The 26S proteasome is ubiquitinated by proteasome-associating ubiquitination enzymes. Proteasome ubiquitination impairs proteasomal degradation and is regulated by deubiquitination, substrate binding, and cellular stress. It is proposed that in situ ubiquitination autoregulates proteasomal activity in cells.
Abstract
The 26S proteasome degrades ubiquitinated proteins, and proteasomal degradation controls various cellular events. Here we report that the human 26S proteasome is ubiquitinated, by which the ubiquitin receptors Adrm1 and S5a, the ATPase subunit Rpt5, and the deubiquitinating enzyme Uch37 are ubiquitinated in situ by proteasome-associating ubiquitination enzymes. Ubiquitination of these subunits significantly impairs the 26S proteasome's ability to bind, deubiquitinate, and degrade ubiquitinated proteins. Moreover, ubiquitination of the 26S proteasome can be antagonized by proteasome-residing deubiquitinating enzymes, by the binding of polyubiquitin chains, and by certain cellular stress, indicating that proteasome ubiquitination is dynamic and regulated in cells. We propose that in situ ubiquitination of the 26S proteasome regulates its activity, which could function to adjust proteasomal activity in response to the alteration of cellular ubiquitination levels.
INTRODUCTION
The ubiquitin (Ub)–proteasome system (UPS) is a vast network dedicated to specifically targeting proteins in the cell for degradation. The UPS plays important roles in many cellular processes, including cell signaling, antigen presentation, DNA damage response, and apoptosis (Glickman and Ciechanover, 2002; Pickart and Cohen, 2004; Finley, 2009). The process of targeting proteins for proteasomal degradation often requires attachment of a Ub chain to a substrate protein using the E1, E2, E3 enzyme cascade, in which Ub is first activated by a Ub-activating enzyme (E1) in an ATP hydrolysis–dependent manner. The activated Ub is then transferred to a Ub-conjugating enzyme (E2). The Ub-charged E2 is recruited to a substrate-bound Ub ligase (E3). Finally, Ub or a Ub chain is attached to the substrate (Deshaies and Joazeiro, 2009; Metzger et al., 2012). The system derives its specificity mainly through hundreds of E3s, each of which targets specific proteins and interacts with one or a subset of E2s.
The ∼2.5-MDa 26S proteasome consists of two large subcomplexes—the 20S proteasome and the 19S regulatory particle (called PA700 in mammals; Glickman and Ciechanover, 2002; Pickart and Cohen, 2004; Finley, 2009). The 20S proteasome is composed of four heteroheptametric rings. The inner two are formed by the β subunits and house the peptidase activities, and the outer two are formed by the α subunits, which gate access to the inner chambers (Groll et al., 1997). The gate can be opened by PA700 (Chu-Ping et al., 1994), which directly binds the α rings (Beck et al., 2012; da Fonseca et al., 2012; Lander et al., 2012; Lasker et al., 2012). PA700 facilitates processing and degradation of mainly polyubiquitinated substrates. It binds ubiquitinated substrates with Ub receptors, including the S5a and Adrm1 subunits (Deveraux et al., 1994; Husnjak et al., 2008; Schreiner et al., 2008). It removes the Ub tag and recycles Ub using the proteasomal deubiquitinating enzymes (DUBs) Uch37, Usp14, and Rpn11 (Lam et al., 1997; Verma et al., 2002; Yao and Cohen, 2002; Hanna et al., 2006). Finally, it unfolds and translocates protein substrates into the degradation chamber of the 20S proteasome using its six ATPases (Thrower et al., 2000; Liu et al., 2006), which form a hexametric ring (Tomko et al., 2010). The 26S proteasome is made up of 33 core proteins. In addition, dozens of proteins associate with the 26S proteasome at substoichiometric levels and are referred to as proteasome-associating proteins (Verma et al., 2000).
Some of the proteasome-associating proteins play important roles in regulating proteasomal activity. For instance, the two proteasome-associating DUBs Usp14 and Uch37 catalyze polyUb chain trimming by shortening polyUb chains (Lam et al., 1997; Hanna et al., 2006), which could either promote or inhibit proteasomal degradation (Liu and Jacobson, 2013). In budding yeast, the Ub ligase Hul5 associates with the 26S proteasome and enhances substrate degradation through elongation of short Ub chains attached on substrate proteins (Crosas et al., 2006). Ub-shuttling proteins such as Rad23/hHR23 and DSK2 can aid in delivering ubiquitinated proteins to the 26S proteasome (Elsasser et al., 2004; Verma et al., 2004). The phosphatase UBLCP1 dephosphorylates proteasomal subunits in the nucleus and functions to inhibit 26S proteasome assembly (Guo et al., 2011). In contrast, ADP-ribosylation of PI31, a 20S proteasome inhibitor (Chu-Ping et al., 1992), reduces its affinity for the 20S proteasome and promotes 26S proteasome assembly (Cho-Park and Steller, 2013). Intriguingly, some of the proteasome-associating proteins may assist the proteasome to cope with environmental changes. For example, NUB1L enhances proteasomal degradation of proteins modified with the Ub-like protein FAT10 in response to interferon signaling (Tanji et al., 2005; Rani et al., 2012). Ecm29 associates with the proteasome in response to oxidative stress and promotes disassembly of the 26S complex into 19S and 20S complexes (Wang et al., 2010). In addition, association of Ubp6/Usp14 on the 26S proteasome can be regulated by levels of free Ub in cells (Hanna et al., 2007). Thus proteasome-associating proteins appear to provide additional means to regulate proteasomal activity.
In this study we find that, while incorporated in the 26S proteasome complex, Adrm1, S5a, Rpt5, and Uch37 are ubiquitinated in situ by proteasome-associating ubiquitination enzymes. This modification is antagonized by the proteasome-associating DUBs, by the binding of polyUb chains onto the proteasome, and by certain cellular stress. Functionally, ubiquitination of the 26S proteasome impairs binding, deubiquitination, and degradation of ubiquitinated proteins. Thus our study unravels a potential mechanism for autoregulating proteasomal activity via in situ ubiquitination, which may function to adjust proteasomal activity in response to the alteration of cellular ubiquitination levels.
RESULTS
Multiple protein ubiquitination enzymes interact with the human 26S proteasome
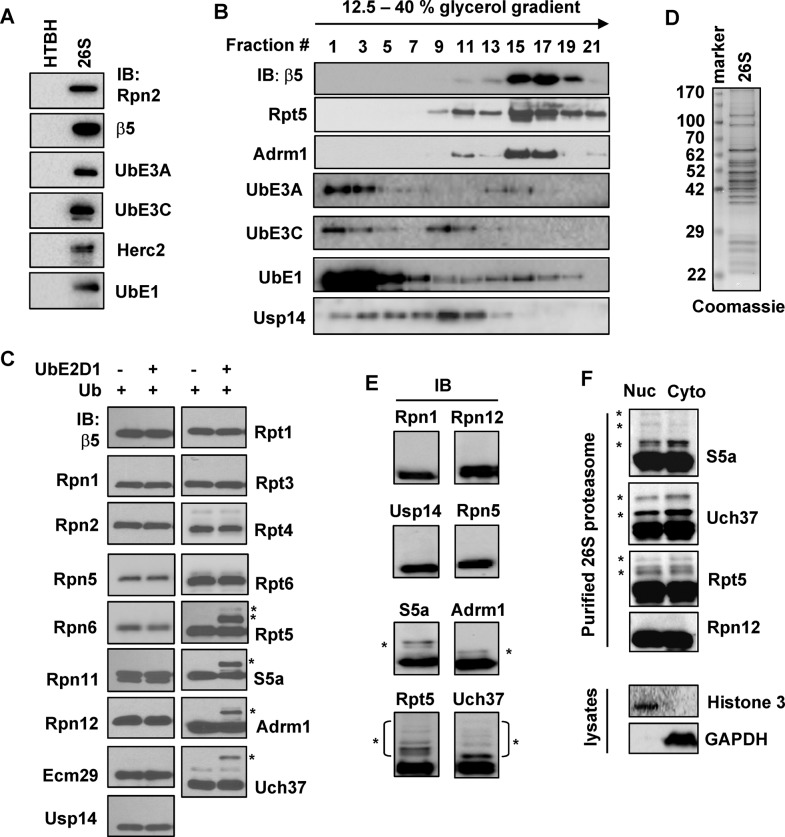
In an effort to identify human 26S proteasome–interacting proteins, we used a previously established method to purify the 26S proteasome from 293T cells stably expressing Rpn11-HTBH. The HTBH tag contains a hexahistidine (6xHis), a TEV protease site, an in vivo biotinylation site, and another 6xHis (Wang et al., 2007; Jacobson et al., 2009). The resulting 26S proteasome showed multiple characteristic bands on SDS–PAGE (Supplemental Figure S1A). Native-PAGE showed that the purified 26S proteasome was resolved mostly as a single catalytically active band (Supplemental Figure S1B). Mass spectrometric analysis identified all subunits of the 26S proteasome, along with several enzymes involved in ubiquitin conjugation, including UbE1, UbE3A, UbE3C, Herc2, Huwe1, and Ubr4 (Supplemental Table S1). The presence of several of these proteins was confirmed by immunoblotting, none of which was identified in the negative control purification using cells expressing the HTBH tag alone (Figure 1A). The finding of several of these E3s in purified 26S proteasome is in agreement with other proteomics studies (Wang et al., 2007; Besche et al., 2009; Tai et al., 2010; Martinez-Noel et al., 2012). A significant portion of each of these proteins dissociated from the 26S proteasome in glycerol gradient sedimentations (Figure 1B). However, this loose association is commonly observed with proteasome-associating proteins, such as the DUB Usp14, which also largely disassociated from the proteasome (Figure 1B). Overall these results suggest that UbE1, UbE3A, UbE3C, and Herc2 are human 26S proteasome-associating proteins.
FIGURE 1:
Four proteins on the human 26S proteasome are ubiquitinated. (A) Samples purified from 293T cells stably expressing the HTBH tag (control) or Rpn11-HTBH (1 μg of 26S proteasome) using streptavidin resin were resolved on SDS–PAGE and immunoblotted against either proteasomal subunits or the protein ubiquitination enzymes identified by mass spectrometry. (B) A 30-μg amount of purified 26S proteasome was subjected to a 12.5–40% glycerol gradient sedimentation (detailed in Supplemental Materials and Methods). Proteins in fractions of the glycerol gradient sedimentation assay were separated on SDS–PAGE and analyzed by immunoblotting. (C) UbE2D1 promotes ubiquitination of Adrm1, S5a, Rpt5, and Uch37 on the 26S proteasome. Purified 26S proteasome (80 nM) was incubated with Ub (50 μM) or Ub plus UbE2D1 (2 μM) for 1 h. Ub aldehyde and epoxomicin were added to inhibit DUB and degradation activities, respectively. Ubiquitination of proteasomal subunits was assayed by immunoblotting against a panel of proteasome subunits or associating proteins. Ubiquitinated species are marked with asterisks (this applies to all other figures). (D) Coomassie-stained SDS–PAGE of 3 μg of 26S proteasome purified from 293T Rpn11-HTBH cells using the protocol that prevents deubiquitination. (E) Immunoblotting the purified human 26S proteasome shown in D. (F) Proteasome ubiquitination occurs in both the cytoplasm and the nucleus. 293T Rpn11-HTBH cells were fractionated into cytoplasmic and nuclear fractions as confirmed by immunoblotting of cell lysates with antibodies against histone 3 (a nucleus marker) and glyceraldehyde-3-phosphate dehydrogenase (a cytoplasm marker). The 26S proteasome was purified by the method that prevents deubiquitination and analyzed by immunoblotting.
S5a, Adrm1, Uch37, and Rpt5 are ubiquitinated on the 26S proteasome
We wondered whether the proteasome-associating E1 and E3s could be acting to ubiquitinate any of the proteasomal subunits. To investigate this, we set up in vitro ubiquitination reactions by incubating purified 26S proteasome with the E2 enzyme UbE2D1, which is known to work with both UbE3A and UbE3C to ubiquitinate proteins (Wang and Pickart, 2005). In a screening using 17 antibodies against different proteasomal proteins, we found that S5a, Adrm1, Uch37, and Rpt5 were the only proteins to be ubiquitinated, predominantly as monoubiquitinated forms based on their molecular weights (marked by asterisks in Figure 1C). No ubiquitination of these proteins in our normally purified 26S proteasome was readily observed (Figure 1C). We reasoned this is likely due to deubiquitination during purification. We therefore modified our purification procedure to prevent deubiquitination by adding protease inhibitors to the cell lysis buffer, shortening the affinity binding time, and directly eluting the streptavidin resin–bound 26S proteasome with SDS sample buffer. Proteasome purified with this method showed characteristic proteasome band patterns when resolved on SDS–PAGE (Figure 1D). Immunoblotting showed that S5a, Adrm1, Uch37, and Rpt5, but not other examined subunits, had slower-migrating populations (Figure 1E), suggesting ubiquitination of these proteins. Comparison of proteasome purified to preserve ubiquitination with the in vitro ubiquitinated proteasome showed nearly identical slower-migrating species for all four proteins, indicating that they are in fact ubiquitinated forms (Supplemental Figure S2). Moreover, cellular fractionation experiments determined that ubiquitinated 26S proteasome existed in both the cytoplasm and the nucleus (Figure 1F). In summary, a population of proteasomal S5a, Adrm1, Uch37, and Rpt5 is ubiquitinated in cells.
The UbE2D family of E2s promotes ubiquitination of the 26S proteasome
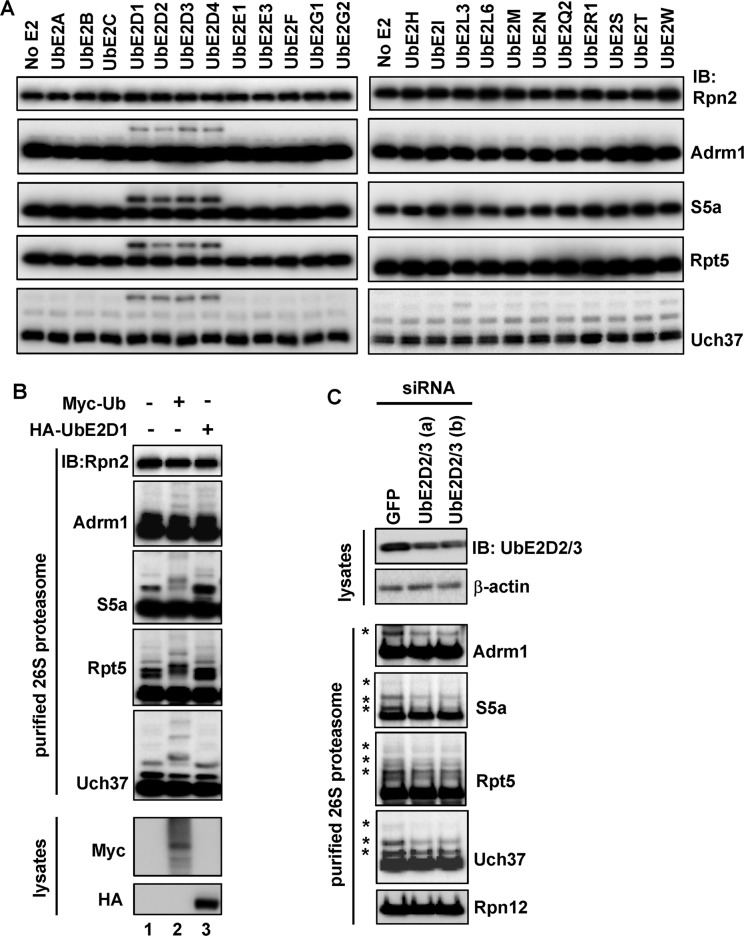
We next sought to identify which E2s mediate ubiquitination of these four proteins. Using similar in vitro ubiquitination reactions as shown in Figure 1C, we screened 23 E2s. Only the four members of the UbE2D family were able to strongly promote ubiquitination of Adrm1, S5a, Rpt5, and Uch37 on the 26S proteasome (Figure 2A). In addition, UbE2L3 and UbE2W slightly enhanced ubiquitination of Uch37 (Figure 2A). Next, we inquired whether UbE2D1 could enhance ubiquitination of the 26S proteasome in cells. To answer this, we transfected 293T Rpn11-HTBH cells with Myc-Ub or HA-UbE2D1 and then purified the proteasome using the method that prevents deubiquitination. In the control assays, overexpression of Myc-Ub promoted ubiquitination of Adrm1, S5a, Rpt5, and Uch37 but not Rpn2 (compare lanes 1 and 2 in Figure 2B). Overexpression of UbE2D1 was also able to promote ubiquitination of all these proteins except Uch37 (compare lanes 1 and 3 in Figure 2B). In agreement with these results, simultaneous knockdown of UbE2D2 and UbE2D3, two highly homologous UbE2D members (Jensen et al., 1995), using small interfering RNA (siRNA) oligos in 293T Rpn11-HTBH cells attenuated ubiquitination of all four proteins on the purified 26S proteasome (Figure 2C). Thus members of the UbE2D family of E2s are capable of promoting ubiquitination of S5a, Adrm1, Uch37, and Rpt5 on the 26S proteasome both in vitro and in cells.
FIGURE 2:
The UbE2D family of E2s promotes ubiquitination of the 26S proteasome. (A) The UbE2D family of E2s promotes ubiquitination of the 26S proteasome in vitro. Reactions were analogous to those in Figure 1C, except that each reaction contained a different E2. (B) UbE2D1 promotes ubiquitination of the 26S proteasome in cells. 293T Rpn11-HTBH cells were transiently transfected with the designated plasmids for expression of Myc-Ub or HA-UbE2D1. The 26S proteasome was purified from these cells and analyzed by immunoblotting. Overexpression of each protein was confirmed by immunoblotting of the whole-cell lysates. (C) Knockdown of UbE2D2 and UbE2D3 impairs ubiquitination of the 26S proteasome. siRNA oligos against GFP or two separate sets of oligos against UbE2D2 and UbE2D3 (a and b) were transfected into 293T Rpn11-HTBH cells, and cells were further cultured for 72 h. The 26S proteasome was then purified from these cells and analyzed by immunoblotting. Knockdown of UbE2D2 and UbE2D3 was confirmed by immunoblotting of the whole-cell lysates using an antibody recognizing both UbE2D2 and UbE2D3.
UbE3A is sufficient for promoting ubiquitination of the 26S proteasome
We next sought to determine whether the proteasome-associated E3s mediate ubiquitination of the 26S proteasome. To this end, we added recombinant UbE3A into the in vitro ubiquitination assay. UbE3A promoted monoubiquitination along with several higher–molecular weight species of all four proteins (compare lanes 4 and 6 in Figure 3A). The higher–molecular weight species are likely polyubiquitinated proteins because the lysine-less Ub(K0), which is only capable of forming monoubiquitination or multi-monoubiquitination, could not support their formation (compare lanes 3 and 4 in Figure 3B). To address whether UbE3A is sufficient for ubiquitination of these proteins on the proteasome, we used purified bovine PA700 to reconstitute the ubiquitination assay. Purified bovine PA700 did not have UbE1, UbE3A, or UbE3C (Figure 3C), likely because they dissociated from PA700 during the multistep purification process (Chu-Ping et al., 1994). When PA700 was used in ubiquitination reactions with UbE1 and UbE2D1, only very faint monoubiquitination was detected for S5a, Rpt5, and Uch37 (Figure 3D, lane 4). Ubiquitination of S5a and Uch37 on PA700 was strongly promoted by addition of UbE3A, whereas Rpt5 ubiquitination was only mildly increased (Figure 3D, lane 6). We were unable to detect bovine Adrm1 ubiquitination using two different anti-Adrm1 antibodies. One antibody did not recognize bovine Adrm1, and the other had cross-reacting bands that migrated near the molecular weight of the ubiquitinated Adrm1 species. Together these results indicate that UbE3A is sufficient to promote ubiquitination of the 26S proteasome in vitro.
FIGURE 3:
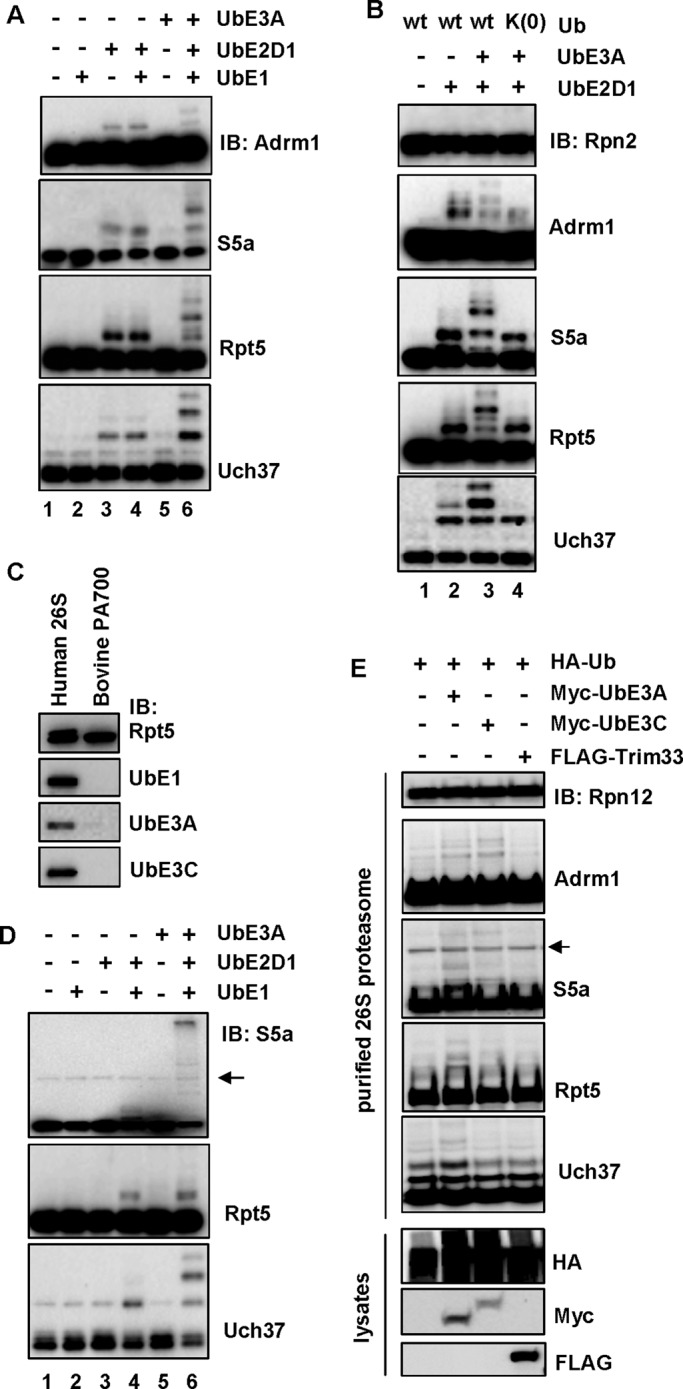
UbE3A is sufficient for ubiquitination of the 26S proteasome. (A) UbE3A stimulates ubiquitination of Adrm1, S5a, Rpt5, and Uch37 on the 26S proteasome in vitro. Purified 26S proteasome was incubated with combinations of UbE1 (100 nM), UbE2D1 (2 μM), and UbE3A (1 μM). Protein ubiquitination was analyzed by immunoblotting. (B) The lysine-less Ub mutant, Ub(K0), supports only monoubiquitination of Adrm1, S5a, Rpt5, and Uch37. Reactions were analogous to those in A, except that Ub(K0) reactions contained Ub(K0) (50 μM) instead of Ub. (C) Purified bovine PA700 does not contain UbE1, UbE3A, and UbE3C. A 1-μg amount of purified human 26S proteasome or 0.5 μg of purified bovine PA700 was resolved on SDS–PAGE and immunoblotted with the indicated antibodies. (D) UbE3A stimulates ubiquitination of S5a, Rpt5, and Uch37 on purified bovine PA700. The assay was analogous to those in A, except that 160 nM PA700 was used instead of the 26S proteasome. The arrow marks a potential nonspecific protein recognized by the anti-S5a antibody (same for E). (E) UbE3A stimulates ubiquitination of the 26S proteasome in cells. 293T Rpn11-HTBH cells were transiently transfected with the designated plasmids for expression of various proteins. The 26S proteasome was purified from these cells and analyzed by immunoblotting. Expression of each protein was confirmed by immunoblotting of the whole-cell lysates.
The proteasome-associating E3s may redundantly mediate ubiquitination of the 26S proteasome
We next examined whether overexpression of proteasome-associated E3s could enhance ubiquitination of the 26S proteasome in cells. We transfected 293T Rpn11-HTBH cells with HA-Ub alone or HA-Ub plus Myc-UbE3A, Myc-UbE3C, or FLAG-Trim33, followed by purification of the 26S proteasome. In comparison with reactions overexpressing HA-Ub, cooverexpression with Myc-UbE3A enhanced ubiquitination of all four proteins, whereas UbE3C increased only Adrm1 and S5a ubiquitination (Figure 3E). In contrast, expression of FLAG-Trim33, an E3 ligase not found on our purified 26S proteasome, had no effect on ubiquitination of any of these four proteins (Figure 3E). We also knocked down UbE3A and UbE3C using siRNA. Although the levels of both proteins were decreased by ∼70%, no effect was observed on ubiquitination of the 26S proteasome (unpublished data). On one hand, this could be caused by low knockdown efficiency. On the other hand, in combination with the overexpression data, the results suggest that the E3s redundantly mediate ubiquitination of the 26S proteasome. Unfortunately, we were not able to purify active UbE3C or overexpress or purify the other three extremely large proteasome-associated E3 ligases (>480 kDa) to examine their effects on ubiquitination of the 26S proteasome.
Ubiquitination of the 26S proteasome impairs processing of ubiquitinated proteins
We next wanted to examine the effect of ubiquitination of the 26S proteasome on proteasomal activities, including binding/deubiquitination of polyUb chains and degradation of ubiquitinated proteins. Before examining these activities, we first wanted to ensure that the ubiquitinated subunits were still incorporated into the 26S complex. Normal and ubiquitinated 26S proteasomes were obtained by setting up reactions as described in Figure 1C. The resulting 26S proteasomes were then separated by glycerol gradient sedimentations. Immunoblotting revealed that the ubiquitinated proteins comigrated with the 26S proteasome and the subunit distribution pattern was very similar between nonubiquitinated and ubiquitinated 26S proteasome (Supplemental Figure S3). Thus ubiquitination of these four proteins does not affect their association with the 26S proteasome.
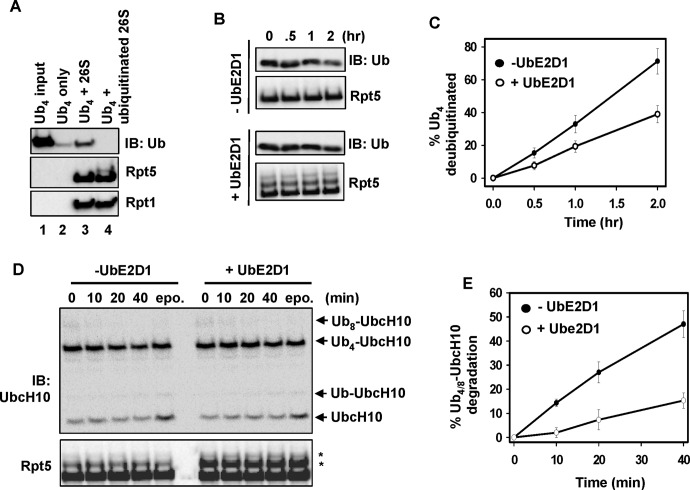
All four ubiquitinatable subunits are implicated in the binding of polyUb chains (Deveraux et al., 1994; Lam et al., 2002; Husnjak et al., 2008; Schreiner et al., 2008). We first examined whether ubiquitination of the 26S proteasome influences its ability to bind polyUb chains. We used a size-exclusion spin column binding assay to monitor polyUb chain binding (Jacobson et al., 2009). Nonubiquitinated and ubiquitinated 26S proteasome were prepared as described, followed by treatment with Ub aldehyde to block the proteasome's polyUb chain–trimming activity. K48 Ub4 chain was then added and incubated with the proteasome, followed by passage through the size-exclusion spin columns. The flowthrough was separated on SDS–PAGE and immunoblotted against Rpt5, Rpt1, and Ub. We found that K48 Ub4 alone was trapped in the spin column and not detected in the flowthrough (Figure 4A, lane 2). Proteasome-bound Ub4 was found in reactions with nonubiquitinated 26S proteasome (Figure 4A, lane 3) but not with ubiquitinated 26S proteasome (Figure 4A, lane 4). Therefore ubiquitination of the 26S proteasome impairs binding of polyUb chains. Accordingly, we would predict 26S proteasome-mediated deubiquitination to be inhibited as well. Indeed, ubiquitination of the 26S proteasome inhibited deubiquitination of K48 Ub4 (Figure 4, B and C; Ub aldehyde was omitted when preparing proteasome for this assay). Moreover, ubiquitination of the 26S proteasome inhibited degradation of Ub4 (K48-linked)-UbcH10 (Figure 4, D and E), an established model substrate for assaying proteasomal degradation in vitro (Liu et al., 2006; Jacobson et al., 2009; Ub aldehyde and epoxomicin were omitted when preparing proteasome for this assay). Together these results indicate that ubiquitination of the 26S proteasome impairs substrate binding, deubiquitination, and degradation.
FIGURE 4:
Ubiquitination of the 26S proteasome impairs processing of ubiquitinated proteins. (A) Ubiquitination of the 26S proteasome impairs binding of K48 Ub4. Ub-chain binding was examined using a size-exclusion spin column assay (detailed in the Supplemental Materials and Methods). The flowthrough was analyzed by immunoblotting. (B) Ubiquitination of the 26S proteasome inhibits deubiquitination of K48 Ub4. The reactions contained 20 nM ubiquitinated or nonubiquitinated 26S proteasome and 300 nM K48-Ub4. Time course–dependent deubiquitination was assayed by immunoblotting of Ub. (C) Quantitation of K48 Ub4 deubiquitination. The Ub4 band intensity in three independent experiments similar to that shown in B was quantitated by densitometry and normalized to the band intensity at time 0. The error bars represent SD. (D) Ubiquitination of the 26S proteasome inhibits degradation of Ub4 (K48)-UbcH10. The reactions contained 30 nM nonubiquitinated or ubiquitinated 26S proteasome and 100 nM Ub4 (K48)-UbcH10. (E) Quantitation of Ub4(K48)-UbcH10 degradation. Both the Ub8 and Ub4 (K48)-UbcH10 bands in two independent experiments similar to that shown in D were quantitated by densitometry and normalized to the band intensity at time 0. The error bars represent SD.
Proteasomal polyUb chain–trimming enzymes antagonize ubiquitination of the proteasome
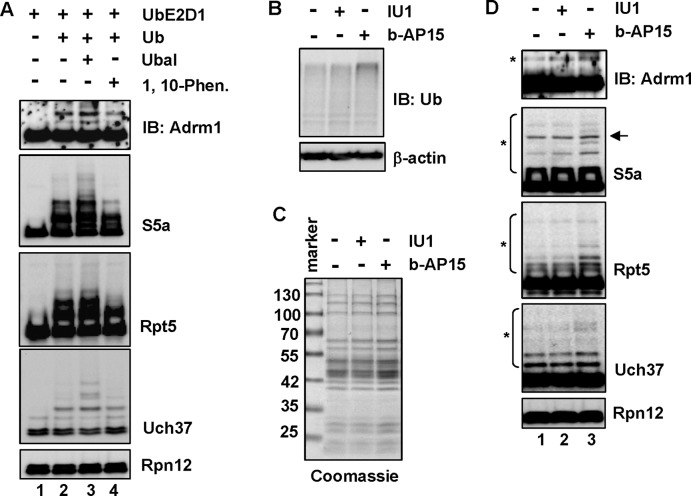
The three known DUBs on the 26S proteasome catalyze two distinct deubiquitinating activities. Uch37 and Usp14 are cysteine proteases that trim polyUb chains, often from the distal end (Lam et al., 1997; Hanna et al., 2006). In contrast, Rpn11 is a Zn2+-dependent metalloprotease that amputates the whole polyUb chain from a substrate by attacking the bond that links Ub to the substrate (Yao and Cohen, 2002). To determine whether DUBs of the 26S proteasome regulate proteasomal ubiquitination, we set up in vitro ubiquitination reactions in which Ub aldehyde (an inhibitor of both Usp14 and Uch37) or 1,10-phenanthroline (an inhibitor of Rpn11) was added in the reactions. Inhibition with Ub aldehyde resulted in a substantial increase of ubiquitination of all four proteins (compare lanes 2 and 3 in Figure 5A), whereas inhibition of Rpn11 with 1,10-phenanthroline had no effect on ubiquitination (compare lanes 2 and 4 in Figure 5A). Thus the DUB activities of Uch37 and/or Usp14 appear to antagonize ubiquitination of the 26S proteasome.
FIGURE 5:
The Ub chain–trimming enzymes on the 26S proteasome mediate in situ deubiquitination of the proteasome. (A) Ub aldehyde (Ubal) treatment increases ubiquitination of the 26S proteasome. Purified human 26S proteasome (80 nM) was incubated for 1 h with UbE2D1 (2 μM), Ub (50 μM), Ubal (2.5 μM), 1,10-phenathroline (5 mM), or their combination as indicated. Ubiquitination of Adrm1, S5a, Rpt5, and Uch37 was assayed by immunoblotting. (B) Immunoblotting of Ub and β-actin in 293T Rpn11-HTBH cells treated with 75 μM IU1 for 6 h or 0.75 μM b-AP15 for 2 h. (C) Coomassie-stained SDS–PAGE of 3 μg of 26S proteasome purified from cells described in B. (D) Immunoblotting of Adrm1, S5a, Rpt5, Uch37, and Rpn12 (loading control) on the purified human 26S proteasome shown in C. The arrow indicates a possible cross-reacting band in the S5a blot.
We next took advantage of the two recently developed cell-permeable inhibitors of proteasomal Ub chain–trimming enzymes to determine their effects on proteasome ubiquitination. IU1 is a Usp14-specific inhibitor (Lee et al., 2010), whereas b-AP15 inhibits both Usp14 and Uch37 (D'Arcy et al., 2011). Treating 293T Rpn11-HTBH cells with either b-AP15 or IU1 caused a mild increase of cellular ubiquitination levels under our experimental conditions (Figure 5B). The 26S proteasome was then purified by the method that prevents deubiquitination (Figure 5C). b-AP15 treatment increased ubiquitination of Adrm1, S5a, and Rpt5, with less effect on Uch37. IU1 treatment also caused mild enhancement of ubiquitination of Adrm1 and S5a (Figure 5D). Together these results indicate that ubiquitination of the 26S proteasome is antagonized in situ by proteasomal polyUb chain trimming enzymes.
Ubiquitination of the 26S proteasome is abolished by the binding of long polyUb chains
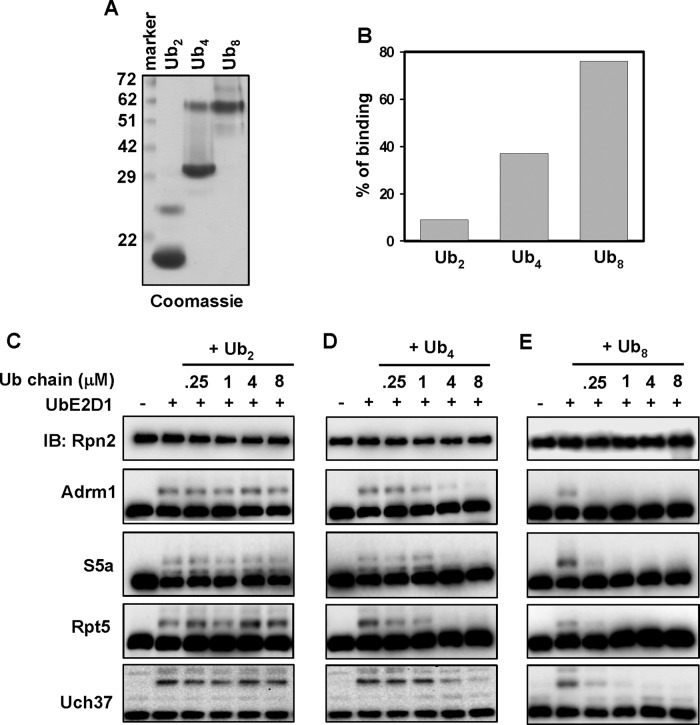
S5a, Adrm1, Uch37, and Rpt5 are all implicated in binding of polyUb chains. We therefore asked whether the binding of polyUb chains on the 26S proteasome influences ubiquitination of these proteins. To test this, we synthesized K48-linked Ub2, Ub4, and Ub8 using a reported method (Figure 6A; Raasi and Pickart, 2005). Consistent with a previous finding (Thrower et al., 2000), purified human 26S proteasome preferred to bind longer polyUb chains. Ub2 had negligible binding, whereas Ub4 and Ub8 bound with increasing affinity (Figure 6B and Supplemental Figure S4).
FIGURE 6:
Ubiquitination of the 26S proteasome is blocked by the binding of polyUb chains. (A) Coomassie-stained SDS–PAGE of 10 μg of synthesized K48 Ub2, Ub4, or Ub8. (B) Quantitation of proteasome binding of various Ub chains. Densitometric quantitation from the size-exclusion spin column assay shown in Supplemental Figure S4. The percentage of each chain bound to the proteasome in relation to the total input. (C–E) The binding of a polyUb chain on the 26S proteasome inhibits proteasome ubiquitination in vitro. Purified 26S proteasome was first preincubated with Ub aldehyde and epoxomicin to inhibit DUB and degradation activities, respectively. Proteasomes were then preincubated with different concentrations of Ub chains as designated. Finally, reactions were initiated by the addition of UbE2D1 and incubated for 1 h at 37°C. Ubiquitination of the 26S proteasome was assayed by immunoblotting.
We next examined the effect of these polyUb chains on S5a, Adrm1, Uch37, and Rpt5 ubiquitination by supplementing increasing concentrations of these chains into in vitro ubiquitination assays. We found that K48-Ub2 had no effect on ubiquitination of any of the four proteins up to the highest concentration of 8 μM (Figure 6C), K48-Ub4 inhibited ubiquitination of all four proteins at 4 μM (Figure 6D), and K48-Ub8 significantly inhibited ubiquitination of all four proteins at 0.25 μM (Figure 6E). Thus the inhibitory effect of these chains increased with their ability to be bound by the proteasome. Taken together, these data show that binding of polyUb chains on the 26S proteasome can block in situ ubiquitination of all four proteins.
Cellular stress affects ubiquitination of the 26S proteasome
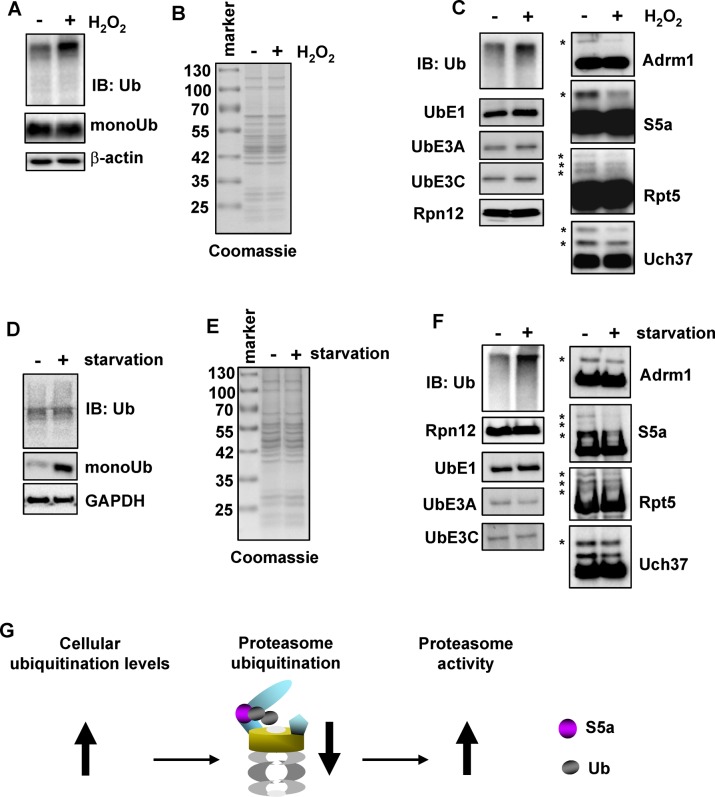
The foregoing result indicates that substrate binding might regulate ubiquitination of the 26S proteasome. Cellular ubiquitination levels are often altered under stress conditions (Fuertes et al., 2003; Shang and Taylor, 2011; Flick and Kaiser, 2012). We therefore investigated the effect of cellular stress on proteasome ubiquitination. To this end, we exposed 293T Rpn11-HTBH cells to oxidative stress by treatment with H2O2, which causes a strong increase in polyubiquitinated proteins (Figure 7A). We then purified the 26S proteasome from these cells using the method that prevents deubiquitination. Coomassie-stained SDS–PAGE of purified 26S proteasome from normal and H2O2-treated cells showed similar band patterns, indicating that our H2O2 treatment did not cause dissociation of the 26S proteasome (Figure 7B). In addition, it did not cause dissociation of UbE1 or the E3 ligases from the 26S proteasome (Figure 7C, left). Immunoblotting revealed that H2O2 treatment increased the amount of proteasome-bound Ub conjugates (Figure 7C, left). Ubiquitination of all four proteasomal proteins was decreased in the H2O2-treated sample (Figure 7C, right). Other cellular stress affected 26S ubiquitination as well. Cells starved in 0.2% fetal bovine serum for 72 h had increased pools of free and conjugated ubiquitin (Figure 7D). Proteasome purified from normal or starved cells looked similar on Coomassie-stained SDS–PAGE (Figure 7E). The proteasome from starved cells had more ubiquitinated substrate bound and showed a significant decrease in S5a ubiquitination along with slight decreases for the other subunits (Figure 7F). We next tested two conditions for proteolytic stress by inhibition of the proteasome. Treatment of cells with the proteasome inhibitor MG132 for 2 h caused a significant reduction in only Rpt5 ubiquitination (Supplemental Figure S5A), whereas a longer treatment of 16 h resulted in a slight increase in ubiquitination of all four proteins (Supplemental Figure S5B). Finally, exposure of cells to heat stress at 42°C for 2 h did not affect ubiquitination of any subunits compared with cells cultured at 37°C (Supplemental Figure S5C). Taken together, these data show that ubiquitination of the 26S proteasome can be influenced by certain cellular stress.
FIGURE 7:
Different types of cellular stress affect ubiquitination of the 26S proteasome. (A) H2O2 treatment increases the cellular ubiquitination levels. 293T Rpn11-HTBH cells were treated with or without 25 μM H2O2 for 5 h. Ub and β-actin in the whole-cell lysates were detected by immunoblotting. (B) Coomassie-stained SDS–PAGE of 3 μg 26S proteasome purified from cells described in A. (C) Immunoblotting of the purified 26S proteasome shown in B. (D) Serum starvation increases cellular Ub and Ub conjugates. 293T Rpn11-HTBH cells were starved in DMEM with 0.2% fetal bovine serum for 72 h. Ubiquitin and glyceraldehyde-3-phosphate dehydrogenase were detected by immunoblotting of whole-cell lysates. (E) Coomassie-stained SDS–PAGE of 3 μg o 26S proteasome purified from cells described in D. (F) Immunoblotting of the purified 26S proteasome shown in E. (G) Model for regulating proteasome ubiquitination by cellular stress. Certain cellular stress causes an increase of the cellular ubiquitination levels and loading of more ubiquitinated proteins onto the 26S proteasome. This could block ubiquitination of the 26S proteasome and maintain the proteasome at an active form to cope with the increased substrate load. Ubiquitination of S5a is shown as an example. The arrow markers ↑ and ↓ represent up- and down-regulation of each designated event, respectively.
DISCUSSION
In this study, we found that the Adrm1, S5a, Rpt5, and Uch37 proteins on the 26S proteasome are subjected to in situ ubiquitination, a process mediated by proteasome-associating ubiquitination enzymes. Of interest, ubiquitination of the 26S proteasome is antagonized by the proteasome-residing DUBs, by the binding of polyUb chains, and by certain cellular stress. Thus proteasome ubiquitination is a dynamic and regulated event in cells. Intriguingly, all four ubiquitinated proteins are implicated in binding or deubiquitinating ubiquitinated substrates. Consequently, ubiquitination of these proteins on the 26S proteasome impairs binding, deubiquitination, and degradation of ubiquitinated proteins in vitro. Taking these results together, we propose a model in which ubiquitination of the 26S proteasome serves to regulate proteasome activity. In a potential case, elevation of cellular ubiquitinated protein levels under certain stress conditions could increase the load of ubiquitinated proteins on the proteasome, which could block ubiquitination of the proteasome and maintain it in an active form (Figure 7G).
In situ ubiquitination of the human 26S proteasome
Recent global proteomic studies reveal that a majority of cellular proteins are substrates for ubiquitination, including the proteasomal components we studied here (Danielsen et al., 2011; Kim et al., 2011; Wagner et al., 2011). Our present study focuses on ubiquitination of proteasomal proteins in the context of the assembled 26S proteasome. In situ ubiquitination of Adrm1, S5a, Rpt5, and Uch37 is accomplished by proteasome-associating UbE1 and E3 Ub ligases together with the UbE2D family of E2s. No E2 enzymes were copurified with our 26S proteasome. This could be due to the fact that association of E2s with E3s is highly transient (Eletr et al., 2005; Yin et al., 2009). Five Ub ligases were identified in our purified human 26S proteasome, in which UbE3A is sufficient for ubiquitinating all four of the identified proteins on the 26S proteasome. Other E3s may also mediate proteasome ubiquitination, as we found that overexpression of UbE3C enhances ubiquitination of Adrm1 and S5a but not Rpt5 and Uch37. The Ub ligases on the 26S proteasome might have other functions. For instance, they could promote ubiquitination of substrates bound on the 26S proteasome, as was shown for Hul5 in yeast (Crosas et al., 2006).
Among the four ubiquitinatable proteins on the proteasome, Adrm1 and S5a are two Ub receptors (Deveraux et al., 1994; Husnjak et al., 2008; Schreiner et al., 2008), Uch37 is a DUB that weakly interacts with Ub chains during deubiquitination, and Rpt5 is an ATPase subunit that may also be involved in Ub binding (Lam et al., 2002), although whether it is a direct interaction requires further investigation. Presumably, the Ub-binding feature is being used to capture Ub-charged E2s and/or E3s, which in turn promotes ubiquitination of these proteins. This phenomenon of regulation of Ub-binding proteins through monoubiquitination has been termed “coupled monoubiquitination” and requires the Ub-binding activity of the modified protein (Hoeller et al., 2007). Of interest, coupled monoubiquitination can be performed in an E3-independent manner (Hoeller et al., 2007). Indeed, UbE1 and UbE2D1 together are able to monoubiquitinate S5a, Uch37, and Rpt5 on purified PA700, which has no E3s. However, UbE3A enhances polyubiquitination of these proteins (Figure 3B). Rpn10, the yeast homologue of human S5a, was found to be monoubiquitinated and/or multiple monoubiquitinated on the yeast 26S proteasome (Crosas et al., 2006; Isasa et al., 2010). Similar modification was also found in Drosophila (Lipinszki et al., 2012). Our finding that human S5a is ubiquitinated on the proteasome indicates that ubiquitination of this subunit is conserved among different species. Our study adds three other proteins to this list and reveals a trend of ubiquitination of subunits that are implicated in Ub binding.
26S proteasome ubiquitination is a regulated event
Ub modification is reversible through the actions of DUBs (Reyes-Turcu et al., 2009). Our results show that the polyUb chain–trimming enzymes of the 26S proteasome mediate in situ deubiquitination of the 26S proteasome. Uch37 and Usp14 are the two known proteasomal DUBs that catalyze polyUb chain trimming. Treating cells with IU1, a Usp14-specific inhibitor (Lee et al., 2010), causes only mild accumulation of ubiquitinated S5a and Adrm1 on the 26S proteasome. Inhibition of both Uch37 and Usp14 with b-AP15 in cells or with Ub aldehyde in vitro results in an increase of ubiquitinated forms of all four proteins. Thus Uch37 and Usp14 may both deubiquitinate the 26S proteasome. It is also possible that other cellular DUBs could counteract ubiquitination of the proteasome as well. Because ubiquitination of the 26S proteasome impairs binding of ubiquitinated proteins, in situ deubiquitination of the 26S proteasome would prevent this inhibitory effect. In this regard, proteasome-residing DUBs may help maintain proteasomes in a more active state.
Our study also shows that binding of polyUb chains on the 26S proteasome inhibits ubiquitination of Adrm1, S5a, Rpt5, and Uch37 in vitro. It is likely that the Ub-interacting capability is required to bind Ub-charged E2s or E3s for ubiquitination of these proteins and thus blocking the Ub-interacting site by polyUb chain binding could abolish their ubiquitination. Because the levels of cellular ubiquitinated proteins are often altered during stress, one implication is that ubiquitination of the 26S proteasome could be regulated by various types of stress. Indeed, we found that mild oxidative stress and serum starvation caused elevation of cellular ubiquitination levels, as well as levels of ubiquitinated proteins bound on the 26S proteasome. Both types of stress also decrease the levels of proteasome ubiquitination, which may maintain the proteasome as an active state to degrade ubiquitinated proteins under mild stress conditions. Certainly, different stresses and the severity of each stress could have different effects on proteasome ubiquitination, as supported by our findings during conditions of acute and prolonged proteolytic stress (Supplemental Figure S5). Our study indicates that ubiquitination of the proteasome can be counteracted by deubiquitination, substrate binding, and cellular stress. Thus proteasome ubiquitination is likely dynamic and regulated. This may explain why such low levels of proteasome ubiquitination are seen in cells.
Ubiquitination of the 26S proteasome impairs processing of ubiquitinated proteins
Our study found that ubiquitination of the proteasome impairs binding of polyUb chains, which might contribute to the observed inhibition of substrate deubiquitination and degradation. A commonly observed function of ubiquitination of Ub-binding proteins is to inhibit their Ub-binding activity. This could be done through intramolecular interactions between the Ub moiety and the Ub-binding domain (Crosas et al., 2006). Alternatively, a Ub-binding protein could bind Ub conjugated on an adjacent protein. In support of this, we found that Adrm1 preferred to bind ubiquitinated Uch37 compared with nonubiquitinated Uch37 (Tian and Liu, unpublished result). Adrm1 recruits Uch37 to the 26S proteasome by a direct interaction between their C-termini (Hamazaki et al., 2006; Jorgensen et al., 2006; Qiu et al., 2006; Yao et al., 2006). Presumably, ubiquitinated Uch37 offers an additional binding event, in which the Ub moieties conjugated on Uch37 directly interact with the N-terminal Pru domain of Adrm1, as the Pru domain is capable of binding monoUb and polyUb chains (Husnjak et al., 2008; Schreiner et al., 2008). The latter interaction may also impair ubiquitination of Adrm1, providing a potential explanation of why the Adrm1 ubiquitination level is the least among the four ubiquitinated proteins. It will be of interest to determine how ubiquitination affects the activity of each of these subunits. For example, it may alter the ATPase activity of Rpt5 or the DUB activity of Uch37. Mutating the ubiquitination sites in combination with reconstitution of the 26S proteasome may help to address their functions. However, recent proteomics studies identified multiple ubiquitination sites on each of these four proteins (Danielsen et al., 2011; Kim et al., 2011; Wagner et al., 2011), which makes reconstitution of the proteasome using the ubiquitination-site mutants very difficult, especially in light of the potential promiscuity of protein ubiquitination. Nevertheless, future investigations are necessary to address these interesting questions and understand whether proteasome ubiquitination regulates proteasome activity and function in cells.
MATERIALS AND METHODS
Proteasome purification
Bovine PA700 was purified according to a previous report (Chu-Ping et al., 1994). Human 26S proteasome was purified from HEK 293T or HeLa cells stably expressing Rpn11-HTBH. The pQCXIP-HTBH and pQCXIP-Rpn11-HTBH plasmids were gifts from Lan Huang (University of California, Irvine, Irvine, CA; Wang et al., 2007). In experiments that require preserving ubiquitination, three modifications were introduced in the regular procedure of proteasome purification that we published (Jacobson et al., 2009). First, 1X EDTA-Free Protease Inhibitor Cocktail (Roche, Basel, Switzerland) was added to the cell lysis buffer. Second, the streptavidin resin (GenScript, Piscataway, NJ) binding time was reduced to 3 h. Third, after washing the 26S proteasome on streptavidin resin, it was directly eluted by boiling in 1× SDS sample buffer.
To determine the effect of overexpression of an E2 or E3 on ubiquitination of the 26S proteasome, four 10-cm plates of 293T Rpn11-HTBH cells (50% confluence) were used for each transfection, in which 10 μg of each plasmid was transfected using calcium phosphate precipitation. In UbE2D2- and UbE2D3-knockdown experiments, predesigned siRNA oligos against UbE2D2 and UbE2D3 (Sigma-Aldrich, St. Louis, MO) were cotransfected onto one 10-cm plate of 293T Rpn11-HTBH cells (30% confluence) using Lipofectamine 2000 (Life Technologies, Grand Island, NY) and following the manufacturer's instructions. An siRNA oligo of green fluorescent protein (GFP) was used as a control. At 6 h posttransfection, the medium was changed to fresh DMEM. At 48 h posttransfection for overexpression experiments or 72 h for knockdown experiments, the cells were harvested and the 26S proteasome was purified as described. In IU1 or b-AP15 treatment experiments, 293T Rpn11-HTBH cells that were 90% confluent were incubated with DMEM with 75 μM IU1 (UBPBio) for 6 h or 0.75 μM b-AP15 (UBPBio) for 2 h. Cells were then harvested for proteasome purification.
For various stress experiments, two 15-cm plates of 293T Rpn11-HTBH cells (90% confluence) cultured in DMEM were used for each condition. For oxidative stress experiments, cells either were treated with 25 μM H2O2 or received no treatment for 5 h. For serum starvation experiments, cells were cultured in DMEM with 0.2% fetal bovine serum (HyClone, Logan, UT) for 72 h. The control cells in medium with 10% fetal bovine serum were harvested at the beginning of starvation. For MG132 treatment, cells were treated with dimethyl sulfoxide (control) or 10 μM MG132 (UBPBio, Aurora, CO) for 2 or 16 h. For heat shock experiments, cells were cultured in CO2 incubator at 37 or 42°C for 2 h before harvest. The 26S proteasome was purified using the method that prevents deubiquitination.
In vitro proteasome ubiquitination
In vitro proteasome ubiquitination assays were performed at 37°C in a buffer consisting of 40 mM Tris, pH 7.2, 40 mM NaCl, 5 mM MgCl2, 2 mM ATP, 2 mM β-mercaptoethanol, and 10% glycerol. Ubiquitination reactions contained either 80 nM purified human 26S proteasome or 160 nM bovine PA700, 50 μM Ub and 2 μM UbE2D1, or other E2s (UBPBio). We also included 100 nM Ub-activating enzyme E1 (UBPBio) and/or 1 μM UbE3A in the ubiquitination assays, as stated in the figure legends. In all ubiquitination assays, 100 μM epoxomicin (UBPBio) and 2.5 μM Ub aldehyde (UBPBio) were included, except that no Ub aldehyde was added for assays in Figure 4C and no Ub aldehyde or epoxomicin was added for assays in Figure 4D. The ubiquitination time was 1 h.
Supplementary Material
Acknowledgments
We thank L. Huang, G. N. DeMartino, and R. E. Cohen for providing valuable reagents. We also thank W. Tian and K-J. Han for generating plasmids and purifying proteins. This work was supported by a grant from the National Institutes of Health (5R01NS72397) and a grant from the American Cancer Society to C.-W. L.
Abbreviations used:
- DUB
deubiquitinating enzyme
- HA
hemagglutinin
- TEV
tobacco etch virus
- Ub
ubiquitin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0585) on April 17, 2014.
REFERENCES
- Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153:614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Ping M, Slaughter CA, DeMartino GN. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain) Biochim Biophys Acta. 1992;1119:303–311. doi: 10.1016/0167-4838(92)90218-3. [DOI] [PubMed] [Google Scholar]
- Chu-Ping M, Vu JH, Proske RJ, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20 S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy P, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K, Kaiser P. Protein degradation and the stress response. Semin Cell Dev Biol. 2012;23:515–522. doi: 10.1016/j.semcdb.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes G, Martin de Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Guo X, Engel JL, Xiao J, Tagliabracci VS, Wang X, Huang L, Dixon JE. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci USA. 2011;108:18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- Jorgensen JP, Lauridsen AM, Kristensen P, Dissing K, Johnsen AH, Hendil KB, Hartmann-Petersen R. Adrm1, a putative cell adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol. 2006;360:1043–1052. doi: 10.1016/j.jmb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinszki Z, Kovacs L, Deak P, Udvardy A. Ubiquitylation of Drosophila p54/Rpn10/S5a regulates its interaction with the UBA-UBL polyubiquitin receptors. Biochemistry. 2012;51:2461–2470. doi: 10.1021/bi3001006. [DOI] [PubMed] [Google Scholar]
- Liu CW, Jacobson AD. Functions of the 19S complex in proteasomal degradation. Trends Biochem Sci. 2013;38:103–110. doi: 10.1016/j.tibs.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32:3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Ubiquitin chain synthesis. Methods Mol Biol. 2005;301:47–55. doi: 10.1385/1-59259-895-1:047. [DOI] [PubMed] [Google Scholar]
- Rani N, Aichem A, Schmidtke G, Kreft SG, Groettrup M. FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat Commun. 2012;3:749. doi: 10.1038/ncomms1752. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 2010;3 doi: 10.3389/fnmol.2010.00012. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji K, Tanaka T, Kamitani T. Interaction of NUB1 with the proteasome subunit S5a. Biochem Biophys Res Commun. 2005;337:116–120. doi: 10.1016/j.bbrc.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pickart CM. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 2005;24:4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;151:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.