Abstract
Cerebral amyloid angiopathy (CAA) related intracerebral hemorrhage (ICH) is a devastating form of stroke with no known therapies. Clinical, neuropathological, and genetic studies have suggested both overlap and divergence between the pathogenesis of CAA and the biologically related condition of Alzheimer's disease (AD). Among the genetic loci associated with AD are APOE and TOMM40, a gene in close proximity to APOE. We investigate here whether variants within TOMM40 are associated with CAA-related ICH and CAA neuropathology. Using cohorts from the Massachusetts General Hospital (MGH) and the Alzheimer's Disease Neuroimaging Initiative (ADNI), we designed a comparative analysis of high-density SNP geno-type data for CAA-related ICH and AD. APOE ε4 was associated with CAA-related ICH and AD, while APOE ε2 was protective in AD but a risk factor for CAA. A total of 14 SNPs within TOMM40 were associated with AD (p< 0.05 after multiple testing correction), but not CAA-related ICH (all p>0.20); as a result, all AD-associated SNPs within TOMM40 showed heterogeneity of effect in CAA-related ICH (BD p<0.001). Analysis of CAA neuropathology in the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP), however, found that neuritic plaque, diffuse plaque burden, and vascular amyloid burden associated with all TOMM40 SNPs (p<0.02). These results suggest that alterations in TOMM40 can promote vascular as well as plaque amyloid deposition, but not the full pathogenic pathway leading to CAA-related ICH.
Keywords: TOMM40, APOE, Cerebral amyloid angiopathy, Alzheimer's disease, Linkage disequilibrium
Introduction
Vascular deposition of β-amyloid or cerebral amyloid angiopathy (CAA) is a common pathological finding in older persons [1, 2] and, in its severest form, is a leading cause of intracerebral hemorrhage (ICH) in the elderly [3–6]. CAA-related ICH accounts for up to 40 % of all nontraumatic ICH and is associated with a 30–50 % rate of mortality, yet lacks any known therapies [7–9]. CAA is a common finding at autopsy among the elderly and advanced CAA is present in roughly 25 % of brains of patients with Alzheimer's disease [1, 10, 11]. Yet, ICH occurs only rarely among these groups. This contrast suggests that the development of ICH may involve a distinct set of biological pathways from those involved in vascular amyloid deposition.
The substantial biological overlap between CAA and AD is reflected in the conditions’ shared genetic risk factors [12–14]. Recent progress in genome-wide association studies (GWAS) has led to the identification of novel risk loci for AD [15, 16]. As expected, these GWAS have consistently yielded an association signal at the 19q13.2 locus, where APOE is located. Accumulating data, however, have raised the possibility that the signal at 19q13.2 may not be entirely due to APOE, but may be the result of association with variants in other genes as well.
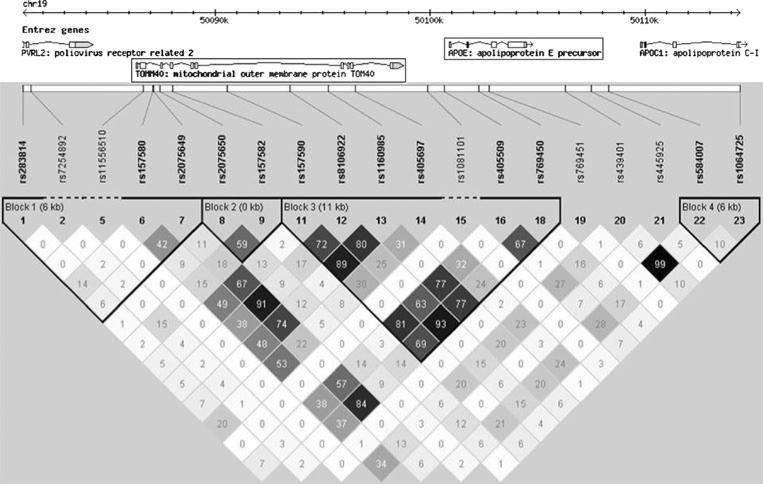
Among the genes at 19q13.2 is TOMM40 (translocase of outer mitochondrial membrane 40), which encodes a membrane-bound mitochondrial protein that occupies the same linkage-disequilibrium (LD) block as APOE ε2 and ε4 (Fig. 1). Some studies have suggested that genetic variation within TOMM40 is associated with AD risk and age at onset [17, 18]. Definitive testing of this association has not been possible, because the strong LD between TOMM40 and APOE prohibits the independent testing of variants within these two genes in standard genetic association studies.
Fig. 1.
Linkage disequilibrium at the 19q13.2 locus. The 19q13.2 locus, and the identified genes that it contains, is represented in the top panel of the figure. The middle panel lists all SNPs identified at the locus in the HapMap Phase 2 project (www.hapmap.org) and their physical position. Numbers in each square in the bottom panel represent the r2 values (i.e., relationships due to linkage disequilibrium) between SNPs, ranging from 0 (no correlation) to 100 (perfect correlation)
We chose to leverage the genetic and biological similarities between CAA-related ICH and AD in order to clarify the relationship between TOMM40 and CAA-related ICH [19, 20]. Genetic variants within APOE, specifically alleles ε2 and ε4, both potently affect risk for both conditions. APOE ε4 increases risk of both AD and CAA, while ε2 reduces AD risk but raises the risk of ICH related to CAA [14, 21]. Strikingly, while the size of the effect of APOE on risk of AD and CAA is roughly similar, GWAS of equal power reveal no association for CAA-related ICH, but significant association for AD (Online Resource 1 and Online Resource 2).
Using genome-wide genotyping data, we performed a comparative genetic analysis of TOMM40 variants in CAA-related ICH and AD to determine whether TOMM40 associates with CAA-related ICH. This analysis was performed in individuals from the Alzheimer Disease Neuroimaging Initiative (ADNI) and Massachusetts General Hospital (MGH-CAA). We used gene-set analysis to further characterize any genetic differences that emerged. Analysis was then extended to related histopathological phenotypes among individuals recruited through the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP).
Methods
Subjects
Cerebral Amyloid Angiopathy Related ICH
Genotype and phenotype data for CAA cases and controls were collected at MGH as part of an ongoing genome-wide association study of intracerebral hemorrhage [14]. Subjects are individuals with ICH aged>55 years, who present to the emergency departments of participating centers and fulfill the criteria for definite or probable CAA-related ICH according to the Boston criteria [10, 22]. In order to minimize inclusion of subjects with undiagnosed AD in the CAA group, we excluded ICH cases with a history of dementia prior to development of the CAA-related ICH, based on interview of the patient or an informant at the time of hemorrhage [14]. Study participation was approved by the Massachusetts General Hospital Institutional Review Board, and all subjects or proxies provided informed consent.
Alzheimer's Disease Neuroimaging Initiative
Participants were selected from the ADNI database (http://www.loni.ucla.edu/ADNI). The ADNI is a large, multisite, collaborative effort launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the US Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations as a public–private partnership aimed at testing whether serial MRI, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of Mild Cognitive Impairment (MCI) and early AD. The ADNI, led by principal investigator Michale Weiner, MD, is the product of many co-investigators from a broad range of academic institutions and private corporations, with patients recruited from more than 50 sites across the United States and Canada (http://www.adni-info.org).
Subjects were prospectively screened and followed according to the ADNI study protocol [23]. The degree of clinical severity for each subject was evaluated by an annual semi-structured interview, which generated an overall Clinical Dementia Rating (CDR) score and the CDR Sum of Boxes [24]. The Mini-Mental State Examination [25] and a neuropsycho-logical battery were also conducted.
AD cases and Cognitively Normal Controls (CNC) were selected from the ADNI database based upon their most recent diagnosis at follow-up. Specifically, participants were selected from the ADNI database if they were clinically classified at follow-up as either: (1) Cognitively Normal Controls (CNC) with CDR=0; or (2) AD individuals who met criteria for probable AD (CDR 1). In order to minimize the chance that clinically symptomatic CAA subjects (i.e., with a past medical history of ICH) could be included in the AD case–control analysis, we excluded all participants with pre-enrollment history of stroke (ischemic or hemorrhagic).
Religious Orders Study and the Rush Memory and Aging Project (ROS and MAP)
The Religious Orders Study (ROS), which began in 1994, enrolled older Catholic priests, nuns, and brothers (aged ≥53 years) from about 40 groups in 12 states [26]. Since January 1994, 1,166 participants, of whom 1,027 were non-Hispanic white, have completed their baseline evaluation. The follow-up rate of survivors exceeds 90 % as does the autopsy rate (546 autopsies of 581 deaths, of whom 516 were non-Hispanic white). Participants were free of known dementia at enrollment.
The Rush Memory and Aging Project (MAP), which began in 1997, enrolled older men and women (aged ≥55 years) free of known dementia from retirement communities in the Chicago area [27]. Since October 1997, 1,533 participants, of whom 1,332 were non-Hispanic white, completed their baseline evaluation. The follow-up rate exceeds 90 %, and the autopsy rate exceeds 80 % (431 autopsies of 527 deaths, of whom 414 were non-Hispanic white).
Participants in both studies agreed to annual clinical evaluations and signed both an informed consent and an Anatomic Gift Act form, donating their brains to Rush investigators at the time of death. All clinical and pathological data were collected, and analyses were performed by study personnel blinded to genotype data. The clinical diagnosis of AD was based on the National Institute of Neurologic and Communicative Disorders and Stroke and the AD and Related Disorders Association [28]. In addition to the diagnosis made at follow-up visits, clinical data were also reviewed at time of death by a neurologist, who did not have access to postmortem pathologic data, and a summary diagnostic opinion was created regarding the most likely clinical diagnosis (AD or normal cognition).
All brains were examined for pathological markers of AD using modified Bielschwoski silver stain. Summary measures of neuritic plaques, diffuse plaques, and neurofibrillary tangles were created from separate counts in five brain regions: midfrontal, middle temporal, inferior parietal, enterhinal cortices, and the hippocampal CA1 sector. A global measure was then made by averaging the five brain regions to create a single standardized summary measure for each pathology [29]. The neuropathologic diagnosis of AD was made by a board-certified neuropathologist without access to any clinical data. Diagnosis was defined as intermediate or high likelihood of AD based on the National Institute on Aging (NIA)-Reagan criteria, which integrates Consortium to Establish a Registry for Alzheimer's Disease (CERAD) semiquantitative estimates of neuritic plaque density and Braak staging of neurofibrillary tangle pathology [30–32].
CAA pathology was measured from tissue in five brain regions on a 5-point scale (0 to 4) regions, using immunohistochemical labeling to amyloid-β [33–36]. Cutoffs for histological CAA/vascular amyloid severity categorization were chosen according to previously published reports, and were also determined without any knowledge of individuals’ genetic data [26]. Additional information pertaining to these studies can be found in previously published literature [26, 27, 37, 38].
Genome-wide SNP Genotyping
Blood samples for the MGH-CAA cases and controls were obtained from each patient. After DNA extraction, samples were genotyped using the Illumina Human 610-Quad Bead-Chip at the Broad Institute. ICH genome-wide SNP genotypes were generated from normalized intensity data using the BeadStudio 3.2 software as previously described, generating a single dataset for 499 individuals [39]. ADNI genome-wide SNP genotyping was performed on the Illumina 660 W SNP array, and individual-level genotype data in the ADNI database [40] were downloaded and merged to form a single dataset containing genome-wide information for 523 individuals. Within the ROS and MAP cohort, DNA was extracted from whole blood lymphocytes or frozen post-mortem brain tissue. Genotype data were generated using the Affymetrix Genechip 6.0 platform at the Broad Institute's Genetic Analysis Platform or the Translational Genomics Research Institute, on self-declared non-Hispanic Caucasians only. APOE alleles ε2 and ε4 were captured via targeted genotyping in all cohorts, according to previously published methods [14, 39].
Statistical Analysis
All genetic analyses for all data sets were performed using PLINK version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/) [41] and SAS software, version 9.1 (SAS Institute, Cary, NC).
Genome-wide Data Quality Control and Population Structure
Quality control (QC) of genotype data for all analyzed individuals was performed according to previously published methods [39]. The QC protocol included filters for missingness, heterozygosity, and concordance between genotype-determined and reported sex. The SNP quality control included filters for minor allele frequency (MAF), missingness, Hardy–Weinberg equilibrium, and differential missingness by case–control status.
Population structure was assessed by performing principal component analysis (PCA) separately in both datasets. PCA was performed in a subset of SNPs selected to minimize genotype missingness using the EIGENSOFT v3.0 package, and confirmed using multidimensional scaling in PLINK v1.07. Principal components 1 and 2 were extracted from the PCA results and entered as additional covariates in separate logistic regression analyses for the genome-wide data of both cohorts (see below) until no additional reduction in genomic inflation factor (GIF) could be achieved.
Imputation and Association Testing
In order to investigate genetic variation within TOMM40, we extracted for association testing all SNPs within the TOMM40 gene (Chromosome 19: kb 50000–50200, HG18 assembly reference), after having performed imputation on the basis of the 1000 Genomes CEU reference panel (release #20101123), using MACHv 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) and excluding SNPs with imputation r2<0.3. APOE alleles ε2 and ε4 were also analyzed for comparison and adjustment purposes.
To test the association of TOMM40 SNPs with both CAA-related ICH and AD, we used logistic regression analyses under the additive genetic model, with the odds ratio (OR) expressing the effect of each copy of the reference (coded) allele. All analyses included age, sex, history of hypertension, years of education, alcohol use, and smoking habits as additional covariates, given their presumed influence on disease risk.
In our biologic extention effort, we tested the association of TOMM40 SNPs with both neuritic and diffuse plaque burden using linear regression analysis in the ROS and MAP cohorts. To test the association of the SNPs with histological CAA (which was coded as a semi-quantitative outcome), we used ordinal logistic regression. All analyses are similarly adjusted for age at death, study (ROS v. MAP), and three principal components from population structure analysis. Imputation was performed similarly to above but was based on the 1000 Genomes European reference panel (release #20100804).
SNP-associated genetic influences on risk of AD vs. CAA-related ICH were compared using the Breslow-Day (BD) test for heterogeneity of effects [42]. This tests the null hypothesis of homogeneity of odds ratio, i.e., identical SNP-related effects in CAA and AD. Statistically significant BD p values reject the null hypothesis, thus supporting the existence of an underlying significant and repeatable difference in effect sizes when comparing different case–control analyses.
TOM Complex Gene-Set Analyses
Gene-set enrichment analysis (GSEA) were performed using PLINK version 1.07 [41]. The implemented gene-set testing method analyzes a pre-defined collection of SNPs, investigating whether the set as a whole contains more associated variants than expected under the null hypothesis. For each gene-set, each SNP is tested for association with case–control status, and p values separately calculated via logistic regression in both CAA and AD (using previously described covariates).
Gene-sets are then refined by excluding SNPs with association p>0.05 and SNPs whose r2 with at least another SNP in the set was >0.60 (for each pair of associated SNPs the one with the lowest p value was retained for analysis). The overall gene-set enrichment for statistically significant associations is summarized by computing the median p value among SNPs satisfying the aforementioned criteria. Gene-set empirical p values were generated via permutation of case–control status (separate permutations for AD and CAA), using 100,000 permutations for each analysis and considering the median p value as the permutation figure of merit.
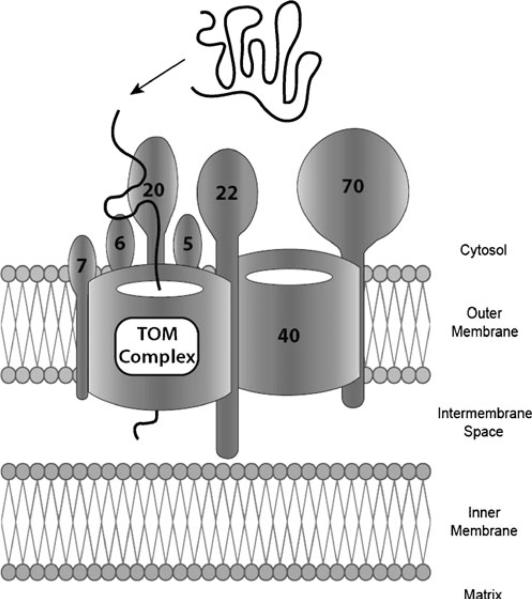
We chose to analyze gene-sets in three stages. In the first stage, each gene encoding a component of the TOM complex (Fig. 2) was defined as our unit of analysis and constituted a single gene-set. For each gene, we extracted all SNPs within the region delimited by the transcription start site and transcription end site (based on coordinates provided by the UCSC Genome Browser, http://genome.ucsc.edu), as well as all SNPs within 500 Kb of these landmarks (in order to include variants potentially in LD with intronic and/or exonic SNPs). SNPs were extracted from the genome-wide dataset after we performed a 1000 Genome Imputation (according to previously described methods). In the second stage, we analyzed all previously considered genes together (thus making the TOM complex our gene-set and unit of analysis), but without including results from TOMM40. This allowed us to test the cumulative association of genetic variants within TOM complex genes, without highly significant results from the primary TOMM40 analysis having undue influence on overall findings. In the third and final stage, we included TOMM40 SNPs to investigate the TOM complex as a whole.
Fig. 2.
The TOM (Translocase of the Outer Mitochondrial membrane) complex and its constituent proteins are shown, along with their localization across the outer mitochondrial membrane. The TOM complex provides a pore through which proteins (solid thick black line) are imported across the mitochondrial outer membrane via a complex series of conformational changes
Results
Analyzed Subjects
We analyzed 1,022 individuals with genotype data from the MGH-CAA and ADNI cohorts (Table 1). Of these, 308 were definite/probable AD cases with 215 cognitively normal ICH-free controls, and 175 were definite/probable CAA cases with 324 controls, also selected to have no prior history of ICH or cognitive impairment.
Table 1.
Subject characteristics
| AD (ADNI) |
CAA-related ICH (MGH) |
ROS/MAP | |||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||
| No. of subjects | 308 | 215 | 175 | 324 | 730 |
| Age (Mean, SD) | 75.5 (7.7) | 75.9 (5.5) | 76.4 (8.2) | 74.1 (8.3) | 81.2 (6.8) |
| Sex (% male) | 42 | 45 | 46 | 45 | 37 |
| History of hypertension (%) | 50 | 45 | 55 | 49 | 58 |
| Ever smoker (%) | 39 | 36 | 41 | 35 | 27 |
| Alcohol abuse (%) | 5 | 2 | 7 | 3 | 3 |
| APOE ε2 (MAF) | 0.21 | 0.07 | 0.16 | 0.07 | 0.082 |
| APOE ε4 (MAF) | 0.32 | 0.14 | 0.25 | 0.12 | 0.151 |
AD=Alzheimer's disease, CAA=cerebral amyloid angiopathy, MAF=minor allele frequency, SD=standard deviation
Controls enrolled in the AD and CAA-related ICH studies did not differ significantly for any of the group characteristics reported on Table 1 (all p>0.20). As expected, the APOE ε2 allele was significantly more frequent in CAA-related ICH cases as opposed to AD cases (p<0.001), consistent with the known discrepancy in genetic effect for this variant. APOE ε4 was more frequent in AD as opposed to CAA-related ICH (MAF 0.32 vs. 0.25, p<0.001).
In our biologic extention cohort, 730 individuals from the ROS and MAP cohorts had genotype date (Table 1). Of these subjects, average summary score, the mean (SD), was 0.78 (0.82) for neuritic plaques and 0.70 (0.78) for diffuse plaques. A total of 723 subjects had CAA as classified by a neuropathological diagnosis, with 21 % assigned an Amyloid Angiopathy Severity of 0, 60 % assigned an Amyloid Angiopathy Severity of 1, and 17 % assigned an Amyloid Angiopathy Severity of 2.
Comparison of Genetic Associations in AD and CAA-Related ICH
Following application of quality control measures, a total of 235 SNPs were successfully imputed at the APOE-TOMM40 locus, and available for analysis. Direct genotyping of the two SNPs that determine APOE alleles revealed that both ε4 and ε2 were associated with AD and CAA-related ICH (Table 2). The direction of the associations was consistent with what had been previously reported in the literature. Of SNPs within TOMM40, a total of 14 were associated with AD after Bonferroni correction for multiple hypothesis testing (p<0.05). We identified no association between TOMM40 SNPs and CAA, and all AD-associated SNPs within TOMM40 showed heterogeneity of effect when compared to CAA (BD p<0.001).
Table 2.
Association results for APOE ε2 / ε4 and SNPs in TOMM40
| SNP | Chrom | Position (BP) | Allele (Coded) | AD |
CAA-related ICH |
BD-p | ||
|---|---|---|---|---|---|---|---|---|
| OR (95 % CI OR) | p | OR (95 % CI OR) | P* | |||||
| APOE E4 | 19 | n/a | ε4 | 3.99 (2.41–7.29) | 3.20 × 10–9 | 3.08 (1.68–5.63) | 2.41 × 10–5 | >0.20 |
| APOE E2 | 19 | n/a | ε2 | 0.44 (0.22–0.89) | 0.024 | 2.89 (1.57–5.33) | 8.72 × 10–5 | <0.0001 |
| rs2075650 | 19 | 50087459 | G | 3.39 (2.40–4.80) | 5.17 × 10–9 | 0.98 (0.69–1.38) | >0.20 | <0.0001 |
| rs34404554 | 19 | 50087749 | G | 3.38 (2.39–4.78) | 6.11 × 10–9 | 1.05 (0.62–1.78) | >0.20 | <0.0001 |
| rs11556505 | 19 | 50087984 | T | 3.38 (2.39–4.78) | 6.34 × 10–9 | 1.06 (0.63–1.75) | >0.20 | <0.0001 |
| rs769449 | 19 | 50101842 | A | 3.59 (2.29–5.62) | 2.38 × 10–5 | 1.24 (0.60–2.58) | >0.20 | 0.0003 |
| rs12972156 | 19 | 50079299 | G | 2.86 (1.92–4.25) | 1.99 × 10–4 | 0.79 (0.43–1.46) | >0.20 | <0.0001 |
| rs12972970 | 19 | 50079436 | A | 2.99 (1.97–4.53) | 0.0002 | 0.86 (0.46–1.63) | >0.20 | <0.0001 |
| rs157582 | 19 | 50088059 | T | 3.02 (2.18–4.18) | 2.74 × 10–8 | 1.08 (0.61–1.90) | >0.20 | <0.0001 |
| rs184017 | 19 | 50086809 | G | 3.00 (2.16–4.15) | 3.87 × 10–8 | 1.08 (0.61–1.88) | >0.20 | <0.0001 |
| rs157581 | 19 | 50087554 | C | 2.82 (2.03–3.92) | 5.86 × 10–7 | 0.89 (0.50–1.57) | >0.20 | <0.0001 |
| rs283815 | 19 | 50082173 | G | 2.53 (1.67–3.83) | 0.010 | 0.91 (0.48–1.72) | >0.20 | <0.0001 |
| rs157580 | 19 | 50087106 | G | 0.48 (0.36–0.64) | 0.0006 | 0.83 (0.60–1.14) | >0.20 | 0.008 |
| rs439401 | 19 | 50106291 | T | 0.52 (0.39–0.69) | 0.005 | 0.86 (0.62–1.20) | >0.20 | 0.010 |
| rs34095326 | 19 | 50087684 | A | 2.79 (1.78–4.37) | 0.007 | 1.00 (0.56–1.80) | >0.20 | 0.001 |
| rs10119 | 19 | 50098513 | A | 2.72 (1.69–4.35) | 0.031 | 0.44 (0.14–1.38) | >0.20 | <0.0001 |
Results of multivariate logistic regression analyses of association of variants in TOMM40 and APOE with AD and CAA-related ICH. Reported p values are adjusted for 235 independent tests using Bonferroni correction. APOE SNPs were directly genotyped, while SNPs within TOMM40 are derived from genome-wide genotyping with imputation as described above. Only SNPs with adjusted p<0.05 in AD case–control analysis are reported. The rightmost column reports results of the heterogeneity of effects test (BD). Significant p values (after Bonferroni adjustment) are consistent with differences in genetic effects between AD and CAA
AD=Alzheimer's disease, BD=Breslow–Day test, BP=base pairs, CAA=cerebral amyloid angiopathy, Chrom=Chromosome, OR=odds ratio, 95 % CI=95 % confidence interval
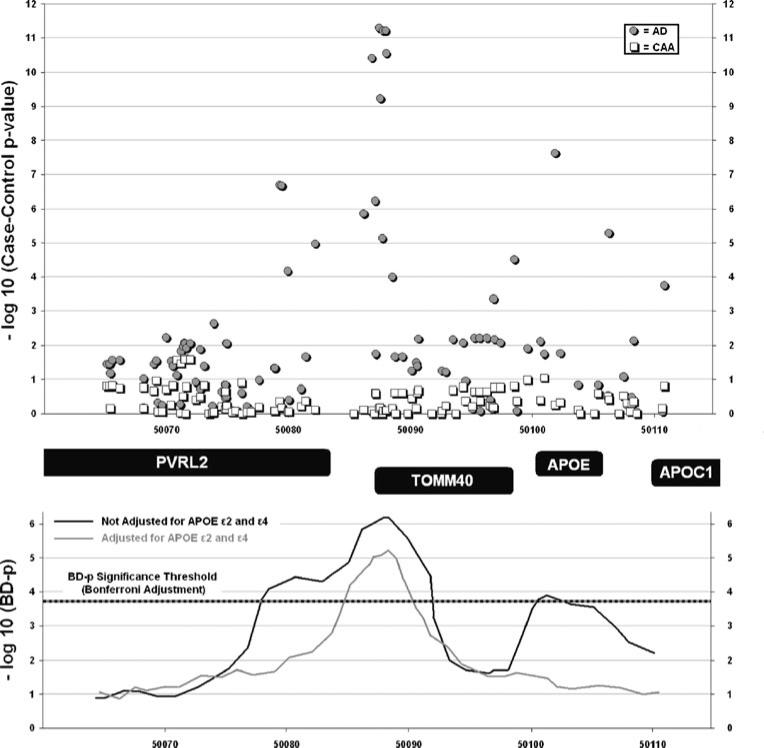
We sought to determine whether the association signal for APOE from genome-wide arrays could arise through linkage disequilibrium with TOMM40. When genome-wide array data were analyzed, SNPs within the APOE locus showed association with AD but not CAA (Fig. 3, top panel). When the significance of the Breslow–Day test for heterogeneity was examined across the extended genetic region, heterogeneity was observed at both TOMM40 and APOE (Fig. 3, bottom panel). We then incorporated direct genotyping of APOE e2 and e4 in order to adjust for the effect of APOE. This eliminated the heterogeneity at the APOE locus, consistent with the inversion of effect for ε2 in the two diseases (AD OR, 0.44; CAA OR, 2.89). Adjustment for APOE, however, did not alter the significant heterogeneity for AD-associated TOMM40 SNPs in CAA.
Fig. 3.
Effect heterogeneity for association of SNPs in the 19q13.2 region. Top panel: negative log of p values for case–control AD/CAA association are plotted against physical position at the 19q13.2 locus using genotypes derived exclusively from genome-wide arrays and imputation as described above. Bottom panel: negative log of p values for the heterogeneity of effects tests (BD) are displayed against physical position at the 19q13.2 locus. The multiple-testing adjusted threshold for BD significance is calculated for 235 independent tests. Reported values refer to BD results for BD-p<0.05 before and after adjustment for APOE ε2 / ε4 genotype. AD=Alzheimer's disease, BD=Breslow–Day, CAA=cerebral amyloid angiopathy
TOM Complex Gene-Set Analysis
Seven major genes of the TOM complex were analyzed via GSEA for association with either AD or CAA (Table 3). The absence of SNPs in TOMM5 and TOMM6 precluded their analysis. Three of the genes, TOMM40, TOMM20, and TOMM7, were independently associated with AD after Bonferroni correction (p<0.05 after multiple testing corrections). SNPs in TOMM22 and TOMM70 demonstrated a trend toward association with AD. We identified no association between TOM complex genes and CAA-related ICH. Finally, the TOM complex as a whole was analyzed both with and without TOMM40, demonstrating an association with AD but not CAA-related ICH. Comparison of Genetic Associations in Diffuse and Neuritic Plaque Burden and Vascular Amyloid Deposition Upon extension, ε4 and ε2 were again found to be associated with AD neuropathology and histological CAA (Table 4). The same 14 SNPs of TOMM40 were associated with diffuse and neuritic plaque burden as well as vascular amyloid burden (p<0.02).
Table 3.
TOM Complex Gene-set Analyses in AD and CAA
| Gene-set | AD p value | CAA p value |
|---|---|---|
| TOMM40 | 0.0001 | 0.23 |
| TOMM20 | 0.012 | 0.33 |
| TOMM22 | 0.082 | 0.15 |
| TOMM5 | n/a | n/a |
| TOMM6 | n/a | n/a |
| TOMM7 | 0.023 | 0.88 |
| TOMM70 | 0.11 | 0.25 |
| TOM Complex (minus TOMM40) | 0.012 | 0.25 |
| TOM Complex (all genes) | 0.003 | 0.32 |
Reported p value are adjusted for multiple testing (Bonferroni) for seven independent tests
AD=Alzheimer's disease, CAA=cerebral amyloid angiopathy, TOM=translocase outer membrane
Table 4.
Association results for APOE ε2 / ε4 and SNPs in TOMM40 using the ROS and MAP cohorts
| SNP | Chrom | Position (BP) | Allele (Coded) | Neuritic plaques |
Diffuse plaques |
Vascular amyloid/CAA |
|||
|---|---|---|---|---|---|---|---|---|---|
| β (95 % CI OR) | p | β (95 % CI OR) | p | OR (95 % CI OR) | P* | ||||
| APOE E4 | 19 | n/a | ε4 | 0.35 (0.27–0.42) | 1.20 × 10–19 | 0.33 (0.26–0.40) | 2.00 × 10–19 | 4.04 (2.93–5.58) | 2.27 × 10–17 |
| APOE E2 | 19 | n/a | ε2 | –0.28 (–0.38–0.19) | 5.94 × 10–9 | –0.26(–0.35–0.17) | 1.04 × 10–8 | 0.42 (0.28–0.61) | –5.57 × 10–6 |
| rs2075650 | 19 | 45395619 | G | 0.48 (0.36–0.61) | 5.83 × 10–14 | 0.47 (0.35–0.58) | 3.03 × 10–14 | 6.79 (4.04–11.4) | 1.36 × 10–12 |
| rs34404554 | 19 | 45395909 | G | 0.49 (0.37–0.62) | 5.42 × 10–14 | 0.47 (0.35–0.59) | 3.06 × 10–14 | 6.95 (4.11–11.8) | 1.29 × 10–12 |
| rs11556505 | 19 | 45396144 | T | 0.49 (0.37–0.62) | 4.81 × 10–14 | 0.47 (0.35–0.59) | 2.69 × 10–14 | 7.00 (4.14–11.9) | 1.15 × 10–12 |
| rs769449 | 19 | 45410002 | A | 0.46 (0.36–0.57) | 9.00 × 10–17 | 0.44 (0.34–0.54) | 9.96 × 10–17 | 6.37 (4.04–10.0) | 6.30 × 10–15 |
| rs12972156 | 19 | 45387459 | G | 0.52 (0.38–0.66) | 7.06 × 10–13 | 0.49 (0.36–0.63) | 8.25 × 10–13 | 7.37 (4.13–13.2) | 3.03 × 10–11 |
| rs12972970 | 19 | 45387596 | A | 0.51 (0.37–0.65) | 4.64 × 10–13 | 0.49 (0.36–0.62) | 5.34 × 10–13 | 7.14 (4.06–12.6) | 2.00 × 10–11 |
| rs157582 | 19 | 45396219 | T | 0.46 (0.35–0.58) | 7.10 × 10–14 | 0.45 (0.33–0.56) | 3.82 × 10–14 | 6.13 (3.73–10.1) | 1.96 × 10–12 |
| rs184017 | 19 | 45394969 | G | 0.47 (0.35–0.59) | 1.11 × 10–13 | 0.46 (0.34–0.57) | 5.60 × 10–14 | 6.38 (3.83–10.6) | 2.51 × 10–12 |
| rs157581 | 19 | 45395714 | C | 0.46 (0.34–0.58) | 8.38 × 10–14 | 0.45 (0.33–0.56) | 4.42 × 10–14 | 6.11 (3.72–10.1) | 2.41 × 10–12 |
| rs283815 | 19 | 45390333 | G | 0.47 (0.34–0.59) | 4.43 × 10–13 | 0.45 (0.33–0.57) | 3.31 × 10–13 | 6.08 (3.62–10.2) | 1.68 × 10–11 |
| rs157580 | 19 | 45395266 | G | –0.08(–0.14––0.01) | 0.030 | –0.05(–0.11–0.02) | 0.147 | 0.83 (0.64–1.07) | 0.166 |
| rs439401 | 19 | 45414451 | T | –0.17 (–0.26––0.08) | 2.34 × 10–4 | –0.13(–0.22––0.05) | 0.002 | 0.55 (0.39–0.78) | 7.21 × 10–4 |
| rs34095326 | 19 | 45395844 | A | 0.51 (0.36–0.66) | 6.63 × 10–11 | 0.50 (0.36–0.64) | 1.22 × 10–11 | 6.41 (3.49–11.8) | 3.30 × 10–9 |
| rs10119 | 19 | 45406673 | A | 0.25 (0.16–0.33) | 1.35 × 10–8 | 0.28 (0.20–0.36) | 1.59 × 10–11 | 3.26 (2.31–4.61) | 3.95 × 10–11 |
Results of multivariate logistic regression and ordinal logistic regression analyses of association of variants in TOMM40 and APOE with neuritic and diffuse plaque count and histological CAA. Reported p values are adjusted for 235 independent tests using Bonferroni correction. APOE SNPs were directly genotyped, while SNPs within TOMM40 are derived from genome-wide genotyping with imputation as described above. Only SNPs with adjusted p<0.05 in AD case–control analysis are reported
AD=Alzheimer's disease, BP=base pairs, CAA=cerebral amyloid angiopathy, Chrom=chromosome, OR=odds ratio, 95 % CI=95 % confidence interval
Discussion
Our data demonstrate that common genetic variants within TOMM40 are associated with risk of clinical AD, histological amyloid burden in the brain parenchyma and vessels, but not with CAA-related ICH. This contrast points to the possibility that the biological pathways involved in amyloid-related vessel rupture may differ from those implicated in the vascular amyloid deposition that must, of necessity, precede it. The Boston Criteria allow diagnosis of CAA-related ICH with a very high degree of accuracy [22, 43]. Virtually all patients with CAA-related ICH in the present study can therefore be assumed to have histopatho-logically severe vascular amyloid deposition. None of the subjects in ROS or MAP, on the other hand, had ICH despite the presence of often extensive amyloid deposition within the cerebral vessels. While vascular amyloid deposition is a common finding in the aging brain, CAA-related ICH, diagnosed through the Boston Criteria, which combines clinical, imaging, and pathological (if available) parameters [22], is substantially less common. Furthermore, our exploratory gene-set analysis raises the hypothesis that additional variants within the TOM complex influence susceptibility to AD, particularly TOMM20 and TOMM7.
While replication of these findings is essential, gene-set analysis raises the possibility that TOMM40 may play a role in clinical expression and pathological changes in AD, including vascular amyloid deposition, but not in the ICH that occurs in CAA. These results raise the hypothesis that the TOMM40-mediated increase in vascular amyloid does not trigger the breakdown in the vessel wall that leads to ICH. Potential explanations include the possibility that the TOMM40-mediated effect is insufficiently severe, or alternatively could be part of a biological pathway that is separate from the pathway that leads to vessel rupture. Vessel rupture is an aspect of CAA-related ICH that appears to involve mechanisms that are distinct from those in AD. The most striking demonstration of this is the divergent role played by APOE e2 in AD and CAA. APOE e2 promotes CAA-related vessel rupture without stimulating either plaque or vascular amyloid deposition [44, 45].
ApoE, the apolipoprotein product of the APOE gene, acts as a transporter of cholesterol and lipids [46]. The gene has three major polymorphic forms (ε2, ε3, and ε4 alleles), and it is these isoforms that modulate amyloid-β metabolism and accumulation in the brain [21]. ApoE plays a critical role in transforming amyloid-β from a monomeric, nontoxic molecule into a toxic, higher-molecular-weight form [21]. Whether TOMM40 might produce its effect through interaction with APOE e2 requires further study, although the LD between the two genes makes it impossible to answer this question within the present study design.
The TOMM40 gene encodes Tom40, a subunit of the translocase of the outer membrane (TOM) complex which lies in the mitochondria and is involved in transporting cytoplasmic peptides and proteins during mitochondrial bio-genesis [18]. The role of Tom40 in regulating protein traffic across the outer mitochondrial membrane appears to be important in the development of AD, perhaps because of the unique bioenergetic requirements of neurons [47, 48]. Mitochondrial dysfunction and oxidative imbalance have been linked to neuronal cell death and AD [18]. Amyloid-β appears to contribute to mitochondrial oxidative stress and dysregulation of internal Ca2+ homeostasis, ultimately leading to impairment of the electron transport chain, reduction in ATP production, and increased production of superoxide anion radicals [48]. TOM is one of two complexes that form the major pathway for mitochondrial import of precursor proteins, the second being the translocase of the inner membrane (TIM) [49]. Experiments in isolated rat mitochondria [49] demonstrate that amyloid-β is selectively transported into mitochondria using the TOM complex. Amyloid-β import into the mitochondria through the TOM complex may facilitate its toxic effects on neurons [49].
The limitations of our study render our findings preliminary. Through the International Stroke Genetics Consortium, we have assembled a substantial proportion of available cases of definite and probable CAA-related ICH with accompanying genotype data. There is no existing CAA-related ICH cohort in which to replicate our findings. An additional important limitation is that there was insufficient coverage on the genome-wide genotyping platforms used of the TOM complex to allow inclusion of all available genes that contribute to the final protein assembly. Thus, for example, TOMM5 and TOMM6 could not be analyzed at all due to an absence of genotyped SNPs in their gene regions. We also did not evaluate any SNPs in the TOMs other than TOMM40 in relation to the pathological expression of vascular or parenchymal amyloid. Nonetheless, while our results are unreplicated, a number of features suggest they may not be false positive. The Breslow–Day p values for assessment of heterogeneity of effects were corrected for multiple comparisons. Furthermore, we detected heterogeneity between AD and CAA-related ICH for genes in the TOM complex that are not part of the 19q13.2 locus and therefore segregate fully independently of the locus.
Conclusion
In summary, through comparisons between association signals for CAA-related ICH and AD as well as histopathological phenotypes, we provide evidence that genetic variation within TOMM40 is implicated in the pathogenesis of AD, parenchymal, and vascular amyloid deposition, but may have little, if any, role in the pathways that lead from CAA to ICH. Further studies are required to replicate our findings and to clarify the functional consequences of TOMM40 variants in AD.
Supplementary Material
Acknowledgement
The authors thank the participants at MGH and within the ROS and the MAP, the Alzheimer's Disease Neuroimaging Initiative (ADNI), the Rush Alzheimer's Disease Center, and the Broad Institute.
Funding/Support This work was supported by the John and Marilyn Keane Stroke Genetics Fund, the Edward and Maybeth Sonn Research Fund, NINDS R01NS059727, the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the National Institutes for Health (NIH) (grant U01 AG024904). The ADNI is funded by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott Laboratories, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation Plc, Genentech Inc, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson Services Inc, Eli Lilly and Company, Medpace Inc, Merck and Co Inc, Novartis International AG, Pfizer Inc, F. Hoffman-La Roche Ltd, and Wyeth Pharmaceuticals, as well as nonprofit partners the Alzheimer's Association and the Alzheimer's Drug Discovery Foundation, with participation from the US Food and Drug Administration. Private sector contributions to the ADNI are facilitated by the Foundation for the NIH. The grantee organization is the Northern California Institute for Research and Education Inc, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angles. This research was also supported by the NIH (grants P30 AG010129 and K01 AG030514) and the Dana Foundation. Drs Biffi and Anderson receive research support from the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research (grant 0775010 N). Dr. Shulman is supported by the Clinical Investigator Training Program: Beth Israel Deaconess Medical Center and Harvard/MIT Health Sciences and Technology, in collaboration with Pfizer Inc. and Merck & Co., Parkinson's Study Group, Harvard Neuro Discovery Center/Massachusetts Alzheimer's Disease Research Center, and Burroughs Wellcome Fund, Career Award for Medical Scientists. Dr Bennett receives research support from Danone Research B.V., the NIH, the Illinois Department of Public Health, and the Robert C. Borwell Endowment Fund. Dr DeJager receives research support from Biogen Idec and the NIH. Dr Rosand receives research support from the National Center for Research Resources. Dr Goldstein (grant K23NS059774) receives research support from the National Institute of Health-National Institute of Neurological Disorders and Stroke.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12975-012-0161-1) contains supplementary material, which is available to authorized users.
Additional Contributions We would like to thank Tammy Gills, PhD, and Marcy MacDonald, PhD, for technical assistance in genotyping APOE variants and the investigators participating in the ADNI study, who contributed to the design and implementation of the ADNI and/or provided data but did participate in the analysis or writing of this report.
Author Contributions Study concept and design: Valant, Rosand and Biffi. Acquisition of data: Valant, Shulman, Devan, Ayres, Schwab, Goldstein, Viswanathan, Greenberg, Bennett, DeJager, Rosand and Biffi. Analysis and interpretation of data: Valant, Keenan, Anderson, Devan, Rosand and Biffi. Drafting of the manuscript: Valant and Biffi. Critical revision of the manuscript for important intellectual comment: Valant, Keenan, Anderson, Shulman, Devan, Ayres, Schwab, Goldstein, Viswanathan, Greenberg, Bennett, De Jager, Rosand and Biffi. Statistical analysis: Keenan, Shulman, Devan and Biffi. Obtained funding: Goldstein, Greenberg, Bennett, De Jager and Rosand. Administrative, technical, and material support: Valant, Ayres and Schwab. Study supervision: Bennett, De Jager, Rosand and Biffi.
Financial Disclosure The authors declare that they have no conflict of interest.
Contributor Information
Valerie Valant, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, CPZN 6818, Boston, MA 02114, USA; Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
Brendan T. Keenan, Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA Program in Translational NeuroPsychiatric Genomics, Institute for Neurosciences, Departments of Neurology and Psychiatry, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Christopher D. Anderson, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, CPZN 6818, Boston, MA 02114, USA; Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
Joshua M. Shulman, Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA Program in Translational NeuroPsychiatric Genomics, Institute for Neurosciences, Departments of Neurology and Psychiatry, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
William J. Devan, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, CPZN 6818, Boston, MA 02114, USA; Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
Alison M. Ayres, Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA
Kristin Schwab, Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA.
Joshua N. Goldstein, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Department of Emergency Medicine, Massachusetts General Hospital, Boston, MA, USA.
Anand Viswanathan, Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA.
Steven M. Greenberg, Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA
David A. Bennett, Rush Alzheimer's Disease Center, Department of Neurological Science, Rush University medical Center, Chicago, IL, USA
Philip L. De Jager, Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA Program in Translational NeuroPsychiatric Genomics, Institute for Neurosciences, Departments of Neurology and Psychiatry, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Jonathan Rosand, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, CPZN 6818, Boston, MA 02114, USA; Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
Alessandro Biffi, Division of Neurocritical Care and Emergency Neurology, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, CPZN 6818, Boston, MA 02114, USA; Hemorrhagic Stroke Research Group, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA.
References
- 1.Vinters HV, Gilbery JJ. Cerebral amyloid angiopathy: incidence and complications in the aging brain. II. The distribution of amyloid vascular changes. Stroke. 1983;14:924–8. doi: 10.1161/01.str.14.6.924. [DOI] [PubMed] [Google Scholar]
- 2.Ellis RJ, Olichney JM, Thal LJ, et al. Cerebral amyloid angiopathy in the brains of patients with Alzheimer's disease: the CERAD experience, Part XV. Neurology. 1996;46:1592–6. doi: 10.1212/wnl.46.6.1592. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi AI, Tuhrim S, Broderick JP, et al. Spontaneous intracerebral hemorrhage. N Engl J Med. 2001;344:1450–60. doi: 10.1056/NEJM200105103441907. [DOI] [PubMed] [Google Scholar]
- 4.Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 2010;9:689–701. doi: 10.1016/S1474-4422(10)70104-6. [DOI] [PubMed] [Google Scholar]
- 5.Vinters HV. Cerebral amyloid angiopathy: a microvascular link between parenchymal and vascular dementia? Ann Neurol. 2001;49:691–3. doi: 10.1002/ana.1055. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg SM, Gurol ME, Rosand J, Smith EE. Amyloid angiopathy-related vascular cognitive impairment. Stroke. 2004;35:2616–9. doi: 10.1161/01.STR.0000143224.36527.44. [DOI] [PubMed] [Google Scholar]
- 7.Van Asch CJ, Luitse MJ, Rinkel GJ, et al. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. 2010;9:167–76. doi: 10.1016/S1474-4422(09)70340-0. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell HC, Rosand J, Knudsen KA, et al. Apolipoprotein E genotype and risk of recurrent lobar intracerebral hemorrhage. N Engl J Med. 2000;342:240–5. doi: 10.1056/NEJM200001273420403. [DOI] [PubMed] [Google Scholar]
- 9.Biffi A, Haplin A, Towfighi A, et al. Aspirin and recurrent intra-cerebral hemorrhage in cerebral amyloid angiopathy. Neurology. 2010;75:693–8. doi: 10.1212/WNL.0b013e3181eee40f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg SM, Vonsattel JP. Diagnosis of cerebral amyloid angiopathy. Sensitivity and specificity of cortical biopsy. Stroke. 1997;28(7):1418–22. doi: 10.1161/01.str.28.7.1418. [DOI] [PubMed] [Google Scholar]
- 11.Ellis RJ, Olichney JM, Thal LJ, et al. Cerebral amyloid angiopathy in the brains of patients with Alzheimer's disease: the CERAD experience, Part XV. Neurology. 1996;46:1592–6. doi: 10.1212/wnl.46.6.1592. [DOI] [PubMed] [Google Scholar]
- 12.Bekris LM, Yu CE, Bird TD, et al. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–27. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Revesz T, Holton JL, Lashley T, et al. Genetics and molecular pathologenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta Neuropathol. 2009;118:115–30. doi: 10.1007/s00401-009-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biffi A, Sonni A, Anderson CD, et al. Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol. 2010;68(6):934–43. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/ MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43(5):429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong MG, Alexeyenko A, Lambert JC, et al. Genome-wide pathway analysis implicates intracellular transmembrane protein transport in Alzheimer's disease. J Hum Genet. 2010;55(10):707–9. doi: 10.1038/jhg.2010.92. [DOI] [PubMed] [Google Scholar]
- 18.Roses AD. An inherited variable Poly-T repeat genotype in TOMM40 in Alzheimer's disease. Arch Neurol. 2010;67(5):536–41. doi: 10.1001/archneurol.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer's disease. Lancet. 2011;377(9770):1019–31. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 20.Revesz T, Ghiso J, Lashley T, et al. Cerebral amyloid angiopathies: a pathologic, biochemical, and genetic view. J Neuropathol Exp Neurol. 2003;62(9):885–98. doi: 10.1093/jnen/62.9.885. [DOI] [PubMed] [Google Scholar]
- 21.Verghese PB, Castellano JM, Holtzman DM. Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol. 2011;10(3):241–52. doi: 10.1016/S1474-4422(10)70325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen KA, Rosand J, Karluk D, Greenberg SM. Clinical diagnosis of cerebral amyloid angiopathy: validation of the Boston criteria. Neurology. 2001;56(4):537–9. doi: 10.1212/wnl.56.4.537. [DOI] [PubMed] [Google Scholar]
- 23.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–9. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–4. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 25.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 26.Arvanitakis Z, Leurgans SE, Wang Z, et al. Cerebral amyloid angiopathy pathology and cognitive decline in older persons. Ann Neurol. 2011;69:320–7. doi: 10.1002/ana.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett DA, Schneider JA, Buchman AS, et al. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–75. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- 28.McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer's disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 29.Bennett DA, Wilson RS, Schneider JA, et al. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer's disease. Neurology. 2003;60:246–52. doi: 10.1212/01.wnl.0000042478.08543.f7. [DOI] [PubMed] [Google Scholar]
- 30.Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18(4 Suppl):S1–2. [PubMed] [Google Scholar]
- 31.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 32.Mirra SS, Heyman A, McKeel D, et al. The consortium to establish a registry for Alzheimer's disease (cerad). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 33.Olichney JM, Hansen LA, Hofstetter CR, et al. Cerebral infarction in Alzheimer's disease is associated with severe amyloid angiopathy and hypertension. Arch Neurol. 1995;52:702–8. doi: 10.1001/archneur.1995.00540310076019. [DOI] [PubMed] [Google Scholar]
- 34.Nakata-Kudo Y, Mizuna T, Yamada K, et al. Microbleeds in Alzheimer disease are more related to cerebral amyloid angiopathy than cerebrovascular disease. Dement Geriatr Cogn Disord. 2006;22:8–14. doi: 10.1159/000092958. [DOI] [PubMed] [Google Scholar]
- 35.Mandybur TI. The incidence of cerebral amyloid angiopahy in Alzheimer's disease. Neurology. 1975;25:120–6. doi: 10.1212/wnl.25.2.120. [DOI] [PubMed] [Google Scholar]
- 36.Attems J, Jellinger KA. Only cerebral capillary amyloid angiopathy correlates with Alzheimer pathology—a pilot study. Acta Neuropathol. 2004;107:83–90. doi: 10.1007/s00401-003-0796-9. [DOI] [PubMed] [Google Scholar]
- 37.Bennett DA, Schneider JA, Arvanitakis Z, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–44. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- 38.Biffi A, Shulman JM, Jagiella JM, et al. Genetic variation at CR1 increases risk of cerebral amyloid angiopathy. Neurology. 2012;78:1–8. doi: 10.1212/WNL.0b013e3182452b40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biffi A, Anderson CD, Desikan RS, et al. Genetic variation and neuroimaging measures in Alzheimer's disease. Arch Neruol. 2010;67(6):677–85. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potkin SG, Guffanti G, Lakatos A, et al. Alzheimer's Disease Neuroimaging Initiative. Hippocampal atrophy as a quantitative trait in genome-wide association study indentifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell SM, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breslow NE, Day NE. Statistical methods in cancer research. Volume I—the analysis of case–control studies. IARC Sci Publ. 1980;32:5–338. [PubMed] [Google Scholar]
- 43.Samarasekera N, Smith C, Al-Shahi Salman R. The association between cerebral amyloid angiopathy and intracerebral haemorrhage: systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2012;83:275–81. doi: 10.1136/jnnp-2011-300371. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg SM, Vonsattel JP, Segal AZ, et al. Association of apolipoprotein E epsilon2 and vasculopathy in cerebral amyloid angiopathy. Neurology. 1998;50(4):961–5. doi: 10.1212/wnl.50.4.961. [DOI] [PubMed] [Google Scholar]
- 45.McCarron MO, Nicoll JA, Stewart J, et al. The apolipoprotein E epsilon2 allele and the pathological features in cerebral amyloid angiopathy-related hemorrhage. J Neuropathol Exp Neurol. 1999;58(7):711–8. doi: 10.1097/00005072-199907000-00005. [DOI] [PubMed] [Google Scholar]
- 46.Grossman I, Lutz MW, Crenshaw DG, et al. Alzheimer's disease: diagnostics, prognosis and the road to prevention. EPMA J. 2010;1(2):293–303. doi: 10.1007/s13167-010-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herculano-Houzel S. Scaling of brain metabolism with a fixed energy budget per neuron: implications for neuronal activity, plasticity and evolution. PLoS One. 2011;6(3):e17514. doi: 10.1371/journal.pone.0017514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430(7000):631–9. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansson-Petersen CA, Alikhani N, Behbahani H, et al. The amyloid β-peptide is imported into mitochondrial via the TOM import machinery and localized to mitochondrial cristae. PNAS. 2008;105(35):13145–50. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell SM, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.