Abstract
The term nanodisk (ND) describes reconstituted high-density lipoprotein particles that contain one or more exogenous bioactive agents. In the present study ND were assembled from apolipoprotein A-I, the zwitterionic glycerophospholipid, dimyristoylphosphatidylcholine (DMPC) and the synthetic cationic lipid, dimyristoyltrimethylaminopropane (DMTAP). ND formulated at a 70:30 weight ratio of DMPC to DMTAP were soluble in aqueous media. The particles generated were polydisperse, with diameters ranging from ~20 to <50 nm. In nucleic acid binding studies, agarose gel retardation assays revealed that a synthetic 23mer double-stranded oligonucleotide (dsOligo) bound to DMTAP containing ND but not to ND formulated with DMPC alone. Sucrose density gradient ultracentrifugation studies provided additional evidence for stable dsOligo binding to DMTAP-ND. Incubation of cultured hepatoma cells with DMTAP-ND complexed with a small interfering (si) RNA directed against glyceraldehyde 3-phosphate dehydrogenase showed 60 % knockdown efficiency. Thus, incorporation of synthetic cationic lipid (i.e. DMTAP) to ND confers an ability to bind siRNA and the resulting complexes possess target gene knockdown activity in a cultured cell model.
Keywords: Nanodisk, dimyristoyltrimethylaminopropane, cationic lipid, siRNA, apolipoprotein, delivery and HepG2 cells
Introduction
Nanodisks (ND) exist as biocompatible, ternary complexes of phospholipid, scaffold protein and an exogenous bioactive agent (Ryan, 2008; 2010). The scaffold component, usually a member of the class of amphipathic apolipoproteins, induces ND self-assembly, ultimately circumscribing the perimeter of a disk-shaped lipid bilayer, shielding otherwise exposed fatty acyl chains at the edge of the particle (Ryan, 2008). ND technology has been applied to solubilization and delivery of small hydrophobic bioactive agents including the polyene antibiotic, amphotericin B (Oda et al., 2006), the isoprenoid, all trans retinoic acid (Singh et al., 2010) and the plant-derived polyphenol, curcumin (Ghosh et al., 2011). In addition, ND technology has been used to solubilize transmembrane spanning proteins in a native-like membrane environment (Bayburt and Sligar, 2010).
In the present study we sought to adapt ND technology to binding and transport of nucleic acid, specifically small interfering (si) RNA. siRNAs are comprised of 21–23mer ribonucleotide duplexes. One of the strands, termed the guide strand, is complementary to that of the target mRNA (Bernstein et al., 2001; Martinez et al., 2002). In the cell cytoplasm siRNA interacts with proteins to assemble a multiprotein-RNA complex, the RNA-induced silencing complex (RISC). The assembled RISC uses the guide strand of siRNA to specifically cleave the target mRNA, thereby preventing its translation. The potential therapeutic utility of RNA interference can be seen by the fact that siRNA-mediated silencing of validated disease targets has been shown to improve outcomes in disease models (Hu-Leiskovan et al., 2005; Landen et al., 2005; Zimmerman et al., 2006). Despite the promise of this technology, significant obstacles related to in vivo delivery of siRNA are yet to be overcome.
siRNA is unstable in vivo due to the ubiquitous presence of RNAses. As such, systemic administration of naked siRNA results in rapid degradation and renal clearance. Numerous strategies (Kanasty et al., 2013) have been pursued to overcome this problem, including chemical modification of siRNA (DePaula et al., 2007; Wada et al., 2012) and development of viral delivery strategies (McCaffrey et al., 2002; Zou et al., 2008). Although viral vectors provide an efficient delivery option, concerns persist regarding host immune response (Miele et al., 2012). Numerous non-viral strategies have been pursued including the use of peptides (Andaloussi et al., 2011), aptamers (Li et al., 2013), antibody-protamine chimeras (Song et al., 2005; Dou et al., 2012), cationic lipids (Morrissey et al., 2005; Sørensen and Sioud, 2010; Semple et al., 2010) and cationic polymers (Urban-Klein et al., 2005; Tachibana et al., 2014) as siRNA carriers. Cationic lipid-based stable nucleic acid particles (Barros and Gollob, 2012) and cyclodextrin nanoparticles (Davis et al., 2010) are currently in clinical trial as potential siRNA carriers.
Given the inherent advantages of ND, which include biocompatibility, self-assembly, nanoscale size, aqueous solubility and intrinsic stability, we formulated ND with a synthetic, bilayer-forming cationic lipid as bioactive agent. Incorporation of cationic lipid into ND conferred double-stranded oligonucleotide (dsOligo) binding activity, thereby generating a potential siRNA binding / transport vehicle. The finding that siRNA containing ND complexes possess target gene knockdown activity in cultured hepatoma cells suggests cationic lipid ND may be useful for in vivo siRNA delivery.
Materials and Methods
Materials
Dimyristoylphosphatidylcholine (DMPC) and dimyristoyltrimethylaminopropane (DMTAP) were obtained from Avanti Polar Lipids Inc. (Alabaster, AL). The ND scaffold protein, recombinant human apolipoprotein (apo) A-I, was expressed in E. coli and isolated as described elsewhere (Ryan et al., 2003). BCA protein assay and enzyme-based phospholipid assay were from Thermo Fisher Scientific (Rockford, IL) and Wako Diagnostics (Richmond, VA) respectively. The KDalert™ glyceraldehyde 3-phosphate dehydrogenase (GAPDH) assay kit and Lipofectamine were from Life Technologies Corp. (Carlsbad, CA).
Nanodisk formulation
Ten mg total lipid, consisting of 100 % DMPC or 70 % DMPC / 30 % DMTAP (w/w), was dissolved in chloroform / methanol (3:1 v/v) and dried under a stream of N2 gas, forming a thin film on a glass vessel wall. Residual organic solvent was removed under vacuum. The prepared lipids were dispersed in phosphate buffered saline (PBS; 20 mM sodium phosphate, 150 mM sodium chloride, pH 7.0) by bath sonication. Subsequently, 4 mg apoA-I was added to the lipid dispersions. Sonication was continued until the turbid mixture clarified, indicating lipid/protein complexes (i.e. ND) had formed. Sample absorbance measurements at 325 nm were made on a Perkin Elmer Lambda 20 UV/VIS spectrophotometer.
Electron miscroscopy
A 4 μl sample of DMTAP-ND was adhered to carbon-coated 400-mesh copper grids previously rendered hydrophilic by glow discharge. The grids were washed for 1 min with three successive drops of deionized water and then exposed to three successive drops of 2% (w/v) uranyl nitrate for 1 min (Ted Pella, Tustin, CA) for negative-staining (Zhang et al., 2013). Images at 19,000× or 80,000× magnification were recorded on 4 K × 4 K Gatan UltraScan CCD under low electron dose conditions using a Tacnai 20 electron microscope (Philips Electron Optics/FEI, Eindhoven, The Netherlands) operating at 200 kV. Each pixel of the micrographs corresponds to 5.5 Å or 1.4 Å at the level of the specimen. Particles in micrographs were semi-automatically selected and boxed using EMAN software (Ludtke, Baldwin and Chu, 1999). Representative nanoparticle images were captured on a grid with 30 nm boxes, 142 pixels/box.
Agarose gel electrophoresis assay
Complementary deoxyribonucleotide strands, 5′-AAC TGG ACT TCC AGA AGA ACA TC-3′ and 5′-GAT GTT CTT CTG GAA GTC CAG TT-3′, were obtained from Eurofins MWG Operon (Huntsville, AL). Equimolar amounts of the complementary strands were mixed in 1xTE buffer, heated at 95 °C for 5 min and slowly cooled to 22 °C to allow annealing. The synthetic 23mer dsOligo was incubated for 30 min at room temperature in PBS with increasing amounts of specified ND. The samples were analyzed by 1% (w/v) agarose gel electrophoresis and stained with ethidium bromide.
Sucrose density gradient ultracentrifugation
Increasing amounts of dsOligo were incubated with DMTAP-ND to achieve +/− charge ratios of 6, 4, 2, 1 and .05 and subjected to 5 to 40% sucrose density gradient centrifugation at 22,000 rpm for 16 h at 10 °C in a Beckman L8-80M ultracentrifuge in a SW41 rotor. Positive charge contributed by DMTAP head group and negative charge from the phosphate backbone of dsOligo were taken into account for +/− charge ratio calculations [e.g. +/− ratio = n(DMTAP) / {n(dsOligo) x 2 x 23} where n represents the number of DMTAP and dsOligo molecules, respectively]. Twelve equal fractions, starting from the top of the gradient, were collected and aliquots of each fraction were assayed for protein, phospholipid and nucleic acid. Protein was quantified by BCA assay and phosphatidylcholine was measured by enzyme-based colorimetric assay. An aliquot from each fraction was electrophoresed on a 1% agarose gel, with or without the addition of 0.1% Triton X-100.
Cell culture
HepG2 cells were obtained from the American Type Culture Collection. Cells were cultured in minimal essential medium supplemented with 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, and 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
GAPDH knockdown
The KDalert™ GAPDH kit includes an optimized GAPDH-specific siRNA and a control siRNA. HepG2 cells were seeded on a 24 well plate at a density of 15,000 cells/well and allowed to attach overnight. The corresponding siRNAs were incubated with DMTAP-ND at a 1:1 (+/−) charge ratio for 15 min at room temperature. Charge ratio was calculated as described above for sucrose density gradient experiments using dsOligo. Cells were incubated with DMTAP-ND harboring GAPDH siRNA or control siRNA and with Lipofectamine-GAPDH siRNA for 48 h. Components provided in the kit were used as per the manufacturer’s protocol for fluorescent readout of GAPDH enzyme activity remaining after treatment. Briefly, the culture medium was aspirated and cells were lysed. Twenty μl of the lysate was transferred onto a 96 well plate and, immediately after adding 180 μl of master mix, the plate was read on a Wallac VICTOR2 1420 multilabel counter (Perkin Elmer Life Science) with excitation and emission filters set at 530/590 nm, respectively. After 4 min a second reading was taken. As per manufacturer guidelines, the initial fluorescence reading was subtracted from the second reading to determine the fluorescence increase (i.e. GAPDH activity remaining in each sample after treatment). Statistical significance between treatment groups was calculated using two-tailed Student’s t-test (GraphPad Prism version 6.0, San Diego, CA). P-values less than 0.05 were considered significant.
Results
DMTAP-ND formulation and characterization
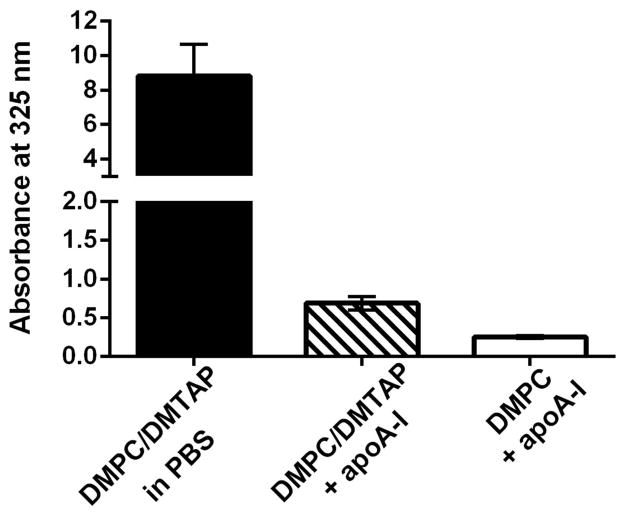
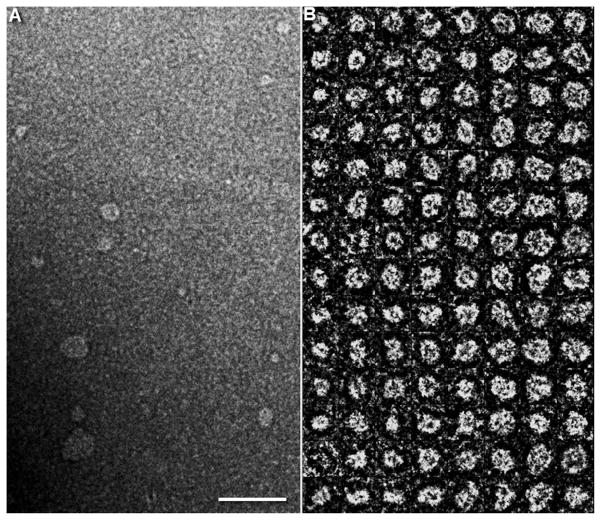
To determine the effect of synthetic cationic lipid (i.e. DMTAP) on ND assembly and structure, lipid dispersions in PBS were incubated with recombinant human apoA-I to induce ND self-assembly. As seen in Figure 1, DMPC/DMTAP dispersed in buffer is turbid with strong light scattering intensity detected at 325 nm. Upon addition of apoA-I, however, both DMPC and DMPC/DMTAP lipid dispersions undergo a marked decline in sample turbidity, consistent with ND particle assembly. Compared to the DMPC only sample, the DMTAP containing sample displayed a slightly higher final absorbance value at 325 nm and this difference was detectable with the naked eye as a faint opacity. Subsequently, the morphology of particles generated upon incubation of apoA-I with the DMPC/DMTAP dispersion was examined by negative stain electron microscopy. Figure 2 depicts examples of the particles generated. In general, DMTAP containing ND displayed particle diameters ~20 to <50 nm. Subsequently, studies were performed to investigate the nucleic acid binding properties of DMTAP-ND.
Figure 1. Effect of apoA-I on aqueous lipid dispersion sample turbidity.
A mixture of DMPC and DMTAP (70:30 w/w; 2.5 mg total lipid) was dispersed in PBS by bath sonication. Sample absorbance was measured at 325 nm after addition of 200 μl PBS or 200 μl PBS containing 1 mg apoA-I. Sample absorbance of ND formulated with DMPC only is shown for comparison.
Figure 2. Negative-stain electron microscopy of DMTAP-ND.
(A) Survey view of DMTAP-ND preparation. (B) Representative views of selected and windowed individual raw DMTAP-ND. Scale bar in panel A is 100 nm and boxes in Panel B are 30 nm.
Interaction of nucleic acid with DMTAP-ND
The binding interaction between a 23 base pair dsOligo and DMTAP-ND was studied by agarose gel electrophoresis (Figure 3). It was hypothesized that, if a stable binding interaction occurs between the dsOligo and DMTAP-ND, dsOligo negative charge character will be neutralized and, thereby, alter its electrophoretic mobility. In the absence of ND, dsOligo migrated to a characteristic position in the gel. When the dsOligo was incubated with DMPC-ND, no change in dsOligo electrophoretic mobility was detected. By contrast, increasing amounts of DMTAP-ND induced corresponding changes in dsOligo mobility. As the intensity of the band corresponding to free dsOligo decreased, a new band appeared with decreased electrophoretic mobility. At a 1:1 charge ratio of dsOligo to DMTAP, the band corresponding to free dsOligo was completely replaced by the slower mobility band. These results indicate that introduction of DMTAP into ND confers dsOligo binding capability.
Figure 3. Effect of ND lipid composition on dsOligo electrophoretic mobility.
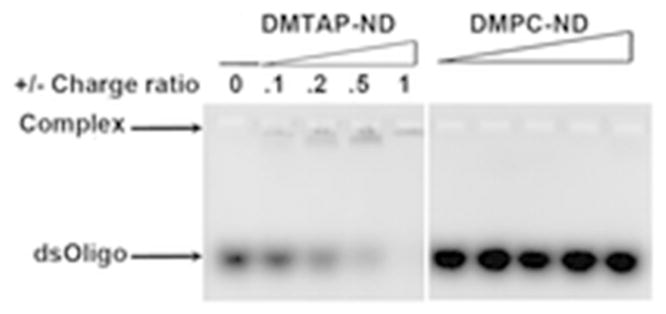
Preformed DMTAP-ND or DMPC-ND were mixed with dsOligo, electrophoresed on an agarose gel (1% w/v) and stained with ethidium bromide. DMTAP-ND and dsOligo were incubated at given charge ratios achieved by adjusting the amount of DMTAP-ND added to a fixed amount of dsOligo.
Sucrose density gradient ultracentrifugation studies
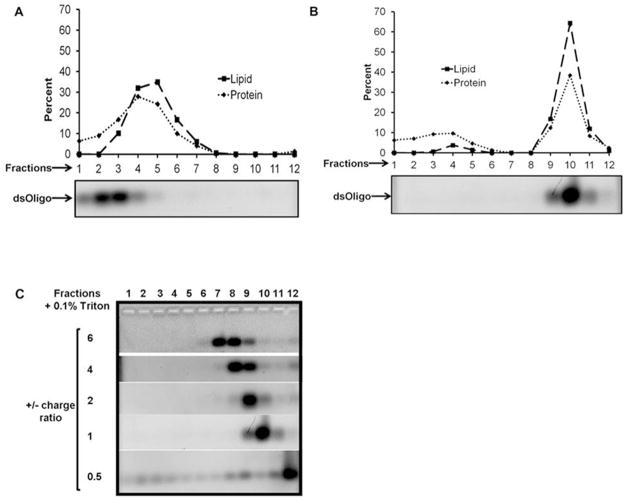
Insofar as the agarose gel electrophoresis assay method employed above detects dsOligo mobility only, a second assay was performed that would allow for detection of ND components in addition to dsOligo. In this experiment a fixed amount of DMTAP-ND was incubated with increasing amounts of dsOligo followed by sucrose density gradient ultracentrifugation. Following centrifugation, twelve fractions, from the top of the gradient to the bottom, were collected. An aliquot of each fraction was subjected to a) agarose gel electrophoresis / ethidium bromide staining and b) lipid and protein analysis. Figure 4 Panel A depicts the migration pattern of the different components when control DMTAP-ND and dsOligo were centrifuged separately. Whereas DMTAP-ND were recovered in gradient fractions 4 and 5, dsOligo was recovered in fractions 2 and 3. Although it may be anticipated that the dsOligo would migrate to a higher density position than DMTAP-ND, the short length of these oligos (i.e. 23mer) influences their migration in this system (Djikeng et al., 2003).
Figure 4. Effect of DMTAP-ND on dsOligo migration in a sucrose gradient.
Specified samples were applied to a 5 to 40 % linear sucrose density gradient and centrifuged at 22,000 rpm for 16 h at 10 °C. Twelve fractions, from the top of the gradient to the bottom, were collected. Protein and phospholipid content in each fraction was determined. In addition, an aliquot of each fraction was electrophoresed on a 1% (w/v) agarose gel and stained with ethidium bromide. (A) Migration pattern for control DMTAP-ND and dsOligo centrifuged separately. (B) Migration pattern of DMTAP-ND and dsOligo following 15 min pre-incubation at a +/− charge ratio of 1. (C) Migration pattern for dsOligo and DMTAP-ND pre-incubated at +/− charge ratios of 6, 4, 2, 1 and 0.5. For Panels B and C, sucrose density gradient fractions were treated with Triton X-100 to disrupt DMTAP-ND and release bound dsOligo.
When DMTAP-ND and dsOligo were pre-incubated at a 1:1 charge ratio prior to sucrose gradient ultracentrifugation, a much different migration pattern was observed (Figure 4, Panel B). In this case, the lipid and protein components of the DMTAP-ND were recovered in gradient fractions 9 – 11. Consistent with complex formation, when aliquots of sucrose gradient fractions were subjected to agarose gel electrophoresis and stained with ethidium bromide, no dsOligo was detected (data not shown). When Triton X-100 was added to the fractions to disrupt the ND, however, dsOligo was detected by agarose gel elecrophoresis (Wheeler et al., 1999; August et al., 2008). Consistent with complex formation, dsOligo co-migrated with ND lipid and protein components in the sucrose gradient. To further characterize this interaction, dsOligo-DMTAP-ND complexes were formulated at +/− charge ratios of 6, 4, 2, 1 and 0.5, subjected to sucrose gradient ultracentrifugation and analyzed (Figure 4C). As the charge ratio decreased, the further the dsOligo migrated into the sucrose gradient. For dsOligo DMTAP-ND complexes formed at a +/− charge ratio of 6, dsOligo was detected in fractions 7 and 8. For complexes with a +/− ratio = 4, they migrated to fractions 8 and 9 while complexes with a +/− ratio = 2 were recovered in fraction 9. At the lowest +/− charge ratio, visible evidence of precipitate formation was observed following centrifugation.
Knockdown experiments with GAPDH siRNA-DMTAP-ND
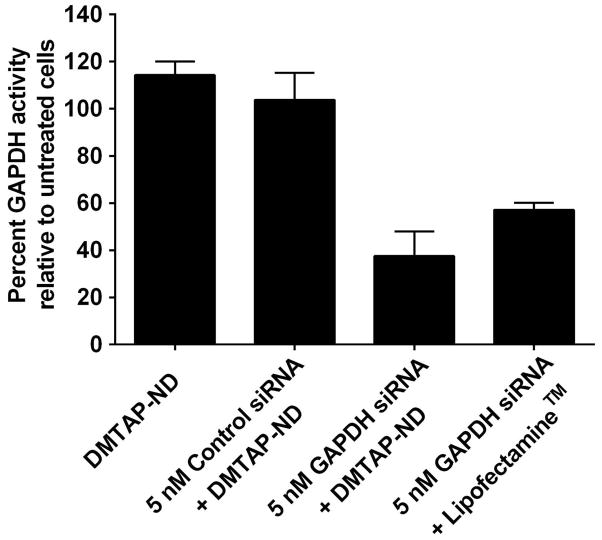
To examine the ability of siRNA bound to DMTAP-ND complexes to knockdown a target gene, DMTAP-ND were mixed with a control siRNA or GAPDH specific siRNA at a +/− charge ratio = 1. Subsequently, HepG2 cells were incubated with the siRNA DMTAP-ND complexes (Figure 5). Whereas cells treated with DMTAP-ND complexed with control siRNA were unaffected (i.e. no knockdown), GAPDH siRNA DMTAP-ND complexes induced a significant 60 % knockdown of GAPDH activity, comparable to the % knockdown observed with the positive control transfection reagent, Lipofectamine.
Figure 5. GAPDH siRNA knockdown studies with DMTAP-ND.
DMTAP-ND with bound GAPDH specific siRNA were formed at a +/− charge ratio of 1:1. HepG2 cells were incubated with the preformed complexes and, after 48 h, cellular GAPDH enzyme activity was measured. The percent GAPDH activity is expressed relative to untreated cells. The commercial transfection reagent, Lipofectamine, was used as positve control while DMTAP-ND with an irrelevant siRNA served as the negative control. Values are mean ± SD (n=3). **P<0.01; ***P<0.001 and n.s. is not significant.
Discussion
It is well known that members of the class of amphipathic apolipoproteins possess potent lipid binding activity capable of inducing formation of discoidal reconstituted high density lipoproteins (rHDL) upon incubation with aqueous phospholipid dispersions. The resulting complexes, comprised solely of phospholipid and apolipoprotein (i.e. rHDL) have been used in studies of cellular cholesterol efflux in vitro (Ma et al., 2012) and in vivo (Besler et al., 2010). These relatively simple complexes can be formed by two different methods, detergent dialysis and, in the case of certain phospholipid substrates, direct solubilization (Ryan, 2008). Over the past ten years rHDL have been repurposed for uses well beyond lipoprotein metabolism. To distinguish between rHDL comprised entirely of phospholipid and apolipoprotein and those particles formulated with an exogenous bioactive agent or transmembrane protein (Ryan, 2010), the term nanodisk (ND) has been coined. Given the ease of formulation and the range of bioactive agents that can be incorporated (i.e. solubilized), ND technology is rapidly expanding and now encompasses drugs, synthetic lipid chelators, fluorescent dyes and proteins. Major advantages of ND include their facile reproducible formulation, product particle stability and biocompatibility. The component parts of ND are interchangeable, thereby creating a versatile platform with myriad potential applications. Given the known ability of synthetic cationic lipids to interact with nucleic acid (Zuhorn et al., 2007), we hypothesized that incorporation of cationic lipid into the ND bilayer would yield a vehicle capable of binding and transport of nucleic acid, specifically, siRNA.
The cationic lipid selected for study (i.e. DMTAP) contains two myristoyl acyl chains bound to a cationic polar head group. Formulation studies revealed that 30 % DMTAP can be incoporporated into ND containing DMPC as co-lipid, with apoA-I functioning as the scaffold component. The particles formed are soluble in aqueous media and of nanoscale size. Two assay methods were employed to evaluate the ability of DMTAP-ND to bind nucleic acid and these studies revealed that, unlike DMPC only particles, introduction of DMTAP into ND confers nucleic acid binding activity. Characterization studies showed that dsOligo and DMTAP-ND co-migrate and that siRNA containing DMTAP-ND possess target mRNA knockdown activity.
Thus, the data presented herein extends ND technology to include a synthetic cationic lipid that confers unique functional properties to the resulting ND. The data support the conclusion that nucleic acid binding to DMTAP-ND occurs via electrostatic attraction. Indeed, in the absence of cationic lipid, no binding occurs. When present, cationic lipid affects the electrophoretic behavior of dsOligo in a concentration dependent manner. Whereas others have employed spherical HDL as a vehicle for siRNA transport, the particles generated herein are discoidal. Chemically modified siRNAs that possess a covalently bound cholesterol moiety bind HDL particles via their lipid anchor (Wolfrum et al., 2007; Nakayama et al., 2012). In other studies, spherical rHDL possessing a nucleic acid-cationic lipid core surounded by anionic lipid and apoA-I have been generated (Rui et al., 2013). In contrast to both these approaches, the present experimental design generates discoidal particles in which the siRNA binds at the particle surface, via electrostatic interaction with cationic lipid molecules intercalated into the ND bilayer. Whereas we have shown that DMTAP-ND are able to bind siRNA and the resulting complexes possess target gene knockdown activity, future studies will be required to assess the potential utility of DMTAP-ND for in vivo delivery of siRNA. At the same time, other cationic lipids as well as different scaffold proteins or bilayer forming lipids can be investigated to optimize the resulting ND in terms of particle size, siRNA binding properties, in vivo stability and cellular uptake of siRNA payload. For example, when apoE was employed in lieu of apoA-I as ND scaffold, enhanced cellular interaction of ND, and bioactive agent uptake, were observed (Ghosh et al., 2014).
Acknowledgments
The authors thank Dr. Todd Sulchek for preliminary DMTAP-ND characterization studies. This work was supported by a grant from NIH (HL-64159) and an American Heart Association Pre-Doctoral Studentship Award (MG). J.B.S. acknowledges support from the Danish VKR and IMK Almene Foundations.
References
- Andaloussi SE, Lehto T, Mäger I, Rosenthal-Aizman K, Oprea II, Simonson OE, et al. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011;39(9):3972–87. doi: 10.1093/nar/gkq1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguste DT, Furman K, Wong A, Fuller J, Armes SP, Deming TJ, et al. Triggered release of siRNA from poly(ethylene glycol)-protected, pH-dependent liposomes. J Control Release. 2008;130:266–274. doi: 10.1016/j.jconrel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros SA, Gollob JA. Safety profile of RNAi nanomedicines. Adv Drug Deliv Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Besler C, Heinrich K, Riwanto M, Lüscher TF, Landmesser U. High-density lipoprotein-mediated anti-atherosclerotic and endothelial-protective effects: a potential novel therapeutic target in cardiovascular disease. Curr Pharm Des. 2010;16:1480–1493. doi: 10.2174/138161210791051013. [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Shen S, Ullu E. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA. 2003;9:802–808. doi: 10.1261/rna.5270203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou S, Yao YD, Yang XZ, Sun TM, Mao CQ, Song EW, et al. Anti-Her2 single-chain antibody mediated DNMTs-siRNA delivery for targeted breast cancer therapy. J Control Release. 2012;161(3):875–83. doi: 10.1016/j.jconrel.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Singh ATK, Xu W, Sulchek T, Gordon LI, Ryan RO. Curcumin nanodisks: formulation and characterization. Nanomedicine. 2011;7:162–167. doi: 10.1016/j.nano.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Ryan RO. ApoE enhances nanodisk-mediated curcumin delivery to glioblastoma multiforme cells. Nanomedicine (Lond) 2014 Jul 24; doi: 10.2217/nnm.13.35. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in murine model of metastatic Ewing’s sarcoma. Cancer Research. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Research. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao Q, Qiu L. Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J Control Release. 2013;171(2):152–62. doi: 10.1016/j.jconrel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ma CI, Beckstead JA, Thompson A, Hafiane A, Wang RH, Ryan RO, et al. Tweaking the cholesterol efflux capacity of reconstituted HDL. Biochem Cell Biol. 2012;90:636–45. doi: 10.1139/o2012-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, et al. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomedicine. 2012;7:3637–57. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotechnology. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Butler JS, Sehgal A, Severgnini M, Racie T, Sharman J, et al. Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Mol Ther. 2012;20:1582–1589. doi: 10.1038/mt.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda MN, Hargreaves P, Beckstead JA, Redmond KA, van Antwerpen R, Ryan RO. Reconstituted high-density lipoprotein enriched with the polyene antibiotic, amphotericin B. J Lipid Res. 2006;47:260–267. doi: 10.1194/jlr.D500033-JLR200. [DOI] [PubMed] [Google Scholar]
- Rui M, Tang H, Li Y, Wei X, Xu Y. Recombinant high density lipoprotein nanoparticles for target-specific delivery of siRNA. Pharm Res. 2013;30:1203–1214. doi: 10.1007/s11095-012-0957-4. [DOI] [PubMed] [Google Scholar]
- Ryan RO. Nanodisks: hydrophobic drug delivery vehicles. Exp Opin Drug Deliv. 2008;5:343–351. doi: 10.1517/17425247.5.3.343. [DOI] [PubMed] [Google Scholar]
- Ryan RO. Nanobiotechnology applications of reconstituted high density lipoprotein. J Nanobiotechnology. 2010;8:28. doi: 10.1186/1477-3155-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RO, Forte TM, Oda MN. Optimized bacterial expression of human apolipoprotein A-I. Protein Expr Purif. 2003;27:98–103. doi: 10.1016/s1046-5928(02)00568-5. [DOI] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- Singh AT, Evens AM, Anderson RJ, Beckstead JA, Sankar N, Sassano A, et al. All trans retinoic acid nanodisks enhance retinoic acid receptor mediated apoptosis and cell cycle arrest in mantle cell lymphoma. Br J Haematol. 2010;150:158–69. doi: 10.1111/j.1365-2141.2010.08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnology. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Sørensen DR, Sioud M. Systemic delivery of synthetic siRNAs. Methods Mol Biol. 2010;629:87–91. doi: 10.1007/978-1-60761-657-3_6. [DOI] [PubMed] [Google Scholar]
- Tachibana Y, Munisso MC, Kamata W, Kitagwa M, Harada-Shiba M, Yamaoka T. Quick nuclear transportation of siRNA and in vivo hepatic ApoB gene silencing with galactose-bearing polymeric carrier. J Biotechnol. 2014 doi: 10.1016/j.jbiotec.2014.01.029. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Wada S, Obika S, Shibata MA, Yamamoto T, Nakatani M, Yamaoka T, et al. Development of a 2′,4′-BNA/LNA-based siRNA for Dyslipidemia and Assessment of the Effects of Its Chemical Modifications In Vivo. Mol Ther Nucleic Acids. 2012;1:e45. doi: 10.1038/mtna.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JJ, Palmer L, Ossanlou M, MacLachlan I, Graham RW, Zhang YP, et al. Stabilized plasmid-lipid particles: construction and characterization. Gene Ther. 1999;6:271–281. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25(10):1149–57. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tong H, Garewal M, Ren G. Optimized negative-staining electron microscopy for lipoprotein studies. Biochim Biophys Acta. 2013;1830:2150–2159. doi: 10.1016/j.bbagen.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Zou X, Qiao H, Jiang X, Dong X, Jiang H, Sun X. Downregulation of developmentally regulated endothelial cell locus-1 inhibits the growth of colon cancer. J Biomed Sci. 2008;16:33. doi: 10.1186/1423-0127-16-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhorn IS, Engberts JB, Hoekstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur Biophys J. 2007;36:349–362. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]