Abstract
Cell migration is a dynamic process that requires the coordinated formation and disassembly of focal adhesions (FAs). Several proteins such as paxillin, focal adhesion kinase (FAK), and G protein–coupled receptor kinase–interacting protein 1 (GIT1) are known to play a regulatory role in FA disassembly and turnover. However, the mechanisms by which this occurs remain to be elucidated. Paxillin has been shown to bind the C-terminal domain of FAK in FAs, and an increasing number of studies have linked paxillin association with GIT1 during focal adhesion disassembly. It has been reported recently that phosphorylation of serine 273 in the LD4 motif of paxillin leads to an increased association with Git1 and focal adhesion turnover. In the present study, we examined the effects of phosphorylation of the LD4 peptide on its binding affinity to the C-terminal domain of FAK. We show that phosphorylation of LD4 results in a reduction of binding affinity to FAK. This reduction in binding affinity is not due to the introduction of electrostatic repulsion or steric effects, but rather by a destabilization of the helical propensity of the LD4 motif. These results further our understanding of the focal adhesion turnover mechanism as well as identify a novel process by which phosphorylation can modulate intracellular signaling.
Focal adhesion kinase (FAK) 1 is an intracellular protein tyrosine kinase whose activity is regulated by integrin-mediated cell adhesion.(1) FAK influences the dynamic regulation of integrin-associated adhesions and the actin cytoskeleton which is tethered there. The continual formation and disassembly of focal adhesions at the leading edge of lamellipodia of migrating cells (focal adhesion turnover) is a process that is crucial to both the control of cell migration(2) and invasion by cancer cells.(3) Observation of elevated FAK expression in a broad variety of tumors suggests that FAK plays an important role in tumorigenesis.(4) It is therefore necessary to understand the mechanisms that regulate the formation and disassembly of complexes between FAK and its binding partners in focal adhesions.
The C-terminal portion of FAK, termed FAK-related nonkinase (FRNK)(5), consists of several proline-rich regions that act as binding sites for many SH3-containing proteins, and a C-terminal focal adhesion targeting (FAT) domain.(5–8) Exogenous expression of FRNK or FAT domain can displace endogenous full-length FAK from focal adhesions resulting in disruption of lamellipodia formation and apoptosis.(9–12) One of the key binding partners of FAK at sites of focal adhesion is paxillin, a multi-domain adaptor protein associated with cell adhesion and growth control.(13–16) Paxillin binds to the C-terminal FAT domain of FAK through two of its five N-terminal LD motifs, termed LD2 and LD4.(17–21) The LD motifs of paxillin are amphipathic alpha helices that can interact with many other proteins,(22) including G protein-coupled receptor kinase-interacting protein 1 (GIT1).(23) Paxillin, GIT1, and FAK are hypothesized to act synergistically to promote cell motility by a mechanism in which paxillin triggers focal adhesion disassembly and turnover by switching its binding partner from FAK to GIT1.(24; 25) This switch was thought likely to be driven by the phosphorylation of paxillin.(24) Mapping of the major paxillin phosphorylation sites by mass spectrometry(26) showed that p21-activated kinase (PAK) phosphorylation of paxillin at S273 within the LD4 motif increased the association between paxillin and GIT1 resulting in an up-regulation of adhesion turnover.(27)
Here we show by chemical shift perturbation and fluorescence anisotropy experiments that phosphorylation at S273 within the LD4 motif of paxillin significantly reduces it’s binding affinity to the FAT domain of FAK. This finding demonstrates that phosphorylation of paxillin at S273 triggers a molecular switch that determines the preferred binding partner of paxillin during the focal adhesion turnover process. In addition, we show that the reduction in binding affinity of phosphorylated LD4 to the FAT domain is due to a shift in the order to disorder transition of the LD4 motif, a novel mechanism by which phosphorylation can regulate selectivity.
EXPERIMENTAL PROCEDURES
Overexpression and purification of the FAT domain of FAK, paxillin, and the FAT-LD2 construct
The cDNA encoding the FAT domain (residues 916 to 1053) was subcloned into a pET28a vector. The N-terminal His-tagged FAT domain was subsequently expressed in E. coli strain BL21(DE3)pLysS. Protein induction, harvest, and purification methods have been described previously.(28; 29) FAT-LD2 was sub-cloned into a pET28a vector in which LD2 oligos were inserted into a BamHI site downstream of FAT. Oligonucleotides that contained BamHI restriction sites and the LD2 sequence flanked 5’ by glycine serine repeats (GGS2GS(GGS2 positive and negative strands) were synthesized at the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. The cDNA encoding chicken paxillin, spanning amino acids 51–313, was subcloned into a pET28a vector. To generate the paxillin amino acid 130–313 construct in pET28a, oligonucleotides containing 5’ NdeI or 3’ EcoRI restriction enzyme sites were designed and used to amplify the sequence through polymerase chain reaction (PCR). Incorporation of paxillin into pET28a was verified by sequencing with T7 terminator primer. Isotope-labeled protein was prepared by using morpholinepropanesulfonic acid–buffered medium containing 15NH4Cl (1 g/) and 13C6-glucose (2.5 g/l).
Peptide synthesis
The Hartwell Center for Bioinformatics and Biotechnology at St Jude chemically synthesized a peptide corresponding to the LD4 motif of chicken paxillin (residues 262–276 of wild type paxillin, SATRELDELMASLSD), and LD4 peptides phosphorylated at the S12 and S14 positions (S273 and S275 of the wild-type paxillin), termed LD4-S273p and LD4-S275p, respectively. For the fluorescence anisotropy experiments, peptides corresponding to LD4, LD4-S273p, and LD4-S275p with a fluorescein tag covalently attached to the N-terminus via a butanoic linker were synthesized. CD experiments confirmed that the fluorescein tag does not inhibit the helical propensity of the individual peptides (data not shown).
NMR spectroscopy
NMR data for structure determination were acquired with Varian Inova 600 MHz and Bruker Avance 800 MHz (with cryoprobe) spectrometers at 37 °C. Sample concentrations for NMR experiments were typically 0.5 to 1.0 mM in 10 mM potassium phosphate buffer, pH 6.5, 10% D2O, and 0.1% sodium azide. Data were processed and displayed by using the programs NMRPipe and NMRDraw(30) on an SGI Octane workstation or PC. Proton chemical shifts were referenced to H2O peaks (4.75 ppm). 13C and 15N chemical shifts were referenced indirectly to DSS using absolute frequency ratios. The program SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) was used for data analysis and assignment. Backbone resonance assignments of the FAT-LD2 construct were made manually on the basis of 3-dimensional HNCA, HNCACB, and CBCA(CO)NH experiments. Aliphatic side-chain resonances were assigned by using 3-dimensional 15N-edited HCCH-COSY and HCCH-TOCSY spectra. Residual gaps and ambiguities were resolved by using nuclear Overhauser effect (NOE) connections measured by 3-dimensional 15N-edited and 13C-edited NOESY experiments. 15N-heteronuclear single quantum correlation (HSQC) experiments were performed to measure the chemical shift perturbations in the FAT-LD2 spectra upon LD4 peptide binding.
Chemical shift perturbation
NMR data for binding affinity studies were acquired on a Varian Inova 600 MHz spectrometer at 37 °C, and processed as described above. 15N and 1H chemical shift values for the displaced peaks in the 15N-HSQC titration experiments were determined for each of the successive titration points by using SPARKY. The average chemical shift perturbation was calculated by using the following equation:(31)
In this equation ΔδN is chemical shift change of the amide nitrogen and ΔδH is the chemical change of the amide proton. The amide nitrogen chemical shift change is divided by 5 to account for the scale difference of the nitrogen and proton dimensions.
The dissociation constant was calculated by nonlinear curve fitting of the plotted chemical shift change versus the molar ratio of the ligand by using the following equation:(32)
In this equation, Δδ is the chemical shift change upon titration, X is the molar ratio of the ligand, and [protein]0 the initial protein concentration.
Determination of binding affinity with fluorescence anisotropy
Fluorescence anisotropy measurements were taken using a Fluorolog fluorometer (Jobin Yvon Horriba) equipped with polarizers. Successive 2- to 20- μl aliquots of 0.5 mM FAT-LD2 were added to a 1-cm cuvette containing 2.0 ml of a 0.5 µM solution of fluorescently labeled LD4 peptide. The experiments were done at pH 8.0 in 10 mM phosphate buffer and either 0% or 15% TFE. Anisotropy was measured at an emission wavelength of 513 nm 3 min after each addition to allow the system to come to equilibrium. Change in anisotropy for binding was plotted and fitted to a standard one-site binding model in order to determine the binding constant Kd and the maximum change in anisotropy, Bmax, using Prism (version 4.0 for Windows, GraphPad Software, San Diego California USA, www.graphapd.com).
Measurements of helix propensity using circular dichroism
CD spectra were obtained with an Aviv 62DS CD spectrometer (Aviv, Lakewood, N.J.) and processed with Igor Pro software (Wavemetrics Inc., Lake Oswego, OR). All experiments were performed at 25 °C using a quartz cuvette with a 0.1-cm path length. The parameters used for the measurements were: 0.5 nm step resolution, 10 s average signaling time, and 1 nm bandwidth. All spectra shown are averages of 3 scans. Concentration of samples ranged from 20 to 40 μM in 10 mM phosphate buffer, pH 6.5. The total volume of each sample was 300 μl. The CD spectra were expressed as molar ellipticity ([θ]). Helix content was calculated as [θ]222 / max[θ]222 where max[θ]222 = −40,000×[1 – (2.5 / n)] and n is the number of amino acid residues.(33)
RESULTS
Covalently tethered LD motifs
The FAT domain of FAK is composed of four helices that form a right-turn elongated bundle maintained by hydrophobic interactions.(17–19) Paxillin binds to two hydrophobic patches on opposite faces of this four-helix bundle through two of its N-terminal helical LD motifs (LD2 and LD4).(21) However, in solution, isolated LD peptides (mainly LD2) can non-specifically interact with the other LD binding site on the FAT domain.(20; 21) To unambiguously study LD binding to the FAT domain, our laboratory uses constructs in which the individual LD motifs are covalently linked to the C-terminal end of the FAT domain via a (GGSx linker (see Methods). Analysis of 15N-HSQC spectra have shown that when tethered to the FAT domain in this manner, covalently linked LD2 or LD4 motifs bind specifically to their preferred binding sites in the FAT domain.(21) Thus, in the FAT-LD2 construct, only the LD4 binding site is open, and in the FAT-LD4 construct, only the LD2 binding site is open. In the present study, we titrated FAT-LD2 with LD4 peptides and could confidently attribute the changes observed in the fluorescence anisotropy and chemical shift perturbation experiments solely to the binding of LD4 peptides to the LD4 binding pocket on the FAT domain.
Binding studies by fluorescence anisotropy
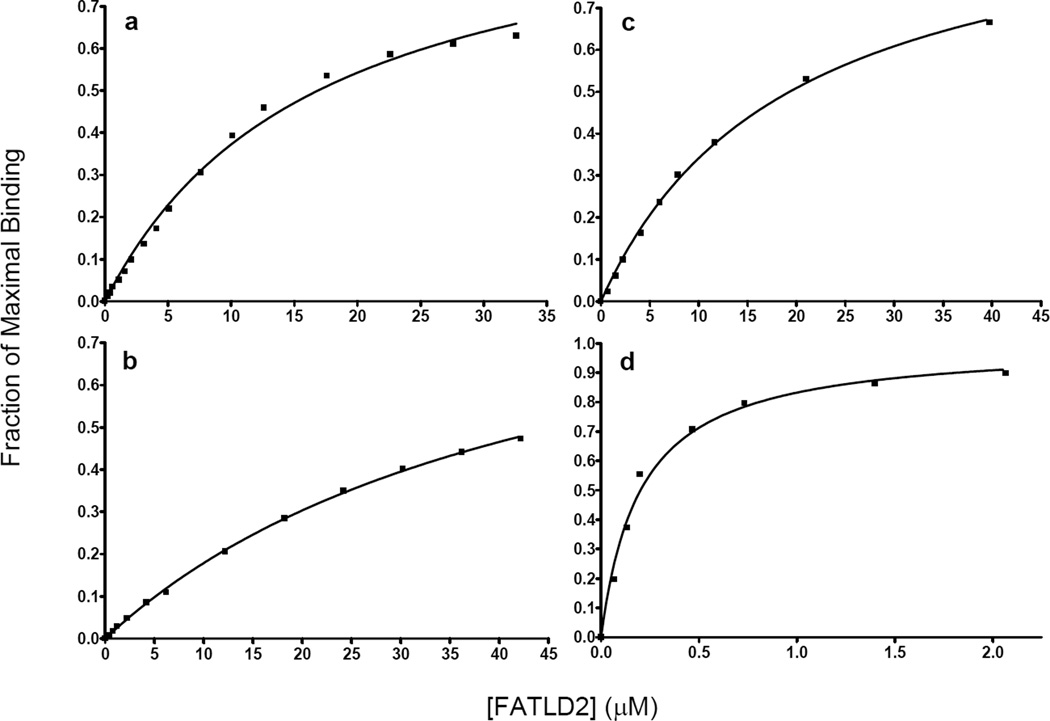
To measure the binding affinity between FAT-LD2 and LD4, we synthesized LD4 peptides tagged at the N-terminus with fluorescein (see Methods) and measured the change in fluorescence anisotropy as a function of titrated protein. The measured Kd values for LD4, LD4-S275p, and LD4-S273p binding to FAT-LD2 were 16.8 ± 1.5 µM, 19.3 ± 1.1 µM, and 45.9 ± 2.4 µM respectively (Fig. 1a-c). Phosphorylation at the S273 position increased dissociate constant by almost 300%, whereas phosphorylation at the S275 position affected binding affinity only slightly over that of unphosphorylated LD4.
Figure 1.
Binding curves of fluorescein-tagged LD4 peptides titrated with increasing concentrations of FAT-LD2, as measured by fluorescence anisotropy in 10-mM phosphate, pH 8.0. The y-axis indicates the fraction of maximal binding. a-c LD4, LD4-S273p, and LD4-S275p, respectively. d LD4 in 15% TFE.
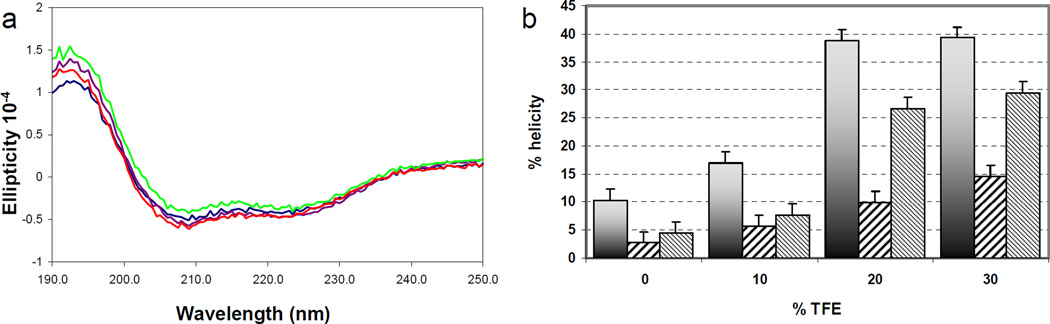
We previously showed that the LD4 motif exists mostly as a random coil in solution and folds into an α-helix upon binding to FAT.(21) We postulated that if we could increase the helicity of unbound LD4, its binding affinity to FAT would increase. To test this hypothesis, we performed a fluorescence anisotropy analysis of the binding of tagged LD4 to FAT-LD2 in the presence of 15% 2,2,2-trifluoroethanol (TFE) (Fig. 1d). The resulting binding curve yielded a Kd of 216 ± 24 nM, demonstrating a 77-fold increase in binding affinity. Circular dichroism (CD) studies of FAT-LD2 titrated with TFE showed it to be stable at the concentrations (5–10 µM) used in the fluorescence experiments (Fig. 2a). These results strongly suggest a link between the binding affinity and the helical configuration of LD4.
Figure 2.
a Circular dichroism scans of 5 µM FAT-LD2 in 0% TFE (blue), 5% TFE (purple), 10% TFE (green), and 15% TFE (red).b Helix-formation propensity of LD4 (shaded), LD4-S273p (wide diagonal), and LD4-S275p (narrow diagonal) as measured by circular dichroism in increasing concentrations of TFE.
Helical propensity of LD4 peptides
To determine the effect of phosphorylation on the propensity of LD4 to form a helix, we performed CD measurements of the fluorescein-tagged peptides titrated with TFE. The effect of increasing amounts of TFE (up to about 20%) on helix formation provides a useful approximation of the helical propensity of non–alanine based peptides in water.(34) At 20% TFE, the LD4-S273p peptide exhibited a helix propensity approximately 75% less than that of unphosphorylated LD4, whereas the LD4-S275p peptide’s helix propensity was reduced only ~30% (Fig. 2b).
Factors that may contribute to the helix-destabilizing effect of phosphorylation at the S273 position include an unfavorable i - i+4 side chain interaction between the negatively charged phosphate and glutamate 269 and the unfavorable peptide solvation effects of phosphoserines at the C-terminal positions of α–helices.(35) The destabilizing effects of C-terminal phosphorylation on an α-helix have also been observed in a monomeric mutant form of phospholamban, in which phosphorylation at serine 16 results in significant unwinding of the cytoplasmic helix and the short loop connecting it to the transmembrane helix.(36)
Effect of LD4 phosphorylation on FAT binding
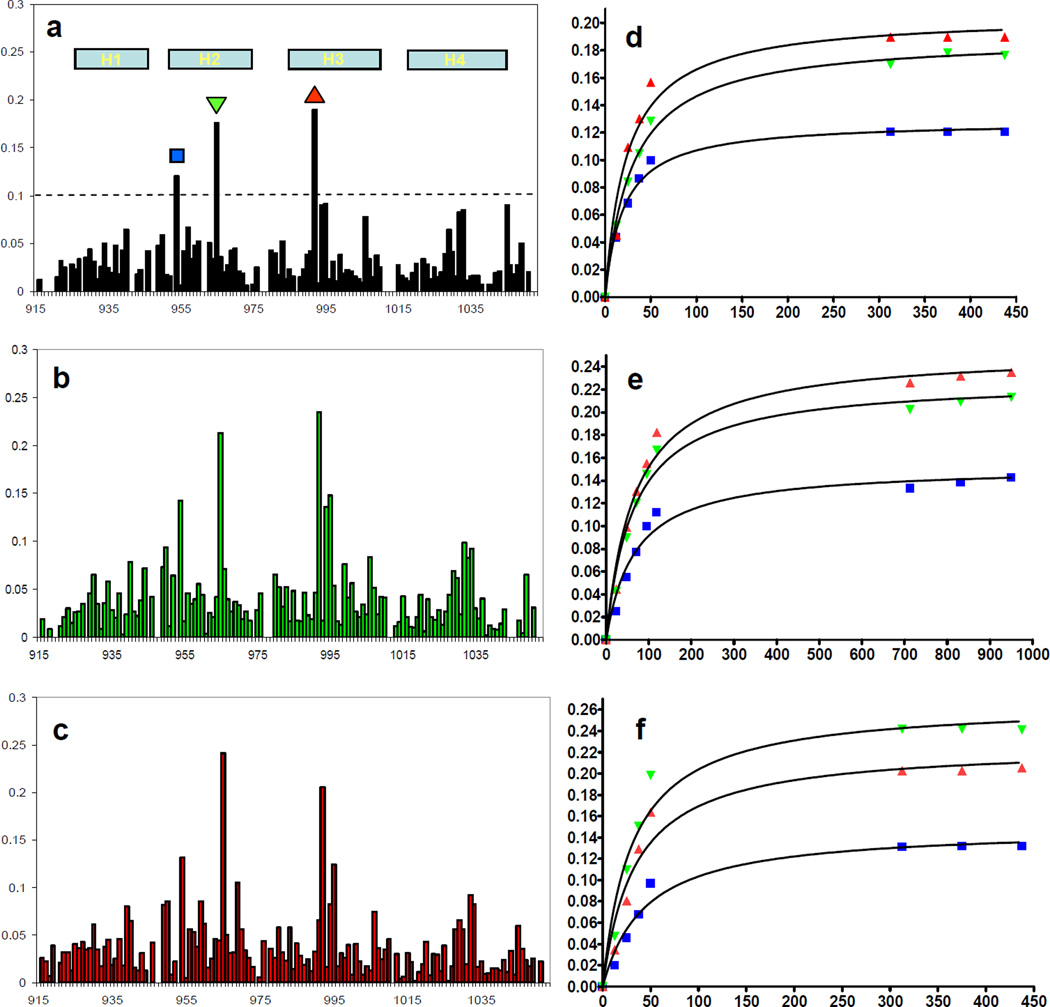
To verify that the phosphorylated LD4 peptides bound similarly to FAT and to the unphosphorylated peptide, we performed titration experiments and monitored the change of chemical shifts in the HSQC spectrum of 15N labeled FAT-LD2. This method can be used to map binding sites and is well suited to detect weak but specific interactions in solution.(37) In the HSQC spectra, unphosphorylated LD4, the LD4-S273p, and LD4-S275p most strongly perturbed the same residues of FAT-LD2 (Fig. 3a-c). We plotted the change in the weighted average of the chemical shift values of the affected residues versus the concentration of ligand to generate binding curves (Fig. 3d-f). Using the chemical shift values from residues M954, N992, and L965 (all within the LD4 binding pocket) and fitting the curves for each peptide to a global Kd, we calculated the binding affinities of LD4, LD4-S275p, and LD4-S273p to be 25.3 ± 2.8 µM, 33.6 ± 5.8 µM, and 65.8 ± 7.2 µM, respectively. As in the fluorescence anisotropy experiments, we observed a slightly reduced binding affinity (within the range of error) for the LD4-S275p peptide and an approximately 3-fold decrease in the binding affinity of the LD4-S273p peptide. Therefore, the binding affinities observed for the non-fluorescently tagged LD4 peptides are consistent with the binding data observed for the fluorescently tagged LD4 peptides.
Figure 3.
a-c Chemical shift perturbation plots of the FAT domain residues in the FAT-LD2 construct titrated with LD4, LD4-S273p, and LD4-S275p, respectively. Helices (H) 1–4 of the FAT domain are indicated in plot A. The largest chemical shifts correspond to residues in the LD4-binding pocket. d-f Binding curves derived from weighted chemical shift values for FAT residues M954 (■), L965 (▼), and N992 (▲).
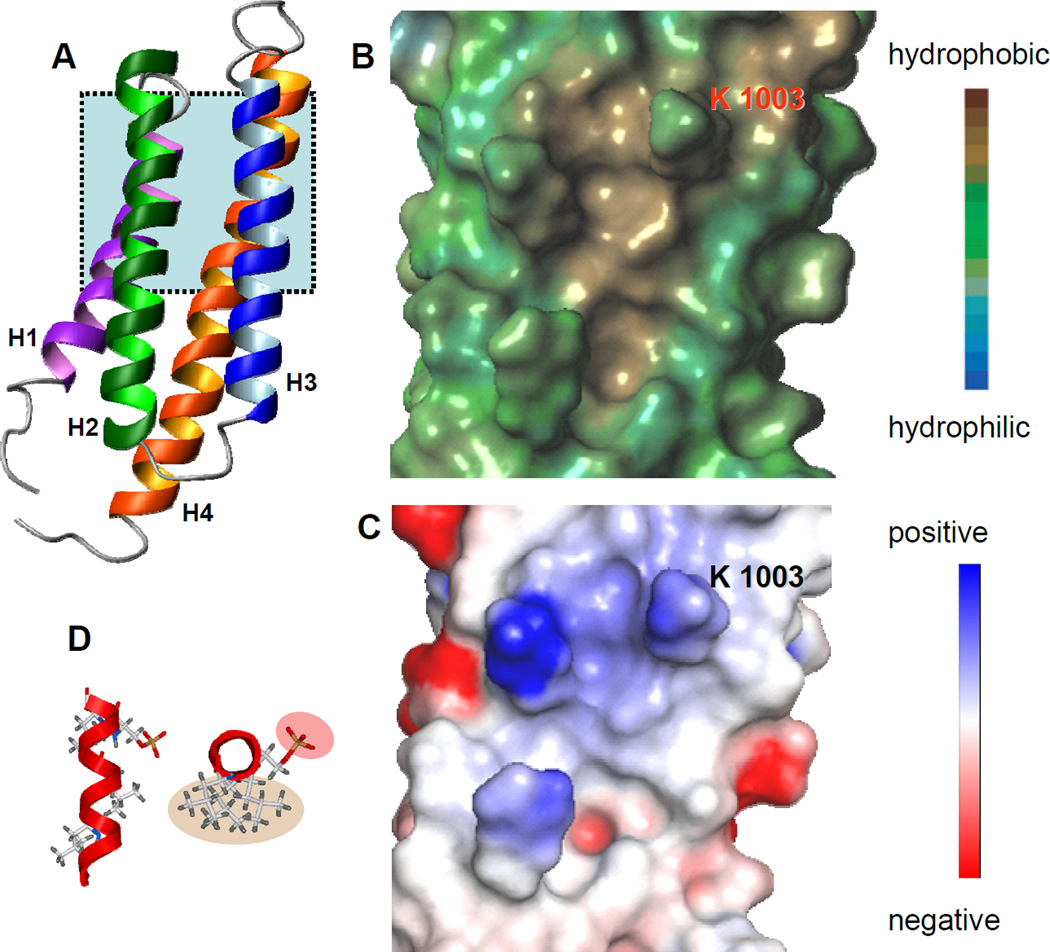
The LD4 motif of paxillin has previously been shown to interact with a hydrophobic pocket on the surface of FAT formed between helices H2 and H3 of the four helix bundle(21) (Fig. 4a). The similarity of the HSQC spectra of LD4-S273p bound to FAT and the HSQC spectra of LD4 bound to FAT strongly suggests that the phosphorylated peptide binds in the same pocket and in the same manner as its unphosphorylated form. When LD4-S273p is modeled as an α-helix, it is clear that the phosphate moiety is oriented away from the hydrophobic binding surface of the LD motif (Fig. 4b). The electrostatic surface of the LD4 binding pocket contains no negatively charged residues that could interact with the phosphate group of LD4-S273p. Indeed, only a positively charged K1003 lies in the vicinity of the phospho group (Fig. 4c). For that reason, it is unlikely that the reduced binding affinity of the LD4-S273p peptide is caused by electrostatic or steric effects. Therefore, destabilization of the α-helix of LD4 upon phosphorylation at S273 is the most likely cause of the reduced binding affinity observed in both fluorescently tagged and untagged peptides.
Figure 4.
a Ribbon diagram of the FAT domain of FAK (Protein Data Bank accession code 1KTM).(17) The LD4 binding pocket (shaded area) is formed between helices H2 and H3 of the four helix bundl. b Lypophilic surface map of the LD4 binding pocket generated using SYBYL v7.2.5 (Tripos Inc.). The hydrophobic surface interacts with hydrophobic leucine residues on LD4. c Electrostatic surface of the LD4 binding pocket generated using Pymol (DeLano Scientific LLC, San Carlos, CA). Only a positively charged lysine, K1003, is in a position to interact with the phosphate moiety on LD4-S273p. d The LD4-S273p peptide modeled as an α-helix by using ICM-Pro v3.4 (Molsoft Inc.). Top view shows leucine residues that form the hydrophobic binding surface (shaded tan) and the phosphorylated Ser 273 (shaded rose). Bottom view shows the relative orientation of the peptide as it binds to the LD4-binding pocket of FAT.
DISCUSSION
Because cell migration is a dynamic process, the mechanisms by which focal adhesions form and disassemble are likely to be regulated in a temporal and spatial manner.(3) Nayal et al. provided an important piece of the puzzle when they observed that phosphorylation of paxillin at S273 increases adhesion turnover in highly motile cells by increasing paxillin-GIT1 binding.(27) Our findings show that phosphorylation of paxillin at S273 also significantly reduces its binding affinity to FAK, causing the disassembly of the FAK-paxillin complex and freeing paxillin to interact with other key binding partners. The modulation of paxillin binding through phosphorylation of its LD4 motif represents a key step in the focal adhesion turnover mechanism.
Although paxillin binds to FAK through both its LD2 and LD4 motifs, the LD4 motif is the key structural element in the formation and disassembly of the FAK-paxillin complex. Exogenous expression of paxillin lacking the LD4 motif in paxillin-null cells results in an 11-fold decrease in focal adhesion disassembly.(38) The importance of LD4 in the interaction between FAK and paxillin can also be seen in western blot analysis of paxillin binding to FAK mutants: mutations that disrupt the LD4 binding site reduce paxillin binding more than mutations that disrupt the LD2 binding site.(20) The results of our binding studies show that when the helical content of LD4 is increased by addition of TFE, its binding affinity to FAK is markedly increased. In contrast, when the helical propensity of LD4 is decreased by phosphorylation at S273 (and, to a lesser extent, at S275), the binding affinity is dramatically reduced. The reduction in the binding affinity of LD4 as the result of phosphorylation at S273 leads to destabilization of the FAK-paxillin interaction and the eventual breakdown of the complex.
Phosphorylation has previously been shown to contribute to complex dissociation.(39) In the case of paxillin binding to FAK, the decrease in the binding affinity of phosphorylated LD4 is likely to reflect perturbation of an order-to-disorder transition rather than the introduction of unfavorable electrostatic or steric effects. Further evidence is provided by immunoblot experiments of a paxillin S273D mutant expressed in CHO-K1 cells that showed only marginal changes in binding to FAK.(27) Although recognized by a phospho-S273-paxillin–specific antibody, the aspartate mutant does not appear to mimic the binding behavior of paxillin-S273p to FAK. This can be explained by the fact that phosphoserine at the C-terminus of α-helices is destabilizing,(35) whereas unphosphorylated serine and aspartate residues have similar effects on helix propensity.(40)
The random coil to helix transition upon binding exhibited by the LD4 motif is a common feature among regulatory proteins classified as intrinsically unstructured proteins (IUPs).(41–43) The LD4 motif of paxillin has all of the characteristics of a set of targeting elements known as alpha-helix–forming molecular recognition elements (α-MoREs).(44) These are specific structural elements that mediate many binding events of disordered regions. They consist of a short region (about 20 residues) that undergoes a disorder-to-order transition that is stabilized upon binding to the partner; this short region is itself located within a segment of disorder. Such disorder-to-order transitions upon binding are common in IUPs involved in regulation, signaling, and control pathways.(45) By contrast, the LD2 motif does not appear to be an α-MoRE as it forms a stable α-helix in solution.(17) The difference between the helix propensities of these two LD motifs may lie in their helix capping motifs. The LD2 helix ends at a proline residue, which, like glycine, is a strong helix breaker.(46; 47) The LD4 sequence contains neither proline nor glycine at the C-terminal end of its helix. Thus, the hydrogen bonds responsible for stabilizing the C-terminal end of the LD4 helix are likely to be provided by interacting residues within its binding partners. Lowering the helix propensity of the LD4 motif by phosphorylation at S273 provides the cell with a subtle yet effective mechanism for modulating the selectivity of paxillin among its various binding partners in focal adhesions.
It has been suggested that FAK, paxillin, and GIT1 can act together to promote focal adhesion turnover.(24), (25; 27) Furthermore, Zhao et al. observed that GIT1 directly associates with FAK, thus linking FAK to PAK signaling, and suggested that both kinases play an important role in the focal adhesion dynamics that underlie cell motility.(24) It is likely that in focal adhesions PAK is brought into proximity with the FAK-paxillin complex by GIT1 and subsequently phosphorylates serine 273 in the LD4 motif of paxillin. Because both GIT1 and FAK interact directly with the LD4 motif of paxillin(16) and phosphorylation of serine 273 differentially alters their binding affinities,(27) this phosphorylation event should facilitate the transition of paxillin from a complex with FAK to a complex with GIT1, thereby initiating focal adhesion turnover.
ACKNOWLEDGEMENTS
We thank Dr. J. Thomas Parsons for kindly providing chicken FRNK cDNA, Dr. Christopher Turner for kindly providing chicken paxillin cDNA, and Dr. Weixing Zhang for technical support.
Footnotes
This work is supported by the Cancer Center Support Grant (CA21765) from the National Cancer Institute, by the American Lebanese Syrian Associated Charities, by grant GM069916 from the National Institutes of Health.
Abbreviations: FA, Focal Adhesion; FAK, focal adhesion kinase; FAT, focal adhesion targeting; FRNK, FAK-related nonkinase; GIT1, G protein–coupled receptor kinase–interacting protein 1; NMR, nuclear magnetic resonance; TFE, 2,2,2-trifluoroethanol; HSQC, Heteronuclear Single Quantum Correlation; IUP, intrinsically unstructured proteins; α-MoREs, alpha-helix–forming molecular recognition elements.
REFERENCES
- 1.Parsons JT. Focal adhesion kinase: the first ten years. J. Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 2.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells -- over and over and over again. Nat. Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 3.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim. Biophys. Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 4.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat. Rev. Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 5.Schaller MD, Borgman CA, Parsons JT. Autonomous expression of a noncatalytic domain of the focal adhesion- associated protein tyrosine kinase pp125FAK. Mol. Cell Biol. 1993;13:785–791. doi: 10.1128/mcb.13.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildebrand JD, Schaller MD, Parsons JT. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J. Cell Biol. 1993;123:993–1005. doi: 10.1083/jcb.123.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cary LA, Guan JL. Focal adhesion kinase in integrin-mediated signaling. Front Biosci. 1999;4:D102–D113. doi: 10.2741/cary. [DOI] [PubMed] [Google Scholar]
- 8.Schlaepfer DD, Hauck CR, Sieg DJ. Signaling through focal adhesion kinase. Prog. Biophys. Mol. Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 9.Xu LH, Yang X, Bradham CA, Brenner DA, Baldwin AS, Jr, Craven RJ, Cance WG. The Focal Adhesion Kinase Suppresses Transformation-associated, Anchorage-independent Apoptosis in Human Breast Cancer Cells. J. Biol. Chem. 2000;275:30597–30604. doi: 10.1074/jbc.M910027199. [DOI] [PubMed] [Google Scholar]
- 10.Golubovskaya V, Beviglia L, Xu LH, Earp HS, III, Craven R, Cance W. Dual inhibition of focal adhesion kinase and epidermal growth factor receptor pathways cooperatively induces death receptor-mediated apoptosis in human breast cancer cells. J. Biol. Chem. 2002;277:38978–38987. doi: 10.1074/jbc.M205002200. [DOI] [PubMed] [Google Scholar]
- 11.Richardson A, Parsons T. A mechanism for regulation of the adhesion-associated proteintyrosine kinase pp125FAK. Nature. 1996;380:538–540. doi: 10.1038/380538a0. [DOI] [PubMed] [Google Scholar]
- 12.Martin KH, Boerner SA, Parsons JT. Regulation of focal adhesion targeting and inhibitory functions of the FAK related protein FRNK using a novel estrogen receptor "switch". Cell Motil. Cytoskeleton. 2002;51:76–88. doi: 10.1002/cm.10018. [DOI] [PubMed] [Google Scholar]
- 13.Tachibana K, Sato T, D'Avirro N, Morimoto C. Direct association of pp125FAK with paxillin, the focal adhesion- targeting mechanism of pp125FAK. J. Exp. Med. 1995;182:1089–1099. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas JW, Cooley MA, Broome JM, Salgia R, Griffin JD, Lombardo CR, Schaller MD. The role of focal adhesion kinase binding in the regulation of tyrosine phosphorylation of paxillin. J. Biol. Chem. 1999;274:36684–36692. doi: 10.1074/jbc.274.51.36684. [DOI] [PubMed] [Google Scholar]
- 16.Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Guibao CD, Zheng J. Structural insight into the mechanisms of targeting and signaling of focal adhesion kinase. Mol. Cell Biol. 2002;22:2751–2760. doi: 10.1128/MCB.22.8.2751-2760.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arold ST, Hoellerer MK, Noble ME. The structural basis of localization and signaling by the focal adhesion targeting domain. Structure. (Camb.) 2002;10:319–327. doi: 10.1016/s0969-2126(02)00717-7. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi I, Vuori K, Liddington RC. The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin. Nat. Struct. Biol. 2002;9:101–106. doi: 10.1038/nsb755. [DOI] [PubMed] [Google Scholar]
- 20.Gao G, Prutzman KC, King ML, Scheswohl DM, DeRose EF, London RE, Schaller MD, Campbell SL. NMR solution structure of the focal adhesion targeting domain of focal adhesion kinase in complex with a paxillin LD peptide: evidence for a two-site binding model. J. Biol. Chem. 2004;279:8441–8451. doi: 10.1074/jbc.M309808200. [DOI] [PubMed] [Google Scholar]
- 21.Bertolucci CM, Guibao CD, Zheng J. Structural features of the focal adhesion kinase-paxillin complex give insight into the dynamics of focal adhesion assembly. Protein Sci. 2005;14:644–652. doi: 10.1110/ps.041107205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbarello DA, Brown MC, Turner CE. The paxillin LD motifs. FEBS Lett. 2002;513:114–118. doi: 10.1016/s0014-5793(01)03244-6. [DOI] [PubMed] [Google Scholar]
- 23.Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J. Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 24.Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikata Y, Birukov KG, Garcia JG. S1P induces FA remodeling in human pulmonary endothelial cells: role of Rac, GIT1, FAK, and paxillin. J. Appl. Physiol. 2003;94:1193–1203. doi: 10.1152/japplphysiol.00690.2002. [DOI] [PubMed] [Google Scholar]
- 26.Webb DJ, Schroeder MJ, Brame CJ, Whitmore L, Shabanowitz J, Hunt DF, Horwitz AR. Paxillin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 2005;118:4925–4929. doi: 10.1242/jcs.02563. [DOI] [PubMed] [Google Scholar]
- 27.Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 2006;173:587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong HC, Mao J, Nguyen JT, Srinivas S, Zhang W, Liu B, Li L, Wu D, Zheng J. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat. Struct. Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong HC, Bourdelas A, Krauss A, Lee H-J, Shao Y-M, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell. 2003;12:1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Worrall JA, Reinle W, Bernhardt R, Ubbink M. Transient protein interactions studied by NMR spectroscopy: the case of cytochrome C and adrenodoxin. Biochemistry. 2003;42:7068–7076. doi: 10.1021/bi0342968. [DOI] [PubMed] [Google Scholar]
- 32.Smet C, Duckert JF, Wieruszeski JM, Landrieu I, Buee L, Lippens G, Deprez B. Control of protein-protein interactions: structure-based discovery of low molecular weight inhibitors of the interactions between Pin1 WW domain and phosphopeptides. J. Med. Chem. 2005;48:4815–4823. doi: 10.1021/jm0500119. [DOI] [PubMed] [Google Scholar]
- 33.Forood B, Feliciano EJ, Nambiar KP. Stabilization of alpha-helical structures in short peptides via end capping. Proc. Natl. Acad. Sci. U.S.A. 1993;90:838–842. doi: 10.1073/pnas.90.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers JK, Pace CN, Scholtz JM. Trifluoroethanol effects on helix propensity and electrostatic interactions in the helical peptide from ribonuclease T1. Protein Sci. 1998;7:383–388. doi: 10.1002/pro.5560070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrew CD, Warwicker J, Jones GR, Doig AJ. Effect of phosphorylation on alpha-helix stability as a function of position. Biochemistry. 2002;41:1897–1905. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]
- 36.Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44:4386–4396. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 37.Wuthrich K. Protein recognition by NMR. Nat. Struct. Biol. 2000;7:188–189. doi: 10.1038/73278. [DOI] [PubMed] [Google Scholar]
- 38.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 39.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem. Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 40.Rohl CA, Doig AJ. Models for the 3(10)-helix/coil, pi-helix/coil, and alpha-helix/ 3(10)-helix/coil transitions in isolated peptides. Protein Sci. 1996;5:1687–1696. doi: 10.1002/pro.5560050822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat. Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 42.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 43.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 45.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 46.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunasekaran K, Nagarajaram HA, Ramakrishnan C, Balaram P. Stereochemical punctuation marks in protein structures: glycine and proline containing helix stop signals. J. Mol. Biol. 1998;275:917–932. doi: 10.1006/jmbi.1997.1505. [DOI] [PubMed] [Google Scholar]