Abstract
Epigenetics is the study of changes in gene expression or cellular phenotype that do not change the DNA sequence. In this review, current methods, both genomic and proteomic, associated with epigenetics research are discussed. Among them, chromatin immunoprecipitation (ChIP) followed by sequencing and other ChIP-based techniques are powerful techniques for genome-wide profiling of DNA-binding proteins, histone post-translational modifications or nucleosome positions. However, mass spectrometry-based proteomics is increasingly being used in functional biological studies and has proved to be an indispensable tool to characterize histone modifications, as well as DNA–protein and protein–protein interactions. With the development of genomic and proteomic approaches, combination of ChIP and mass spectrometry has the potential to expand our knowledge of epigenetics research to a higher level.
Keywords: approach, chromatin immunoprecipitation, DNA methylation, epigenetic, genomic, histone modification, mass spectrometry, proteomic
Epigenetics was first introduced by Waddington in 1939 to name “the causal interactions between genes and their products, which bring the phenotype into being” [1]. Despite decades of debate and research, a consensus definition of epigenetics remains both contentious and ambiguous [2]. The term ‘epigenetics’ has most commonly evolved to include any process that alters gene activity without changing the DNA sequence, and epigenetic marks are somatically inherited and, therefore, can be passed on as disease cells replicate. The field of epigenetics was given its name and a vague definition only 70 years ago, but it is now a dynamic and rapidly expanding discipline, challenging and revising traditional paradigms of potential inheritance [1,3]. This increasingly important research area examines how the covalent attachment of chemical groups to DNA and its associated histone proteins can influence phenotype without alteration of the DNA sequence [4].
Common types of epigenetic regulation include DNA methylation, DNA hydroxy-methylation, histone variants and modifications, nucleosome remodeling, and small and large noncoding regulatory RNAs [5]. Chromatin is composed of chromosomal DNA wrapped with histones, nonhistone proteins (including structural and transcription factors) and RNA [6]. The nucleosome is the repeating subunit of chromatin, in which 147 bps of DNA are wrapped around an octamer of core histones formed by four histone partners (an H3–H4 tetramer and two H2A–H2B dimers). Nucleosomes have an active role in regulating processes such as transcription, DNA repair and apoptosis [7,8].
Perhaps DNA methylation is the best known of epigenetic process, in part, because it has been the easiest to study with the existing technology [9]. Understanding the functions of DNA methylation requires consideration of the distribution of methylation across the genome. Most work in animals has focused on 5-methylcytosine in the context of CpG sequences [10]. In mammals, CpG methylation is an important mechanism to ensure the repression of transcription of repeat elements and transposons [11–13], and has been directly implicated in genomic imprinting and X-chromosome inactivation [14,15]. Alterations in DNA methylation are associated with many human diseases and are a hallmark of cancer, while the relationship between DNA methylation and gene silencing has proved to be challenging to unravel [10,16].
Another significant epigenetic process is histone modification, which is an indicator of active or repressed chromatin [1,9]. Usually, tightly folded chromatin tends to be shut down or not expressed, while more open chromatin is functional or expressed [9]. Increasing evidence indicates that post-translationally modified histones serve as extremely selective binding platforms for specific regulatory proteins that drive distinct nuclear processes [17]. Histones can be modified in many ways, including by methylation, acetylation, propionylation, butyrylation, formylation, phosphorylation, ubiquitylation, sumoylation, citrullination, proline isomerization and ADP ribosylation [18–20]. Post-translational modifications of histones, along with deposition of histone variants, form a ‘histone code’ which are ‘written’ by specific chromatin-modifying enzymes, and then ‘read’ by downstream effector proteins and protein complexes to signal the transcriptional ‘on-and-off’ status of target genes [21,22]. For example, methylation of lysines H3K4 and H3K36 is correlated with transcriptional activation, whereas methylation of lysines H3K9 and H3K27 occurs primarily in association with transcriptional repression [23].
Both DNA methylation and histone modification are involved in establishing patterns of gene repression during development [24]. DNA modifications typically correspond to longterm epigenetic memory: once methylated, genomic DNA remains methylated through the generations. By contrast, histone modifications typically provide short-term epigenetic memory and can be reversed after a few cell division cycles [25,26]. These epigenetic modifications do not work alone; DNA methylation is linked to histone modifications in a mutually dependent relationship, which has implications for understanding normal development as well as somatic cell reprogramming and tumorigenesis [27]. Thus, all the histone modifications in a nucleosome or region, together with the DNA methylation pattern, specifies chromatin structure and, therefore, transcriptional activity [28].
Many methods have been developed to investigate DNA methylation, histone modifications and protein–chromatin interactions. Some of them are extremely powerful for performing experiments to determine what epigenetic mechanisms are involved and the importance of epigenetics in different aspects of gene expression and cell biology [29]. In this review, current methods, both genomic and proteomic, associated with epigenetics research will be discussed.
Genome-scale approaches to studying DNA methylation
Several methods have been developed over the past few years to map DNA methylation on a genomic scale [30]. Most of these are based on one of three techniques: digestion with methylation-sensitive restriction enzymes, affinity enrichment of methylated DNA or chemical conversion with sodium bisulfite [31,32]. For example:
■ Methylation-sensitive digestion uses prokaryotic restriction enzymes to selectively fractionate only methylated or only unmethylated DNA [33,34];
■ Methyl-DNA immunoprecipitation (MeDIP) is a large-scale antibody-based technique for the unbiased detection of methylated DNA [35,36];
■ Methylated DNA capture by affinity purification (MethylCap®; Diagenode, Liège, Belgium) uses a methyl-binding domain protein to obtain DNA fractions with similar methylation levels [37,38];
■ The Infinium® (Illumina, CA, USA) approach is based on the chemical reaction of unmethylated single-stranded DNA with sodium bisulfite, which introduced methylation-specific SNPs into the DNA sequence [39,40].
Genomic DNA is first treated with one of the methylation-dependent steps and then analyzed by various molecular biology techniques, including probe hybridization and sequencing, which can be applied to reveal the location of the 5-methylcytosine residues. The combination of different types of pretreatment and different analytical steps has resulted in a plethora of techniques for determining DNA methylation patterns and profiles [31,41,42], for example, MeDIP profiles generated using next-generation sequencing or microarray analysis following MeDIP [43,44], and bisulfite conversion of either the entire genome (methylC-sequencing) [45] or a CpG island-enriched partition (reduced representation bisulfite sequencing) [46,47], or shotgun bisulfite sequencing of bisulfite-treated DNA [48]. As DNA sequencing becomes faster, cheaper and more capable of genomic applications it is becoming feasible for routine whole-genome sequencing of bisulfite-converted or MeDIPprecipitated DNA to detect genome-wide DNA methylation profiles [1,3].
Genome-scale approaches to studying histone modifications
Although histone modifications have been studied for several decades now, for many histone modifications their functional role is not yet fully understood. In the past few years, there has been a dramatic increase in the amount of information available about the functions of these covalent histone modifications [49,50]. Major contributions to this knowledge are the rapidly advancing tools that are available to perform high-throughput genomic screenings for histone modifications [51]. At approximately the same time that a histone code was proposed to exist, genome-scale methods for mapping histone modifications were introduced [52,53], and now there are dozens of studies that profile histone modifications [54,55]. Among those, chromatin immunoprecipitation (ChIP) is the most wildly used method for profiling histone modifications. In this method, a chromatin extract is prepared and antibodies specific for each histone modification are used for immunoprecipitation of chromatin that carries the modification in question.
Most of the existing methods for studying histone modifications on a genomic scale combine the use of ChIP with serial analysis of gene expression [56], including the serial analysis of chromatin occupancy [57], the genome-wide mapping technique [58], ChIP combined with paired-end ditag sequencing [59], genome wide by hybridization to a microarray (ChIP-chip) or by direct next-generation sequencing of the immunoprecipitated chromatin (ChIP-seq) [60,61]. These technologies provide new insights into the target genomic regions, which harbor various histone modifications during a specific physiological state of the cell.
With ChIP technology, information can be obtained about precise mapping of histone methylation patterns at specific promoters, genes or other genomic regions [62]. The most prevalent technique used to map histone modifications at a genomic scale has been ChIP-chip. Previous ChIP-chip studies reported that H3K4 methylation was a hallmark of active genes [63] and that, inversely, H3K27 methylation was a hallmark of repressed genes [64–66]. However, ChIP-chipbased whole-genome analyses are comparatively expensive, while ChIP-seq is a nascent technology and it is anticipated that costs will decrease since sequence throughput and access to the technology will increase in future [61,67]. The first reported use of ChIP-seq was to identify the genome-wide locations of 20 different histone lysine and arginine methylations in addition to H2A.Z, RNA polymerase II and CTCF in human CD4+ T cells [68]. To date, ChIP-seq has been used to produce genome-wide maps for a variety of histone modifications in developing tissue or cancer models [17].
Proteomic approaches for epigenetics research
Mass spectrometry (MS)-based proteomics is increasingly used in functional biological studies and has proved to be a powerful analytical strategy for understanding the role of histone modifications [69,70]. In addition, for mapping DNA–protein or protein–protein interactions, MS is a sensitive and highly versatile analytical technique that enables the rapid identification of proteins from complex biological samples [71]. However, a major technical obstacle for the identification of native protein complexes by MS is the isolation of a sufficient amount of purified material from a cell lysate [72]. Affinity purification strategies employing epitope tags (also known as fusion or affinity tags) and mild buffer conditions have been widely used for both largescale and targeted protein interaction mapping efforts [73].
Affinity purification is an applicable technique used to purify proteins of interest based on its specific binding affinity to a particular ligand [74]. Traditionally, affinity purifications have been performed using antibodies directed against a specific protein, which is often referred to as immunopurification or immunoprecipitation [75]. In this approach, the experimental conditions must be optimized for every antibody–protein pair studied, and successful binding of an antibody to an endogenous protein must first be obtained, which can be a long and costly process. Since large-scale studies are limited by this approach, a variety of strategies have been developed for purifying proteins based on the addition of specific tag(s) to the bait proteins.
Tandem affinity purification is a generic approach for the purification of protein complexes, which was developed in yeast some 10 years ago by the group of Séraphin [76]. It involves incorporation of a dual-affinity tag into the protein of interest and introduction of the construct into desired cell lines or organisms. Although the tandem affinity purification–MS strategy has been widely used for mapping yeast protein–protein interactions, it has limitations in higher eukaryotes [77]. With modifications to this method, many variations of the original tag are currently available: influenza hemagglutinin or Myc; proteins that bind molecules with high affinity, such as avidin (biotin); short peptide tags, such as the widely used FLAG tag; and fluorescent protein tags, such as green fluorescent protein [75,78]. The size of the affinity tag is important because small affinity tags decrease the possibility of interference with the biological functions of the tagged protein, such as protein folding, recruitment into protein complexes or subcellular localization. While other affinity tags also have some advantages, green fluorescent protein offers benefits in addition to an efficient purification handle, which permits the direct comparison of imaging and proteomics data [79]. Tandem affinity purification–MS methods have limitations in capturing more transient interactions, so shorter protocols have been designed with a single step of purification instead of two [80]. For example, single FLAG or hemagglutinin tags have been recently applied by Breitkreutz et al. to characterize networks of transient interactions between yeast kinases, phosphatases and their substrates [81]. However, other approaches have emerged, for example, the high-throughput method termed BAC TransgeneOmics, which allows transgenes to be expressed in cultured mammalian cells under the control of their endogenous promoters and native regulatory elements, is one of the most promising technologies for the elucidation of gene expression and protein function [82].
To ensure the stability of the protein complex isolated for MS analysis, crosslinkers can be another effective tool by effectively ‘freezing’ a series of concurrently formed heterogeneous protein subcomplex species or DNA–protein complexes in their in vivo state and stabilizing complexes for subsequent purification [83]. Over recent years, a large number of chemical crosslinking reagents have been developed. Broadly, they may be classified into several categories according to their reactivity (e.g., amineor thiolreactive, and homoand hetero-bifunctional), their hydrophobicity or hydrophilicity, and the length of the spacer between the reactive groups. A balanced level of chemical crosslinking is required to preserve the native chromatin state during purification, while still allowing for solubility and interaction with affinity reagents [84,85]. In one strategy reported, live cells are treated with formaldehyde, which rapidly permeates the cell membrane and generates crosslinks between interacting proteins in the cell.
One issue for mapping nucleosome distribution is whether the chromatin should be crosslinked with formaldehyde before micrococcal nuclease digestion. The aim of crosslinking is to fix the antigen of interest to its chromatin binding site. In general, histones themselves do not need to be crosslinked as they are already tightly associated with DNA. However, to prevent histone exchange, chemical crosslinking is required. An isotopic labeling technique combining affinity purification and MS called transient isotopic differentiation of interactions as random or targeted is used for optimizing the levels of chemical crosslinking for affinity purification of cognate chromatin sections [86,87]. Besides, other DNA binding proteins that have a weaker affinity for DNA or histones may also need to be crosslinked. In some cases, crosslinking may further stabilize fragile nucleosomes at promoter and enhancer regions. Unfortunately, crosslinking with formaldehyde may also stabilize non-nucleosome protein complexes on chromatin and lead to difficulty in enzymatically digesting the crosslinked DNA. This can result in difficulties in analyzing data of some protected regions. Combination of sonication can be useful when enzymatic digestion is ineffective on fully crosslinked samples [88–90].
With the development of highly specific epitope tags and crosslinkers, it is feasible to pursue large-scale interaction mapping efforts in human cells that parallel those already performed in yeast [91]. However, it should be noted that conventional biochemical and affinity purification approaches are prone to false positive identification of interaction partners. One way to overcome the issue of ‘dirty’ pull-downs is the use of quantitative approaches using chemical crosslinking, isotope labeling and affinity purification, which offer the possibility to distinguish bona fide interaction partners from background contaminants [71]. By using epitope tagging, multiple proteins can be tagged with the same epitope and purified in an identical manner. Thus, background contaminants will be present in equal amounts in light and heavy forms across all purifications, while specific interactions will have skewed intensity ratios for heavy versus light peptides. The development of labeling methodologies, such as isotope-coded affinity tags, isobaric tags for relative and absolute quantification, and stable isotope labeling by amino acids in cell culture (SILAC), have allowed greater provision of quantitative measurements [92,93].
The major technical hurdle on the MS side for answering emerging questions is the task of examining all modification sites present simultaneously on a given histone protein [94]. Various MS techniques have been applied to histone post-translational modifications (PTMs) study [95–100]. The classical method for structural analysis of proteins by MS, now referred to as the ‘bottom-up’ approach, first digests protein samples with proteases to create peptides suitable in size for sequence determination prior to analysis by MS/MS. However, once the proteins are diced into bite-sized pieces, it is impossible to tell if two modifications are physically linked or not. However, new methodologies that use a top-down proteomics approach ensure that all detected modifications coexist. There is hope that we may, in the future, be able to look at the intact modification pattern of different histones in a given nucleosome [101,102]. Top-down methods directly measure the masses of full-length histones, and the modification level and their relative stoichiometry can be obtained by MS profiling and protein fragmentation [103,104]. Sometimes full top-down treatment is not needed because a significant number of PTMs are located in the N-terminal region of the core histones. Therefore, an alternative version to the top-down method, a ‘middle-down’ approach, has been applied to characterize large peptides that usually contain less than 50 N-terminal amino acid residues of histone tails [62]. It would be attractive to use middle and top down approaches to study longer histone pep tides (>20 amino acids), up to intact proteins, so to distinguish cis and trans, and inter/intra cooccurrence of PTMs on the same histones at specific chromatin regions [105,106].
Combined genomic & proteomic approaches for epigenetics
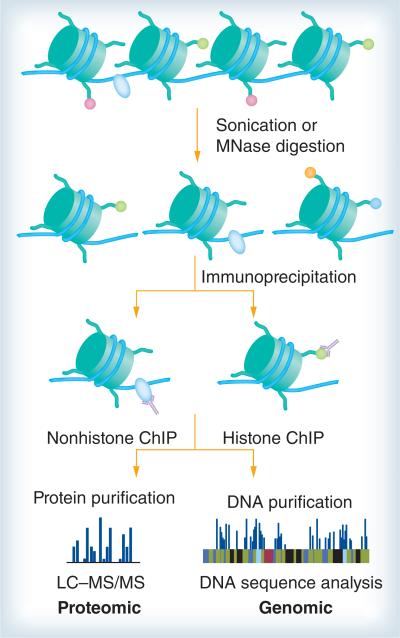
For investigation in epigenetic research, ChIP and MS are popular and highly complementary strategies (figure 1) [107]. ChIP is a reliable method for purifying DNA that is in close contact with a particular protein in animal cells, as long as the interactions are sufficiently stable and a highly specific antibody is available [108,109]. In addition, ChIP experiments provide information on the localization of modifications within the genome [110]. There are mainly two types of ChIP, crosslinked ChIP (XChIP) and native ChIP (NChIP). XChIP is used for mapping target sites of transcription factors and other chromatin-associated proteins, while NChIP is suited for mapping the DNA target of histone modifiers. The basic steps of XChIP are as follows. First, the living cells are treated with a crosslinking agent, usually formaldehyde. Next, the chromatin is sheared by sonication or enzymatic digestion, and specific DNA sequences associated with a particular protein are subjected to precipitation with specific antibodies. After the crosslinks are reversed, the purified DNA products are then analyzed by various molecular techniques to determine the genomic localization of DNA. When the target protein interacts with this locus, more DNA is pulled down and amplified, resulting in a higher signal [111,112]. Unlike XChIP, NChIP uses native chromatin sheared by micrococcal nuclease digestion without crosslinkers.
Figure 1. Combination of chromatin immunoprecipitation and mass spectrometry.
After cell lysis, the chromatin is sheared by sonication or enzymatic digestion, and specific DNA sequences associated with a particular protein are subjected to precipitation with specfic antibodies. Subsequently, the purified DNA products are analyzed by various molecular techniques to determine the genomic localization of DNA. In addition, proteins are separated by affinity purification, then analyzed by MS.
ChIP: Chromatin immunoprecipitation; LC-MS: Liquid chromotography-mass spectrometry; MS: Mass spectrometry.
The drawbacks of conventional ChIP assays have for a long time been the requirement for large numbers of cells, which limits applicability to rare cell samples (such as cells from small tissue biopsies, rare stem cell populations or cells from embryos), and/or lengthy procedures with limited applications. This has been necessary to compensate for the loss of cells upon recovery after crosslinking, for the overall inefficiency of ChIP and the relative insensitivity of ChIPenriched DNA detection. Several emerging approaches aiming to reduce cell numbers are listed in Table 1 [113,114].
Table 1.
Summary of several approaches aiming at reducing cell numbers.
| Method name | Description | Number of cells | Application | Ref. |
|---|---|---|---|---|
| CChIP | Based on conventional NChIP but using ‘carrier’ chromatin, which allows detailed and reproducible epigenetic analysis of small numbers of cells | Procedure allows ChIP assays on as few as 100 cells and it can generate consistent results from 1000 cells | The procedure has been validated with primary mouse embryo material, but should be applicable to cells from various sources, including tissue biopsies and FACS-sorted cell populations. It may also be applicable to formaldehyde XChIP, thereby allowing the analysis of nonhistone proteins | [142] |
| Q2ChIP | As an alternative to CChIP, Q2ChIP involves a chromatin preparation from a larger number of cells than CChIP, but includes chromatin dilution and aliquoting steps. In addition, Q2ChIP involves a crosslinking step, enabling the analysis of immunoprecipitation of transcription factors or other nonhistone DNA-bound proteins | 100,000 cells are used as starting material | Q2ChIP is suitable for analysis of both histone modifications and transcription factor binding from greatly reduced amounts of chromatin relative to conventional ChIP | [143] |
| μChIP | The basis of the μChIP assay was the Q2ChIP assay with modifications. This 1D μChIP assay enables the analysis of histone or RNAPII binding throughout the human genome using high-density oligonucleotide arrays (ChIP-chip) | Chromatin is usually prepared from 1000 cells. | μChIP is applicable to small fresh tissue biopsies, and a crosslink-while-thawing procedure makes the assay suitable for frozen biopsies | [144,145] |
| Microplate-based assay to enhance throughput: matrix ChIP | To increase the throughput and to simplify the assay, matrix ChIP, which utilizes surface-immobilized antibodies in a 96-well plate, was developed, where all steps from chromatin precipitation to PCR-ready DNA purification are carried out in microplate wells without sample transfers | Microplate-based number of cells | Application of all steps, from immunoprecipitation to DNA purification, is carried out in microplate wells without sample transfers, potentially enabling automation. A total of 96 ChIP assays for histone and various DNA-bound proteins can be conducted in a single day | [146] |
μChIP: Micro-chromatin immunoprecipitation; CChIP: Carrier chromatin immunoprecipitation; ChIP: Chromatin immunoprecipitation; FACS: Fluorescence-activated cell sorting; NChIP: Native chromatin immunoprecipitation; Q2ChIP: Quick and quantitative chromatin immunoprecipitation; RNAPII: RNA polymerase II; XChIP: Crosslinked chromatin.
However, ChIP only provides information on the DNA interactions of the protein chosen for precipitation, while not providing information on some region-specific modifications, variants and nonhistone-associated proteins. The only way to address this issue is to use MS [49]. At the level of individual histones, MS has emerged as a powerful analytical strategy to detect all PTMs in a detailed, unbiased and global fashion, and to reveal interplay between them [115–117]. With the development of genomic and proteomic approaches, combination of ChIP and MS has the potential to expand our knowledge of epigenetics research to a higher level.
The Garcia laboratory combined ChIP with MS technology to characterize the local chromatin make-up of histone PTM-binding proteins, alongside the characterization of proteogenomic mapping of histone PTM readers by using human HEK293 cell lines [118]. Histone PTMs were quantified by MS (ChIP–MS), and their associated DNAs were mapped using deep sequencing. The results showed that Brdand HP1-bound nucleosomes were enriched with patterns of histone PTMs, which is consistent with actively transcribed euchromatin and silent heterochromatin, respectively. More recently, ChIP–MS has been successfully used for the study of the male-specific lethal interactions on crosslinked chromatin using Drosophila S2 cells (109–1010 cells), and the success supports the general applicability of this method for unbiased analysis of chromatin-associated proteins [119]. Improvements such as development of a dual tag that will function well on crosslinked material makes it a notable possibility that ChIP–MS can be adapted to crude tissues and developmental stages in the future.
Proteins associated with DNA have the dual nature of participating in conventional soluble protein–protein interactions as well as participating in larger DNA–protein macrocomplexes [120]. The analysis of biomolecular macrocomplexes requires certain preconditions to be fulfilled. First, macrocomplexes also often show reduced stability. Furthermore, macromolecular complexes are usually composed by means of noncovalent interactions. By using conventional immunopurification protocols, these macrocomplexes can be easily lost during the purification steps as they pellet along with the DNA/chromatin and hence are not available for the downstream immunopurification step [121]. Hence, a holistic view of cellular protein–protein interactions occurring on chromatin requires the development of new purification techniques (i.e., peptide affinity purifications, chromatin templates and oligonucleotide-based affinity purifications).
Lambert et al. recently described a method for affinity purification, termed modified ChIP (mChIP) performed in Saccharomyces cerevisiae cells [121]. mChIP permits the efficient enrichment of DNA-bound proteins along with their associated protein, enabling subsequent protein identification by MS. The mChIP method relies on chromatin solubilization, with a single affinity purification step, whereby chromatinbound protein networks are isolated [122]. To date, mChIP purifications coupled to MS have been performed on more than 100 different chromatin-related baits, including histone proteins and their chaperones. Another modified, preparative version of ChIP called chromatin proteomics is a global, quantitative proteomic strategy designed by Bonaldi to analyze the protein component characterizing distinct chromatin regions [107]. Chromatin proteomics isolated native mononuclesomes up to crosslinked 500-bp oligo nucleosomal stretches derived from distinct chromatin domains in HeLa S3 cells (108–109 cells) [107]. The results revealed unique functional interactions among various chromatin modifiers, thus, suggesting potential novel roles for the identified proteins and indicating new regulatory pathways, such as a heterochromatin-specific modulation of DDR involving H2A.X and WICH, both enriched in silent domains. The strategy emerges as an additional and valuable tool, which offers the possibility of characterizing histone variants enriched at specific chromatin domains [107,123].
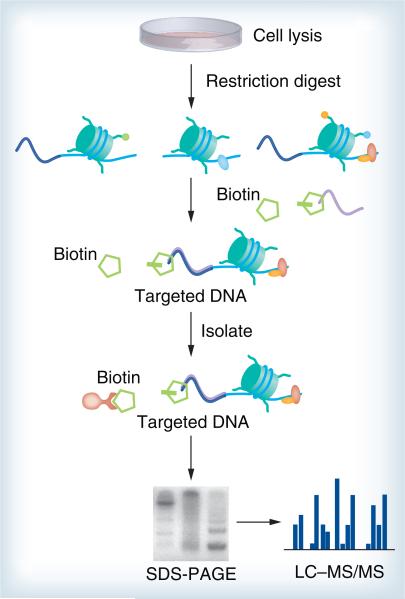
Affinity purification approaches have been devised for the isolation of a chromatin region [124]. More recently, elegant approaches that, in principle, allow the targeted enrichment of a defined genomic region are proteomics of isolated chromatin (PICh), global exonucleasebased enrichment of chromatin-associated proteins for proteomics (GENECAPP), chromatin affinity purification with MS (ChAP–MS) and insertional ChIP (iChIP) [125–129]. PICh (Figure 2) is to use a DNA probe to hybridize to a specific chromatin locus and isolate it together with all associated proteins, which are then identified by MS. PICh is opposite to ChIP, which uses protein antigens to capture the associated DNA. A global identification of telomere-associated proteins in HeLa S3 cells (109–1010 cells) was achieved by Déjardin and Kingston with PICh, where crosslinked telomeric regions enriched in repetitive DNA sequences are captured and purified with complementary probes immobilized on beads [127]. Yet, a drawback of PICh is the limited applicability to regions rich in repetitive DNA sequences. It remains to be seen if this method can be multiplexed for parallel analysis of multiple gene sequences. Furthermore, none of the methods have demonstrated sensitivity for identification of in vivo bound DNA-binding proteins under the condition of the sequence of interest being present at only a single copy per cell. To combat this problem, a new strategy named GENECAPP that is amenable to multiplexing and may offer single-copy sensitivity was developed. GENECAPP is one of the presently existing tools able to identify proteins that interact with the genome at locations of interest. Key steps in this process include crosslinking, exonuclease digestion, sequence-specific capture and MS protein identification. The success of specific hybridization capture of FoxO1 proved that it is a powerful tool for studies of protein–DNA and protein–protein interactions in in vitro model systems [128].
Figure 2. Proteomics of isolated chromatin.
After cell lysis and restriction digestion, the targeted loci are captured by primers that bind to a specific genomic region. An enzymatic step incorporates biotin labels only to chromosomal fragments that contain the targeted sequence and streptavidin-coated magnetic particles isolate the targeted chromatin. Proteins associated with the isolated regions are separated by SDS-PAGE and then analyzed by MS.
LC–MS: Liquid chromotography–mass spectrometry; MS: Mass spectrometry.
ChAP–MS is an unbiased approach whereby a unique native genomic locus is isolated, and high-resolution proteomic identification of specifically associated proteins and histone PTMs is carried out. The ChAP–MS procedure involves formaldehyde crosslinking, cell lysis, sonication, affinity purification, gel electrophoresis of eluants, trypsinization and high-resolution MS. Using ChAP–MS, an approximately 1000-bp section of GAL1 chromatin, as well as the bound proteins and histone PTMs, was specifically enriched. Compared with ChIP, ChAP–MS is a more cost-effective option for characterizing specifically bound proteins and histone PTMs. However, adaptation of this technology in mammalian cell lines may be difficult unless any advances emerge that permit ChAP–MS analysis of in vivo untagged or unaltered samples [126].
PICh has been successfully applied on telomeric sequences, of which there are 92 in a normal diploid cells, and ChAP–MS has suffered from a high level of contaminating proteins so far. It remains to be seen if any of these methods can successfully analyze proteins at single genomic locations [95]. To perform biochemical analyses of specific genomic regions retaining molecular interaction, iChIP was developed [129]. It is noteworthy that only 4 × 107 cells containing 24 copies of cHS4-core per genome are sufficient to identify p68 and Matrin-3 as protein components of the insulator complex, demonstrating that proteins bound to low-copy number loci can be analyzed by iChIP–MS [130]. However, iChIP also has some disadvantages, such as it requires insertion of LexA binding element into the target loci and expression of a tagged LexA DNAbinding domain, which may affect chromatin structure, such as nucleosome positioning, and abrogate normal genome activities, such as gene expression. Regarding the future application, iChIP is not restricted to cultured cell lines but can also be easily extended to organisms in vivo. In fact, iChIP was recently applied to cells of the entire body of a fruit fly [131]. To increase the utility of iChIP, it would be promising to compare samples prepared in different conditions, for example, different cell types, in the absence or presence of stimulation, and by combination of iChIP with isobaric tags for relative and absolute quantification or SILAC.
Large-scale affinity-based pull-down experiments have high sensitivity in protein complex identification, yet they are prone to generating numerous false positive identifications of protein–protein interactions because of overexpressed or ‘sticky’ nonspecific bait. To circumvent this problem, affinity purifications of complex subunits can be combined with endogenous tagging methods and SILAC labeling techniques [95,132,133]. SILAC provides an in vivo strategy to label the proteins with different stable isotopic forms of the amino acids, which makes it possible to quantitatively distinguish the differences at the protein level between different conditions. By comparing the ratio between the labeled and unlabeled form, the quantitative readout distinguishes specifically interacting proteins from contaminants and also identifies low-affinity interactions that might not be apparent in conventional purifications [95].
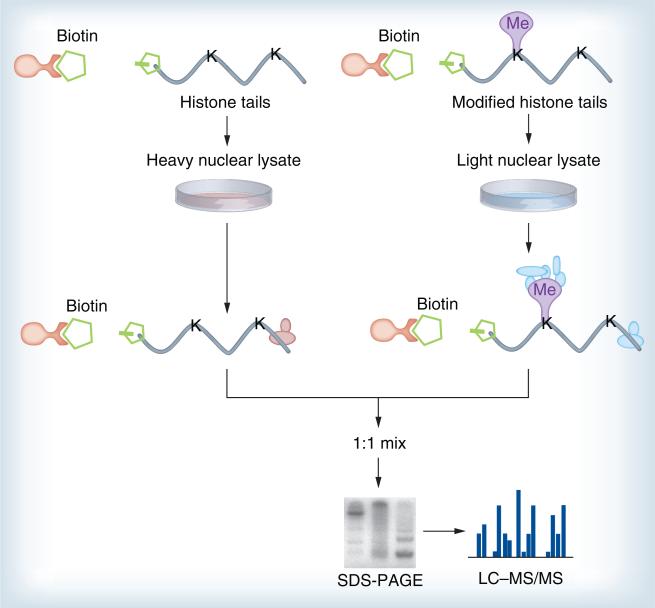
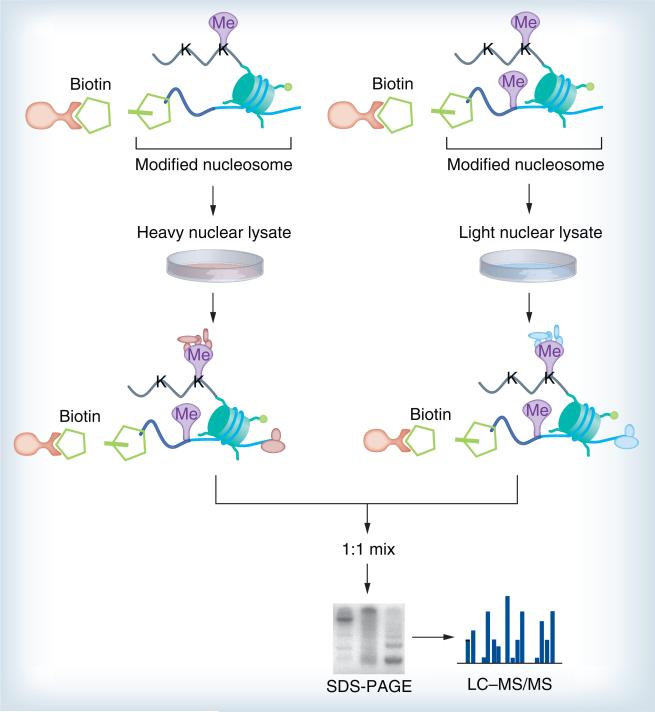
Affinity-interaction assays using histone peptide baits (Figure 3) in conjunction with SILAC-based proteomics were recently developed to pull-down and identify proteins specifically interacting with distinct H3 and H4 trimethyl lysines [70,134]. Similarly, in S. cerevisiae, analysis using inducible expression of tagged histone H3 revealed that there was a predominantly conservative inheritance of whole nucleosomes, but actively transcribed genes did contain both new and old H3–H4 dimers [135,136]. The approach has since been modified into the SILAC nucleosome affinity purification (SNAP) (Figure 4) method for capturing proteins that recognize differentially modified nucleosomes [137]. This approach has been successfully used to identify proteins (HeLa S3 cells) that are sensitive to nucleosomes methylated on histones and DNA, thus, defining categories of ‘crosstalk’ between these two distinct classes of modification. Again, isotopic labels are inverted to increase confidence in the association of a protein with a particular nucleosome. SNAP has some powerful advantages, such as nucleosomes providing a more physiological substrate and allowing the identification of proteins whose affinity may be too weak to be detected by the current methods, as well as proteins that recognize multiple independent modifications on chromatin.
Figure 3. Affinity purification of histone tails.
Histone tails can be used as baits for affinity purification, enabling the identification of proteins binding to particular histone modifications. The unmodified or modified peptides were immobilized on beads and treated with nuclear extracts from metabolically labeled cells. The beads were then washed to remove nonspecifically associated proteins and pooled. Associated proteins were eluted and mixed in a 1:1 ratio, resolved with SDS-PAGE gel, then the entire lane was in-gel digested and analyzed by LC–MS/MS.
LC–MS: Liquid chromotography–mass spectrometry; MS: Mass spectrometry.
Figure 4. Stable isotope labeling by amino acids in cell culture nucleosome affinity purification.
First, chemically modified histones are prepared, assembled into nucleosomes with biotinylated DNA and finally immobilized on streptavidin beads. The recombinant nucleosomes containing methylated or unmethylated DNA are treated with nuclear extracts from metabolically labeled cells. The beads are washed and combined, and the bound proteins are eluted and mixed in a 1:1 ratio, resolved with SDS-PAGE gel, in-gel digested and analyzed by MS.
LC–MS: Liquid chromotography–mass spectrometry; MS: Mass spectrometry.
Similarly, crosslinking-assisted and SILAC-based protein identification combines SILAC-based quantitative MS with photo-crosslinking-based histone peptide probes to identify PTM-dependent protein–protein interactions, as reported recently [138]. This probe is based on the unstructured N-terminal ‘tail’ of histone H3, with K4 trimethylated, and enables covalent capture of proteins that recognize methylated histone tails. It is possible that crosslinking assisted and SILAC-based protein identification can be extended to analyze protein–protein interactions in complex proteomes that depend on other PTMs (e.g., phosphorylation).
Methylated DNA is specifically bound by methyl-CpG binding proteins. By using biotinylated double-stranded oligonucleotides (wild-type and control baits) that have been immobilized on streptavidin magnetic beads that promote methylation-specific binding, MeCP2 and MBD3 were pulled down from Xenopus oocyte extracts [139]. In addition, there is great potential in combining methylated DNA pull-downs with MS approaches to aid the identification of proteins with DNA methylation-specific binding. A methyl-CpG pull-down assay combined with SILAC was developed for the purpose of identifying novel ‘readers’. The result proved this combination to be a generic and scalable strategy to uncover such DNA protein interactions by SILAC that uses a fast and simple one-step affinity capture of transcription factors from crude nuclear extracts (108 HeLa-S3 cells) [140]. Recently, by using this method, dynamic readers for 5-hydroxymethylcytosine and its oxidized derivatives were identified in mouse embryonic stem cells [141]. These methylated DNA affinity precipitation assays have a broad application potential as they can combine with a number of biochemical techniques and can be used in different model systems.
Future perspective
The power and scope of the genome-wide data sets generated with ChIP-seq and related techniques, in particular, now that they are combined with other technologies, such as MS, are expanding our knowledge of epigenetics research. By using ChIP-seq and related techniques, DNA sequences that are directly or indirectly bound to proteins of interest throughout the genome can be identified. In addition, recent gains in sensitivity of the proteomic techniques have enabled the detection of subtle changes in the protein composition of chromatin caused by various stimuli. But no study can yet claim to have ‘fully’ characterized all physical interactions of proteins within chromatin, even combining techniques. There are still a lot of difficulties with these techniques: for example, enriching chromatin samples from specific regions at a quantity and purity sufficient for MS analysis; identification of proteins at single genomic loci; and the vast amount of cells required for most of the combined techniques. But we believe that with the development of the combination of genomic and proteomic technologies, we can get a systems biology outlook on epigenetic processes that will lay the foundation for the development of drug treatments for human diseases and conditions that are believed to be of epigenetic origin.
Executive summary.
Background
■ The term ‘epigenetics’ has most commonly evolved to include any process that alters gene activity without changing the DNA sequence.
■ The best known epigenetic processes are DNA methylation and histone modifications.
■ Epigenetic modifications do not work alone; DNA methylation is linked to histone modifications in a mutually dependent relationship.
Genome-scale approaches to studying DNA methylation
■ Most of these are based on one of three techniques: digestion with methylation-sensitive restriction enzymes, affinity enrichment of methylated DNA or chemical conversion with sodium bisulfite.
■ The combination of different types of pretreatment followed by different analytical steps has resulted in a plethora of techniques for determining DNA methylation patterns and profiles (e.g., methyl-DNA immunoprecipitation-sequencing, methyl-DNA immunoprecipitation-chip, methylC-sequencing, reduced representation bisulfite sequencing and shotgun bisulfite sequencing of bisulfite-treated DNA).
Genome-scale approaches to studying histone modifications
■ Chromatin immunoprecipitation (ChIP) is the most wildly used method for profiling histone modifications.
■ The most prevalent technique used to map histone modifications at a genomic scale has been the combination of ChIP with DNA microarrays and next-generation sequencing.
Proteomic approaches for epigenetics research
■ Mass spectrometry (MS)-based proteomics is increasingly used in functional biological studies and has proved to be a powerful tool to characterize histone modifications as well as DNA–protein interactions.
■ Mapping DNA–protein/protein–protein interactions by MS remains a significant challenge that requires the use of affinity purification strategies or other complementary techniques such as chemical crosslinking.
■ Various MS techniques including ‘bottom-up’, ‘top-down’ and ‘middle-down’ have been applied to histone post-translational modification studies.
Combining genomic & proteomic approaches for epigenetics
■ ChIP and MS are popular and complementary strategies to investigate the epigenetic components of chromatin.
■ Recent developments in the ChIP field have led to the emergence of protocols aiming at reducing cell numbers.
■ Combining affinity purifications of complex subunits with endogenous tagging methods and stable isotope labeling by amino acids in cell culture labeling techniques allows specifically interacting proteins to be distinguished from contaminants, and also identifies low-affinity interactions that might not be apparent in conventional purifications.
Future perspective
■ The combination of genomic and proteomic technologies is expanding our knowledge of epigenetics research.
Acknowledgments
BA Garcia acknowledges funding from a National Science Foundation Early Faculty CAREER award grant, and the NIH New Innovator award grant (DP2OD007447) from the Office of the Director.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Gene Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Nakao M. Epigenetics: interaction of DNA methylation and chromatin. Gene. 2001;278:25–31. doi: 10.1016/s0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 6.Sajan SA, Hawkins RD. Methods for identification of higher‑order chromatin structure. Annu. Rev. Genomics Hum. Genet. 2012;13:59–82. doi: 10.1146/annurev-genom-090711-163818. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 8.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 9.Weinhold B. Epigenetics: the science of change. Environ. Health Persp. 2006;114:A160. doi: 10.1289/ehp.114-a160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10■.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [Describes how the function of DNA methylation is intrinsically linked to the mechanisms for establishing, maintaining and removing the methyl group.] [DOI] [PubMed] [Google Scholar]
- 11.Bird AP, Wolffe AP. Methylation-induced repression – belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 12.Dodge JE, Okano M, Dick F, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 13.Hammoud SS, Cairns BR, Carrell DT. Analysis of gene-specific and genome-wide sperm DNA methylation. Methods Mol. Biol. 2013;927:451–458. doi: 10.1007/978-1-62703-038-0_39. [DOI] [PubMed] [Google Scholar]
- 14.Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 15.Ng HH, Adrian B. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Wang L, Wang Q, Li W. Histone modifications and chromatin organization in prostate cancer. Epigenomics. 2010;2:551–560. doi: 10.2217/epi.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 22.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 23.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 25.Bird A. DNA methylation patterns and epigenetic memory. Gene Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 26.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 28.Alelu-Paz R, Ashour N, Gonzalez-Corpas A, Ropero S. DNA methylation, histone modifications, and signal transduction pathways: a close relationship in malignant gliomas pathophysiology. J. Signal. Trans. 2012;2012:956–958. doi: 10.1155/2012/956958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30■.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [Describes several methods that have been developed to map DNA methylation on a genomic scale. Most of these methods combine DNA analysis by microarrays or high-throughput sequencing with one of four ways of translating DNA methylation patterns into DNA sequence information or library enrichment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 32.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 33.Brunner AL, Johnson DS, Kim SW, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oda M, Glass JL, Thompson RF, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–3839. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coverdale LE, Martin CC. Epigenomics – genome wide modifications of cytosine and new dimensions in our understanding of differentiation and disease. Curr. Genomics. 2005;6:491–500. [Google Scholar]
- 37.Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using methylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 42.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 43.Sørensen AL, Collas P. Immunoprecipitation of methylated DNA. Methods Mol. Biol. 2009;567:249–262. doi: 10.1007/978-1-60327-414-2_16. [DOI] [PubMed] [Google Scholar]
- 44.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 48.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 51.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 53.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vega VB, Cheung E, Palanisamy N, Sung WK. Inherent signals in sequencing-based chromatin-immunoprecipitation control libraries. PLoS One. 2009;4:e5241. doi: 10.1371/journal.pone.0005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roh T, Zhao K. High-resolution, genome-wide mapping of chromatin modifications by GMAT. Methods Mol. Biol. 2008;387:95. doi: 10.1007/978-1-59745-454-4_7. [DOI] [PubMed] [Google Scholar]
- 59.Ng P, Tan JJ, Ooi HS, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84–e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li KK, Luo C, Wang D, Jiang H, Zheng YG. Chemical and biochemical approaches in the study of histone methylation and demethylation. Med. Res. Rev. 2012;32:815–867. doi: 10.1002/mrr.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 64.Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Gene Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viré E, Brenner C, Deplus R, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2005;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 66.Peters AH, Schübeler D. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 2005;17:230–238. doi: 10.1016/j.ceb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Irvine RA, Hsieh CL. Q-PCR in combination with ChIP assays to detect changes in chromatin acetylation. Methods Mol. Biol. 2004;287:45–52. doi: 10.1385/1-59259-828-5:045. [DOI] [PubMed] [Google Scholar]
- 68.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 70.Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 71■■.Vasilescu J, Figeys D. Mapping protein–protein interactions by mass spectrometry. Curr. Opin. Biotech. 2006;17:394. doi: 10.1016/j.copbio.2006.06.008. [Overview of developments in protein interaction mapping by mass spectrometry (MS) and MS-based methods that employ novel affinity purification strategies and affinity tags, as well as a description of in vivo and in vitro chemical crosslinking.] [DOI] [PubMed] [Google Scholar]
- 72.Ethier M, Lambert JP, Vasilescu J, Figeys D. Analysis of protein interaction networks using mass spectrometry compatible techniques. Anal. Chim. Acta. 2006;564:10–18. doi: 10.1016/j.aca.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 73.Bauer A, Kuster B. Affinity purification- mass spectrometry. Eur. J. Biochem. 2003;270:570–578. doi: 10.1046/j.1432-1033.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 74.Roque A, Lowe C. Affinity chromatography: history, perspectives, limitations and prospects. Methods Mol. Biol. 2008;421:1–21. [PubMed] [Google Scholar]
- 75.Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 76.Puig O, Caspary F, Rigaut G, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 77.Li Y. The tandem affinity purification technology: an overview. Biotechnol. Lett. 2011;33:1487–1499. doi: 10.1007/s10529-011-0592-x. [DOI] [PubMed] [Google Scholar]
- 78.Trinkle-Mulcahy L. Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics. 2012;12:1623–1638. doi: 10.1002/pmic.201100438. [DOI] [PubMed] [Google Scholar]
- 79.Galan JA, Paris LL, Zhang HJ, Adler J, Geahlen RL, Tao WA. Proteomic studies of Syk-interacting proteins using a novel amine-specific isotope tag and GFP nanotrap. J. Am. Soc. Mass Spectrom. 2011;22:319–328. doi: 10.1007/s13361-010-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oeffinger M. Two steps forward – one step back: advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics. 2012;12:1591–1608. doi: 10.1002/pmic.201100509. [DOI] [PubMed] [Google Scholar]
- 81.Breitkreutz A, Choi H, Sharom JR, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poser I, Sarov M, Hutchins JR, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang X, Bruce JE. Chemical crosslinking for protein–protein interaction studies. Methods Mol. Biol. 2009;492:283–293. doi: 10.1007/978-1-59745-493-3_17. [DOI] [PubMed] [Google Scholar]
- 84.Sinz A. Chemical crosslinking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 85.Leitner A, Walzthoeni T, Kahraman A, et al. Probing native protein structures by chemical crosslinking, mass spectrometry, and bioinformatics. Mol. Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Byrum SD, Taverna SD, Tackett AJ. Quantitative analysis of histone exchange for transcriptionally active chromatin. J. Clin. Bioinforma. 2011;1:17. doi: 10.1186/2043-9113-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Byrum S, Mackintosh SG, Edmondson RD, Cheung WL, Taverna SD, Tackett AJ. Analysis of histone exchange during chromatin purification. J. Integr. OMICS. 2011;1:61–65. doi: 10.5584/jiomics.v1i1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Northrup DL, Zhao K. Application of ChIP-seq and related techniques to the study of immune function. Immunity. 2011;34:830–842. doi: 10.1016/j.immuni.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Das PM, Ramachandran K, Vanwert J, Singal R. Chromatin immunoprecipitation assay. Biotechniques. 2004;37:961–969. doi: 10.2144/04376RV01. [DOI] [PubMed] [Google Scholar]
- 90.O'Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 91.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 92.Stunnenberg HG, Vermeulen M. Towards cracking the epigenetic code using a combination of high-throughput epigenomics and quantitative mass spectrometry-based proteomics. Bioessays. 2011;33:547–551. doi: 10.1002/bies.201100044. [DOI] [PubMed] [Google Scholar]
- 93.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Ueberheide BM, Mollah S. Deciphering the histone code using mass spectrometry. Int. J. Mass Spectrom. 2007;259:46–56. [Google Scholar]
- 95■■.Bartke T, Borgel J, Dimaggio PA. Proteomics in epigenetics: new perspectives for cancer research. Brief Funct. Genomics. 2013;12(3):205–218. doi: 10.1093/bfgp/elt002. [Overview of the applications of MS-based proteomics in studying various aspects of chromatin biology, the use of MS in the discovery and mapping of histone modifications and how novel proteomic approaches are being utilized to identify and study chromatin-associated proteins and multisubunit complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Young NL, Dimaggio PA, Garcia BA. The significance, development and progress of high-throughput combinatorial histone code analysis. Cell Mol. Life Sci. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 98.Zee BM, Garcia BA. Validation of protein acetylation by mass spectrometry. Methods Mol. Biol. 2013;981:1–11. doi: 10.1007/978-1-62703-305-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J. Proteomics. 2012;75:3419–3433. doi: 10.1016/j.jprot.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 100.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol. Cell Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 103.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 105.Young NL, Dimaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol. Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 107■.Soldi M, Bonaldi T. The proteomic investigation of chromatin functional domains reveals novel synergisms among distinct heterochromatin components. Mol. Cell Proteomics. 2013;12:764–780. doi: 10.1074/mcp.M112.024307. [Overview of affinity-interaction assays using different baits in conjunction with SILAC-based proteomics and the development of a global, quantitative proteomic strategy named ‘chromatin proteomics’ to analyze the protein component characterizing distinct chromatin regions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 109.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 110.Voigt P, Leroy G, Drury WJ, 3rd, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vinckevicius A, Chakravarti D. Chromatin immunoprecipitation: advancing analysis of nuclear hormone signaling. J. Mol. Endocrinol. 2012;49:R113–R123. doi: 10.1530/JME-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 113.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front. Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- 114■.Collas P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010;45:87–100. doi: 10.1007/s12033-009-9239-8. [Describes recent developments in the chromatin immunoprecipitation field that have led to the emergence of protocols aiming to reduce cell numbers.] [DOI] [PubMed] [Google Scholar]
- 115.Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2007;11(1):66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 116.Beck HC. Mass spectrometry in epigenetic research. Methods Mol. Biol. 2010;593:263–282. doi: 10.1007/978-1-60327-194-3_13. [DOI] [PubMed] [Google Scholar]
- 117.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat. Rev. Genet. 2012;13:469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leroy G, Chepelev I, Dimaggio PA, et al. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang CI, Alekseyenko AA, Leroy G, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol. 2013;20:202–209. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Price CM, Cech TR. Telomeric DNA–protein interactions of oxytricha macronuclear DNA. Gene Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- 121.Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell Proteomics. 2009;8:870–882. doi: 10.1074/mcp.M800447-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 123.Chu DS, Liu H, Nix P, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol. Cell Biol. 2003;23:9275–9282. doi: 10.1128/MCB.23.24.9275-9282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rusk N. Reverse ChIP. Nat. Methods. 2009;6:187–187. [Google Scholar]
- 126.Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP–MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Reports. 2012;2(1):198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Déjardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu CH, Chen S, Shortreed MR, et al. Sequence-specific capture of protein–DNA complexes for mass spectrometric protein identification. PLoS One. 2011;6:e26217. doi: 10.1371/journal.pone.0026217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fujita T, Fujii H. Direct identification of insulator components by insertional chromatin immunoprecipitation. PLoS One. 2011;6:e26109. doi: 10.1371/journal.pone.0026109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fujita T, Fujii H. Locus-specific biochemical epigenetics/chromatin biochemistry by insertional chromatin immunoprecipitation. ISRN Biochem. 2013;2013:913273. doi: 10.1155/2013/913273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Agelopoulos M, Mckay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012;1:350–359. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eberl HC, Mann M, Vermeulen M. Quantitative proteomics for epigenetics. Chembiochem. 2011;12:224–234. doi: 10.1002/cbic.201000429. [DOI] [PubMed] [Google Scholar]
- 133.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 134.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 135.Katan-Khaykovich Y, Struhl K. Splitting of H3–H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc. Natl Acad. Sci. USA. 2011;108:1296–1301. doi: 10.1073/pnas.1018308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein–protein interactions. J. Am. Chem. Soc. 2012;134:1982–1985. doi: 10.1021/ja210528v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bogdanovic O, Veenstra GJ. Affinity-based enrichment strategies to assay methyl-CpG binding activity and DNA methylation in early Xenopus embryos. BMC Res. Notes. 2011;4:300. doi: 10.1186/1756-0500-4-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Spruijt CG, Gnerlich F, Smits AH, et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 142.O'Neill LP, Vermilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 143.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 144.Dahl JA, Collas P. MicroChIP – a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 146.Flanagin S, Nelson JD, Castner DG, Denisenko O, Bomsztyk K. Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events. Nucleic Acids Res. 2008;36:e17. doi: 10.1093/nar/gkn001. [DOI] [PMC free article] [PubMed] [Google Scholar]