are similar to those of the WT complex. Although the D30N/N88D mutations do not cause a dramatic change in the total vdW energy, residue 30's interactions, mainly electrostatic and H bonding interactions with Gag R452, are substantially reduced. We observed that the R452 side chain reoriented from its position in the WT complex and in previous modeling predictions (43) and shifted toward protease D29 to at least partially compensate for lost favorable electrostatic interactions with residue 30. In the case of coevolution, the larger side chains introduced by L449F or S451N mutation in the p1-p6 cleavage site increase the overall contacts of the substrate. This interdependency of interactions at different sites within the p1-p6 cleavage site across complexes with the D30N/N88D PR suggests that, rather than reinstating the specific lost contact, preserving the overall interaction potential by coevolution is essential for recognition and cleavage.
Previously, we showed that L449 and S451 within the p1-p6 cleavage site do not mutate simultaneously (15). Our structures show that having both mutations at the same time does not offer any advantages in altering substrate-PR interactions (H bonds and vdW contacts) and, furthermore, worsens the fit within the substrate envelope. The overall van der Waals interaction energies of the double mutant substrate were similar to those with single mutations, and Vin was reduced for D30N/N88D PR. Corresponding with two large side chains located close to each other, the double mutant p1-p6 also had a relatively larger Vout that may affect PR binding and cleavage. Thus, two mutations do not provide any additional advantage in the context of the D30N/N88D PR mutations, in agreement with their very infrequent observation together (15).
In our previous study (14) we found that within the p1-p6 cleavage site, L449 and S451, along with P453, which is just outside the cleavage site, vary, with a significantly higher rate in the presence of D30N/N88D (32, 30, and 66%, respectively) than in the absence of these protease mutations (15, 17, and 30%, respectively). However, unlike with other cleavage site mutations, the fold change in resistance to nelfinavir was not altered. Although we might expect the resistant protease to less efficiently cleave the p1-p6 site and consequently the virus to have reduced fitness in the absence of these compensatory mutations, we did not observe a difference in the replicative capacity in patient-derived viral sequences in the presence or absence of cleavage site mutations. However, within these viral strains there are likely other mutations elsewhere either in the protease or in Gag to modulate the fitness of the virus. Only by engineering these changes into an otherwise unaltered system can the level of compensation conferred by these coevolving sites be properly assessed.
The WT p1-p6 cleavage site, in a manner analogous to that for the NC-p1 cleavage site, may not be the optimum sequence for efficient cleavage (11), and this suboptimal amino acid sequence may regulate the sequential processing of Gag (6, 44) in the viral life cycle. Compared to the WT sequence, the p1-p6 peptide with the L449F mutation is cleaved more efficiently even by the WT protease (11, 16). Gag processing by WT protease is enhanced in the presence of L449F (11, 16, 45, 46). The S451N mutation may have similar effects on cleavage of the p1-p6 substrate. Similar to the more efficient processing of Gag A431V at the NC-p1 cleavage site (24, 27), enhanced contacts with the protease due to bulkier side chain substitutions by F449 and N451 may increase the kinetics of cleavage at the p1-p6 site. Such a more efficient cleavage at the p1-p6 site by WT PR could affect Gag processing by altering the relative rate of p1-p6 cleavage and/or by changing the order of cleavage within Gag. Therefore, the F449 and N451 in the context of the WT PR would likely be detrimental to this regulated processing of Gag and inadvertently affect viral fitness. However, in the context of the NFV-resistant PR, which is expected to be less efficient at cleaving the WT p1-p6 junction, these enhanced contacts with the coevolved p1-p6 substrate improve recognition and cleavage, which may help restore the correct order of Gag processing.
Our structural analysis demonstrates that the interactions between PR and the p1-p6 cleavage site are influenced by mutations in either the protease or the substrate. The loss of interactions at one site due to NFV resistance mutations is compensated for by improved interactions at other sites in a correlated manner. This interdependency in PR–p1-p6 interactions influences the overall fit of the substrate within the substrate envelope and is likely to affect substrate recognition and cleavage in the presence of PR resistance mutations. Coevolution enables the virus to maintain a fine balance of substrate recognition and cleavage while avoiding inhibition by PIs.
ACKNOWLEDGMENTS
Nelfinavir was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This research was supported by National Institutes of Health (NIH) grant R01-GM65347.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Flexner C. 1998. HIV-protease inhibitors. N. Engl. J. Med. 338:1281–1293. 10.1056/NEJM199804303381808 [DOI] [PubMed] [Google Scholar]
- 2.Thompson MA, Aberg JA, Cahn P, Montaner JSG, Rizzardini G, Telenti A, Gatell JM, Günthard HF, Hammer SM, Hirsch MS, Jacobsen DM, Reiss P, Richman DD, Volberding PA, Yeni P, Schooley RT. 2010. Antiretroviral treatment of adult HIV infection. JAMA 304:321–333. 10.1001/jama.2010.1004 [DOI] [PubMed] [Google Scholar]
- 3.Volberding PA, Deeks SG. 2010. Antiretroviral therapy and management of HIV infection. Lancet 376:49–62. 10.1016/S0140-6736(10)60676-9 [DOI] [PubMed] [Google Scholar]
- 4.Shafer RW, Schapiro JM. 2008. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 10:67–84 http://www.aidsreviews.com/resumen.asp?id=998&indice=2008102&u=unp [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe DE, Tritch R, Hutchison CA, III, Loeb DD, Swanstrom R. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retroviruses 5:577–591. 10.1089/aid.1989.5.577 [DOI] [PubMed] [Google Scholar]
- 6.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. 2005. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 2:66. 10.1186/1742-4690-2-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual gag poly protein cleavage sites. J. Virol. 72:2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369–381. 10.1016/S0969-2126(02)00720-7 [DOI] [PubMed] [Google Scholar]
- 9.Clemente JC, Moose RE, Hemrajani R, Whitford LR, Govindasamy L, Reutzel R, McKenna R, Agbandje-McKenna M, Goodenow MM, Dunn BM. 2004. Comparing the accumulation of active- and nonactive-site mutations in the HIV-1 protease. Biochemistry 43:12141–12151. 10.1021/bi049459m [DOI] [PubMed] [Google Scholar]
- 10.Bally F, Martinez R, Peters S, Sudre P, Telenti A. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retroviruses 16:1209–1213. 10.1089/08892220050116970 [DOI] [PubMed] [Google Scholar]
- 11.Feher A, Weber IT, Bagossi P, Baross P, Mahalingam B, Louis JM, Copeland TD, Yorshin IY, Harrison RW, Tozser J. 2002. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. J. Biochem. 269:4114–4120. 10.1046/j.1432-1033.2002.03105.x [DOI] [PubMed] [Google Scholar]
- 12.Mammano F, Petit C, Clavel F. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. 2004. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 11:1333–1338. 10.1016/j.chembiol.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 14.Kolli M, Stawiski E, Chappey C, Schiffer CA. 2009. Human immunodeficiency virus type 1 protease-correlated cleavage site mutations enhance inhibitor resistance. J. Virol. 83:11027–11042. 10.1128/JVI.00628-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolli M, Lastere S, Schiffer CA. 2006. Co-evolution of nelfinavir-resistant HIV-1 protease and the p1-p6 substrate. Virology 347:405–409. 10.1016/j.virol.2005.11.049 [DOI] [PubMed] [Google Scholar]
- 16.Maguire MF, Guinea R, Griffin P, Macmanus S, Elston RC, Wolfram J, Richards N, Hanlon MH, Porter DJ, Wrin T, Parkin N, Tisdale M, Furfine E, Petropoulos C, Snowden BW, Kleim JP. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398–7406. 10.1128/JVI.76.15.7398-7406.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, Heilek-Snyder G, Dulude D, Cammack N, Brakier-Gingras L, Konvalinka J, Parkin N, Krausslich HG, Brun-Vezinet F, Boucher CA. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 4:e36. 10.1371/journal.pmed.0040036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. 1997. Characterization of the backbone dynamics of folded and denatured states of an SH3 domain. Biochemistry 36:2390–2402. 10.1021/bi962548h [DOI] [PubMed] [Google Scholar]
- 19.Patick AK, Duran M, Cao Y, Shugarts D, Keller MR, Mazabel E, Knowles M, Chapman S, Kuritzkes DR, Markowitz M. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali A, Bandaranayake RM, Cai Y, King NM, Kolli M, Mittal S, Murzycki JF, Nalam MN, Nalivaika EA, Ozen A, Prabu-Jeyabalan MM, Thayer K, Schiffer CA. 2010. Molecular basis for drug resistance in HIV-1 protease. Viruses 2:2509–2535. 10.3390/v2112509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandaranayake RM, Kolli M, King NM, Nalivaika EA, Heroux A, Kakizawa J, Sugiura W, Schiffer CA. 2010. The effect of clade-specific sequence polymorphisms on HIV-1 protease activity and inhibitor resistance pathways. J. Virol. 84:9995–10003. 10.1128/JVI.00505-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandaranayake RM, Prabu-Jeyabalan M, Kakizawa J, Sugiura W, Schiffer CA. 2008. Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6. J. Virol. 82:6762–6766. 10.1128/JVI.00018-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozisek M, Bray J, Rezacova P, Saskova K, Brynda J, Pokorna J, Mammano F, Rulisek L, Konvalinka J. 2007. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J. Mol. Biol. 374:1005–1016. 10.1016/j.jmb.2007.09.083 [DOI] [PubMed] [Google Scholar]
- 24.Ozen A, Haliloglu T, Schiffer CA. 14 February 2012. HIV-1 protease and substrate coevolution validates the substrate envelope as the substrate recognition pattern. J. Chem. Theory Comput. 10.1021/ct200668a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose JR, Salto R, Craik CS. 1993. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J. Biol. Chem. 268:11939–11945 [PubMed] [Google Scholar]
- 26.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. 2008. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 283:13459–13470. 10.1074/jbc.M708506200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prabu-Jeyabalan M, Nalivaika EA, King NM, Schiffer CA. 2004. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 78:12446–12454. 10.1128/JVI.78.22.12446-12454.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 29.Prabu-Jeyabalan M, Nalivaika EA, Romano K, Schiffer CA. 2006. Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate. J. Virol. 80:3607–3616. 10.1128/JVI.80.7.3607-3616.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navaza J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. A50:157–163 [Google Scholar]
- 32.Surleraux DL, Tahri A, Verschueren WG, Pille GM, de Kock HA, Jonckers TH, Peeters A, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. 2005. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 48:1813–1822. 10.1021/jm049560p [DOI] [PubMed] [Google Scholar]
- 33.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3:1171–1179. 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn MD, Isupov MN, Murshudov GN. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57:122–133. 10.1107/S0907444900014736 [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. D53:240–255 [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 37.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383. 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrödinger, LLC. 2011. Suite 2011: Prime, version 3.0. Schrödinger, LLC, New York, NY [Google Scholar]
- 39.Schrödinger, LLC. 2011. Suite 2011: Maestro, version 9.2. Schrödinger, LLC, Portland, OR [Google Scholar]
- 40.DeLano WL. 2002. The PyMol molecular graphics system. Delano Scientific, San Carlos, CA [Google Scholar]
- 41.Ozen A, Haliloglu T, Schiffer CA. 2011. Dynamics of preferential substrate recognition in HIV-1 protease: redefining the substrate envelope. J. Mol. Biol. 410:726–744. 10.1016/j.jmb.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. 2000. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 301:1207–1220. 10.1006/jmbi.2000.4018 [DOI] [PubMed] [Google Scholar]
- 43.Shibata J, Sugiura W, Ode H, Iwatani Y, Sato H, Tsang H, Matsuda M, Hasegawa N, Ren F, Tanaka H. 2011. Within-host co-evolution of Gag P453L and protease D30N/N88D demonstrates virological advantage in a highly protease inhibitor-exposed HIV-1 case. Antiviral Res. 90:33–41. 10.1016/j.antiviral.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 44.Pettit SC, Sheng N, Tritch R, Erickson-Vitanen S, Swanstrom R. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15–25. 10.1007/978-1-4615-5373-1_2 [DOI] [PubMed] [Google Scholar]
- 45.Maguire MF, MacManus S, Griffin P, Guinea C, Harris W, Richards N, Wolfram J, Tisdale M, Snowden W, Kleim JP. 2001. Interaction of HIV-1 protease and gag gene mutations in response to amprenavir-selective pressure exerted in amprenavir-treated subjects—contribution of Gag p6 changes L449F and P453L. Antivir. Ther. 6(Suppl 1):48 [Google Scholar]
- 46.Myint L, Matsuda M, Matsuda Z, Yokomaku Y, Chiba T, Okano A, Yamada K, Sugiura W. 2004. Gag non-cleavage site mutations contribute to full recovery of viral fitness in protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:444–452. 10.1128/AAC.48.2.444-452.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]