Abstract
PDZ domains are important interaction modules in many intracellular pathways and abnormal activations of many of those pathways lead to diseases, including several types of cancer. The domains are characterized by the ability to recognize the extreme COOH-terminus of target proteins, such as G protein-coupled receptors and ion channels. Because PDZ protein-protein interaction is a key factor in the function of cellular pathways and signal transmission in those pathways, developing small-molecule inhibitors to compete with PDZ targets is very attractive in dissecting molecular mechanisms and formulating pharmaceutical agents. Moreover, there is a growing interest in developing small-molecule drugs to block signaling within cells. The modulation of PDZ-involved interactions in cells might be an approach to target the G protein-coupled receptors and ion channels, which are among the most important classes of drug targets in the pharmaceutical industry today. Here, we review recent progress in the development of small-molecule PDZ inhibitors, and especially focus on two PDZ domain-containing target proteins, postsynaptic density 95 and dishevelled.
Introduction
PDZ [postsynaptic density 95 (PSD-95), disc large (Dlg), and zonula occludens 1 (ZO-1) homology] domains are protein-protein interaction modules that are found in bacteria, yeast, plants, and animals.1 They exist in single or multiple copies and in combination with other interaction modules, such as SH2, SH3, PTB, and WW domains. Most PDZ proteins are scaffold proteins, and only a few of them have catalytic functions.2,3 PDZ domains are ~90 residues long and are folded into a compact globular structure comprising six β-strands (βA ~ βF) flanked by two α-helices (αA and αB).4 PDZ domains recognize the extreme C-terminal and/or internal sequence of their target proteins.5 The C-terminal region of target proteins forms an additional β-strand in the groove between the βB-strand and the αB-helix structure of the PDZ domain, and the ligand carboxylate is hydrogen-bonded with the backbone amide of the first loop region of the PDZ domain.6 These interactions play a critical role in many cellular and biological processes, such as cell-cell junctions, signaling pathways, and subcellular membrane trafficking.3,7,8 Several studies have provided solid evidence that PDZ domains are involved in human congenital diseases and in regulating aspects of cytoarchitecture in mice.9,10 The development of a modulator for targeting PDZ protein-protein interactions in complex diseases will therefore give a promising opportunity to understand and to control many cellular and biological processes that lead to cancers and other disorders.11
Another reason that PDZ domain are drawing more attention is because PDZ domain proteins can recognize important drug targets, including G protein-coupled receptors (GPCRs) and ion channels inside cells.12,13 For example, PICK1, which is one of the well-characterized PDZ-containing proteins, interacts with several subtypes and subunits of the glutamate (Glu) receptor family, a well known GPCR protein. This GPCR protein has a key role in a number of neurological disorders, such as stroke. GPCR drugs accounted for ~30% of the small-molecule drugs on the market 5 years ago.14 Most of them focus on outside-in signaling by targeting GPCRs. However, these drugs vary in effectiveness due to well-recognized off-target effects. Recent efforts to develop GPCR drugs have extended to block signaling inside cells by targeting downstream components in cellular pathways.15 Several small-molecule inhibitors for targeting downstream signaling have been tested in clinical trials and look promising.15 Thus, blocking the interaction between the PDZ domain and the C-terminal region of GPCRs specifically might be a way to develop new therapeutics.11 Several groups have begun the search for novel antagonists that target PDZ protein-protein interactions.16–27 Recent studies have shown a bright future for therapeutic use of PDZ protein-protein interaction antagonism. There are many excellent reviews of the structure basis and bioactivities of PDZ domain,3,10,11 and in this review, will recent progress in antagonist development for the PDZ domain.
Most recent progress in antagonist development for PDZ domains have been focused on two important PDZ-containing proteins, postsynaptic density-95(PSD-95) and dishevelld(Dvl). The former couples N-methyl d-aspartate (NMDA) receptor activity to the production of nitric oxide and then mediates NMDA receptor-dependent excitotoxicity and the latter interacting with Frizzled(Fz) protein is critical for transmitting the Wnt signal downward into cell.28,29,41 Since these two important PDZ protein-protein interactions are involved in the several diseases including several type of cancers, the identification of small-molecule inhibitors for blocking these interactions provides a potential therapeutic approach to cure them.
Antagonists of PSD-95 PDZ/NMDA and PSD-95/nNOS interaction
The N-methyl d-aspartate (NMDA) receptor, one of the major excitatory neurotransmitters in mammalian central nervous systems, plays an essential role in the normal physiology of neurons. Overstimulation of NMDA causes glutamate-induced excitotoxicity, including stroke, brain trauma, and epilepsy.9 Many treatments of glutamate-induced excitotoxicity by NMDA receptor inhibitors have been tested. However, most of them failed in clinical trials due to adverse effects.38–40 Thus, the search for alternative approaches to mediate NMDA receptors has become quite active in recent years. One potential approach is to block downstream excitotoxic signaling instead of inhibiting NMDA receptors.9 The most attractive target is the interaction between NMDA receptors and PSD-95, a target protein downstream of the NMDA receptor signaling pathways.
The PSD-95 protein contains three non-identical PDZ domains, an SH3 domain, and an inactive guanyl kinase (GK) domain (Figure 1). Of the three PDZ domains of PSD-95, the second (PDZ2) was extensively investigated.39,41–47 PDZ2 binds to the COOH-terminus of NMDA receptor NR2 subunits as well as the internal sequence of neuronal nitric oxide synthase (nNOS).42,46 This binding couples NMDA receptor activity to the production of nitric oxide, which plays important roles in the central nervous system as a messenger molecule.48 The effect of blocking PSD-95/NMDA (or nNOS) protein-protein interaction on ischemic brain damage in rats was investigated using a Tat peptide consisting of the nine COOH terminal residues of the NR2B subunit, Tat-NR2B9c.43 Remarkably, the antagonist of NMDA receptor/PSD-95 interaction suppressed excitotoxicity and ischemic brain damage in rats effectively without affecting NMDA receptor activity.43 Very recently, Cui et al.42 conducted the proteomic analysis of the interactions of neuronal signaling proteins with human PDZ domains (> 6,500 interactions) using an ELISA-based assay. They found that the Tat-NR2B9c peptide binds specifically to PSD-95 family members (PSD-95, PSD-93, SAP97, and SAP102) and Tax interaction protein 1 (TIP1). As they suppressed the Tat-NR2B9c-binding proteins in primary murine neuron culture by RNA interference, notably, only neurons lacking PSD-95 or nNOS, but not other PDZ domains, exhibited reduced excitotoxic vulnerability. The results suggested that PSD-95 inhibition might be a target for treatment for stroke. A phase I clinical trial is ongoing for a drug based on the Tat-NR2B9c peptide by Arbor Vita Corporation and NoNO Inc.42 Besides the PDZ2 domain, other PDZ domains (PDZ1 and PDZ3) in PSD-95 were also studied.23,49 Spaller et al.21,26 designed and investigated monovalent linear and cyclic peptides to target the first and third PDZ domains of PSD-95. However, the role of those domains in disease remains unclear.50
Figure 1.
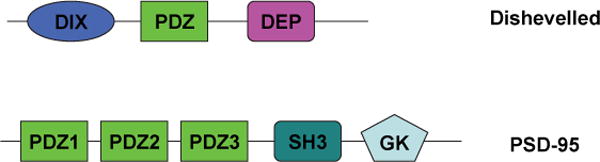
The schematic structure of dishevelled and PSD-95 proteins.
Non-peptide inhibitors for the PSD-95 PDZ2 domain were also identified by separating the effective components from traditional Chinese medicine preparations used in stroke therapy, such as extracting flavonoids from Scutellaria baicalensis extracts.44 Four flavonoid-family natural products, baicalin, norwogonoside, oroxylin A-glucuronide, and wogonoside, were found to bind to PDZ2, as shown by the nuclear magnetic resonance heteronuclear single quantum correlation technique. Their binding motif was distinct from that of the NR2B-PDZ2 binding complex. It was characterized by minimal chemical shift changes in the GLGL-loop, which are involved in key binding interactions in the PSD-95 PDZ2/NR2B peptide complex. The lack of key binding interactions directly resulted in weak binding affinity (Kd ~10 mM) for these extracts. However, this binding motif is very helpful in identifying new leads for developing drugs to target NMDA receptor signaling pathways.
Antagonists of Dishevelled/Frizzled interaction
Dishevelled (Dvl) proteins, consisting of ~700 amino acid residues, are a key component in Wnt signaling pathways, which are involved in embryo development and in malignant diseases.28,29 At least three such pathways, including Wnt/β-catenin, Wnt/planar-cell polarity, and Wnt/Ca2+ pathways, have been identified.28 Three homologues, Dvl1, Dvl2, and Dvl3, were identified in mammalian systems. The Dvl protein contains three highly conserved domains: an N-terminal DIX domain that binds to Axin, a central PDZ domain that is involved in protein-protein interactions, and a C-terminal DEP domain that functions in membrane relocation and protein-protein interactions (Figure 1).30–32
In Wnt/β-catenin signaling, Wnt ligands bind to the co-receptors of LRP5/6 and Frizzled (Fz), a 7-transmembrane domain protein, and activate the phosphoprotein Dvl. The activated Dvl protein relays the Wnt signals to the downstream components. Although the mechanism involving Dvl remains unclear, the GSK-3–dependent phosphorylation of β-catenin is suppressed. The β-catenin accumulates and enters the nucleus to activate the Wnt target gene. Although the role of Dvl in Wnt pathway is not completely understood yet, we demonstrated that the protein-protein interaction between the membrane-bound Fz receptor and the Dvl protein in the cytoplasm plays a key role in conveying Wnt signals downward.31 Using nuclear magnetic resonance spectroscopy we showed that the Dvl PDZ domain directely interacted to a conserved sequence (KTXXXW) in Fz, localized two residues after the seventh transmembrane domain.31,33 We also found that a short Fz7 peptide from the internal sequence motif attenuated Wnt1-induced β-catenin signaling, implying that the inhibitors of Dvl PDZ domain block Wnt signaling effectively at the Dvl level. Moreover, the importance of the Dvl PDZ domain is emphasized by the observation that Dvl protein is overexpressed in several cancer cells and tissues.34–36 Dvl overexpression is critical to Wnt signaling activation in these cancer cell lines. Uematsu et al.34 underlined the importance of the Dvl PDZ domain in malignant pleural mesothelioma, showing that a PDZ domain deletion mutant (ΔPDZ-Dvl) cells inhibits endogenous Dvl and stabilizes cytosolic β-catenin in mesothelioma and downregulates the downstream target genes. They also showed that transfection of ΔPDZ-Dvl inhibits tumorigenicity of mesothelioma cell lines in soft agar and in athymic mice. In non-small cell lung cancer cell lines, they found that Dvl-3 was overexpressed in 75% of freshly microdissected cancer samples compared with autologous matched normal tissues.35 Small interfering RNA designed to inhibit the Dvl protein reduced β-catenin expression and its TCF-dependent transcriptional activity. The results confirm that Dvl is a possible therapeutic target for the development of novel molecular treatments in several cancers.
To date, two successful antagonists targeting Dvl/Fz interaction have been reported.16,37 Shan et al.37 discovered a peptidomimetic compound (NSC668036) that can bind to the Dvl PDZ domain with a binding affinity of ~237 μM using structure-based virtual ligand screening and nuclear magnetic resonance experimental tools. The National Cancer Institute (NCI) three-dimensional small-molecule database that includes the coordinates of more than 250,000 drug-like chemical compounds was virtually screened for the best hits. In Xenopus embryos, NSC668036 inhibited Wnt3A-induced, but not β-catenin-induced, canonical Wnt/β-catenin signaling, indicating that the compound may block Wnt signaling at the Dvl level. Fujii et al.16 reported that FJ9, another interesting non-peptide antagonist targeting Dvl/Fz interaction, showed potential therapeutic usage. The compound suppressed β-catenin–dependent tumor cell growth in human cancer cells and in a mouse xenograft model. Both NSC668036 and FJ9 provide a structural basis for the rational design of high-affinity inhibitors of the Dvl/Fz7 interaction. Additional studies are in progress to find more efficient compounds for clinical use.
Perspective
The development of inhibitors for targeting PDZ protein-protein interaction is now in progress and the initial successes have been achieved so far are very exciting.16–18,20,25,27 However, there are several challenges to using them for therapeutic treatments.9,11,42 First of all, due to the diversity of PDZ domains (~500 PDZ domains- in the human genome) and the fact that many of them have similar binding sites, developing small-molecule inhibitors targeting specific PDZ/target interactions is – challenging task.1 Further, one commonly used approach is to use a peptide inhibitor that mimics the binding site of the target protein, as shown in the example of the antagonist for targeting the PSD-95/NMDA protein-protein interaction42,43 However, the peptide inhibitor might suffer from bioavailability limitations, such as metabolic stability and unfavorable physiochemical properties, which could reduce its therapeutic benefit. To overcome such limitations, modification of peptide inhibitors, such as β-strand peptidomimetic and cyclic peptides, for PDZ protein-protein interaction was suggested.22,51 The development of small organic molecules is also a promising approach. However, structural information about PDZ-ligand complexes is limited, which may prevent the further optimization of PDZ/target antagonists.27 More thorough understanding of the structural basis of PDZ domains with distinct binding sites and more efficient methods to select proper templates to design inhibitors with high specificity are urgently needed to develop therapeutics targeting PDZ protein-protein interaction.37
Table 1.
Antagonists of PDZ protein-protein interactions.
| Target | Antagonist | Phase of development | Roles/Functions |
|---|---|---|---|
| PSD-95 PDZ2/NMDA (N-methyl d-aspartate) or nNOS | NA-1 | Phase I (Ending March 2007) | Treatment of ischemic brain damage; in animal toxicity studies, NA-1 showed safety and tolerability after intravenous administration.42 |
| PSD-95 PDZ2/NMDA (N-methyl d-aspartate) or nNOS | Flavonoids | Lead | Flavonoids bind to the PSD-95 PDZ2 domain.44 |
| Dishevelled PDZ/Fz7 Wnt receptor | Fz7peptide (GSKTLQSWRRYH) Dapper peptide (SGSLKLMTTV) |
Biological study | In Xenopus embryos, Fz7(or Dapper) peptide attenuates Wnt3A-induced canonical Wnt signaling.31 |
| Dishevelled PDZ/Fz7 Wnt receptor | NSC668036 | Biological study HIT | In Xenopus embryos, NSC668036 inhibits the canonical Wnt signaling induced by Wnt3A.37 |
| Dishevelled PDZ/Fz7 Wnt receptor | FJ9 | Lead | Induction of apoptosis in human cancer cell lines and tumor growth inhibition in a mouse xenograft model.16 |
References
- 1.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998 May 26;95:5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan JS, Zhang M. Signaling complex organization by PDZ domain proteins. Neurosignals. 2002 Nov;11:315–21. doi: 10.1159/000068256. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Wang W. Organization of signaling complexes by PDZ-domain scaffold proteins. Acc Chem Res. 2003 Jul;36:530–8. doi: 10.1021/ar020210b. [DOI] [PubMed] [Google Scholar]
- 4.Morais Cabral JH, Petosa C, Sutcliffe MJ, Raza S, Byron O, Poy F, Marfatia SM, Chishti AH, Liddington RC. Crystal structure of a PDZ domain. Nature. 1996 Aug 15;382:649–52. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 5.Bockaert J, Fagni L, Dumuis A, Marin P. GPCR interacting proteins (GIP) Pharmacol Ther. 2004 Sep;103:203–21. doi: 10.1016/j.pharmthera.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997 Jan 3;275:73–7. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 7.Fanning AS, Anderson JM. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996 Nov 1;6:1385–8. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 8.Kurakin A, Swistowski A, Wu SC, Bredesen DE. The PDZ domain as a complex adaptive system. PLoS ONE. 2007;2:e953. doi: 10.1371/journal.pone.0000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen W, Wang W, Zhang M. Targeting PDZ domain proteins for treating NMDA receptor-mediated excitotoxicity. Curr Top Med Chem. 2006;6:711–21. doi: 10.2174/156802606776894474. [DOI] [PubMed] [Google Scholar]
- 10.Dev KK. PDZ domain protein-protein interactions: a case study with PICK1. Curr Top Med Chem. 2007;7:3–20. doi: 10.2174/156802607779318343. [DOI] [PubMed] [Google Scholar]
- 11.Dev KK. Making protein interactions druggable: targeting PDZ domains. Nat Rev Drug Discov. 2004 Dec;3:1047–56. doi: 10.1038/nrd1578. [DOI] [PubMed] [Google Scholar]
- 12.Day P, Kobilka B. PDZ-domain arrays for identifying components of GPCR signaling complexes. Trends Pharmacol Sci. 2006 Oct;27:509–11. doi: 10.1016/j.tips.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007 Oct;28:518–25. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002 Sep;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 15.McKinsey TA, Kass DA. Small-molecule therapies for cardiac hypertrophy: moving beneath the cell surface. Nat Rev Drug Discov. 2007 Aug;6:617–35. doi: 10.1038/nrd2193. [DOI] [PubMed] [Google Scholar]
- 16.Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, Guy RK, Jablons DM. An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res. 2007 Jan 15;67:573–9. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]
- 17.Fujii N, Shelat A, Hall RA, Guy RK. Design of a selective chemical probe for class I PDZ domains. Bioorg Med Chem Lett. 2007 Jan 15;17:546–8. doi: 10.1016/j.bmcl.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Fujii N, Haresco JJ, Novak KA, Gage RM, Pedemonte N, Stokoe D, Kuntz ID, Guy RK. Rational design of a nonpeptide general chemical scaffold for reversible inhibition of PDZ domain interactions. Bioorg Med Chem Lett. 2007 Jan 15;17:549–52. doi: 10.1016/j.bmcl.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Fujii N, Haresco JJ, Novak KA, Stokoe D, Kuntz ID, Guy RK. A selective irreversible inhibitor targeting a PDZ protein interaction domain. J Am Chem Soc. 2003 Oct 8;125:12074–5. doi: 10.1021/ja035540l. [DOI] [PubMed] [Google Scholar]
- 20.Klosi E, Saro D, Spaller MR. Bivalent peptides as PDZ domain ligands. Bioorg Med Chem Lett. 2007 Nov 15;17:6147–50. doi: 10.1016/j.bmcl.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Saro D, Spaller MR. Thermodynamic profiling of conformationally constrained cyclic ligands for the PDZ domain. Bioorg Med Chem Lett. 2004 Mar 22;14:1385–8. doi: 10.1016/j.bmcl.2003.09.103. [DOI] [PubMed] [Google Scholar]
- 22.Piserchio A, Salinas GD, Li T, Marshall J, Spaller MR, Mierke DF. Targeting specific PDZ domains of PSD-95; structural basis for enhanced affinity and enzymatic stability of a cyclic peptide. Chem Biol. 2004 Apr;11:469–73. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Saro D, Li T, Rupasinghe C, Paredes A, Caspers N, Spaller MR. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 2007 May 29;46:6340–52. doi: 10.1021/bi062088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saro D, Klosi E, Paredes A, Spaller MR. Thermodynamic analysis of a hydrophobic binding site: probing the PDZ domain with nonproteinogenic peptide ligands. Org Lett. 2004 Sep 30;6:3429–32. doi: 10.1021/ol049181q. [DOI] [PubMed] [Google Scholar]
- 25.Sharma SC, Rupasinghe CN, Parisien RB, Spaller MR. Design, Synthesis, and Evaluation of Linear and Cyclic Peptide Ligands for PDZ10 of the Multi-PDZ Domain Protein MUPP1. Biochemistry. 2007 Nov 6;46:12709–20. doi: 10.1021/bi7008135. [DOI] [PubMed] [Google Scholar]
- 26.Udugamasooriya G, Saro D, Spaller MR. Bridged peptide macrocycles as ligands for PDZ domain proteins. Org Lett. 2005 Mar 31;7:1203–6. doi: 10.1021/ol0475966. [DOI] [PubMed] [Google Scholar]
- 27.Joshi M, Vargas C, Boisguerin P, Diehl A, Krause G, Schmieder P, Moelling K, Hagen V, Schade M, Oschkinat H. Discovery of low-molecular-weight ligands for the AF6 PDZ domain. Angew Chem Int Ed Engl. 2006 Jun 2;45:3790–5. doi: 10.1002/anie.200503965. [DOI] [PubMed] [Google Scholar]
- 28.Malbon CC, Wang HY. Dishevelled: a mobile scaffold catalyzing development. Curr Top Dev Biol. 2006;72:153–66. doi: 10.1016/S0070-2153(05)72002-0. [DOI] [PubMed] [Google Scholar]
- 29.Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005 Oct;132:4421–36. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 30.Wharton KA., Jr Runnin’ with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003 Jan 1;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 31.Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell. 2003 Nov;12:1251–60. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007 Jun;14:484–92. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 33.Umbhauer M, Djiane A, Goisset C, Penzo-Mendez A, Riou JF, Boucaut JC, Shi DL. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signalling. EMBO J. 2000 Sep 15;19:4944–54. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uematsu K, Kanazawa S, You L, He B, Xu Z, Li K, Peterlin BM, McCormick F, Jablons DM. Wnt pathway activation in mesothelioma: evidence of Dishevelled overexpression and transcriptional activity of beta-catenin. Cancer Res. 2003 Aug 1;63:4547–51. [PubMed] [Google Scholar]
- 35.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003 Oct 16;22:7218–21. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani K, Miyamoto S, Nagahata T, Konishi N, Emi M, Onda M. Upregulation and overexpression of DVL1, the human counterpart of the Drosophila dishevelled gene, in prostate cancer. Tumori. 2005 Nov;91:546–51. doi: 10.1177/030089160509100616. [DOI] [PubMed] [Google Scholar]
- 37.Shan J, Shi DL, Wang J, Zheng J. Identification of a specific inhibitor of the dishevelled PDZ domain. Biochemistry. 2005 Nov 29;44:15495–503. doi: 10.1021/bi0512602. [DOI] [PubMed] [Google Scholar]
- 38.Kemp JA, McKernan RM. NMDA receptor pathways as drug targets. Nat Neurosci. 2002 Nov;5(Suppl):1039–42. doi: 10.1038/nn936. [DOI] [PubMed] [Google Scholar]
- 39.Aarts MM, Tymianski M. Molecular mechanisms underlying specificity of excitotoxic signaling in neurons. Curr Mol Med. 2004 Mar;4:137–47. doi: 10.2174/1566524043479202. [DOI] [PubMed] [Google Scholar]
- 40.Ikonomidou C, Turski L. Why did NMDA receptor antagonists fail clinical trials for stroke and traumatic brain injury? Lancet Neurol. 2002 Oct;1:383–6. doi: 10.1016/s1474-4422(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 41.Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999 Jun 11;284:1845–8. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- 42.Cui H, Hayashi A, Sun HS, Belmares MP, Cobey C, Phan T, Schweizer J, Salter MW, Wang YT, Tasker RA, Garman D, Rabinowitz J, Lu PS, Tymianski M. PDZ protein interactions underlying NMDA receptor-mediated excitotoxicity and neuroprotection by PSD-95 inhibitors. J Neurosci. 2007 Sep 12;27:9901–15. doi: 10.1523/JNEUROSCI.1464-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aarts M, Liu Y, Liu L, Besshoh S, Arundine M, Gurd JW, Wang YT, Salter MW, Tymianski M. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science. 2002 Oct 25;298:846–50. doi: 10.1126/science.1072873. [DOI] [PubMed] [Google Scholar]
- 44.Tang W, Sun X, Fang JS, Zhang M, Sucher NJ. Flavonoids from Radix Scutellariae as potential stroke therapeutic agents by targeting the second postsynaptic density 95 (PSD-95)/disc large/zonula occludens-1 (PDZ) domain of PSD-95. Phytomedicine. 2004;11:277–84. doi: 10.1078/0944711041495173. [DOI] [PubMed] [Google Scholar]
- 45.Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol. 2003 Mar 14;327:203–14. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 46.Tochio H, Mok YK, Zhang Q, Kan HM, Bredt DS, Zhang M. Formation of nNOS/PSD-95 PDZ dimer requires a preformed beta-finger structure from the nNOS PDZ domain. J Mol Biol. 2000 Oct 27;303:359–70. doi: 10.1006/jmbi.2000.4148. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999 Jan 1;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc Natl Acad Sci U S A. 1991 Jul 15;88:6368–71. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passafaro M, Sala C, Niethammer M, Sheng M. Microtubule binding by CRIPT and its potential role in the synaptic clustering of PSD-95. Nat Neurosci. 1999 Dec;2:1063–9. doi: 10.1038/15990. [DOI] [PubMed] [Google Scholar]
- 50.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg RJ, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998 Apr;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 51.Hammond MC, Harris BZ, Lim WA, Bartlett PA. Beta strand peptidomimetics as potent PDZ domain ligands. Chem Biol. 2006 Dec;13:1247–51. doi: 10.1016/j.chembiol.2006.11.010. [DOI] [PubMed] [Google Scholar]


