Abstract
Lipids are a major class of biological molecules and play many key roles in different processes. The diversity of lipids is on the same order of magnitude as that of proteins: cells express tens of thousands of different lipids and hundreds of proteins to regulate their metabolism and transport. Despite their clear importance and essential functions, lipids have not been as well studied as proteins. We discuss here some of the reasons why it has been challenging to study lipids and outline technological developments that are allowing us to begin lifting lipids out of their “Cinderella” status. We focus on recent advances in lipid identification, visualization, and investigation of their biophysics and perturbations and suggest that the field has sufficiently advanced to encourage broader investigation into these intriguing molecules.
Lipids are fundamental building blocks of all cells and play many important and varied roles. They are key components of the plasma membrane and other cellular compartments, including the nuclear membrane, the endoplasmic reticulum, the Golgi apparatus, and trafficking vesicles such as endosomes and lysosomes. The lipid composition of different organelles, cell types, and ultimately tissues can vary substantially, suggesting that different lipids are required for different functions (Saghatelian et al., 2006; Klose et al., 2013). Mammalian cells express tens of thousands of different lipid species and use hundreds of proteins to synthesize, metabolize, and transport them. Although the complexity and diversity of lipids approach those of proteins, we have a much poorer understanding of their functions, making lipids in many ways the “Cinderellas” of cell biology. We discuss here recent advances, and challenges, in investigating how these molecules contribute to the many biological processes in which they participate.
Like proteins, lipids can have structural (e.g., by stabilizing different membrane curvatures) or signaling roles. Posttranslational lipidation of proteins (e.g., palmitoylation or farnesylation) and carbohydrate-linked lipids (glycolipids) are also important examples of cellular lipid pools. It is clear that higher-order organization is key to most lipid functions, and they are believed to assemble into signaling platforms that contain both lipids and proteins (Kusumi et al., 2012). Some lipid microdomains are termed lipid rafts, and much ongoing effort is being focused on defining their parameters (Simons and Sampaio, 2011; Suzuki et al., 2012; Klotzsch and Schutz, 2013). Because of this focus on lipid rafts, we have a better understanding of the properties of proposed raft lipids (e.g., sphingomyelins and cholesterol) than of many other lipid species. Much of what we know about lipids has come from studying synthetic membranes with specific lipid compositions. Although model membranes usually consist of very few (<10) lipid species, they have been useful for understanding the biophysical properties of lipids. At the other end of the spectrum, lipids have been implicated in various diseases, with cholesterol metabolism being a prominent example. We are now getting to a point at which we can address the roles of lipids in the middle of the spectrum—in cells.
WHY DON'T WE KNOW AS MUCH ABOUT LIPIDS AS ABOUT OTHER BIOMOLECULES?
The answers to the preceding question are threefold: a perception among many scientists that lipids are too difficult to work with, the nature of lipids and how they function in ensembles, and a lack of techniques comparable to protein analysis to visualize lipids and manipulate their levels, both globally and locally. We argue here that recent advances are now allowing us to begin to address these challenges.
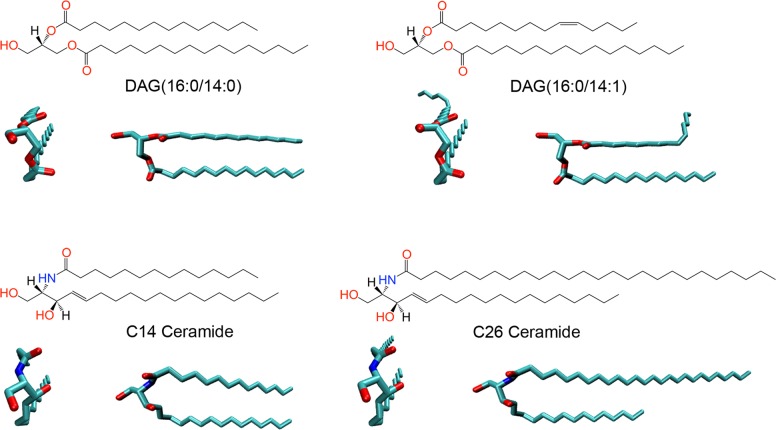
Traditionally, lipid biology has been dominated by chemists, biochemists, and biophysicists. This has resulted in a nomenclature based on chemical scaffolds and connectivities, unlike proteins, which are primarily classified according to their biological function (e.g., kinase or acyl transferase). Confusingly, some lipids with similar chemical structures have different names (e.g., diacyl glycerol and ceramide), and other lipids with similar names have quite different structures (C14 and C26 ceramides) and therefore likely different functions (Figure 1). Perhaps it would be easier to appreciate lipid complexities if we thought about them based on their chemical structures, which is not unfamiliar to biologists accustomed to looking at molecular interactions, for example, in enzymes’ active sites.
FIGURE 1:
Chemical and predicted three-dimensional (3D) structures of illustrative lipids. Chemical structures and top and side views are shown for each lipid. C14 and C26 ceramide and a diacyl glycerol (DAG) with and without a double bond are shown to illustrate differences between lipids within the same species. The major variations are in the length of the fatty acid side chain (14 vs. 26 carbons) or the degree of saturation (C14 side chain with zero or one double bond). C14 ceramide vs. DAG (16:0/14:0) is shown to illustrate similar structures that have been assigned to different lipid families. The side chains are the same in both lipids; parts of the head groups vary. The 3D structures of these lipids within membranes, especially the side chain arrangements, will likely adjust depending on the local cellular environment and lipid packing. The 3D structures were obtained using the molefacture tool of VMD 1.9.1 (www.ks.uiuc.edu/Research/vmd/allversions/cite.html) and minimized with Molecular Operating Environment 2013.08 (Chemical Computing Group, Montreal, Canada) with Amber 99 force field (Cornell et al., 1995).
Most lipids (other than sterols) contain hydrophobic side chains and polar head groups, and different combinations of the side chains and the head groups account for much of lipid diversity (Figure 1). It is well known that modifications to lipid head groups are essential for some lipid functions (e.g., phosphorylation of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2, or PIP2] into phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3, or PIP3]). However, it has only recently become clear that the identity of the hydrophobic side chains is also important (Zhang and Wakelam, 2014). For example, although it has been known that some lipids participate in cell division (Atilla-Gokcumen et al., 2010), we recently showed that dividing HeLa cells display exquisite chemical specificity in the regulation of their lipid content, including their side chains (Atilla-Gokcumen et al., 2014). Looking at chemical structures, it becomes immediately clear that the physical properties—for example, interactions within and between different leaflets of a membrane—of, say, a lipid with a 14-carbon (C14) side chain would be quite different from those of a lipid with a C26 side chain or with significant spatial perturbations due to double bonds in the side chains (Figure 1). Therefore, when investigating lipid function, it is important to consider the contributions both of side chains and of head groups.
The amphipathic nature of lipids, with the functional importance of both side chains and head groups, leads to a major reason why it has been difficult to make inroads into lipid function: lipids very often function in heterogeneous ensembles of lipids and proteins, and therefore there are few linear relationships that can be systematically dissected, analogous, for example, to kinase A phosphorylating kinase B. As we appreciate more that many proteins also function in dynamic and complex relationships, some of the systems biology approaches to studying protein networks should become applicable to lipids, especially when coupled with the techniques discussed in the next sections.
WHAT IS BEING DONE TO INVESTIGATE LIPID FUNCTION, AND WHERE DO WE STILL NEED TO GO?
There are many challenges to understanding the roles of lipids in biological processes. We need to 1) identify which lipid families and species participate in the process; 2) visualize these lipids in relevant cellular compartments or structures, preferably in live cells so that turnover can be evaluated; 3) measure physical and mechanical properties of relevant lipids and membranes, including identification of interaction partners; and 4) perturb lipid levels for phenotypic and therefore functional analyses. We highlight some strategies that are beginning to address these challenges.
Lipid identification
Proteins and nucleic acids consist of different subunits that are repeatedly linked by the same type of chemical bond, making them amenable to iterative sequencing. In contrast, different subunits within lipids have many different chemical connectivities made by complex networks of biosynthetic enzymes, which has made systematic lipid identification more challenging. While it is possible to detect some lipid species by nuclear magnetic resonance (NMR), mass spectrometry (MS) is the most general method to assign the chemical structures of different lipids, and advances over the past decade now allow the detection of small quantities in complex mixtures such as cell extracts. Relevant lipids can be identified in global or targeted lipid-profiling analyses, and we can determine which lipids are enhanced or depleted across multiple samples using software to identify compositional changes. The chemical detective work needed to propose a chemical structure for a lipid of interest based on MS fragmentation patterns has been greatly aided by the recent emergence of databases such as Lipidmaps (Fahy et al., 2007) and Metlin (Smith et al., 2005; Tautenhahn et al., 2012). Lipid MS is now getting to a stage where it can be used by a nonspecialist chemical or cell biology lab such as ours. For example, we were able to show that dividing cells regulate their lipid content and identified which lipids change with the cell cycle (Atilla-Gokcumen et al., 2014).
What's next? Next-generation MS; toward spatial resolution
Although the compositional analysis of lipids, especially in tissues, has been informative, it is not yet at the resolution that would allow systematic mechanistic insights. Usually analyses are conducted with total lipid extracts, meaning that they contain lipids from all cellular lipid compartments. To analyze specifically the composition of a region or membrane of interest, we need either to improve our ability to isolate this region biochemically for subsequent MS or to couple MS with techniques providing spatial resolution. Both strategies are gaining traction. As we learn more about membrane-based processes, we identify the proteins involved and can use these proteins as markers in biochemical isolations.
Combining MS with imaging is a promising way to assess lipid distribution in two-dimensional space. Secondary ion mass spectrometry (SIMS) can visualize the organization of isotope-labeled lipids in very high lateral resolution (∼50 nm; Klitzing et al., 2013). Matrix-assisted laser desorption ionization (MALDI) imaging also allows visualization of the spatial distribution of different biomolecules, especially large ones. It has been mostly used to characterize phospholipids in tissues at 10- to 50-μm lateral resolution (reviewed in Berry et al., 2011). Combining the detailed chemical information of MS measurements with spatial resolution will become even more important as our understanding of the localization of different lipids and their interactions with different partners during biological processes improves.
Lipid visualization
With most biological molecules, function depends on localization, and this is truer for lipids because they cannot diffuse freely across the cytoplasm. Lipids can be visualized by fluorescently tagging either lipid-binding proteins or lipids directly. Although both approaches have furthered our understanding, neither is ideal because both can perturb local interactions and packing within ensembles. Lipids can be chemically linked to a fluorophore, and fluorescent lipid derivatives are commercially available. For solubility and ease of introduction into cells, fluorescent lipids often have short hydrophobic side chains. Because it is becoming clear that cells carefully and specifically regulate their lipid side chains, suggesting that they have important functions, fluorescent lipids with short side chains have limited applications. Fluorescent fusion proteins are extensively used to investigate the localization of lipids in live and fixed cells. For example, the tagged PH domain of PLCδ1 coupled to a fluorescent protein is used to study PIP2 lipids (Stauffer et al., 1998), important regulators of the actin cytoskeleton (Varnai et al., 1999; Schultz et al., 2010).
What's next? Functional fluorescent tags visualized by superresolution microscopy
As we better understand lipid function, it will be possible to design markers with minimal functional disruption—for example, by identifying new lipid binding domains. Caged lipids (discussed later under Lipid perturbation/functional studies) are also promising, as is click chemistry to synthesize tagged lipids in situ. A bio-orthogonal chemical functionality (meaning that it does not occur in nature—e.g., alkyne or azide) can be introduced into a lipid of interest, which is then reacted or “clicked” with a corresponding probe such as a fluorophore or an affinity tag, ideally after it is in the right cellular context and bound to its interaction partners (Prescher and Bertozzi, 2005). Model lipids with “clicked” tags have been synthesized and visualized in cells (Neef and Schultz, 2009; Haberkant et al., 2013). Once the specific introduction of bio-orthogonally derivatized lipids into cells, preferably at physiological levels, has been optimized, this approach will be applicable to many different lipids because it is less disruptive of lipid packing (and therefore potentially function) due to the small size and minimal local perturbation of the “clickable” azide or alkyne groups (Grammel and Hang, 2013).
As lipid tagging improves, better imaging techniques are also needed. A major limitation of visualizing cellular lipids has been that they often localize to vesicles or small structures below the detection limit of conventional fluorescence microscopy. Recent advances in superresolution microscopy (e.g., stochastic optical reconstruction microscopy [STORM] and photoactivated localization microscopy [PALM]) are promising and can decrease the resolution limit to ∼20 nm (Sengupta et al., 2012; Owen and Gaus, 2013). Because few lipid markers exist and not all dyes are suitable for this technique, there are not yet many examples of direct lipid visualization. The development of new lipid tags suitable for PALM/STORM will be key. Other superresolution approaches are also options (Eggeling et al., 2009).
Lipid biophysical properties
The physical properties of lipids are essential because they dictate how lipids interact with other lipid and protein partners. In most protein–protein interactions relatively small parts of each protein participate in the interaction. Lipids are much smaller than proteins, and therefore a larger percentage of their total surface engages with binding partners. This makes small changes in the structure and therefore biophysical properties of individual lipids and their ensemble proportionally more relevant. Much excellent work has been done in model membranes, and this work is being translated into more complex biological systems.
The lateral organization of lipids in a membrane can give some clues about their functions and can be measured by atomic force microscopy (AFM) at nanoscale resolution. Much has been learned by AFM in model membranes (Garcia-Manyes and Sanz, 2010), and we used this technique to show that, surprisingly, lipids isolated from dividing cells form different and more rigid domains than lipids isolated from nondividing cells, even though the total change in cellular lipids was quite small (Atilla-Gokcumen et al., 2014). AFM is beginning to be used to investigate mechanical properties directly in cells—for example, at the plasma membrane. Our study showed that the force needed to break the plasma membrane in dividing versus nondividing cells was higher, consistent with the observations we made with isolated lipids (Atilla-Gokcumen et al., 2014).
What's next? Combining current techniques to address increased complexity in cells
We are only just beginning to correlate the biophysical properties of cellular lipids with function in complex systems (i.e., live cells), and it has only recently become possible to use approaches such as AFM to measure these. Other physical and analytical techniques, such as Raman spectroscopy, for example, have the potential to add valuable complementary information. Coupling some of the different techniques we discuss will help us to understand the roles of lipids and link them with protein function in specific cellular compartments. We expect that this will become possible soon: AFM and SIMS have already successfully been used together, as has been superresolution microscopy with AFM (Anderton et al., 2011; Hodges et al., 2013).
To investigate how the physical properties of lipids affect membrane organization, different dyes to sense the membrane environment are being developed. For example, Laurdan and molecular rotor dyes can be used to assess quantitatively the lipid organization state in membranes (Owen et al., 2012; Lopez-Duarte et al., 2014). Both dye families report on the plasma membrane and internal membrane compartments. It will be very informative to develop environmentally sensitive dyes compatible with superresolution microscopy.
Lipid perturbation/functional studies
To understand the function of lipids, we need to be able to manipulate their cellular and ideally subcellular levels. Unlike biomolecules for which synthesis is streamlined, for example, by the ribosome, lipids are synthesized by many different enzymes, making systematic lipid deletions analogous to RNA interference (RNAi) impossible. This leaves us with three options to modify the amounts of a lipid of interest: chemical sequestration (deplete), perturbation of biosynthetic enzymes (increase or deplete), and lipid addback (increase). For example, methyl-β-cyclodextrin is commonly used to remove cholesterol from membranes, but this approach is not general, and the lipid specificities are poorly understood. Altering the lipid composition by inhibiting lipid biosynthetic enzymes via small molecules or RNAi is more promising if some caveats are kept in mind. Lipid biosynthesis is tightly regulated, with many possible synthetic routes and interconnected feedback loops, which will likely result in different lipid profiles if short small-molecule or longer RNAi experiments are used (Eggert et al., 2006; Atilla-Gokcumen et al., 2011; Castoreno and Eggert, 2011). It is not straightforward to predict the lipid composition of cells in cases in which biosynthesis has been inhibited. For example, when we analyzed cells where we had perturbed biosynthesis, we found that their lipids were not as expected by traditional biochemistry (i.e., depletion of substrate and accumulation of product; Atilla-Gokcumen et al., 2011, 2014). However, substrate/product specificities are a secondary consideration when enzymes are used for lipid perturbation because phenotypes can be directly correlated with lipid composition as determined by MS. At present, perturbing biosynthetic enzymes or lipid-transport/binding proteins is the most systematic and comprehensive way of changing lipid levels, but it is imprecise because we do not know the specificities or regulation, including localization, of most of these proteins. As our understanding grows, we should be able to perturb lipid levels and therefore functions more accurately.
What's next? Targeted delivery of caged lipids
The major next challenges in manipulating lipid levels are the need to do this with spatial, temporal, and quantitative control in different membrane structures. A direct strategy is to simply add lipids to cells, which is not as straightforward as it may seem because many lipids are not commercially available or have limited solubility or it cannot be predicted if/where a lipid will be transported into cells. Photoactivation of chemically cloaked (or caged) lipids involves the (not trivial) synthesis of a lipid derivative with a chemical tag that can be removed upon irradiation. This approach not only allows the generation of endogenous lipids at specific times and locations in cells, but it also serves as a delivery tool because the cellular permeability of the lipid is enhanced. Further optimization may also allow control over how much lipid is delivered to a desired location. Caged lipids are promising and have been successfully used to study several lipid-mediated signaling pathways (Subramanian et al., 2010; Mentel et al., 2011).
OUTLOOK
Lipid research in cell biology is at an exciting juncture because we are finally at a stage at which we know enough to appreciate what we do not yet understand. Lipid biochemistry and biophysics have made major contributions in model membranes that can now be coupled with modern analytical approaches to translate these findings into cells and organisms. While there are too many outstanding questions to list here, one major next challenge, for example, is to understand how or whether specific lipids interact with specific membrane proteins and whether this is necessary for function. We have some clues that this is the case. For example, MS analysis of an ATPase complex found specific lipids bound to the complex (Zhou et al., 2011). Another study found that lipids move between organelles through a series of specific protein-binding events (Mesmin et al., 2013). We can address questions like these only with modern interdisciplinary science. This requires a combination of approaches that draw on biology, chemistry, physics, and engineering, including the ones we discuss here. Using such integrated strategies, we are now at a point at which we can start to uncover the many important and varied roles of lipids in cell biology.
Acknowledgments
We thank Francesca Collu and Franca Fraternali for help with the three-dimensional structures shown in Figure 1 and Roger Morris for comments on the manuscript. Our work was funded by National Institutes of Health Grant R01 GM082834, a Human Frontier Science Program Young Investigator Award, Marie Curie Career Integration Grant 304137, and European Research Council Starting Grant 306659.
Abbreviations used:
- AFM
atomic force microscopy
- C14
side chain with 14 carbons
- C26
side chain with 26 carbons
- DAG
diacyl glycerol
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
- PALM
photoactivated localization microscopy
- PH domain
pleckstrin homology domain
- PIP2 or PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PIP3 or PI(3,4,5)P3
phosphatidylinositol 3,4,5-trisphosphate
- PLCδ1
phospholipase C δ1
- RNAi
RNA interference
- SIMS
secondary ion mass spectrometry
- STORM
stochastic optical reconstruction microscopy
Footnotes
*These authors contributed equally to this work.
REFERENCES
- Anderton CR, Lou K, Weber PK, Hutcheon ID, Kraft ML. Correlated AFM and NanoSIMS imaging to probe cholesterol-induced changes in phase behavior and non-ideal mixing in ternary lipid membranes. Biochim Biophys Acta. 2011;1808:307–315. doi: 10.1016/j.bbamem.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Atilla-Gokcumen GE, Bedigian AV, Sasse S, Eggert US. Inhibition of glycosphingolipid biosynthesis induces cytokinesis failure. J Am Chem Soc. 2011;133:10010–10013. doi: 10.1021/ja202804b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilla-Gokcumen GE, Castoreno AB, Sasse S, Eggert US. Making the cut: the chemical biology of cytokinesis. ACS Chem Biol. 2010;5:79–90. doi: 10.1021/cb900256m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. Dividing cells regulate their lipid composition and localization. Cell. 2014;156:428–439. doi: 10.1016/j.cell.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev. 2011;111:6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoreno AB, Eggert US. Small molecule probes of cellular pathways and networks. ACS Chem Biol. 2011;6:86–94. doi: 10.1021/cb1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Eggert US, Field CM, Mitchison TJ. Small molecules in an RNAi world. Mol BioSyst. 2006;2:93–96. doi: 10.1039/b515335b. [DOI] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manyes S, Sanz F. Nanomechanics of lipid bilayers by force spectroscopy with AFM: a perspective. Biochim Biophys Acta. 2010;1798:741–749. doi: 10.1016/j.bbamem.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Grammel M, Hang HC. Chemical reporters for biological discovery. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberkant P, et al. In vivo profiling and visualization of cellular protein-lipid interactions using bifunctional fatty acids. Angew Chem Int Ed Engl. 2013;52:4033–4038. doi: 10.1002/anie.201210178. [DOI] [PubMed] [Google Scholar]
- Hodges J, Tang X, Landesman MB, Ruedas JB, Ghimire A, Gudheti MV, Perrault J, Jorgensen EM, Gerton JM, Saffarian S. Asymmetric packaging of polymerases within vesicular stomatitis virus. Biochem Biophys Res Commun. 2013;440:271–276. doi: 10.1016/j.bbrc.2013.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzing HA, Weber PK, Kraft ML. Secondary ion mass spectrometry imaging of biological membranes at high spatial resolution. Methods Mol Biol. 2013;950:483–501. doi: 10.1007/978-1-62703-137-0_26. [DOI] [PubMed] [Google Scholar]
- Klose C, Surma MA, Simons K. Organellar lipidomics—background and perspectives. Curr Opin Cell Biol. 2013;25:406–413. doi: 10.1016/j.ceb.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Klotzsch E, Schutz GJ. A critical survey of methods to detect plasma membrane rafts. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120033. doi: 10.1098/rstb.2012.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- Lopez-Duarte I, Vu TT, Izquierdo MA, Bull JA, Kuimova MK. A molecular rotor for measuring viscosity in plasma membranes of live cells. Chem Commun (Camb) 2014;50:5282–5284. doi: 10.1039/c3cc47530a. [DOI] [PubMed] [Google Scholar]
- Mentel M, Laketa V, Subramanian D, Gillandt H, Schultz C. Photoactivatable and cell-membrane-permeable phosphatidylinositol 3,4,5-trisphosphate. Angew Chem Int Ed Engl. 2011;50:3811–3814. doi: 10.1002/anie.201007796. [DOI] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Neef AB, Schultz C. Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed Engl. 2009;48:1498–1500. doi: 10.1002/anie.200805507. [DOI] [PubMed] [Google Scholar]
- Owen DM, Gaus K. Imaging lipid domains in cell membranes: the advent of super-resolution fluorescence microscopy. Front Plant Sci. 2013;4:503. doi: 10.3389/fpls.2013.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun. 2012;3:1256. doi: 10.1038/ncomms2273. [DOI] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry. 2006;45:9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- Schultz C, Neef AB, Gadella TW, Jr, Goedhart J. Labeling lipids for imaging in live cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5459. pdb.prot5459. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Van Engelenburg S, Lippincott-Schwartz J. Visualizing cell structure and function with point-localization superresolution imaging. Dev Cell. 2012;23:1092–1102. doi: 10.1016/j.devcel.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Subramanian D, Laketa V, Muller R, Tischer C, Zarbakhsh S, Pepperkok R, Schultz C. Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat Chem Biol. 2010;6:324–326. doi: 10.1038/nchembio.348. [DOI] [PubMed] [Google Scholar]
- Suzuki KG, Kasai RS, Hirosawa KM, Nemoto YL, Ishibashi M, Miwa Y, Fujiwara TK, Kusumi A. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat Chem Biol. 2012;8:774–783. doi: 10.1038/nchembio.1028. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wakelam MJ. Lipidomics in the analysis of malignancy. Adv Biol Regul. 2014;54C:93–98. doi: 10.1016/j.jbior.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinkovic D, Murata T, Bernal RA, Stock D, Robinson CV. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]