Summary
Dynamic instability, the stochastic switching between growth and shrinkage, is essential for microtubule function. This behavior is driven by GTP hydrolysis in the microtubule lattice, and is inhibited by anticancer agents like Taxol. We provide new insight into the mechanism of dynamic instability, based on high-resolution cryo-EM structures (4.7–5.6 Å) of dynamic microtubules and microtubules stabilized by GMPCPP or Taxol. We infer that hydrolysis leads to a compaction around the E-site nucleotide at longitudinal interfaces, as well as movement of the α–tubulin intermediate domain and H7 helix. Displacement of the C-terminal helices in both α– and β–tubulin subunits suggests an effect on interactions with binding partners that contact this region. Taxol inhibits most of these conformational changes, allosterically inducing a GMPCPP-like state. Lateral interactions are similar in all conditions we examined, suggesting that microtubule lattice stability is primarily modulated at longitudinal interfaces.
Introduction
Microtubules are ubiquitous cytoskeletal filaments critical for multiple cellular processes, including intracellular trafficking, establishment and maintenance of cell morphology, and cell division (Hyams and Lloyd, 1993). For many microtubule-dependent processes, the underlying dynamics of the polymer play a pivotal role. Perhaps the most striking example is mitosis, when chromosome motions are driven by microtubule dynamics and segregation is primarily powered by microtubule depolymerization (Desai and Mitchison, 1997; McIntosh et al., 2010; Rieder and Salmon, 1994). Highlighting this fact, many successful anti-proliferative drugs bind to tubulin and interfere with microtubule dynamics (Dumontet and Jordan, 2010). Describing the conformational cycle accompanying tubulin polymerization, nucleotide hydrolysis, and microtubule depolymerization is essential for our understanding of microtubule dynamics and would significantly aid in improving existing anticancer drugs, as well as facilitating the development of novel agents.
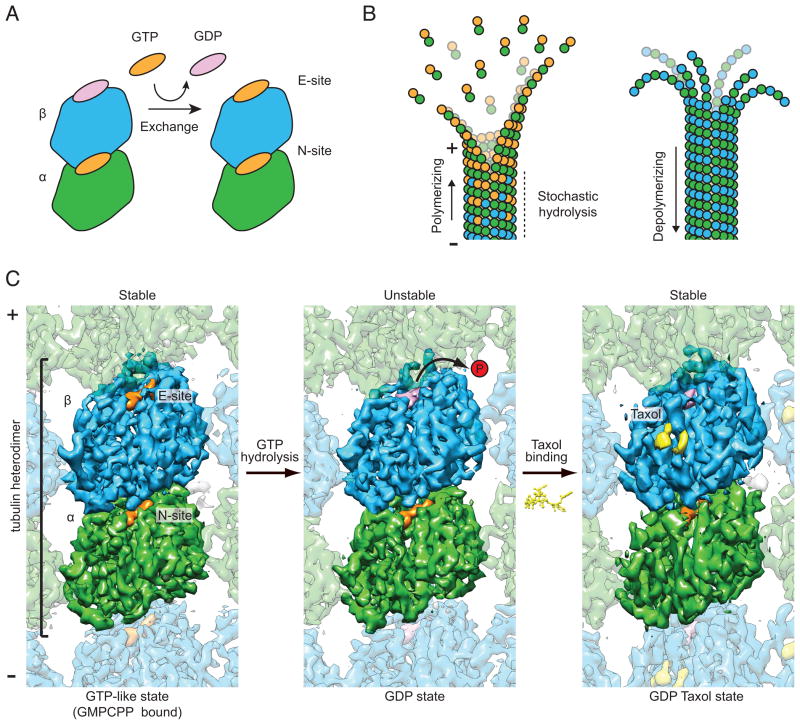
Dynamic instability, the stochastic switching between phases of microtubule growth and shrinkage, is driven by the binding and hydrolysis of GTP by the αβ-tubulin dimer (Mitchison and Kirschner, 1984). Tubulin dimers associate longitudinally to form polar protofilaments, which associate laterally to form a tube. Subunit addition occurs preferentially at the end of the microtubule capped by β-tubulin subunits, termed the “plus-end”. αβ-tubulin contains two GTP-binding sites (Fig. 1A). The N-site (non-exchangeable) in α-tubulin is buried within the tubulin dimer at a longitudinal monomer-monomer (or intradimer) interface (Nogales et al., 1998). This site is constitutively occupied by GTP and has been ascribed a structural role (Menendez et al., 1998). The nucleotide at the E-site (exchangeable) in β-tubulin is exposed on the surface of an unpolymerized dimer and the terminal subunits of a microtubule plus-end (Mitchison, 1993; Nogales, 2000). Free αβ-tubulin dimers exchange bound GDP for GTP at the E-site, rendering them competent for polymerization (Fig, 1A). Upon addition of a tubulin dimer to a growing microtubule plus end, the α-tubulin subunit in the incoming dimer contacts the E-site GTP of the terminal β-tubulin subunit, completing the binding pocket that enables hydrolysis (Nogales et al., 1999). Thus, microtubule growth and GTP hydrolysis are coupled, giving rise to the metastable character of this polymer. While a lattice of GTP-tubulin is stable and promotes polymerization, the GDP-tubulin lattice is unstable and prone to depolymerization, or ‘catastrophe’ (Desai and Mitchison, 1997). In the long-standing ‘GTP-cap’ model (Mitchison and Kirschner, 1984), a microtubule will continue to grow as long as it contains GTP-tubulin subunits at its plus end (i.e. subunit addition outpaces hydrolysis). When this GTP cap is lost, rapid depolymerization ensues (Fig. 1B).
Figure 1. High-resolution cryo-EM structures of dynamic and stabilized microtubules.
A) Cartoon of the αβ–tubulin dimer, which spontaneously exchanges bound GDP for GTP in solution. B) Cartoon illustrating structural intermediates of microtubule polymerization and depolymerization. C) Cryo-EM maps of GMPCPP (left panel, 4.7 Å resolution), GDP (middle panel, 4.9 Å resolution) and taxol-stabilized (right panel, 5.6 Å resolution) microtubules, viewed from inside the microtubule lumen. α-tubulin, green, β-tubulin, blue, GMPCPP/GTP, orange, GDP, pink, taxol, yellow. Maps are contoured at 1.1 σ. (See also Table S1a, Figures S1 and S2 and Movies S1, S2 and S3)
The detailed molecular mechanism by which tubulin GTP binding and hydrolysis controls microtubule dynamics remains elusive despite decades of intensive study. Structural studies have led to the consensus view that conformational changes in tubulin must be correlated with the transition from polymerization to depolymerization. A straight tubulin conformation is found within the body of the microtubule (Li et al., 2002b; Nogales et al., 1999), and all high-resolution structural analyses of this state to date have been limited to electron crystallography of zinc-induced 2D sheets, which contain protofilament-like head-to-tail assemblies of straight αβ-tubulin (Nettles et al., 2004; Nogales et al., 1998). A curved conformation is found in microtubule depolymerization peels (Mandelkow et al., 1991), head-to-tail arrays of bent tubulin heterodimers wherein longitudinal contacts are maintained after lateral contacts are broken (Fig. 1B). The existence of peels as a depolymerization intermediate, together with the fact that contacts between protofilaments are mediated by a series of loop-loop interactions (Li et al., 2002b; Sui and Downing, 2010), has led to the assumption that lateral contacts are the most labile of the microtubule lattice interactions. In this model, hydrolysis-induced destabilization of the distant lateral contacts would occur through an unknown allosteric mechanism.
Despite the apparently clear link between tubulin conformation and polymerization state, a robust connection between tubulin nucleotide state and tubulin dimer conformation has been difficult to establish. There is a continuing controversy over whether different nucleotide-dependent curvature states exist for tubulin dimers in solution (Barbier et al., 2010; Elie-Caille et al., 2007; Müller-Reichert et al., 1998; Nogales and Wang, 2006). An intermediate curvature state in alternative polymers formed at low temperatures with the non-hydrolysable analog GMPCPP (Wang and Nogales, 2005) has been proposed to correspond structurally to an assembly sheet intermediate observed during fast microtubule growth (Fig. 1B) (Chrétien et al., 1995). This led to the speculation that exchange of GDP for GTP causes unassembled tubulin to adopt a straighter conformation, compatible with polymerization. However, all high-resolution X-ray crystal structures of tubulin to date have captured the molecule in a bent conformation, irrespective of nucleotide state (Nawrotek et al., 2011), supporting a model wherein lattice contacts, rather than nucleotide, controls tubulin conformation (Aldaz et al., 2005). However, in all such structures, polymerization was suppressed by binding tubulin to a microtubule depolymerizer (Ayaz et al., 2012; Gigant et al., 2000; Prota et al., 2013) and/or to microtubule destabilizing drugs (Gigant et al., 2005; Ravelli et al., 2004), which are likely to intrinsically bias tubulin towards a bent conformation. The most direct information on unassembled tubulin in different nucleotide states has come from low resolution SAXS data obtained under extremely low magnesium conditions to inhibit polymerization, which was most consistent with a constitutively bent conformation (Rice et al., 2008). Otherwise, the solution conformation of individual tubulin dimers has been extrapolated from that of individual protofilaments (Hyman et al., 1992; Vale et al., 1994), aberrant assemblies, or inhibited states, none of which is devoid of shortcomings, highlighting the need for improved techniques to probe native tubulin and microtubule structure.
In order to probe the link between nucleotide state, tubulin conformation, and the stability of the microtubule lattice, we have generated cryo-EM reconstructions of microtubules composed of tubulin bound to GMPCPP, GDP, and GDP in the presence of taxol, at around 5 Å resolution, sufficient to generate pseudo-atomic structures in conjunction with Rosetta modeling. In addition to providing insight into how GTP hydrolysis leads to microtubule destabilization, our new methodology should be broadly applicable to future structural studies of microtubule-drug complexes and the interactions between Microtubule Associated Proteins (MAPs) and microtubules.
Results
High-resolution cryo-EM structures of dynamic and stabilized microtubules
In order to gain insight into the role of nucleotide state in microtubule structure and stability, we used cryo-electron microscopy (cryo-EM) (Figure S1, S2) to obtain structures of microtubules with significantly improved resolution from those previously reported. Prior cryo-EM studies of microtubules have been unable to achieve better than ~8 Å resolution, probably due to a mixture of technical issues related to the quality of the microscope, limitations in the image processing, or difficulties with microtubule structure preservation. To improve data quality, we collected our images on a latest-generation FEI 300 kV Titan electron microscope, where 3.7 Å resolution diffraction rings were visible in the power spectra of micrographs containing ice contamination (Figure S1B), and where reference-free class averages of microtubule segments showed signal to ~6.7 Å resolution (Figure S1C).
An additional limitation in prior cryo-EM reconstructions of naked microtubules (Sui and Downing, 2010), or microtubules decorated with the kinetochore complex Ndc80 (Alushin et al., 2010), which binds microtubules with a tubulin monomer repeat, was the inability of the image processing algorithms to distinguish the highly similar α- and β-tubulin subunits. This resulted in reconstructions stalling at about 8 Å resolution, at which point α- and β-tubulin become structurally distinct. To address this problem, we decorated microtubules with kinesin motor domain, which provides a low-resolution feature marking each tubulin dimer and aids in identifying the microtubule seam (the special lateral contact breaking helical symmetry with heterotypic interactions). While the kinesin signal drove initial alignments for our modified IHRSR (Iterative Helical Real Space Refinement) approach (Methods, Figure S2), the kinesin density was ultimately less ordered than tubulin and is excluded from display items.
Using this experimental strategy we have obtained significantly improved reconstructions of microtubules in two distinct nucleotide states: GTP-like (bound to GMPCPP) (Fig. 1, left) and GDP (dynamic microtubules in which GTP is hydrolyzed during assembly) (Fig. 1, center). Both structures are resolved to better than 5 Å (Fourier Shell Correlation (FSC) 0.143 criterion, Figure S2B), sufficient to visualize the bound nucleotides, all secondary structural elements (including individual β strands), and large aromatic side chains (Fig. 2B–C, Figure S4). We also report the structure of microtubules stabilized with taxol at 5.6 Å resolution (Fig. 1, right).
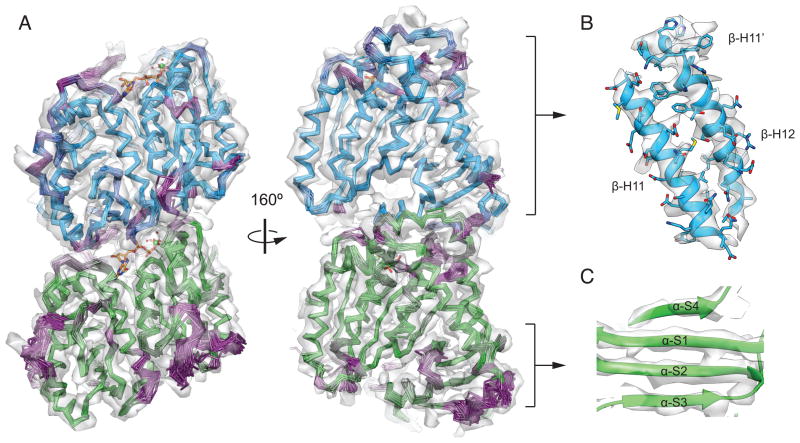
Figure 2. Rosetta Modeling of the GMPCPP Microtubule.
A) The low energy 1% GMPCPP ensemble is shown in cartoon representation, colored as in Fig. 1. Bound nucleotides are shown in stick representation and colored by heteroatom, as are magnesium ions (green) and coordinating water molecules. The map is displayed as a transparent grey isosurface. Regions of high-variability in the Rosetta ensemble (above an RMSF threshold of 0.89) are colored in shades of purple. B) β-tubulin C-terminal helices from the energy minimized consensus, all-atom Rosetta model, colored as in Fig. 1. Map is displayed as in A. C) Individual beta strands in the α-tubulin intermediate domain. (See also Tables S1b and S1c and Figures S3 and S4.)
Comparison of the GMPCPP and GDP reconstructions indicates a remodeling of the longitudinal interface between dimers corresponding to an axial dimer repeat change of 2.4% (from 83 to 81 Å) (Movie S1). A longer monomer repeat (averaging together α– and β–tubulin) for the GMPCPP vs. GDP lattice in the context of naked microtubules was reported almost twenty years ago (Hyman et al., 1995). Our study now demonstrates that the change corresponds to a compaction at the dimer interface, rather than a compression of the structure of each monomer. Importantly, we do not observe this dimer interface compaction when comparing the GMPCPP and GDP-Taxol reconstructions (Movie S2), suggesting that taxol binding allosterically affects remodeling of the interdimer interface. This finding is again in agreement with early studies showing that the monomer axial repeat in microtubules formed in the presence of taxol is increased with respect to GDP microtubules (Arnal and Wade, 1995). Reconstructions of kinesin-free microtubules demonstrated that the effect that we see for GMPCPP is also present in the absence of kinesin (although in this case α– and β-tubulin are indistinguishable) (Movie S3), in accord with the earlier results (Arnal and Wade, 1995; Hyman et al., 1995).
Rosetta full-atom modeling of the microtubule lattice
Previous atomistic models of microtubule structure have generally relied on rigid-body docking of the electron-crystallographic structure of tubulin into cryo-EM maps of the microtubule lattice that never surpassed 8 Å resolution (Li et al., 2002a; Maurer et al., 2012; Sindelar and Downing, 2010). While the structures reported here (4.7–5.6 Å) still falls short of true atomic resolution, tubulin features appear in much greater detail than ever reported before within a microtubule. To take full advantage of these features and produce a biochemically consistent atomic model of microtubule structure, we used a Rosetta protocol (DiMaio et al., 2009; Song et al., 2013) that utilizes the cryo-EM density map in structure refinement through the addition of an energy term representing model-map agreement to the standard Rosetta energy function (see Expanded Experimental Procedures). We used symmetry in energy calculations to model neighboring interactions in a 3 × 3 lattice segment. The final ensemble of microtubule models for each state, consisting of the 1% lowest energy models, is well-converged in terms of total energy and fit to the map (Fig. 2A and Figure S3A). For example, the average pair-wise RMSD within the GMPCPP ensemble is 0.52 Å. Regions of higher variability within any given ensemble (> 0.89 Å root mean squared fluctuation, RMSF, purple in Figure 2) correspond to loops or regions where density is weak and thus indicative of flexibility. The ensembles for the three microtubule states are distinct from each other, with average pair-wise RMSDs ranging from 0.86 Å to 1 Å (Figure S3B). To facilitate analysis, “consensus” models for each ensemble were computed by averaging the coordinates of ensemble members, followed by side-chain optimization and energy minimization.
Cross-correlation scores for each consensus model against each map had the expected pattern, with each model fitting its associated map best (Table S1b). We also compared each consensus model to both the 14pf GMPCPP map and an independently determined 13pf map of GMPCPP not used in the Rosetta refinement (Table S1c). The GMPCPP model fits both the 14pf GMPCPP map used for refinement and the 13pf GMPCPP map better than the consensus models for the other states (Table S1c), providing additional support that the refinement procedure captures structural features corresponding to nucleotide state rather than noise in the maps.
Remodeling of E-site structural elements
To gain further insight into the conformational differences between the GMPCPP and GDP models, and by extension, a GTP-like and GDP state of the microtubule lattice, we analyzed a single protofilament of two dimers from each model, superimposed at the indicated β-tubulin subunit (Fig. 3A). The overall RMSD between the GMPCPP and GDP models, computed from optimally aligned dimers, is 0.75 Å. Larger RMSD changes are observed within the α-subunit (0.85 Å) than within the β-subunit (0.63 Å). Interestingly, larger structural changes occur in the alpha subunit C-terminal half compared to the N-terminal half: 1 Å vs. 0.48 Å, whereas the difference between the N and C-terminal portions of the beta subunit exhibit similar differences of 0.62 Å RMSD. We next calculated difference vectors for the Cα positions, which represent the average differences between the two ensembles (Fig. 3B). Calculation of control vectors demonstrates that the pattern of displacements we observe are robust to the chosen frame of reference for superposition (i.e. α- or β-tubulin) (Fig. S5). The pattern of conformational changes is subtle but complex, and is most clearly visualized in morphs between the representative models of each state (Supplementary Movies 4–7).
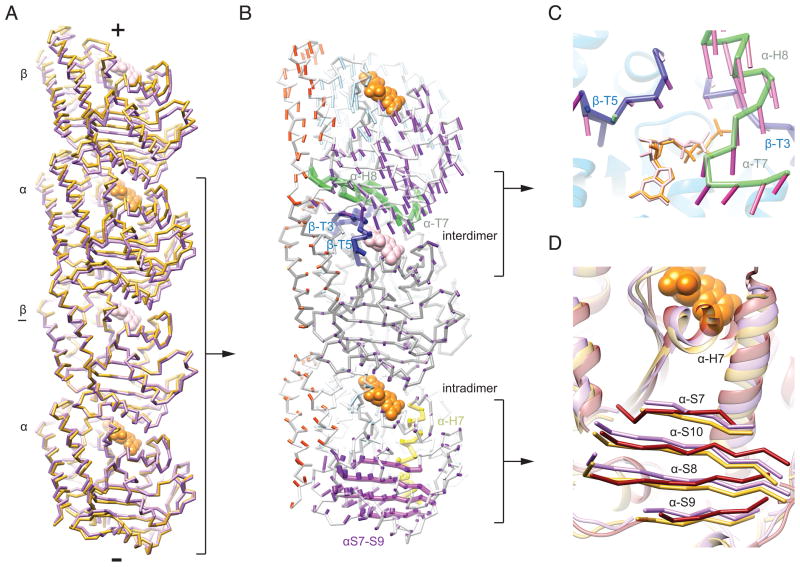
Figure 3. Hydrolysis results in a compression of the E-site at the interdimer interface.
A) Cα traces of two adjacent tubulin dimers from the GMPCPP (gold) and GDP (light purple) consensus models, superimposed at the underlined β tubulin. View is tangential to the microtubule lumen. Nucleotides from the consensus models are shown in orange (GTP) and pink (GDP). B) Displacement vectors between Cα coordinates from the consensus models of the GMPCPP state to the GDP state, superimposed as in A, are displayed as narrow cylinders. The chain-trace displayed corresponds to the GMPCPP consensus model: α-tubulin is light grey and β-tubulin dark grey. For clarity, vectors are only displayed for every other Cα pair. Vectors are colored by subdomain unless otherwise noted: N-terminal domain, light blue, intermediate domain, purple, C-terminal domain, red; vector length has been scaled 1.5 fold to aid visualization throughout. Selected structural elements along with associated vectors are highlighted: β-tubulin nucleotide binding loops, dark blue, α-tubulin T7-H8, green, α-tubulin intermediate domain beta sheet, purple, α-tubulin H7, yellow. Nucleotides are displayed as in A. C) View of the E-site structural unit, colored as in B. The RGB values of vector colors correspond to angular displacements relative to a Cartesian coordinate system, i.e. vectors of similar color point in a similar direction. D) View of the α-tubulin intermediate domain and H7 in the GMPCPP model (gold), GDP model (light purple), and 1SA0 (dark red), superimposed on the beta sheet of the α-tubulin N-terminal domain. (See also Figure S5 and Movies S1, S3, S4 and S5)
The overall compaction of the interdimer interface is clearly illustrated by the displacement vectors of the α-tubulin subunit at that interface (Fig. 3B). Although there are few internal rearrangements within the β-subunit, we observe a substantial remodeling of the T3, and T5 loops, which engage the E-site nucleotide at the interdimer interface (Fig. 3C; Movie S4). These β-tubulin loops move downwards towards the rest of the subunit, as well as inwards towards the microtubule lumen. Such motions had been predicted to accompany repositioning of the nucleotide following hydrolysis, based on comparisons of available tubulin and FtsZ structures (Aylett et al., 2011). Notably, our modeling shows that loop T3 fills in the cavity created upon loss of the γ-phosphate. We infer that this structural transition is coupled to the movement we observe for the T7 loop and helix H8 of the adjacent α subunit, which contains the presumed catalytic residue Glu254 (Lowe et al., 2001), as this region translates nearly 2 Å in a direction that matches the motion of the β-tubulin loops (Fig. 3C; Movie S4). Our vector analysis suggests that this α-T7-H8 region moves fairly independently from the remainder of the α-subunit (Fig. 3B), which, nevertheless, shows an overall translation towards the minus end of the lattice. We therefore propose that the β-tubulin T3 and T5 loops and the α-tubulin T7-H8 region act as a cohesive structural unit across the interdimer interface (Fig. 3B, blue), presumably coordinated by the E-site nucleotide.
Remodeling of longitudinal dimer contacts is coupled to conformational changes in α-tubulin
The observed changes at the interdimer contact, around the E-site, are accompanied by internal rearrangements of the tubulin dimer involving the intermediate and C-terminal domains of α-tubulin (Fig. 3B, bottom subunit). The RMSD between the intermediate domains from the GMPCPP and GDP consensus models (comprising strands S7–S10 and helices H9–10) when aligned on the N-terminal strands (S1–S5) shows a significant difference of 1.64 Å RMSD. The structural change is dominated by a translation of 1.28 Å towards the plus end, moving as a unit with the C-terminal base of H7, effectively tucking up into the dimer (Fig. 3D).
Conformational differences between the electron crystallographic structure of tubulin, corresponding to a straight, protofilament-like state, and X-ray crystallographic structures of curved, peel-like tubulin states, differ most significantly by a rotation of the intermediate domain and a translation of H7 (Ravelli et al., 2004;(Gigant et al., 2000; Nogales and Wang, 2006). Comparing α-tubulin from the crystal structure of bent tubulin bound to a stathmin-like domain (PDB:1SA0, (Ravelli et al., 2004)) with the GMPCPP and GDP models, indicates that upon hydrolysis the α-tubulin intermediate domain within the microtubule undergoes a shift similar to that reported for the straight to bent transition, but no rotation, likely due to the lattice constraints (Fig. 3D and Movie S5). This result is consistent with a model wherein reorganization of the E-site following GTP hydrolysis generates internal strain within the tubulin dimer that would subsequently relax during depolymerization, a long-standing concept in the tubulin field (Caplow et al., 1994). Interestingly, while in the stathmin-bound structure the structural changes within the β-subunit are even larger than those observed in the α-subunit, within the microtubule the nucleotide has a much smaller effect on this region of β-tubulin, with no appreciable translation or rotation.
Interplay between nucleotide state and MAP binding sites
We additionally observe hydrolysis-induced changes that propagate outwards to the microtubule surface, where the vast majority of MAPs and motors interact with tubulin (Al-Bassam et al., 2002; Alushin et al., 2012; Redwine et al., 2012; Sindelar and Downing, 2010) (Fig. 4 and Movie S6). Helices H11 and H12 in α and β subunits translate 1.1 Å and 0.53 Å, respectively, away from the lumen upon GTP hydrolysis. For the α subunit, where changes are most substantial, vector analysis demonstrates that the movement of the C-terminal helices parallels the rearrangement we observe in the intermediate domain, suggesting the intermediate domain transmits rearrangements in the E-site to the microtubule surface (Figure 3B, Movie S6). The rearrangement of αH12 is of particular importance, as this helix constitutes part of the binding site for the kinesin and cytoplasmic dynein motors (Figure 4) (Al-Bassam et al., 2002; Redwine et al., 2012; Sindelar and Downing, 2010). This finding may explain the observation that kinesin preferentially binds to GMPCPP microtubules over destabilized GDP microtubules (Nakata et al., 2011).
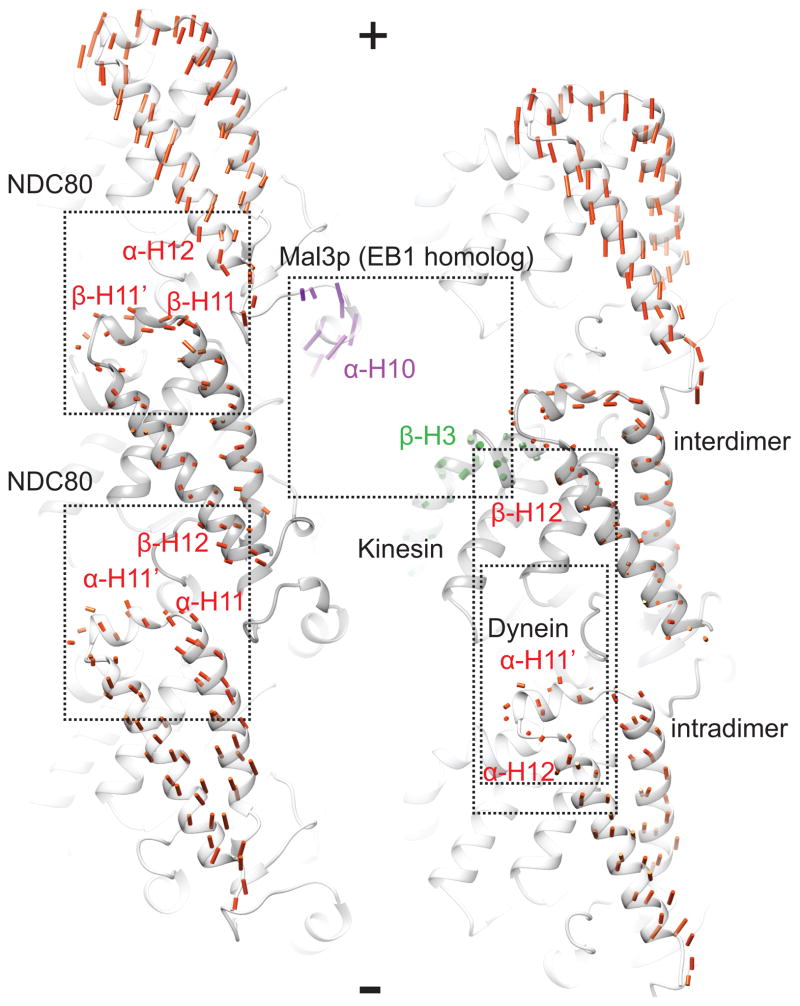
Figure 4. Rearrangements upon hydrolysis alter MAP binding sites on the microtubule surface.
Rearrangements on the microtubule surface. Vectors are colored by subdomain as in Fig. 3. Binding sites for key MAPs are indicated. (See also Movie S6)
Recent reports indicate that GTPγS stabilized microtubules are an optimal structural mimic of the cap recognized by the plus-end tracking factor end-binding protein 1 (EB1, Mal3p in fission yeast) (Maurer et al., 2011; Maurer et al., 2012). Maurer et al. reported an additional lateral contact present in the GTPγS microtubule with respect to others previously visualized, involving the β-tubulin H3 helix, a region closely apposed to the E-site γ-phosphate. As this helix was found to be a component of the Mal3p binding site on the microtubule, they proposed that this extra contact could be a key mechanism for plus-tip recognition (Maurer et al., 2012). In our studies we observe minimal rearrangements in this region (Fig. 4B, green), which is never involved in lateral contacts. We do not believe that this structural disparity is due to a difference in non-hydrolyzable nucleotide, but likely to the difference in resolution of the two studies. On the other hand, we observe a substantial nucleotide state-dependent displacement of α-tubulin H10 and the S9-H10 loop in the intermediate domain (Fig. 4B, purple; Movie S6), which were also identified as a major component of the Mal3p binding site. We therefore propose that rearrangement of the α-tubulin intermediate domain could play a significant role in plus-tip recognition.
Taxol binding restores the GDP lattice to a GMPCPP-like state
Superposition of our GMPCPP and GDP-taxol models demonstrates that, remarkably, taxol binding globally reverses the majority of the conformational changes we observe when comparing the GMPCPP and GDP states (Fig. 5A). We do not observe any major conformational differences between the GMPCPP and GDP-taxol states, such as rearrangement of tubulin subdomains or formation of a second layer of lateral contacts in the GMPCPP state, as was reported in a recent cryo-EM study (Yajima et al., 2012). Possible explanations for these discrepancies include the different symmetries of the microtubules chosen for processing (15 PF 4-start helical microtubules vs. 14 PF 3-start microtubules in this study), image analysis procedures, the presence of bound kinesin in our work, and the different resolutions. The restoration of a GMPCPP-like state by taxol extends to the MAP-interacting regions of the C-terminal domain (Fig. 5A), suggesting that caution should be employed when considering taxol-bound microtubules to be in a GDP-like state in studies of MAP-microtubule interactions.
Figure 5. Taxol binding restores the GDP lattice to a GMPCPP-like extended state.
A) Analogous to Fig. 2A, but comparing the GMPCPP (gold) to the GDP–taxol (light blue) state. B) View from the microtubule lumen of the superimposed GDP (light purple) and GDP–taxol (light blue) models. Note the swelling of the taxol binding site (1), opening of the E-site interface (2), and reversal of the longitudinal displacement of the α-tubulin intermediate domain (3). Nucleotides from the GDP-taxol consensus models are displayed and colored as in Fig. 3. Taxol is yellow. (See also Figure S6 and Movies S2 and S7)
Taxol binds distal to the E-site nucleotide, at the interface between the nucleotide binding and intermediate domains on the luminal face of β-tubulin (Nogales et al., 1998) (Fig. 1C). The global remodeling we observe with respect to the GDP structure suggests that taxol is capable of allosterically reversing the conformational transition accompanying GTP hydrolysis, thus leading to an extended state at the E-site and a reversal in the movement of the intermediate domain in α-tubulin (Fig. 5B and Movie S7). The T3 and T5 nucleotide binding loops do not return to a GTP-like conformation, suggesting that the E-site is opened by an alternative mechanism. In order to be accommodated in the β-subunit, taxol acts as a wedge to increase the volume of the taxol-binding pocket, which swells from 883 Å3 in the GDP model to 996 Å3 in the GDP-taxol model (Figure 5B). The volume of the pocket is 914 Å3 in the GMPCPP model, suggesting that most of the volume increase is a specific effect of taxol binding. This expansion results from slight rearrangements around the taxol binding site (Fig. 5B), which we infer to be allosterically linked to expansion of the E-site and reversal of the longitudinal displacement of the α-tubulin intermediate domain. Vector analysis comparing GDP vs. GDP-taxol and GMPCPP vs. GDP-taxol suggests that these rearrangements are specifically generated by taxol binding (Figure S6).
The electron crystallographic structure of zinc-induced sheets, 1JFF, was obtained in the stabilizing presence of taxol. Intriguingly, the axial repeat in 1JFF is similar to that of the GDP microtubule (81.2 vs. 81.3 Å). On the other hand, comparison of the tubulin dimer structures shows that both α– and β-tubulin subunits in 1JFF are more dissimilar to any of the three microtubule-based structures we described here, than the three structures are to each other (Table S1d). The volume of the taxol binding pocket is also slightly smaller in the zinc sheets (956 Å3), an observation that, taken together with the different lateral contacts present in this polymer, suggests that taxol may stabilize sheets by an alternative mechanism (there are no available sheet structures in the absence of a stabilizing drug). These differences indicate that physiological lateral contacts are necessary for the allosteric effect of Taxol at the E-site, highlighting the importance of studying the mechanisms of microtubule stabilizing agents in a microtubule context. Future studies of microtubules at higher resolution will be required to uncover the detailed allosteric mechanism by which taxol restores the microtubule lattice to a GTP-like state.
Lateral contacts are not significantly affected by nucleotide state or drug binding
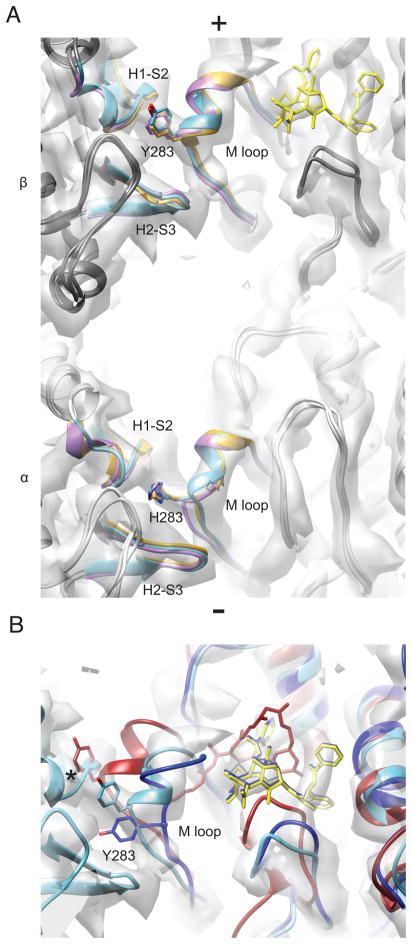
There is a single major point of interaction that defines the lateral interface between protofilaments. The H1-S2 and H2-S3 loops, components of the nucleotide binding domain, interact with a component of the intermediate domain known as the M-loop (S7-H9 loop) across the interface (Fig. 6A). This segment adopts a helical conformation in both α- and β-tubulin in all three states we examined, resembling that of a crystallographic study of tubulin bound to the microtubule stabilizing agent zampanolide (Prota et al., 2013). The position of β-tubulin Tyrosine 283 is well-defined and consistently buried at the interface, suggesting that this residue plays a key role in the lateral interaction. Although tyrosine is also found at the equivalent position in α-tubulin, the next residue in the sequence, Histidine 271, occupies the bridging density.
Figure 6. Lateral contacts are highly similar between stable and unstable states.
View from the microtubule lumen of homotypic inter-protofilament lateral contacts. The GDP–taxol map is displayed as a transparent grey isosurface. A) α-tubulin is colored light grey, β-tubulin, dark grey. Consensus models are displayed with lateral contacts colored (GMPCPP, gold, GDP, light purple, and GDP–taxol, light blue). Key residues mediating lateral contacts are displayed in stick representation, as is taxol. Taxol and its associated density are colored yellow. B) Superpositions of β-tubulin subunits from the electron crystallographic structure of taxol-bound tubulin (PDB 1JFF, dark blue)(Lowe et al., 2001) the structure of unassembled, inhibited zampanolide-bound tubulin (PDB 4I4T, dark red) (Prota et al., 2013), and the GDP-taxol model from this study (light blue, taxol is yellow). Note that Y283 is only in position to mediate the lateral contact in the GDP-taxol model, and that a clash is present with the laterally adjacent subunit in the zampanolide model (asterisk).
The peeling of protofilaments at depolymerizing ends (Mandelkow et al., 1991), and the fact that lateral contacts are mediated by loop-loop interactions burying a small surface area (Li et al., 2002b; Sui and Downing, 2010), led to the assumption that lateral interactions are labile and thus most likely to be affected by nucleotide hydrolysis. Previous computational studies (Grafmuller et al., 2013) supported this hypothesis, proposing that GTP hydrolysis results in an ordered-to-disordered transition in the M-loop.
In contradiction with this model, we observe minimal differences in the lateral interfaces between the GMPCPP, GDP, and GDP-taxol representative models (Fig. 6A). This finding argues against a substantial weakening of lateral contacts upon nucleotide hydrolysis being the major determinant of microtubule destabilization. Additionally, our observation also suggests that stabilization of a particular M-loop conformation may not be the major mechanism by which drugs that bind to the taxol site stabilize the microtubule lattice as originally proposed (Nogales et al., 1999). This conclusion is further supported by the considerable difference in the position and conformation of the M-loop in our microtubule structures and the extended conformation present in the electron crystallographic structure of GDP taxol zinc-induced sheets (Lowe et al., 2001), where taxol also had a stabilizing effect (Nogales et al., 1995). This difference is even more pronounced when comparing our present microtubule structures with that of the recent X-ray crystallography atomic model of zampanolide-bound, unassembled tubulin (PDB:4I4T, (Prota et al., 2013)) where the helical M loop is in a position that would clash with the laterally adjacent protofilament (Fig. 6B). Rather, our data are most consistent with a model wherein the microtubule stabilizing effect of taxol concerns the structure and strength of longitudinal interfaces linked to a GTP-like conformation of the tubulin dimer, as originally envisioned by Amos and Lowe (Amos and Lowe, 1999).
Discussion
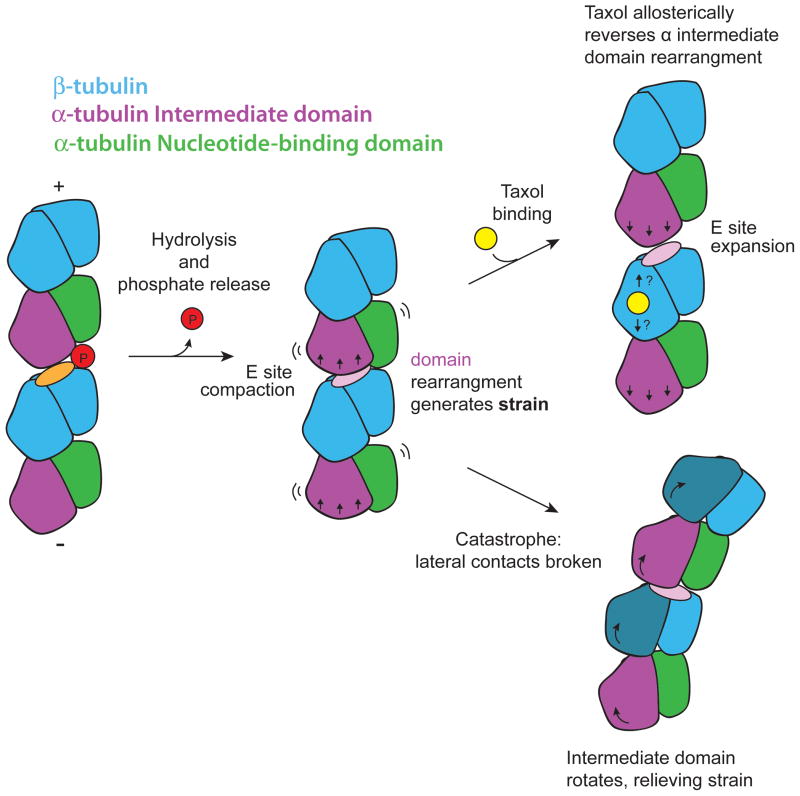
Our results suggest the mechanism by which nucleotide hydrolysis is linked to destabilization of the microtubule lattice (Fig. 7). We propose that upon hydrolysis and phosphate release, compaction of the E-site leads to an energetically unfavorable longitudinal translation of the α-tubulin intermediate domain, including the H7 helix. The observed rearrangements of tubulin upon hydrolysis are reminiscent of the conformational trajectory towards a depolymerized, bent tubulin structure (Ravelli et al., 2004). In the context of the microtubule, only the translational component of that trajectory is present, while the rotational component is not apparent. This difference is likely due to lattice constraints that maintain a strained, straight conformation compatible with the microtubule lattice. Thus, we propose that hydrolysis leads to conformational strain that would be released by bending during depolymerization. This model is consistent with the changes we observe upon taxol binding, a stabilizer of the GDP lattice. The predominant effect of taxol is to restore the longitudinal interface and the α-tubulin intermediate domain to a GTP-like state, suggesting that these structural transitions are responsible for destabilization of the microtubule lattice upon nucleotide hydrolysis.
Figure 7. Proposed model of destabilizing and stabilizing structural transitions in the microtubule lattice.
Cartoon of conformational transitions colored as in Fig. 1, except the α-tubulin intermediate domain is purple. Left, nucleotide hydrolysis and phosphate release leads to compaction of the E-site and rearrangement of the α-tubulin intermediate domain (middle), generating destabilizing strain while tubulin remains within the constraints of the microtubule lattice. Taxol binding (right, top), allosterically leads to a reversal of E-site compaction and the α-tubulin rearrangement; unraveling the detailed mechanism of this transition will require structural analysis at near-atomic resolution. Subtle structural changes could be propagated across the E-site interdimer interface (up arrow), within the dimer (down arrow), or both. In the absence of binding by a stabilizing agent, strain would be dissipated by tubulin bending during catastrophe (right, bottom), when the α-tubulin intermediate domain (and β-tubulin intermediate domain, dark blue) are capable of undergoing rotation due to the relief of steric constraints imposed by lateral contacts.
The substantial changes we have visualized by comparing GMPCPP, GDP, and GDP-taxol microtubules contrast with the minimal effect of nucleotide state on the inhibited form of tubulin previously described by X-ray crystallographic studies. In the context of an RB3-bound, bent tubulin dimer, the presence of a gamma phosphate at the E-site affected the position of merely two amino acids adjacent to the site, rearrangements which were not further propagated throughout the structure (Nawrotek et al., 2011). This suggests that the inhibition of tubulin for crystallization purposes by cellular factors that bind and stabilize its bent conformation can override the conformational effects of nucleotide hydrolysis and stabilizing drugs that otherwise occur within a microtubule.
More recently, the structure of the RB3-inhibited tubulin bound to epothilone A or zampanolide showed the ordering of the adjacent M-loop region (otherwise unresolved in previous crystal structures) into a small helix, without any other significant changes. This structure lead to the proposal that inducing the folded conformation of the β-tubulin M-loop was the primary mechanism by which antimitotic agents that bind at the taxol site stabilize the microtubule lattice, i.e. by facilitating lateral contacts (Prota et al., 2013). However, the position of the M-loop region in the context of a rotated intermediate domain clashes with the neighboring lateral subunit when this crystal structure is placed in the microtubule lattice. Our finding that lateral contacts are not substantially altered in different nucleotide states or upon taxol binding suggests that this set of tubulin polymerization interfaces plays a more passive role in microtubule stability than anticipated. Our results instead support a model in which the energy contribution of lateral contacts may be essentially constant, while longitudinal contacts and conformational strain within the tubulin monomers (particularly α-tubulin) are modulated.
It is possible that the folding of the M-loop observed in the crystal structure of zampanolide/epothilone A-bound tubulin could facilitate the incorporation of tubulin dimers into the microtubule, and thus be a component of the assembly-driving action of taxol-like compounds. Once assembled, however, the stability of the microtubule lattice is likely governed by the total energy of lateral and longitudinal contacts in the GTP state vs. the GDP state. Our analysis supports a model in which microtubule-stabilizing agents like taxol primarily exert a stabilizing influence on the microtubule lattice by affecting internal rearrangements of the tubulin dimer that modulate conformational strain and longitudinal contacts. This concept is consistent with a study demonstrating that taxol induces the formation of straight individual protofilaments from GDP-tubulin peels (Elie-Caille et al., 2007). In particular, our studies suggest that modulating longitudinal interfaces by altering the conformation of β-tubulin is a robust mechanism for affecting microtubule stability, as previously predicted (Amos and Lowe, 1999), a model that should aid future design efforts for drugs that target the taxol binding site.
Future Prospects
Our study demonstrates that microtubules are structurally tractable by cryo-EM at resolutions approaching those obtained by X-ray crystallography, thus enabling the detailed study of nucleotide and drug effects and the interaction of microtubule binding proteins, in the context of the native tubulin polymer. The three microtubule models we present here should provide an important resource for computational modeling studies of dynamic instability, effects of drug binding, and the physical properties of microtubules.
Several outstanding questions and concerns remain to be addressed. The presence of kinesin, currently a technical necessity, may alter certain structural transitions occurring between different states. Indeed, our reconstructions of kinesin-free microtubules suggest that the presence of kinesin slightly dampens the magnitude of E-site remodeling (Movie S3). We optimistically anticipate that future methodological advances in image processing and instrumentation will allow similar and hopefully even superior resolution to be achieved for undecorated microtubule specimens.
Additionally, caution must be employed when interpreting the structural details of GMPCPP-bound microtubules as the bona fide “GTP state” of the microtubule lattice, in light of the differences observed with different GTP analogs. One potential explanation for such differences is that there are additional structural stages in the microtubule polymerization-depolymerization cycle, such as one corresponding to a post-hydrolysis state, but before Pi release, that may be mimicked by one of these GTP analogs. Tubulin could also adopt several distinct conformations prior to GTP hydrolysis (related perhaps to the transition from an open growth sheet to the closed microtubule cylinder), which different analogs may selectively stabilize. Future high-resolution studies of microtubules bound to GTPγS and other nucleotide state mimics, like the GDP-Pi analog GDP-BeF3, will be important to investigate and address these issues.
Finally, although our present structural analysis leads to the unexpected conclusion that lateral contacts are not substantially altered upon nucleotide hydrolysis, we cannot rule out the possibility of more subtle effects that could nevertheless contribute significantly to energetics, and which would only be revealed at higher, i.e. atomic, resolution. Additionally, heterotypic lateral contacts across the seam, where tubulin subunits within a single dimer on one side of the interface are contacting subunits contributed by two dimers on the other, are more likely to be affected by the change in dimer repeat upon remodeling of the E-site with GTP hydrolysis. The seam could therefore constitute a “weak point” to initiate disruption of the lattice at the onset of catastrophe, consistent with early observations of microtubule conversion to sheets when depolymerization is induced by cold treatment (Simon and Salmon, 1990). A mismatch of axial repeats will not only occur at the seam of microtubule segments with a homogeneous nucleotide state, but would also be present at homotypic lateral contacts in a transition zone along the microtubule lattice where hydrolysis is taking place and therefore containing a mixture of GTP and GDP states. While a mixed nucleotide lattice is not amenable to experimental analysis by current structural methods, which require averaging of microtubule segments, computational modeling of these transitions, guided by the structural findings described here, could contribute to our molecular understanding of the catastrophe process. In particular, it would be interesting to explore potential cooperativity in the structural transitions we have described, which seems likely given the steric constraints imposed by the tightly packed microtubule lattice.
Experimental Procedures
Full details of experimental procedures are presented in the Extended Experimental Procedures.
Kinesin and microtubule preparation for cryo-EM
Kinesin was diluted into EM buffer (for buffer compositions see Extended Experimental Procedures) and desalted. ATP was added and aggregates removed by ultracentrifugation. Taxol-stabilized GDP MTs were prepared as previously described (Alushin et al., 2010). GMPCPP microtubules were prepared by polymerizing porcine tubulin in CB1 buffer. The MTs were pelleted and resuspended in cold EM buffer, generating a solution of GDP-tubulin. After clarification, GMPCPP was exchanged for bound GDP for one hour on ice. GMPCPP-MTs were then polymerized in EM buffer at 37°C. Dynamic (primarily GDP) microtubules were polymerized in CB1 buffer at 37°C for 30 minutes, then pelleted.
All cryo-EM samples were prepared on glow-discharged C-flat grids (Protochips, Inc) using a Vitrobot (Maastricht Instruments). Taxol and GMPCPP microtubules were diluted to 0.25 mg/ml in EM buffer and 4 μl was applied to the grid. The MTs were allowed to adsorb and kinesin was then added to the grid. After a short incubation the sample was blotted and plunged into liquid ethane. Pelleted dynamic GDP-microtubules were resuspended into warm kinesin solution. 4 μl of this MT-kinesin mixture was added to the EM grid and allowed to incubate for 1 minute before blotting and vitrification.
Electron Microscopy and Image processing
Cryo grids were loaded into a 626 single tilt cryo-transfer system (Gatan) and imaged in a Titan electron microscope (FEI) operated at 300 keV. Data was collected using the Leginon software (Suloway et al., 2005) for exposure targeting, and the FEI Low-dose kit for focusing and image acquisition. Low-dose exposures (25 e−/Å2) were acquired at 72,000X magnification on Kodak SO163 film, with underfocus ranging from 1.4 to 3.5 μm. Films were digitized at 0.87 Å/pixel using a Nikon CoolScan 8000 (see Table S1a for number of films digitized for each sample). Image processing was performed within the Appion processing environment (Lander et al., 2009). Contrast transfer function (CTF) was estimated and the best quality micrographs selected for further processing. Microtubules were manually selected and overlapping segments were extracted with a spacing of 80Å, phase-flipped and binned by a factor of 2. The particle stacks were subjected to iterative multivariate statistical analysis (MSA) and multi-reference alignment (MRA). Particles in classes that did not clearly show kinesin density (for kinesin-bound samples), or whose power spectra did not exhibit layer lines beyond 10 Å, were excluded from further processing, as were classes that did not have either 13 or 14 protofilaments.
Three-dimensional reconstruction
Undecorated 13 and 14 pf MT densities were used as initial models for a preliminary reconstruction of the kinesin-bound taxol MT dataset. The protofilament with the best apparent kinesin density was extracted and used to generate 13 and 14 pf microtubule maps with seams. These preliminary densities served as starting models for refinement of complete datasets using IHRSR (Egelman, 2007). Our modified IHRSR reconstruction schema (Figure S2) allows us to take advantage of the helical character of the microtubule lattice, without artifacts due to seam symmetrization. A final refinement of the alignment parameters was performed only for the 14pf MT particles using FREALIGN (Grigorieff, 2007) with a modified script that incorporated the symmetrization and MT seam regeneration after each round. The final resolution for each reconstruction was estimated by calculating the Fourier Shell Correlation (FSC) of a single dimer (Supplementary Figure 2B). Resolution-dependent negative b-factors were applied to the three reconstructions using BFACTOR (http://grigoriefflab.janelia.org/bfactor).
Atomic model building and refinement with Rosetta
Initial models of the microtubule lattice were obtained through rigid-body docking the electron crystallographic structure of tubulin (PDB:1JFF (Lowe et al., 2001)) into the cryo-EM density maps using Chimera. Ligand conformations were copied from the corresponding crystal structures and were held fixed during refinement. The lattice was modeled in a 3 × 3 arrangement using symmetry operations as previously described (DiMaio et al., 2011).
Structure refinement was carried out using the RosettaCM protocol (Song et al., 2013) supplemented with an electron density term (DiMaio et al, 2009). Regions in the starting 1JFF structure which fit the density poorly were rebuilt using short fragments from proteins of known structure with similar local sequences, and recombination with X-ray crystallographic tubulin structures at higher resolution in different nucleotide states was used to increase accuracy in the vicinity of the nucleotide binding sites (see Supplemental Materials for details). Following loop region rebuilding and structure hybridization, models were relaxed in the Rosetta all atom forcefield supplemented with the electron density term. Analyses in the text are based on the lowest 1% total energy (Rosetta energy + electron density fit) models for each map. The spread of energies and RMSDs of these low-energy ensembles was small (average 0.56 Å RMSD).
Molecular graphics
All structural figures and movies were generated with UCSF Chimera (Goddard et al., 2007; Pettersen et al., 2004). Displacement vectors were generated with a Python program that creates marker (.cmm) files that are viewable and editable in Chimera (available upon request).
Supplementary Material
Research Highlights.
Microtubule structures at 5Å illustrate the effect of hydrolysis and drug binding.
The longitudinal interface by the E-site nucleotide compacts upon GTP hydrolysis.
α-tubulin’s intermediate domain and helix H7 move, generating strain in the dimer.
Taxol inhibits these changes, allosterically inducing a GTP-like state.
Acknowledgments
We thank Patricia Grob and Tom Houweling for electron microscopy and computer support, respectively, Tom Goddard for help with UCSF Chimera, Yifan Song and Frank Dimaio for making code available for use prior to publication, and Stuart Howes for assistance with kinesin purification. We are grateful to Robert Glaeser for assistance with data collection and many discussions. The kinesin expression construct was the gift of Erik Jonsson and Ron Vale. This work was funded by NIGMS (GM051487 to E.N), and by a Damon Runyon Cancer Research Foundation fellowship (to G.C.L., DRG 2055-10). E.N. and D.B. are Howard Hughes Medical Institute Investigators.
Footnotes
Accession Numbers
The cryo-EM maps and coordinates of the energy minimized consensus Rosetta models have been deposited for the three microtubules studies we describe: GMPCPP-stabilized microtubule (EMDB:5895; PDB:3J6E), GDP, dynamic microtubule (EMDB:5896; PDB:36JF) and taxol stabilized microtubule (EMDB:5897; PDB:3J6G)
Author contributions
G.M.A. and G.C.L. performed sample preparation, electron microscopy experiments and data processing. E.H.K. performed Rosetta modeling analysis. R.Z. performed data processing. G.M.A. wrote the initial draft of the paper and E.H.K. generated supplemental movies. All authors contributed to data analysis and assembling the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol. 2002;157:1187–1196. doi: 10.1083/jcb.200201048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaz H, Rice LM, Stearns T, Agard DA. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005;435:523–527. doi: 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA, Lowe J. How Taxol (R) stabilises microtubule structure. Chemistry & Biology. 1999;V6:R65–R69. doi: 10.1016/s1074-5521(99)89002-4. [DOI] [PubMed] [Google Scholar]
- Arnal I, Wade RH. How does taxol stabilize microtubules? Curr Biol. 1995;5:900–908. doi: 10.1016/s0960-9822(95)00180-1. [DOI] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Lowe J, Amos LA. New insights into the mechanisms of cytomotive actin and tubulin filaments. Int Rev Cell Mol Biol. 2011;292:1–71. doi: 10.1016/B978-0-12-386033-0.00001-3. [DOI] [PubMed] [Google Scholar]
- Barbier P, Dorleans A, Devred F, Sanz L, Allegro D, Alfonso C, Knossow M, Peyrot V, Andreu JM. Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers. J Biol Chem. 2010 doi: 10.1074/jbc.M110.141929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplow M, Ruhlen RL, Shanks J. The free energy of hydrolysis of a microtubule-bound nucleotide triphosphate is near zero: all of the free energy for hydrolysis is stored in the microtubule lattice. J Cell Biol. 1994;127:779–788. doi: 10.1083/jcb.127.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Ann Rev Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Leaver-Fay A, Bradley P, Baker D, Andre I. Modeling symmetric macromolecular structures in Rosetta3. PLoS One. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Elie-Caille C, Severin F, Helenius J, Howard J, Muller DJ, Hyman AA. Straight GDP-tubulin protofilaments form in the presence of taxol. Curr Biol. 2007;17:1765–1770. doi: 10.1016/j.cub.2007.08.063. [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M. The 4 angstrom X-ray structure of a tubulin: stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Grafmuller A, Noya EG, Voth GA. Nucleotide-dependent lateral and longitudinal interactions in microtubules. J Mol Biol. 2013;425:2232–2246. doi: 10.1016/j.jmb.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Hyams JS, Lloyd CW. Microtubules. New York: Wiley-Liss; 1993. [Google Scholar]
- Hyman AA, Chretien D, Arnal I, Wade RH. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, DeRosier D, Nogales E, Downing KH. Structure of the microtubule at 8 Å resolution. Structure. 2002a;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 A resolution. Structure. 2002b;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta- tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubules dynamics and microtubules caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci U S A. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Volkov V, Ataullakhanov FI, Grishchuk EL. Tubulin depolymerization may be an ancient biological motor. J Cell Sci. 2010;123:3425–3434. doi: 10.1242/jcs.067611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez M, Rivas G, Diaz JF, Andreu JM. Control of the structural stability of the tubulin dimer by one high affinity bound magnesium ion at nucleotide N-site. J Biol Chem. 1998;273:167–176. doi: 10.1074/jbc.273.1.167. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Localization of an exchangeable GTP binding site at the plus end of microtubules. Science. 1993;261:1044–1047. doi: 10.1126/science.8102497. [DOI] [PubMed] [Google Scholar]
- Müller-Reichert T, Chrétien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: Information froma slowly hydrolyzable analogue of GTP, guanylyl (α,β)methylenediphosphonate. Proc Natl Acad Sci USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Niwa S, Okada Y, Perez F, Hirokawa N. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J Cell Biol. 2011;194:245–255. doi: 10.1083/jcb.201104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek A, Knossow M, Gigant B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J Mol Biol. 2011;412:35–42. doi: 10.1016/j.jmb.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Nettles JH, Li H, Cornett B, Krahn JM, Snyder JP, Downing KH. The binding mode of epothilone A on alpha,beta-tubulin by electron crystallography. Science. 2004;305:866–869. doi: 10.1126/science.1099190. [DOI] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Ann Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wang HW. Structural mechanisms underlying nucleotide- dependent self-assembly of tubulin and its relatives. Curr Opin Struct Biol. 2006 doi: 10.1016/j.sbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High resolution structure of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Zhang SX, Downing KH. Preservation of 2-D crystals of tubulin for electron crystallography. J Struc Biol. 1995;115:199–208. doi: 10.1006/jsbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Zurwerra D, Field JJ, Diaz JF, Altmann KH, Steinmetz MO. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci U S A. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JR, Salmon ED. The structure of microtubule ends during the elongation and shortening phases of dynamic instability examined by negative-stain electron microscopy. J Cell Sci. 1990;96(Pt 4):571–582. doi: 10.1242/jcs.96.4.571. [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH. Structural Basis of Interprotofilament Interaction and Lateral Deformation of Microtubules. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Vale RD, Coppin CM, Malik F, Kull FJ, Milligan RA. Tubulin GTP hydrolysis influences the structure, mechanical properties, and kinesin-driven transport of microtubules. J Biol Chem. 1994;269:23769–23775. [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H, Ogura T, Nitta R, Okada Y, Sato C, Hirokawa N. Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J Cell Biol. 2012;198:315–322. doi: 10.1083/jcb.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.