Abstract
Objectives
Multidrug-resistant Enterobacteriaceae pose a significant threat to public health. We aimed to study the impact of sewage treatment effluent on antibiotic resistance reservoirs in a river.
Methods
River sediment samples were taken from downstream and upstream of a waste water treatment plant (WWTP) in 2009 and 2011. Third-generation cephalosporin (3GC)-resistant Enterobacteriaceae were enumerated. PCR-based techniques were used to elucidate mechanisms of resistance, with a new two-step PCR-based assay developed to investigate blaCTX-M-15 mobilization. Conjugation experiments and incompatibility replicon typing were used to investigate plasmid ecology.
Results
We report the first examples of blaCTX-M-15 in UK river sediment; the prevalence of blaCTX-M-15 was dramatically increased downstream of the WWTP. Ten novel genetic contexts for this gene were identified, carried in pathogens such as Escherichia coli ST131 as well as indigenous aquatic bacteria such as Aeromonas media. The blaCTX-M-15 gene was readily transferable to other Gram-negative bacteria. We also report the first finding of an imipenem-resistant E. coli in a UK river.
Conclusions
The high diversity and host range of novel genetic contexts proves that evolution of novel combinations of resistance genes is occurring at high frequency and has to date been significantly underestimated. We have identified a worrying reservoir of highly resistant enteric bacteria in the environment that poses a threat to human and animal health.
Keywords: antibiotic resistance, β-lactamases, CTX-M, environmental pathogens, carbapenem resistance
Introduction
Growing evidence suggests anthropogenic activities such as agriculture contribute to environmental reservoirs of resistant bacteria that can directly or indirectly transfer to humans.1,2 Waste water treatment plants (WWTPs) process waste from several sources, including human, animal and industrial waste, providing a hotspot for horizontal gene transfer to occur between bacteria from many origins. Few studies have demonstrated the impacts of liquid WWTP effluent on antibiotic resistance loads in rivers, particularly with reference to third-generation cephalosporin (3GC) resistance.3 The most common mechanism conferring resistance to 3GCs is the production of plasmid-mediated extended-spectrum β-lactamases (ESBLs), of which the most prevalent are the CTX-M enzymes encoded by blaCTX-M.4 Evidence suggests insertion sequence elements ISEcp1 and IS26 mobilized progenitors of blaCTX-M onto plasmids from the chromosome of Kluyvera species, a common rhizosphere organism.5 Subsequently, plasmid-borne blaCTX-M genes have disseminated throughout the Enterobacteriaceae and Gammaproteobacteria.6 Currently, there are >145 different genotypes of blaCTX-M (http://www.lahey.org/studies), which are often region specific, with blaCTX-M-15 being the most prevalent in humans worldwide and in the UK.4 Rivers are routinely used for the release of WWTP effluent and are a repository for sewage through storm drain overflow. A surveillance study found blaCTX-M-14 in UK rivers; however, to date, the most clinically important ESBL blaCTX-M-15 has not been found.7 Environmental reservoirs of antibiotic-resistant bacteria are likely to represent a significant exposure risk to humans, through direct contact or indirectly through contaminated drinking water or irrigation of crops. We hypothesize that waste water disposal methods are a contributing factor in resistance gene dissemination in rivers. The current study involves a comparative analysis of 3GC-resistant Enterobacteriaceae upstream and downstream of a WWTP effluent point.
Materials and methods
Sampling
Sampling took place in December 2009 and January 2011. Sediment core samples were taken from a river in the UK midlands at three sites in triplicate, 300, 600 and 900 m upstream of a WWTP and three sites in triplicate, 300, 600 and 900 m downstream of a WWTP. The treatment plant served ∼500 000 people and processed >120 million litres of raw sewage per day using a primary settlement tank, secondary activated sludge treatment and final tertiary filtration. Upstream of the treatment plant, geospatial analyses had indicated no other WWTP for ≥10 km. All samples were immediately stored at 4°C and processed within 24 h.
Viable counts
Sediment from one of each downstream sample site (300, 600 and 900 m) was pooled in equal parts (1 g total) and resuspended in 1 mL of PBS buffer. This was repeated for upstream sediment samples. In total, each of the triplicate sediment samples was pooled to form three downstream samples and three upstream samples. Chromocult Coliform Agar (Merck) was prepared in accordance with the manufacturer's instructions and amended with cefotaxime (2 mg/L) or ceftazidime (16 mg/L). Downstream and upstream samples were plated (200 μL) in triplicate for each antibiotic and unamended Chromocult, before incubating for 24 h at 30°C. Viable plate counts were taken; blue colonies indicated presumptive Escherichia coli and pink colonies indicated other coliforms termed presumptive coliforms excluding E. coli (PCEs). Reference strains of E. coli, Klebsiella oxytoca, Citrobacter freundii, Pseudomonas fluorescens and Aeromonas media were used to evaluate the performance of Chromocult at 30°C.
Bacterial isolation
PCEs and E. coli were picked and streaked to obtain pure cultures. The number of isolates obtained for each site differed due to different resistance gene prevalences between sample sites.
Antimicrobial susceptibility determination
MICs of cefotaxime (1–2048 mg/L) and imipenem (1–32 mg/L) were determined using a broth microdilution method based on CLSI and EUCAST standards as previously described.8
DNA extractions
Isolates were incubated overnight at 30°C in Luria broth (LB) and DNA was extracted using a Nucleospin Blood Kit (Macherey-Nagel) in accordance with the manufacturer's instructions.
Identification of bacteria
Bacteria were first identified by sequencing PCR products obtained using the universal 27F and 1525R 16S rRNA primers.9 Further identification of Enterobacteriaceae was performed by partial sequencing of dnaJ as previously described.10 Aeromonas spp. were identified using partial sequencing of gyrB.11 E. coli strains were typed using the Achtman scheme.12
Detection of 3GC resistance genes in isolates
PCR amplification of bla genes, including blaTEM, blaSHV and blaCTX-M, was performed as previously described.13
Analysis of blaCTX-M-15 flanking regions
Characterization of the regions flanking blaCTX-M was performed with PCR as previously described.14
Further analysis of flanking regions unidentified by conventional PCR was done by modifying the two-step gene-walking method (please see the Supplementary data at JAC Online)15 and newly designed primers (CTXD-F, 5′-TCACCCAGCCTCAACCTAAG-3′; and CTXD-R, 5′-CGCTCATCAGCACGATAAAG-3′) were used to detect duplications (please see the Supplementary data at JAC Online).
Conjugation assays
E. coli DH10B (StrR) with induced rifampicin resistance was used as a recipient strain for solid conjugal mating assays with positive blaCTX-M-15 strains as donors. Transconjugants were selected using LB plates amended with streptomycin (100 mg/L), rifampicin (100 mg/L) and cefotaxime (2 mg/L). Positive transconjugants were confirmed using the PCR primer pair CTX-F and CTX-R.
Plasmid replicon typing
Plasmid replicon types in blaCTX-M-15-positive strains were identified using a PCR-based method.16 Strains with identical replicon types were further analysed using restriction fragment length polymorphism (RFLP) (please see the Supplementary data at JAC Online).
Statistical analysis
All statistics were performed using Genstat 15th edition SP1 (VSN International). For comparison of means, log counts were checked for normal distribution using the Shapiro–Wilk test followed by analysis using a paired-sample t-test. Proportions were compared using Fisher's exact test.
Results
Viable plate counts
There was a significant increase in the numbers of 3GC-resistant presumptive E. coli and PCEs in the river sediment downstream of effluent discharge in both 2009 and 2011(Figure 1) (t-test P < 0.0001 in all cases). No significant difference was recorded in numbers of 3GC-resistant E. coli in downstream samples between 2009 and 2011; however, there was a significant increase in numbers of 3GC-resistant PCEs in upstream samples between 2009 and 2011 (t-test P < 0.0001 in all cases). The mean average number of total coliforms in 2011 was 4 × 105/g of wet sediment downstream and 2 × 105/g of wet sediment upstream. The mean average number of E. coli in 2011 was 8 × 103/g of wet sediment downstream and 4 × 103/g of wet sediment upstream. From this, we can calculate that 0.95% of E. coli were resistant to 3GCs downstream compared with 0.13% of E. coli upstream. Similarly, for coliforms there were 0.079% resistant downstream compared with 0.042% upstream.
Figure 1.
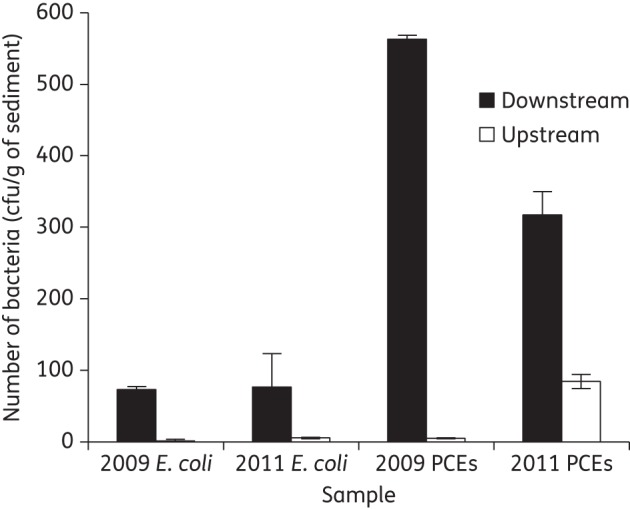
Counts of 3GC-resistant presumptive E. coli and PCEs from samples collected downstream and upstream of a WWTP in 2009 and 2011. Error bars are ± standard errors of biological replicates.
Identification of bacteria
All isolated 3GC-resistant presumptive E. coli (n = 41) were confirmed as E. coli by sequencing dnaJ PCR products (Table 1). Isolated 3GC-resistant PCE isolates (n = 19) were also identified using dnaJ (Table 1). In both downstream and upstream samples, a proportion of PCEs (18.2% downstream and 50% upstream) were identified as members of the Aeromonas genus, species A. media. Multilocus sequence typing (MLST) analysis of E. coli isolates revealed uncharacterized sequence types (STs) particularly from upstream samples (80%), indicating the existence in this environment of novel STs. Downstream of the WWTP, the human-associated ST3103 and ST38 were codominant in 2009, but neither of these STs was detected in 2011 samples, which were dominated by the well-recognized human disease-associated types ST131 (20%) and ST167 (25%) [Table 2 and Table S1 (available as Supplementary data at JAC Online)].
Table 1.
Prevalence of different β-lactamases determined by PCR screening
| Sample site | Organism | Number isolated | blaCTX-M prevalence (%) | blaTEM prevalence (%) | blaSHV prevalence (%) |
|---|---|---|---|---|---|
| Downstream 2009 | E. coli | 11 | 100 | 100 | 0 |
| Downstream 2011 | E. coli | 20 | 100 | 100 | 0 |
| Downstream 2011 | K. oxytoca | 3 | 100 | 100 | 0 |
| Downstream 2011 | C. freundii | 3 | 100 | 100 | 0 |
| Downstream 2011 | C. braakii | 1 | 100 | 100 | 0 |
| Downstream 2011 | Raoultella ornithinolytica | 1 | 0 | 100 | 0 |
| Downstream 2011 | A. media | 2 | 0 | 0 | 0 |
| Downstream 2011 | P. fluorescens | 1 | 100 | 100 | 0 |
| Upstream 2011 | E. coli | 10 | 100 | 100 | 0 |
| Upstream 2011 | K. oxytoca | 1 | 0 | 0 | 0 |
| Upstream 2011 | C. freundii | 3 | 33.3 | 66.6 | 33.3 |
| Upstream 2011 | A. media | 4 | 50 | 100 | 0 |
Table 2.
Molecular characterization of 52 blaCTX-M-positive isolates
| CTX-M genotype (genetic context group) | Composition of isolates in each genetic context | Associated plasmid Inc replicon types | Cefotaxime MIC (mg/L) | Transfer through conjugation |
|---|---|---|---|---|
| 2009 isolates | ||||
| CTX-M-1 | downstream: E. coli ST38 (5) | FIB | 1024–2048 | yes |
| CTX-M-15 international environment (Group A) | downstream: E. coli ST3103 (4) | F, K, IL/IY | >2048 | yes |
| CTX-M-15 Group I | downstream: E. coli ST New (1) | FIB, I1/IY | 64 | yes |
| CTX-M-15 Group J | downstream: E. coli ST3103 (1) | FIB, I1/IY | >2048 | yes |
| 2011 isolates | ||||
| CTX-M-15 international environment (Group A) | downstream: C. braakii (1), C. freundii (1), E. coli incl. ST131 (3), ST167 (1) and ST New (2) | FIB, K, HI2, A/C, FIIA | 16–2048 | yes |
| upstream: E. coli incl. ST131 (1) and ST New (3) | ||||
| CTX-M-15 Group I | downstream: K. oxytoca (2) | FIIA, HI2 | >2048 | yes |
| CTX-M-15 Group K | downstream: C. freundii (2), E. coli incl. ST1060 (1) and ST167 (1) | FIA, FIB, K, IL/IY, A/C | 1024–2048 | yes |
| upstream: A. media (1) and E. coli ST New (1) | ||||
| CTX-M-15 Group L | upstream: C. freundii (1) | FIA | 64 | yes |
| CTX-M-15 Group M | downstream: E. coli ST New (1) | HI2 | 16 | yes |
| CTX-M-15 Group N | downstream: E. coli ST167 (3) and K. oxytoca (1) | FIB, K | 1024–2048 | yes |
| upstream: A. media (1) | ||||
| CTX-M-15 Group O | downstream: E. coli ST1421 (1) | FIB, IL/IY | >2048 | yes |
| CTX-M-15 Group P | downstream: E. coli ST New (1) | F, K | 128 | yes |
| CTX-M-15 Group Q | downstream: E. coli incl. ST131 (1) and ST New (2) | FIB, FIIA, HI2, K | >2048 | yes |
| CTX-M-15 Group R | upstream: E. coli ST New (1) | FIB, HI2 | 16 | no |
| CTX-M-15 unidentified groups | downstream: E. coli ST New (3) and P. fluorescens (1) | F, FIA, FIIA, FIB, K | 32–512 | yes |
| upstream: E. coli incl. ST410 (1) and ST New (3) | ||||
Downstream: isolates recovered downstream of WWTP. Upstream: isolates found upstream of WWTP. ST is the result from MLST with New referring to an isolate with no MLST type matching the MLST database. GenBank accession numbers: Group I, KF155153; Group J, KF155154; Group K, KF155155; Group L, KF155156; Group M, KF155157; Group N, KF155158; Group O, KF155159; Group P, KF155160; and Group Q, KF155161.
Detection and characterization of β-lactamases in resistant isolates
All E. coli were positive for blaCTX-M and blaTEM, but negative for blaSHV (Table 1). Sequencing revealed all blaCTX-M-bearing isolates in 2011 carried blaCTX-M-15 and 54.5% of isolates in 2009 carried blaCTX-M-15 with the remainder carrying blaCTX-M-1.
Analysis of genetic variation in blaCTX-M flanking regions
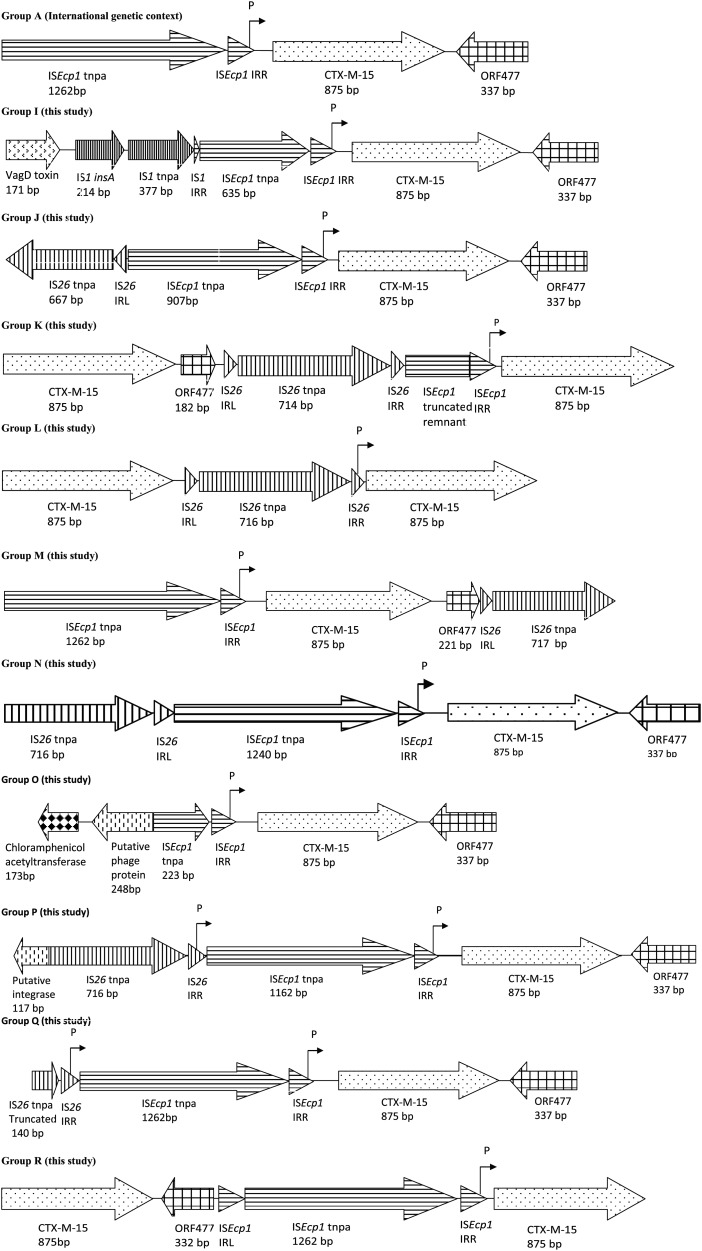
A total of 11 different genetic contexts were found in association with blaCTX-M-15 (Figure 2, Table 2 and Table S1), 10 of which were novel and denoted I–R in keeping with the nomenclature as previously described.14 A total of five genetic contexts were found upstream of the WWTP and nine genetic contexts were found downstream of the WWTP. Three of the genetic contexts were downstream and upstream of the WWTP simultaneously: Group A was the most prevalent and accounted for 67% of the total context type in 2009 and 29% in 2011; Group N was found in A. media in upstream samples, but in E. coli and K. oxytoca in downstream samples; and Group K was found in A. media and E. coli in upstream samples, but in E. coli and C. freundii in downstream samples. Three groups carried multiple copies of blaCTX-M-15: Group K was the only group recovered downstream of the WWTP that consisted of a repeated blaCTX-M-15; and the other two contexts (Groups L and R) both came from upstream of the WWTP. The CTXD-F and CTXD-R primers allowed for detection of blaCTX-M-15 repeats found in Groups K, L and R; however, several groups were unresolved even after two-step gene walking, due to multiple copies of blaCTX-M-15 and repeat regions. Aside from Group A, Group I was the only other group recorded in both 2009 and 2011, but in E. coli in 2009 and K. oxytoca in 2011.
Figure 2.
Flanking regions of blaCTX-M-15 recovered in isolates obtained during this study as confirmed by two-step PCR and sequencing. Nomenclature is an extension of a previously defined typing system.14
Several of the groups contained new elements not previously associated with blaCTX-M-15 flanking regions, such as IS1, putative phage proteins, toxin genes and resistance genes to other antibiotics (Figure 2).
Relationship between genetic context and MIC
Isolates containing seven of the new genetic contexts had MICs of cefotaxime >1024 mg/L. Significantly higher MICs were characteristic of isolates found in downstream samples compared with upstream samples (t-test P = 0.024); however, promoter analysis (Figure S1, available as Supplementary data at JAC Online) revealed that all but one of the contexts shared the same promoter region of blaCTX-M-15 as Group A. One E. coli strain had a high level of resistance to imipenem (>32 mg/L), though this was not conferred through blaCTX-M-15 (Table S1).
Plasmid diversity and relationships to genetic contexts
In both years, a number of different replicon types were associated with CTX-M-carrying strains (Figure 3, Table 2 and Table S1). IncF was the most prevalent replicon type both downstream and upstream of the WWTP. The same genetic context was regularly recorded in strains carrying different replicon types, with the Group A context being associated with the most diverse range of replicons (seven types; Table 2). PFGE analysis was used to compare isolates with the same blaCTX-M-15 context and plasmid replicons (Table S1). Group N was carried by the same plasmid upstream and downstream of the WWTP across different families; initially upstream in A. media and downstream in K. oxytoca and E. coli.
Figure 3.
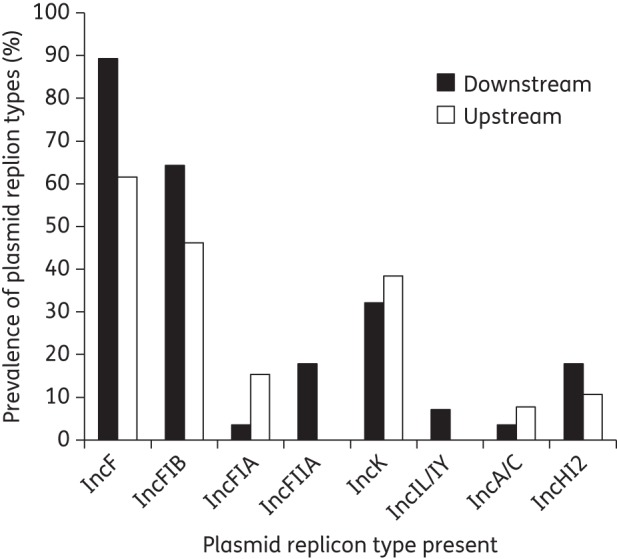
Prevalence of different plasmid replicon types as validated by PCR for 41 blaCTX-M-15-bearing isolates from river sediment downstream of WWTP effluent and upstream of WWTP effluent in 2011.
Conjugation experiments revealed all but two of the blaCTX-M-bearing isolates could transfer this gene at frequencies between 10−3 and 10−7, with expression of cefotaxime resistance ≥2 mg/L in transconjugants. Conjugation rates varied depending on the host background and the plasmid composition (Table S1).
Discussion
Tertiary treatment by WWTPs is the most rigorous level of waste water treatment in the UK as set out by the Water Services Regulation Authority (Ofwat).17 We demonstrated that even with this level of treatment, WWTP effluent has a significant impact on numbers of 3GC-resistant bacteria in river sediment communities. As well as an increase in the total numbers of 3GC-resistant bacteria, the WWTP had an impact on the prevalence of 3GC resistance in bacteria with a 7-fold increase in the prevalence of 3GC-resistant E. coli. The cause of 3GC resistance predominantly resulted from the dissemination of blaCTX-M-15. This is the first report of blaCTX-M-15 in UK river waters and represents a worrying trend as this gene is the most common ESBL in E. coli and Klebsiella spp. causing clinical disease.4 Novel hosts were isolated across the Gammaproteobacteria, including Citrobacter braakii, A. media and P. fluorescens, none of which has previously been reported as a carrier of blaCTX-M-15. Many of the resistant bacteria were pathogens, such as E. coli ST131, ST167 and ST38, C. freundii and K. oxytoca. In particular, the finding of blaCTX-M-15 in the pandemic pathogen E. coli ST131 as a viable and significant reservoir in environmental samples represents a serious threat to human health. This supports recent findings of the threat that rivers pose to human health highlighted by a study in which one-third of people swimming in areas of the River Thames suffered gastrointestinal illness.18 In addition, we report the first finding of an imipenem-resistant E. coli in a UK river, an indication of the emerging spread of carbapenem resistance in the environment, which is a great cause for concern.
We demonstrated that there was high genetic diversity in blaCTX-M-15 carriage and hypothesize that such an unprecedented diversity can be attributed to the direct introduction of bacteria by WWTP effluent possibly combined with in situ selection either in the river or WWTP. Selection is likely to be aided by antibiotic and detergent residues that have previously been detected in WWTP effluent as well as the high density of bacteria present in WWTPs, which will facilitate cell-to-cell contact.3,19 This hypothesis is supported by plasmid analyses as replicon typing revealed eight types present in isolates carrying blaCTX-M-15. Of particular concern is the frequency (46%) at which multiple plasmid replicons were colocalized in one isolate. This would allow for interplasmid transfer of blaCTX-M-15 through transposition and homologous recombination and each different genetic context of blaCTX-M-15 may be indicative of a transfer event.20
The carriage of multiple plasmid incompatibility groups will contribute to higher conjugation rates. This resulted in conjugation frequencies that were higher than reported for similar studies conducted with clinical strains and plasmids.21 The extensive mobility of plasmids was further emphasized by the recovery of identical plasmids in diverse backgrounds. Of concern was the pool of plasmids shared between hosts regarded as clinical bacteria and those regarded as indigenous to the river environment.
We have demonstrated repeated evidence of the significant introduction of clinically relevant ESBL-producing bacteria by WWTP effluent into a UK river. Many of the pathogens had novel blaCTX-M-15 flanking regions, including E. coli ST131-carrying Group Q. The prevalence of human-associated bacteria with a high diversity of blaCTX-M-15 flanking regions downstream of the WWTP supports the hypothesis that community carriage is more extensive than currently thought.22 The change in prevalence of E. coli STs between 2009 and 2011, with ST131 becoming one of the most dominant STs, is likely a reflection of the clonal spread of this ST in the human population.22
An increase in the number of 3GC-resistant coliforms upstream between 2009 and 2011 is potentially a consequence of faecal contamination from surrounding farm environments. This timescale coincides with the detection of the ESBL gene blaCTX-M-1 in UK cattle, chickens, turkeys and most recently dogs.23 The gene blaCTX-M-15 has been detected throughout Europe in companion animals and a diverse range of wild birds.24 Whilst it is not possible to determine the direction of spread from humans to animals, the significant environmental reservoir in rivers will impact both. In conclusion, we report a reservoir of blaCTX-M-15 in a UK river with clear evidence of extensive recombination of the gene within plasmid populations. A growing environmental reservoir presents a risk to human health, with evidence implicating WWTP effluent as a major contributor to the formation of this reservoir.
Further research is needed into sewage treatment systems that result in minimal introduction of resistant bacteria and selecting agents such as antibiotic residues and quaternary ammonium compounds. Stricter regulations and higher levels of treatment are needed if we are to halt the rise of antibiotic resistance in the environment.
Funding
This work was supported by the Natural Environment Research Council (grant NE/E004482/1). G. C. A. A. was supported by a BBSRC studentship. W. H. G. has been supported by the ERDF and ESF since moving to the University of Exeter.
Transparency declarations
None to declare.
Author contributions
G. C. A. A., W. H. G. and E. M. W. designed the research, G. C. A. A. performed the research, all authors analysed the data and all authors contributed to writing the paper.
Supplementary data
References
- 1.Smith DL, Dushoff J, Morris JG. Agricultural antibiotics and human health. PLoS Med. 2005;2:e232. doi: 10.1371/journal.pmed.0020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jechalke S, Kopmann C, Rosendahl I, et al. Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl Environ Microbiol. 2013;79:1704–11. doi: 10.1128/AEM.03172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tennstedt T, Szczepanowski R, Braun S, et al. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol Ecol. 2003;45:239–52. doi: 10.1016/S0168-6496(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 4.Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64(Suppl 1):i3–10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 5.Humeniuk C, Arlet G, Gautier V, et al. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother. 2002;46:3045–9. doi: 10.1128/AAC.46.9.3045-3049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu SY, Zhang YL, Geng SN, et al. High diversity of extended-spectrum β-lactamase-producing bacteria in an urban river sediment habitat. Appl Environ Microbiol. 2010;76:5972–6. doi: 10.1128/AEM.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanji H, Murphy NM, Akhigbe C, et al. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J Antimicrob Chemother. 2011;66:512–6. doi: 10.1093/jac/dkq472. [DOI] [PubMed] [Google Scholar]
- 8.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–75. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 9.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrant E, Goodfellov M, editors. Nucleic Acids Techniques in Bacterial Systematics. Wiley: Chichester; 1991. pp. 115–48. [Google Scholar]
- 10.Pham HN, Ohkusu K, Mishima N, et al. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn Micr Infec Dis. 2007;58:153–61. doi: 10.1016/j.diagmicrobio.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Yanez MA, Catalan V, Apraiz D, et al. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol. 2003;53:875–83. doi: 10.1099/ijs.0.02443-0. [DOI] [PubMed] [Google Scholar]
- 12.Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sturenburg E, Kuhn A, Mack D, et al. A novel extended-spectrum β-lactamase CTX-M-23 with a P167T substitution in the active-site omega loop associated with ceftazidime resistance. J Antimicrob Chemother. 2004;54:406–9. doi: 10.1093/jac/dkh334. [DOI] [PubMed] [Google Scholar]
- 14.Dhanji H, Patel R, Wall R, et al. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother. 2011;66:1005–12. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 15.Pilhofer M, Bauer AP, Schrallhammer M, et al. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res. 2007;35:e135. doi: 10.1093/nar/gkm836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carattoli A, Bertini A, Villa L, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Environment Agency. Catchment Risk Assessment of Steroid Oestrogens from Sewage Treatment Works. 2008. http://nora.nerc.ac.uk/2810/1/SCHO0308BNVO-e-e.pdf. (26 February 2014, date last accessed )
- 18.Public Health England. Epidemiological Investigation of an Outbreak of Gastrointestinal Illness Following a Mass-participation Swim in the River Thames London October 2012. http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317139088329. (26 February 2014, date last accessed)
- 19.Gaze WH, Abdouslam N, Hawket PM, et al. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob Agents Chemother. 2005;49:1802–7. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge SR, Zong Z, Iredell JR. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob Agents Chemother. 2011;55:4971–8. doi: 10.1128/AAC.00025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harajly M, Khairallah MT, Corkill JE, et al. Frequency of conjugative transfer of plasmid-encoded ISEcp1-blaCTX-M-15 and aac(6′)-lb-cr genes in Enterobacteriaceae at a tertiary care center in Lebanon—role of transferases. Ann Clin Microbiol Antimicrob. 2010;9:19. doi: 10.1186/1476-0711-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickramasinghe NH, Xu L, Eustace A, et al. High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J Antimicrob Chemother. 2012;67:1108–13. doi: 10.1093/jac/dks018. [DOI] [PubMed] [Google Scholar]
- 23.Randall LP, Clouting C, Horton RA, et al. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother. 2011;66:86–95. doi: 10.1093/jac/dkq396. [DOI] [PubMed] [Google Scholar]
- 24.Ewers C, Grobbel M, Stamm I, et al. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. J Antimicrob Chemother. 2010;65:651–60. doi: 10.1093/jac/dkq004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.