Abstract
Background
Alzheimer's disease is a common debilitating dementia with known heritability, for which 20 late onset susceptibility loci have been identified, but more remain to be discovered. This study sought to identify new susceptibility genes, using an alternative gene-wide analytical approach which tests for patterns of association within genes, in the powerful genome-wide association dataset of the International Genomics of Alzheimer's Project Consortium, comprising over 7 m genotypes from 25,580 Alzheimer's cases and 48,466 controls.
Principal Findings
In addition to earlier reported genes, we detected genome-wide significant loci on chromosomes 8 (TP53INP1, p = 1.4×10−6) and 14 (IGHV1-67 p = 7.9×10−8) which indexed novel susceptibility loci.
Significance
The additional genes identified in this study, have an array of functions previously implicated in Alzheimer's disease, including aspects of energy metabolism, protein degradation and the immune system and add further weight to these pathways as potential therapeutic targets in Alzheimer's disease.
Introduction
The prevalence of Alzheimer's disease (AD) is increasing as more people live into old age. Hope for finding preventative and clinical therapies lies in the ability to gain a better understanding of the underlying biology of the disease, and genetics will provide a valuable starting point for advancement. Rare monogenic forms of AD, the majority of which are attributable to mutations in one of three genes, APP, PSEN1 and PSEN2, exist, but common, late-onset AD is genetically complex with heritability estimated to be between 56–79%[1], [2]. Along with the APOE polymorphism[3], 20 common susceptibility loci have been identified associated with AD[4]–[9]. (This figure does not include CD33 as it did not show genome-wide significance in the original report[9].) Recently, a moderately rare variant in TREM2 has also shown evidence for association[10]. However, new variants remain to be found. This study sought to identify new susceptibility genes, using an alternative gene-wide analytical approach, which focuses on the pattern of association within gene regions.
Genome-wide association (GWA) studies to date have focused on single nucleotide polymorphisms (SNPs) as the unit of analysis. Single locus tests are the simplest to generate and to interpret, but have limitations. For example, if susceptibility is conferred by multiple variants within a locus[11], [12], this gives rise to complex patterns of association that might not be reflected by association to the same SNPs in different samples, despite apparently reasonably powered tests[13], [14]. In addition, rare risk-increasing variants may not be tagged by single SNPs, as is e.g. the case for CLU in which significant enrichment of rare variants in patients was observed independent of the single locus GWA signal[15]. It is therefore likely that the power to detect association might be enhanced by exploiting information from multiple signals within genes encompassed by gene-wide statistical approaches[12]. Disease risk may reflect the co-action of several loci but the number of loci involved at the individual or the population levels are unknown, as is the spectrum of allele frequencies and effect sizes[16]. The observations of multiple genome-wide significant or suggestive linkage signals for disorders, that do not readily replicate between studies but which are not randomly distributed across the genome[17], [18] is compatible with the existence of multiple risk alleles of moderate effect that would implicate a locus in disease risk, when analysed together. Thus the first aim of this study is to test for gene-wide association with AD, using a powerful mega-meta analysis of genome-wide datasets as part of the International Genomics of Alzheimer's Project (IGAP) Consortium comprising four AD genetic consortia (see the full list of consortia members in Materials S1): Genetic and Environmental Risk in Alzheimer's Disease (GERAD), European Alzheimer's Disease Initiative (EADI), Cohorts for Heart and Aging in Genomic Epidemiology (CHARGE) and Alzheimer's Disease Genetics Consortium (ADGC) (see full IGAP datasets description in Materials S2). A two stage study was undertaken. In Stage 1 the combined sample included 17,008 AD cases and 37,154 controls. In Stage 2 loci with p-values (combined over all SNPs at the locus) less than 10−4 were selected for replication for 8,572 AD cases and 11,312 controls of European ancestry. We observed evidence for gene-wide association at loci which implicate genes which already show genome-wide significant association from single SNP analysis (CR1, BIN1, HLA-DRB5/HLA-DRB1, CD2AP, EPHA1, PTK2B, CLU, MS4A6A, PICALM, SORL1, SLC24A4, ABCA7, APOE), three new genes in the vicinity of lately reported single SNP hits[9] (ZNF3, NDUFS3, MTCH2) and two novel loci (TP53INP1, combined p = 1.4×10−6 and IGHV1-67 combined p = 7.9×10−8).
Results
Initially, we tested for excess genetic signal revealed by the Stage 1 IGAP SNP GWAS study. We observed more SNPs at all significance intervals, and more genes at multiple significance thresholds, than expected by chance (Table S1). This is unlikely to be due to uncorrected stratification, since each of the individual GWAS samples in the IGAP Stage 1 analysis was corrected for ethnic variation. Thus it is likely that the sample contains novel genetic signals, in addition to those detected by the primary analysis[9], [19].
Next, we looked at overrepresentation of significant genes in the Stage 1 data. Table 1 gives the observed and expected numbers of significant genes at significance levels 10−4, 10−5, 10−6 when all genes are counted in the analyses and when the known genes (Table S1) and genes within 500kb of them are excluded, the observed numbers of genes are much larger than expected at all significance levels (all p≤0.001). Thus there are more loci associated with AD to find.
Table 1. Overrepresentation of replication of significant genes/loci available at Stage 2, excluding all loci of 0.5 Mb around genes previously reported[4]–[8] and Stage 1 IGAP genes[9], [19] containing genome-wide significant SNPs.
| GENES | LOCI | ||||
| Stage 1 significance level | Significant at Stage 1 | Replicated (p≤0.05) at Stage 2 | Significant at Stage 1 | Replicated (p≤0.05) at Stage 2 | Over-representation p-value |
| p≤10−4 | 27 | 9 (33%) | 9 | 3 (33%) | 0.109 |
| p≤10−3 | 74 | 17 (23%) | 36 | 8 (22%) | 0.125 |
| p≤0.01 | 229 | 49 (21%) | 102 | 26 (25%) | 0.0001 |
| p≤0.05 | 390 | 77 (20%) | 171 | 33 (19%) | 0.007 |
| Total (p≤1) | 887 | 124 (14%) | 444 | 60 (13.5%) | 4.6×10−12 |
Over-representation p-values were calculated with chi-square/Fisher's exact tests counting the genes within 0.5 Mb as one locus.
Furthermore, the number of independent nominally significant loci at Stage 2 (N = 60, (13.5%)) was significantly greater than expected by chance (p = 4.6×10−12). The percentage of replicated loci increased with the decrease of the gene-wise significance threshold at Stage 1 (see Table 2 for details).
Table 2. Overrepresentation of significant loci, excluding regions of 0.5[4]–[8] and Stage 1 IGAP genes[9], [19] containing genome-wide significant SNPs.
| Numbers of loci (genes) | |||
| p≤10−4 | p≤10−5 | p≤10−6 | |
| Observed | 9(27) | 4(8) | 2(2) |
| Expected | 2.5 | 0.25 | 0.025 |
| p-value | 0.001 | 0.00013 | 0.0003 |
The observed number of genes is calculated by combining significant loci within 0.5 Mb into one signal. The APOE region is excluded (CHR19; 44,411,940–46,411,945bp). The total number of genes after exclusions is 24,849.
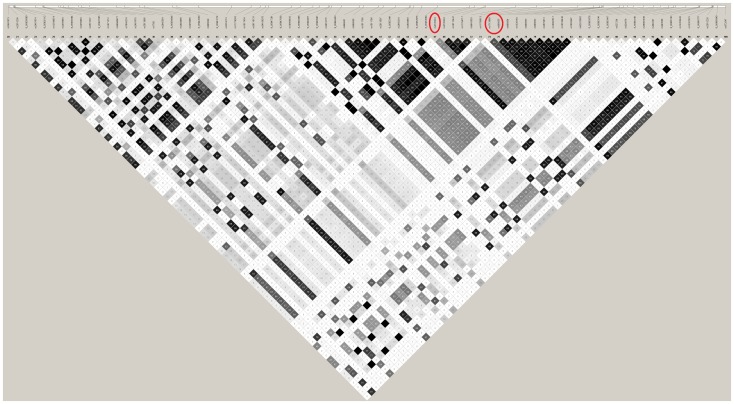
Combining the gene-wide p-values in both stages 1 and 2, using Fisher's method revealed two new gene-based genome-wide significant (p<2.5×10−6) loci TP53INP1 and IGHV1-67. The TP53INP1 gene is located on chromosome 8∶95,938,200–95,961,615 and its combined gene-based p-value = 1.4×10−6 (Table 3). Table S3 provides details for each SNP contributing to the gene-based result. Out of 45 SNPs in the gene, three SNPs (rs4735333, rs1713669, rs896855) have p-value≤10−4. Figure 1 shows the LD plot of this gene and suggests that there are at least two partially independent signals in the TP53INP1 gene (r2 between the pairs of most significant SNPs rs4735333-rs1713669 and rs1713669- rs896855 are 0.65 and 0.6 respectively).
Table 3. New genome-wide significant genes associated with AD.
| Gene Name | Chr | Position | Stage 1 gene-wide p-value | Stage 2 gene-wide p-value | N of SNPs per gene | Combined gene-wide p-value | Combined best SNP p-value | Biological function |
| TP53INP1 | 8 | 95,938,200–95,961,615 | 1.7×10−2 | 4.5×10−3 | 45 | 1.4×10−6 | 1.5×10−7 | Regulation of autophagy, cell cycle arrest |
| IGHV1-67 | 14 | 107,136,620–107,137,059 | 2.3×10−4 | 3.2×10−5 | 2 | 7.9×10−8 | 3.9×10−5 | Immunoglobulin heavy chain region: adaptive immunity |
| New genes in the vicinity of recently reported single SNP genome-wide significant hits[9], [19]: | ||||||||
| ZNF3 | 7 | 99,661,653–99,679,371 | 2.7×10−2 | 1.8×10−6 | 27 | 8.6×10−7 | 3.1×10−7 | Transcription factor, leucocyte activation |
| NDUFS3 | 11 | 47,600,632–47,606,114 | 1.2×10−6 | 2.2×10−2 | 5 | 4.8×10−7 | 2.9×10−6 | Mitochondrial electron transport, NADH to ubiquinone |
| MTCH2 | 11 | 47,638,858–47,664,206 | 1.7×10−5 | 8.7×10−3 | 34 | 2.5×10−6 | 7.2×10−8 | Mitochondrial inner membrane |
Gene-wide p-values are shown for those genes with p<2.5×10−6 for which the best single-SNP p-value in that gene is greater than 5×10−8 in the combined Stage 1 and Stage 2 sample. Previously reported genes[4]–[8] ± 0.5 Mb around them are excluded.
Gene-wide p-values in the combined Stage 1 and Stage 2 sample obtained by combining the p-values from the Stage 1 with those from the Stage 2 using Fisher's method.
Figure 1. Linkage disequilibrium structure of TP53INP1 gene.
The SNPs which are significant at 10−4 level are circled in red.
The IGHV1-67 gene on chromosome 14∶107,136,620–107,137,059 has combined p-value = 7.9×10−8 (Tables 3). This gene is covered by two SNPs (rs2011167, rs1961901), both are significant at 10−4 level. LD plot in Figure 2 and Table S4 indicate that the two most significant SNPs in IGHV1-67 gene represent almost the same signal (r2 = 0.92, calculated with SNAP software[20], 1000 genomes Pilot 1 dataset, CEU population panel, (http://www.broadinstitute.org/mpg/snap)).
Figure 2. Linkage disequilibrium structure of IGHV1-67 gene ±5 kb.
The SNPs which are significant at 10−4 level are circled in red.
To look at the gene expression patterns in these novel genes, we used the Webster-Myers expression dataset[21], available at http://labs.med.miami.edu/myers/LFuN/data%20ajhg.html. Comparing 137 AD vs 176 controls with temporal or frontal cortex expression values by t-test, t showed significantly higher TP53INP1 expression in cases compared to controls (p = 0.0128). Further examination in the BRAINEAC database[22] (www.braineac.org) from the UK Brain Expression Consortium showed TP53INP1 to have a best cis-eQTL p-value of 6.8×10−6 (for rs4582532 SNP, which is about 7.6 kb upstream of the gene). The three SNPs with association p≤10−4 mentioned above (rs4735333, rs1713669, rs896855) had significant cis-eQTL p-values of 8.2×10−6, 7.8×10−5 and 1.1×10−5 respectively in BRAINEAC brain expression data. The r2 between the cis-eQTL and the three associated SNPs were 0.80, 0.65, and 0.81, respectively). Further analysis of additional independent brain expression and methylation datasets (see Methods S1) indicated significant cis eQTLs and meQTLs for TP53INP1 (Tables S10 and S11). The probe for the meQTL is in a CpG island region that corresponds well with ENCODE DNAse/ChIP-seq/Histone marks and is located upstream (∼1.5 kb) of the TP53INP1 transcription start site. In combination these results suggest a possible epigenetic mechanism whereby the associated variants in the region influence TP53INP1 expression in several brain regions. These expression data provide further evidence supporting the functional relevance of TP53INP1 to AD susceptibility. The IGHV1-67 gene was not found in those databases.
In addition we detected two genome-wide significant loci 1) ZNF3 (chr7: 99,661,653–99,679,371; p = 8.6×10−7) and 2) two closely located genes on chromosome 11 MTCH2 (47,638,858–47,664,206, combined p = 2.5×10−6) and NDUFS3 (47,600,632–47,606,114, combined p = 4.8×10−7) (Table 4). None of these genes harbour genome-wide significant SNPs in the SNP GWAS analysis on its own (see Tables S5-S7). Figures S1-S3 show LD plots of these additional genes.
Table 4. New genome-wide significant genes associated with AD in the vicinity of recently reported single SNP genome-wide significant hits[9], [19].
| Gene Name | Chr | Position | Stage 1 gene-wide p-value | Stage 2 gene-wide p-value | N of SNPs per gene | Combined gene-wide p-value | Combined best SNP p-value | Biological function |
| ZNF3 | 7 | 99,661,653–99,679,371 | 2.7×10−2 | 1.8×10−6 | 27 | 8.6×10−7 | 3.1×10−7 | Transcription factor, leucocyte activation |
| NDUFS3 | 11 | 47,600,632–47,606,114 | 1.2×10−6 | 2.2×10−2 | 5 | 4.8×10−7 | 2.9×10−6 | Mitochondrial electron transport, NADH to ubiquinone |
| MTCH2 | 11 | 47,638,858–47,664,206 | 1.7×10−5 | 8.7×10−3 | 34 | 2.5×10−6 | 7.2×10−8 | Mitochondrial inner membrane |
Gene-wide p-values are shown for those genes with p<2.5×10−6 for which the best single-SNP p-value in that gene is greater than 5×10−8 in the combined Stage 1 and Stage 2 sample. Previously reported genes[4]–[8] ± 0.5 Mb around them are excluded.
Gene-wide p-values in the combined Stage 1 and Stage 2 sample obtained by combining the p-values from the Stage 1 with those from the Stage 2 using Fisher's method. The LD between rs1476679 (chr7∶100,004,446) reported by IGAP [9] and the best SNP in ZNF3 is r2 = 0.16. The LD between rs10838725 (chr11: 47,557,871) reported by IGAP [9] and the best SNPs in the region on chr 11 in the table are r2 = 0.3 and 0.88 for NDUFS3 and MTCH2 respectively.
ZNF3 and NDUFS3, MTCH2 genes on chromosomes 7 and 11, respectively, lie close to rs1476679 (chr7∶100,004,446; ZCWPW1) and rs1083872 (chr11∶47,557,871; CELF1) SNPs, which are shown to be genome-wide significant in the IGAP study, when combining Stage 1 and Stage 2 data. Figures S1-S3 show LD structure of these genes in relation to the IGAP singe genome-wide significant hits. (Note that the NDUFS3 gene on chromosome 11 was gene-based genome-wide significant already at Stage 1.) Although none of these SNPs actually lie within the genes mentioned above, it is possible that they may account for the gene-based signals through linkage disequilibrium. In order to test whether the gene-based signals are independent of these strongly-associated SNPs, we performed single-SNP association for each SNP annotated to these genes by regression, adjusting for the significant SNPs mentioned above, along with the other study covariates. The resulting p-values were combined into gene-based tests, as described previously. Under this conditional analysis ZNF3 gene does not show significant association, however NDUFS3 still shows a trend towards significance (p = 0.081) (see Table S8 for details). Furthermore, five genes in chr11∶47,593,749–47,615,961 (KBTBD4, NDUFS3, LOC100287127, FAM180B, C1QTNF4) all have p<0.05 with gene-based analysis ±10 kb, when conditioning by the genome-wide significant hit rs10838725 in this region. This may partially be explained by the SNP rs10838731 (p = 1.2×10−3 after conditioning by rs10838725) which is shared by all latter five genes.
Gene-based analysis with ±10 kb around genes did not reveal additional genome-wide significant loci in the Stage 1 data set. Moreover, the significance of the genes identified above did not improve in general, indicating that adding 10 kb flanking regions to genes introduces more noise to the gene-based signal. The combined Stage 1 and Stage 2 gene-based analysis provided further evidence for significant signals in the loci on chr 11 with 8 genes (SPI1, SLC39A13, LOC100287086, PTPMT1, KBTBD4, NDUFS3, LOC100287127, FAM180B) and on chr 7 with 6 genes (LOC100128334, MCM7, PILRB, PILRA, LOC100289298, C7orf51), all reaching genome-wide significance. This is likely to be due to the fact that including genes' flanking regions captures a greater number of the same SNPs or SNPs in high LD showing significant association.
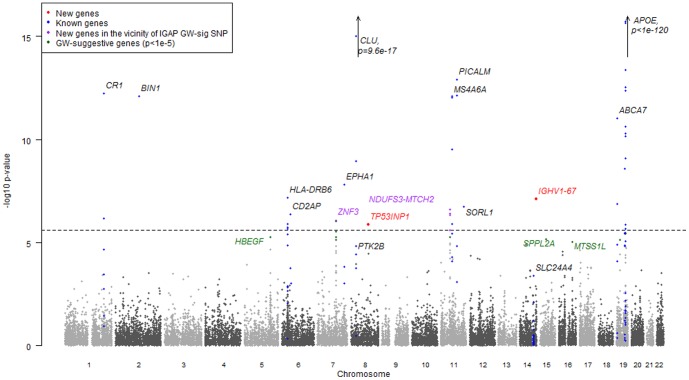
The Manhattan plot of the gene-based p-values (Figure 3) gives a general overview of the gene-based results and shows the new loci in relation to previously reported genes (see also QQ-plots in Figure S4). The results of gene-wide analysis for the genes, which were previously reported as associated with AD[4]-[8] and those which are GWAS significant in the Stage 1 analysis are presented in Table S9. Out of 16 reported susceptibility genes, 15 are nominally significant with gene-wide analysis (almost all p-values are smaller than 10−4), however not all of them reach the gene-based genome-wide significance level (2.5×10−6) when the number of SNPs per gene and LD structure of the gene is taken into account.
Figure 3. Manhattan plot of gene-wide p-values in the Stage 1 dataset and combined gene-wide p-values where Stage 2 data are available.
Each dot represents a gene, genes in blue lie within the previously reported[4]–[8] associated regions.
We did not observe genome-wide significance for CD33 gene. This gene was genome-wide significant in Stage 1 (p = 1.9×10−6), but the association was attenuated when combining Stage 1 and Stage 2 data (p = 1.79×10−5), similar to the single SNP association result in the SNP GWAS study[9], [19].
Discussion
In this study we show that there are more signals in the GWAS imputed data at SNP- and gene-based levels than revealed by single SNP analysis. A gene-based analysis is a next logical step after the single SNP analyses in any attempt to combine possible several signals in genes and thus enhance the power of the association analyses.
The first new gene TP53INP1 (chromosome 8) encodes a protein that is involved in mediating autophagy-dependent cell death via apoptosis through altering the phosphorylation state of p53[23] and in modulating cell-extracellular matrix adhesion and cell migration[24]. TP53INP1 encodes a pro-apoptotic tumor suppressor and its antisense oligonucleotide has been used as potential treatment for castration-resistant prostate cancer[25]. This association is notable, given the potential inverse association between cancer and AD that has previously been reported [26], [27].
The second new gene IGHV1-67 (chromosome 14) is a pseudogene in the immunoglobulin (IgG) variable heavy chain region of chromosome 14: its function is unknown but all genes in this region are most likely to be involved in IgG heavy chain VDJ recombinations that lead to the full repertoire of antigen-detecting immune cell clones[28].
The gene-based analysis in this study has shown its utility to enhance the information provided by single SNP analysis (i.e. NDUFS3 gene was genome-wide significant from Stage 1 using gene-based analysis whereas this gene was only genome-wide significant after combining the two stages of single SNP analysis).
ZNF3 is a zinc-finger protein at the same locus on chromosome 7 as ZCWPW1 thus rendering it a candidate as the gene that contains the functional signal in this region. Although we can not identify which gene actually confers the risk to AD, it is interesting that ZNF3 function is unknown though it interacts with BAG3 which is involved in ubiquitin/proteasomal functions in protein degradation[29] and ZNF3 is regulated by upstream binding of BACH1 whose target genes have roles in the oxidative stress response and control of the cell cycle[30].
In the cluster of genes on chromosome 11, MTCH2 encodes one of the large family of inner mitochondrial membrane transporters[31] which is associated with mitochondrially-mediated cell death[32], adipocyte differentiation[33], insulin sensitivity[34] and has a genetic association with increased BMI[35]. NDUFS3 also has functions in the mitochondria as it encodes an iron-sulphur component of complex 1 (mitochondrial NADH:ubiquinone oxidoreductase) of the electron transport chain. A deficiency causes a form of Leigh syndrome[36] an early-onset progressive neurodegenerative disorder with a characteristic neuropathology consisting of focal lesions including areas of demyelination and gliosis[37].
In summary, we report two novel genes TP53INP1 (chr8: 95,938,200–95,961,615; combined p = 1.4×10−6) and IGHV1-67 (chr14: 107,136,620–107,137,059; combined p = 7.9×10−8), which were not reported as genome-wide significant before. We also report ZNF3 gene on chromosome 7 and a cluster of genes on chromosome 11 (SPI1-MTCH2), showing gene-based genome-wide significant association with Alzheimer's disease. These genes are in proximity with, but not the same as, those detected by genome-wide significant SNPs, demonstrating support for the signals identified by IGAP[9], [19]. They have an array of functions previously implicated in AD including aspects of energy metabolism, protein degradation and the immune system and add further weight to these pathways as potential therapeutic targets in AD.
Materials and Methods
Stage 1 data
The main dataset was reported by the IGAP consortium[9], [19] and consists in total of 17,008 cases and 37,154 controls. This sample of AD cases and controls comprises 4 data sets taken from genome-wide association studies performed by GERAD, EADI, CHARGE and ADGC (see primary IGAP manuscript[9], [19] for more details). The full details of the samples and methods for conduct of the GWA studies are provided in the respective manuscripts[4]-[8].
Each of these datasets was imputed with Impute2[38] or MACH[39] software using the 1000 genomes data (release Dec2010) as a reference panel. In total 11,863,202 SNPs were included in the SNPs allelic association result file. To make our analysis as conservative as possible, we only included autosomal SNPs which passed stringent quality control criteria, i.e. we included only SNPs with minor allele frequencies (MAF) ≥0.01 and imputation quality score greater than or equal to 0.3 in each individual study, resulting in 7,055,881 SNPs which are present in at least 40% of the AD cases and 40% of the controls in the analysis. The summary statistics across datasets were combined using fixed-effects inverse variance-weighted meta-analysis. We corrected all individual SNPs p-values for genomic control (GC) λ = 1.087. These SNPs are well imputed on a large proportion of the sample, which increases confidence in the accuracy of the association analysis upon which gene-wide analysis is based.
Stage 2 data
11,632 SNPs with p-values <10−3 in the IGAP meta-analysis were successfully genotyped in a Stage 2 sample comprising 8,572 cases and 11,312 controls (see primary IGAP manuscript[9], [19] for more details). An additional 771 SNPs were successfully genotyped to test all genes with gene-wide p-values <10-4 in the IGAP Stage 1 analysis, excluding genes reported prior to IGAP[4]–[8], the four loci reaching genome-wide significance in the Stage 1 IGAP meta-analysis[9], [19] and the 0.5Mb regions around them (Table S2). These SNPs cover 887 genes and correspond to 444 independent loci where all genes within 0.5 Mb are counted as one locus.
Assignment of SNPs to genes
SNPs were assigned to genes if they were located within the genomic sequence lying between the start of the first and the end of the last exon of any transcript corresponding to that gene. The chromosome and location for all currently known human SNPs were taken from the dbSNP132 database, as was their assignment to genes (using build 37.1). In total, we retained 2,804,431 (39.7% of the total) SNPs which annotated 28,636 unique genes with 1–16,514 SNPs per gene. For the gene-wide analysis we have excluded genes which contain only one SNP in the IGAP Stage 1 analysis, leaving a total of 25,310 genes. If a SNP belongs to more than one gene, it was assigned to each of these genes. In order to account for possible signals which are correlated with those in a gene, gene-wide analysis was also performed using a 10 kb window around genes to assign SNPs to genes.
Gene-wide analysis
The gene-wide analysis was performed based on the summary p-values while controlling for LD and different number of markers per gene using an approximate statistical approach[40] adopted for set-based analysis of genetic data[41]. This is a method for calculating the significance of a set of SNPs in the absence of individual genotype data based on a theoretical approximation to Fisher's statistic for combining p-values. Fisher's statistic (-∑ln(pi)) combines probabilities and under the null hypothesis has a chi-square distribution with 2N degrees of freedom, where N is the number of markers, and the summation above is for i = 1,…,N). If Fisher's statistic combines the results of several tests when the tests are independent, the approximate method combines non-independent tests and requires only the list of p-values for each SNP and knowledge of correlations between SNPs. Then the value of Fisher's statistic and the number of degrees of freedom is corrected by the coefficient which depends upon the number of SNPs and correlations (LD) between them. This approximation was applied to the Stage 1 and Stage 2 samples separately, and the resulting gene-wide p-values combined using Fisher's method (since these are independent). LD between markers was computed using 1000 genomes data. The gene-based genome-wide significant level was set to 2.5×10−6 to account for the number of tested genes[42].
Test for excess of associated SNPs/loci
The effective number N of independent SNPs in the whole genome (excluding genes with SNPs that are genome-wide significant in the Stage 1 IGAP dataset ± 0.5 Mb was estimated by the method described in [43] taking LD into account, as were the observed number of independent SNPs significant at each p-value criterion (adjusting individual SNP p-values for genomic control λ = 1.087 before hand). LD was computed from the 1000 Genomes database (http://www.1000genomes.org/). In the absence of excess association, the expected number of independent SNPs significant at significance level α is a normally distributed random variable whose mean and standard deviation (SD) can be calculated as αN and √Nα(1-α) (mean and SD for a binomial distribution). The number of independent SNPs (and thus statistical tests) in the whole genome were estimated as ∼3.7×106, ∼3.6×106 and ∼3.5×106 at significance levels below 0.1, between 0.05 and 0.1, and 0.2 and above respectively (see [43] for details on the dependence between the significance levels and the estimated number of independent tests). We then calculated mean of the expected number of significant SNPs in intervals α 1 < p ≤ α 2, (α 1, α 2 = 0, 10−6, 10−5, …, 0.5) as difference between the expected numbers of independent SNPs at α 2 and α 1 significance levels and SD as the square root of sum of the corresponding variances.
We calculated the significance of the excess number of genes attaining the specified thresholds based upon the assumption that, under the null hypothesis of no association, the number of significant genes at a significance level of α in a scan is distributed as a binomial (N,α), where N is the total number of genes, assuming that genes are independent. Genes within 0.5 Mb of each other are counted as one signal when calculating the observed number of significant genes. This prevents significance being inflated by LD between genes, where a single association signal gives rise to several significantly-associated genes. The total number of genes was not corrected for LD in this way, making the estimate of significance of the excess number of genes conservative.
Supporting Information
Overrepresentation of significant SNPs excluding previously reported [4]-[8] genes ±0.5Mb and the APOE region as above.
(DOCX)
List of genes that are genome-wide significant in the IGAP stage 1 dataset and the flanking regions which included SNPs either in r2≥0.3 or association p-value≤10-3 whichever covers the largest region.
(DOCX)
Detailed SNP information for TP53INP1 gene.
(XLS)
Detailed SNP information for IGHV1-67 gene.
(XLS)
Detailed SNP information for ZNF3 gene.
(XLS)
Detailed SNP information for NDUFS3 gene.
(XLS)
Detailed SNP information for MTCH2 gene.
(XLS)
Gene-based analysis results, when single SNPs p-values, contributing to the gene-based p-value were adjusted for the best genome-wide significant SNP in the nearby location.
(DOCX)
Gene-wide analysis for genes which show GWAS significant association with AD in the stage 1 IGAP dataset.
(DOCX)
Brain eQTL Tissues.
(XLSX)
Brain Meth QTLs.
(XLSX)
ZNF3 gene with rs1476679 (ZCWPW1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs1476679 is highlighted in blue.
(TIF)
NDUFS3 gene rs10838725 (CELF1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs10838725 is highlighted in blue.
(TIF)
MTCH2 gene with rs10838725 (CELF1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs10838725 is highlighted in blue.
(TIF)
QQ-plot of gene-wide p-values for all genes (A) and excluding previously reported [4]-[8] GWAS significantly associated genes ±0.5Mb (B) in the discovery dataset. Genomic control λ = 1.08 and 1.07 respectively.
(TIFF)
Expression quantitative trait loci (eQTL) and Methylation quantitative trait loci (meQTL) analyses.
(DOCX)
Full IGAP datasets description.
(DOCX)
List of IGAP consortium members.
(DOC)
Acknowledgements.
(DOCX)
Acknowledgments
This work was made possible by the generous participation of the control subjects, the patients and their families. Complete acknowledgments are detailed in the Materials S3.
Funding Statement
The i-Select chips was funded by the French National Foundation on Alzheimer's disease and related disorders. The French National Fondation on Alzheimer's disease and related disorders supported several I-GAP meetings and communications. Data management involved the Centre National de Génotypage,and was supported by the Institut Pasteur de Lille, Inserm, FRC (fondation pour la recherche sur le cerveau) and Rotary. This work has been developed and supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant (Development of Innovative Strategies for a Transdisciplinary approach to ALZheimer's disease) and by the LABEX GENMED grant (Medical Genomics). The French National Foundation on Alzheimer's disease and related disorders and the Alzheimer's Association (Chicago, Illinois) grant supported IGAP in-person meetings, communication and the Alzheimer's Association (Chicago, Illinois) grant provided some funds to each consortium for analyses. EADI The authors thank Dr. Anne Boland (CNG) for her technical help in preparing the DNA samples for analyses. This work was supported by the National Foundation for Alzheimer's disease and related disorders, the Institut Pasteur de Lille and the Centre National de Génotypage. The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Agence Nationale de la Recherche, ANR supported the COGINUT and COVADIS projects. Fondation de France and the joint French Ministry of Research/INSERM «Cohortes et collections de données biologiques» programme. Lille Génopôle received an unconditional grant from Eisai. The Three-city biological bank was developed and maintained by the laboratory for genomic analysis LAG-BRC - Institut Pasteur de Lille. Belgium sample collection: The patients were clinically and pathological characterized by the neurologists Sebastiaan Engelborghs, Rik Vandenberghe and Peter P. De Deyn, and in part genetically by Caroline Van Cauwenberghe, Karolien Bettens and Kristel Sleegers. Research at the Antwerp site is funded in part by the Belgian Science Policy Office Interuniversity Attraction Poles program, the Foundation Alzheimer Research (SAO-FRA), the Flemish Government initiated Methusalem Excellence Program, the Research Foundation Flanders (FWO) and the University of Antwerp Research Fund, Belgium. Karolien Bettens is a postdoctoral fellow of the FWO. The Antwerp site authors thank the personnel of the VIB Genetic Service Facility, the Biobank of the Institute Born-Bunge and the Departments of Neurology and Memory Clinics at the Hospital Network Antwerp and the University Hospitals Leuven. Finish sample collection: Financial support for this project was provided by the Health Research Council of the Academy of Finland, EVO grant 5772708 of Kuopio University Hospital, and the Nordic Centre of Excellence in Neurodegeneration. Italian sample collections: the Bologna site (FL) obtained funds from the Italian Ministry of research and University as well as Carimonte Foundation. The Florence site was supported by grant RF-2010-2319722, grant from the the Cassa di Risparmio di Pistoia e Pescia (Grant 2012) and the Cassa di Risparmio di Firenze (Grant 2012). The Milan site was supported by a grant from the «fondazione Monzino». The authors thank the expert contribution of Mr. Carmelo Romano. The Roma site received financial support from Italian Ministry of Health, Grant RF07-08 and RC08-09-10-11-12. The Pisa site is grateful to Dr. Annalisa LoGerfo for her technical assistance in the DNA purification studies. Spanish sample collection: the Madrid site (MB) was supported by grants of the Ministerio de Educación y Ciencia and the Ministerio de Sanidad y Consumo (Instituto de Salud Carlos III), and an institutional grant of the Fundación Ramón Areces to the CBMSO. The authors thank I. Sastre and Dr. A. Martínez-García for the preparation and control of the DNA collection, and Drs. P. Gil and P. Coria for their cooperation in the cases/controls recruitment. The authors are grateful to the Asociación de Familiares de Alzheimer de Madrid (AFAL) for continuous encouragement and help. Swedish sample collection: Financially supported in part by the Swedish Brain Power network, the Marianne and Marcus Wallenberg Foundation, the Swedish Research Council (521-2010-3134), the King Gustaf V and Queen Victoria's Foundation of Freemasons, the Regional Agreement on Medical Training and Clinical Research (ALF) between Stockholm County Council and the Karolinska Institutet, the Swedish Brain Foundation and the Swedish Alzheimer Foundation. CHARGE AGES: The AGES-Reykjavik Study is funded by National Institutes of Health (NIH) contract N01-AG-12100 (National Institute on Aging (NIA) with contributions from the National Eye Institute, National Institute on Deafness and Other Communication Disorders and National Heart, Lung, and Blood Institute (NHLBI)), the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). ASPS/PRODEM: The Austrian Stroke Prevention Study and The Prospective Dementia Register of the Austrian Alzheimer Society was supported by The Austrian Science Fond (FWF) grant number P20545-P05 (H. Schmidt) and P13180; The Austrian Alzheimer Society; The Medical University of Graz. Cardiovascular Health Study (CHS): This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and HHSN268200960009C; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629, AG15928, AG20098, AG027058 and AG033193 (Seshadri) from the NIA. A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Framingham Heart Study (FHS): This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with A_ymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study as also supported by grants from the National Institute on Aging: AG08122 and AG033193 (Seshadri). Drs. Seshadri and DeStefano were also supported by additional grants from the National Institute on Aging: (R01 AG16495; AG031287, AG033040), the National Institute of Neurological Disorders and Stroke (R01 NS17950), and the National Heart, Lung and Blood Institute (U01 HL096917, HL093029 and K24HL038444, RC2-HL102419 and UC2 HL103010. Fundació ACE would like to thank patients and controls who participated in this project. This work has been funded by the Fundación Alzheimur (Murcia), the Ministerio de Educación y Ciencia (PCT-010000-2007-18), (DEX-580000-2008-4), (Gobierno de España), Corporación Tecnológica de Andalucía (08/211) and Agencia IDEA (841318) (Consejería de Innovación, Junta de Andalucía). The authors thank to Ms. Trinitat Port-Carbó and her family for their generous support of Fundació ACE research programs. The Rotterdam Study: The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Health Research and Development; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission;and the Municipality of Rotterdam; by grants from the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), Internationale Stichting Alzheimer Onderzoek, Hersenstichting Nederland, the Netherlands Genomics Initiative–Netherlands Organization for Scientific Research (Center for Medical Systems Biology and the Netherlands Consortium for Healthy Aging), the Seventh Framework Program (FP7/2007-2013), the ENGAGE project (grant agreement HEALTH-F4-2007-201413), MRACE-grant from the Erasmus Medical Center, the Netherlands Organization for Health Research and Development (ZonMW Veni-grant no. 916.13.054). ARIC: The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01- HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022 and grants R01-HL087641, RC2-HL102419 (Boerwinkle, CHARGE-S), UC2 HL103010, U01-HL096917 (Mosley) and R01-HL093029; NHGRI contract U01- HG004402; and NIH contract HHSN268200625226C and NIA: R01 AG033193 (Seshadri). Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. GERAD Cardiff University was supported by the Wellcome Trust, Medical Research Council (MRC), Alzheimer's Research United Kingdom (ARUK) and the Welsh Government. ARUK supported sample collections at the Kings College London, the South West Dementia Bank, Universities of Cambridge, Nottingham, Manchester and Belfast. The Belfast group acknowledges support from the Alzheimer's Society, Ulster Garden Villages, N. Ireland R & D Office and the Royal College of Physicians/Dunhill Medical Trust. The MRC and Mercer's Institute for Research on Ageing supported the Trinity College group. DCR is a Wellcome Trust Principal Research fellow. The South West Dementia Brain Bank acknowledges support from Bristol Research into Alzheimer's and Care of the Elderly. The Charles Wolfson Charitable Trust supported the OPTIMA group. Washington University was funded by NIH grants, Barnes Jewish Foundation and the Charles and Joanne Knight Alzheimer's Research Initiative. Patient recruitment for the MRC Prion Unit/UCL Department of Neurodegenerative Disease collection was supported by the UCLH/UCL Biomedical Centre and their work was supported by the NIHR Queen Square Dementia BRU. LASER-AD was funded by Lundbeck SA. The Bonn group would like to thank Dr. Heike Koelsch for her scientific support. The Bonn group was funded by the German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant number 01GI0102, 01GI0711, 01GI0420. The AgeCoDe study group was supported by the German Federal Ministry for Education and Research grants 01 GI 0710, 01 GI 0712, 01 GI 0713, 01 GI 0714, 01 GI 0715, 01 GI 0716, 01 GI 0717. The Homburg group was funded by the German Federal Ministry of Education and Research (BMBF): German National Genome Research Network (NGFN); Alzheimer's disease Integrated Genome Research Network; AD-IG: 01GS0465. Genotyping of the Bonn case-control sample was funded by the German centre for Neurodegenerative Diseases (DZNE), Germany. The GERAD Consortium also used samples ascertained by the NIMH AD Genetics Initiative. Harald Hampel was supported by a grant of the Katharina-Hardt-Foundation, Bad Homburg vor der Höhe, Germany. The KORA F4 studies were financed by Helmholtz Zentrum München; German Research Center for Environmental Health; BMBF; German National Genome Research Network and the Munich Center of Health Sciences. The Heinz Nixdorf Recall cohort was funded by the Heinz Nixdorf Foundation (Dr. Jur. G.Schmidt, Chairman) and BMBF. Coriell Cell Repositories is supported by NINDS and the Intramural Research Program of the National Institute on Aging. The authors acknowledge use of genotype data from the 1958 Birth Cohort collection, funded by the MRC and the Wellcome Trust which was genotyped by the Wellcome Trust Case Control Consortium and the Type-1 Diabetes Genetics Consortium, sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development and Juvenile Diabetes Research Foundation International. The Nottingham Group (KM) are supported by the Big Lottery. MRC CFAS is part of the consortium and data will be included in future analyses. ADGC The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, R01 AG041797; MIRAGE R01 AG025259; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG06781, UO1 HG004610; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, MO1RR00096, and UL1 RR029893; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582, UL1RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50, P50 AG016575, P50 AG016576, P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG034504 to AJM, The Banner Alzheimer's Foundation, The Johnnie B. Byrd Sr. Alzheimer's Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council), South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer's Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. Marcelle Morrison-Bogorad, PhD., Tony Phelps, PhD and Walter Kukull PhD are thanked for helping to co-ordinate this collection. ADNI Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, Glaxo-SmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., Alzheimer's Association, Alzheimer's Drug Discovery Foundation, the Dana Foundation, and by the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, K01 AG030514. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. The authors thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-o_cio ADGC members. Support was also from the Alzheimer's Association (LAF, IIRG-08-89720; MP-V, IIRG-05-14147) and the United States Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. Peter St George-Hyslop is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, et al. (2006) Role of genes and environments for explaining Alzheimer disease. Archives of General Psychiatry 63: 168–174. [DOI] [PubMed] [Google Scholar]
- 2. Bettens K, Sleegers K, Van Broeckhoven C (2013) Genetic insights in Alzheimer's disease. Lancet neurology 12: 92–104. [DOI] [PubMed] [Google Scholar]
- 3. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 4. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics 41: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, et al. (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature Genetics 43: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature Genetics 41: 1094–U1068. [DOI] [PubMed] [Google Scholar]
- 7. Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nature Genetics 43: 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, et al. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA: the journal of the American Medical Association 303: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guerreiro RJ, Hardy J (2011) Alzheimer's disease genetics: lessons to improve disease modelling. Biochemical Society transactions 39: 910–916. [DOI] [PubMed] [Google Scholar]
- 11. Ioannidis JP (2007) Non-replication and inconsistency in the genome-wide association setting. Human heredity 64: 203–213. [DOI] [PubMed] [Google Scholar]
- 12. Neale BM, Sham PC (2004) The future of association studies: gene-based analysis and replication. American journal of human genetics 75: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moskvina V, O'Donovan MC (2007) Detailed analysis of the relative power of direct and indirect association studies and the implications for their interpretation. Human heredity 64: 63–73. [DOI] [PubMed] [Google Scholar]
- 14. Terwilliger JD, Hiekkalinna T (2006) An utter refutation of the "fundamental theorem of the HapMap". European journal of human genetics: EJHG 14: 426–437. [DOI] [PubMed] [Google Scholar]
- 15. Bettens K, Brouwers N, Engelborghs S, Lambert JC, Rogaeva E, et al. (2012) Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Molecular neurodegeneration 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. American journal of human genetics 46: 222–228. [PMC free article] [PubMed] [Google Scholar]
- 17. Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, et al. (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. American journal of human genetics 73: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, et al. (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. American journal of human genetics 73: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert JCea (2013) Extended meta-analysis of 74,538 individuals identifies 11 new susceptibility loci for Alzheimer's disease. [DOI] [PMC free article] [PubMed]
- 20. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webster JA, Gibbs JR, Clarke J, Ray M, Zhang WX, et al. (2009) Genetic Control of Human Brain Transcript Expression in Alzheimer Disease. American Journal of Human Genetics 84: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trabzuni D, Ryten M, Walker R, Smith C, Imran S, et al. (2011) Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 119: 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seux M, Peuget S, Montero MP, Siret C, Rigot V, et al. (2011) TP53INP1 decreases pancreatic cancer cell migration by regulating SPARC expression. Oncogene 30: 3049–3061. [DOI] [PubMed] [Google Scholar]
- 24. Seillier M, Peuget S, Gayet O, Gauthier C, N'Guessan P, et al. (2012) TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell death and differentiation 19: 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giusiano S, Baylot V, Andrieu C, Fazli L, Gleave M, et al. (2012) TP53INP1 as new therapeutic target in castration-resistant prostate cancer. Prostate 72: 1286–1294. [DOI] [PubMed] [Google Scholar]
- 26.Driver JA, Beiser A, Au R, Kreger BE, Splansky GL, et al.. (2012) Inverse association between cancer and Alzheimer's disease: results from the Framingham Heart Study. British Medical Journal 344. [DOI] [PMC free article] [PubMed]
- 27. Roe CM, Fitzpatrick AL, Xiong C, Sieh W, Kuller L, et al. (2010) Cancer linked to Alzheimer disease but not vascular dementia. Neurology 74: 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watson CT, Breden F (2012) The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes and immunity 13: 363–373. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Yang LN, Cheng L, Tu S, Guo SJ, et al.. (2013) BAG3 Interactome Analysis Reveals a New Role in Modulating Proteasome Activity. Molecular & cellular proteomics: MCP. [DOI] [PMC free article] [PubMed]
- 30. Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, et al. (2011) The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. The Journal of biological chemistry 286: 23521–23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmieri F (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Molecular aspects of medicine 34: 465–484. [DOI] [PubMed] [Google Scholar]
- 32. Katz C, Zaltsman-Amir Y, Mostizky Y, Kollet N, Gross A, et al. (2012) Molecular basis of the interaction between proapoptotic truncated BID (tBID) protein and mitochondrial carrier homologue 2 (MTCH2) protein: key players in mitochondrial death pathway. The Journal of biological chemistry 287: 15016–15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernhard F, Landgraf K, Kloting N, Berthold A, Buttner P, et al. (2013) Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 56: 311–322. [DOI] [PubMed] [Google Scholar]
- 34. Fall T, Arnlov J, Berne C, Ingelsson E (2012) The role of obesity-related genetic loci in insulin sensitivity. Diabetic medicine: a journal of the British Diabetic Association 29: e62–66. [DOI] [PubMed] [Google Scholar]
- 35. Haupt A, Thamer C, Heni M, Machicao F, Machann J, et al. (2010) Novel obesity risk loci do not determine distribution of body fat depots: a whole-body MRI/MRS study. Obesity 18: 1212–1217. [DOI] [PubMed] [Google Scholar]
- 36. Benit P, Slama A, Cartault F, Giurgea I, Chretien D, et al. (2004) Mutant NDUFS3 subunit of mitochondrial complex I causes Leigh syndrome. Journal of medical genetics 41: 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dahl HH (1998) Getting to the nucleus of mitochondrial disorders: identification of respiratory chain-enzyme genes causing Leigh syndrome. American journal of human genetics 63: 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown MB (1975) A method for combining non-independent, one-sided tests of significance. Biometrics 31: 978–992. [Google Scholar]
- 41. Moskvina V, O'Dushlaine C, Purcell S, Craddock N, Holmans P, et al. (2011) Evaluation of an approximation method for assessment of overall significance of multiple-dependent tests in a genomewide association study. Genetic epidemiology 35: 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, et al. (2012) Exome sequencing and the genetic basis of complex traits. Nature genetics 44: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moskvina V, Schmidt KM (2008) On multiple-testing correction in genome-wide association studies. Genetic epidemiology 32: 567–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overrepresentation of significant SNPs excluding previously reported [4]-[8] genes ±0.5Mb and the APOE region as above.
(DOCX)
List of genes that are genome-wide significant in the IGAP stage 1 dataset and the flanking regions which included SNPs either in r2≥0.3 or association p-value≤10-3 whichever covers the largest region.
(DOCX)
Detailed SNP information for TP53INP1 gene.
(XLS)
Detailed SNP information for IGHV1-67 gene.
(XLS)
Detailed SNP information for ZNF3 gene.
(XLS)
Detailed SNP information for NDUFS3 gene.
(XLS)
Detailed SNP information for MTCH2 gene.
(XLS)
Gene-based analysis results, when single SNPs p-values, contributing to the gene-based p-value were adjusted for the best genome-wide significant SNP in the nearby location.
(DOCX)
Gene-wide analysis for genes which show GWAS significant association with AD in the stage 1 IGAP dataset.
(DOCX)
Brain eQTL Tissues.
(XLSX)
Brain Meth QTLs.
(XLSX)
ZNF3 gene with rs1476679 (ZCWPW1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs1476679 is highlighted in blue.
(TIF)
NDUFS3 gene rs10838725 (CELF1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs10838725 is highlighted in blue.
(TIF)
MTCH2 gene with rs10838725 (CELF1) reported by Lambert et al (2013) study. SNPs which are significant at 1e-3 level are circled in red, rs10838725 is highlighted in blue.
(TIF)
QQ-plot of gene-wide p-values for all genes (A) and excluding previously reported [4]-[8] GWAS significantly associated genes ±0.5Mb (B) in the discovery dataset. Genomic control λ = 1.08 and 1.07 respectively.
(TIFF)
Expression quantitative trait loci (eQTL) and Methylation quantitative trait loci (meQTL) analyses.
(DOCX)
Full IGAP datasets description.
(DOCX)
List of IGAP consortium members.
(DOC)
Acknowledgements.
(DOCX)