Abstract
Background
Epidemiologic studies of respiratory infections frequently rely on separate sample collections for the detection of bacteria and viruses. The requirement for two specimens presents cost, logistical, and acceptability challenges.
Objectives
To determine the agreement in detection of respiratory viruses using RT-PCR between two different types of samples collected on the same day: nasal swabs preserved in viral transport medium (NS) and nasopharyngeal swabs preserved in skim milk-tryptone-glucose-glycerol [STGG] medium (NP), the current standard for pneumococcal colonization studies.
Study design
Paired NS and NP samples were collected between May 2009 and September 2011 as part of the RESPIRA-PERU study, a large prospective cohort of Andean children <3 years of age. NS samples used polyester swabs and viral transport medium whereas NP samples used rayon wire-handled swabs and STGG medium. Samples were tested for influenza, human metapneumovirus (MPV), respiratory syncytial virus (RSV), human rhinovirus (HRV), parainfluenza virus 3 (PIV3) and adenovirus (ADV) using real-time RT-PCR. We calculated the agreement, and compared cycle thresholds (CT) between NP and NS samples.
Results
Among 226 paired NP-NS samples, we observed very high agreement with a Kappa statistic ranging from 0.71 for ADV to 0.97 for MPV. CT values were similar for both strategies.
Conclusions
NP samples preserved in STGG provide a simple and reliable strategy for identification of both pneumococcus and respiratory viruses. This single specimen collection strategy could be used for epidemiologic studies, especially in resource-limited settings. Furthermore, archived NP-STGG specimens from previous studies could be reliably tested by RT-PCR for viruses.
Keywords: respiratory virus, children, epidemiology, nasal swab, nasopharyngeal swab
BACKGROUND
Epidemiologic studies of respiratory infections frequently rely on separate sample collections for the detection of bacteria and viruses. For example, the standard method for detection of pneumococcal nasopharyngeal colonization uses a deep nasopharyngeal swab (NP) preserved in skim milk-tryptone-glucose-glycerol (STGG).1, 2 In contrast, many recent studies of respiratory viruses used nasal mid-turbinate swabs (NS) preserved in viral transport medium. The requirement for separate collection procedures, different media, and different swab types presents barriers to studying the interaction of viruses with bacteria in clinical and field studies, including increased cost and complexity, increased storage needs, and subject dissatisfaction with multiple procedures.
OBJECTIVES
To determine the agreement in RT-PCR-based detection of six common respiratory viruses between NS and NP samples. Furthermore, we also used cycle thresholds (CT) to indirectly compare the amount of viral genetic material detected by each sampling strategy.
STUDY DESIGN
Paired NS and NP samples were collected between May 2009 and September 2011 as part of the RESPIRA-PERU study, a large prospective cohort of Andean children <3 years of age.3 Signed informed consent was obtained from parents of participating children. Study children were under observation through weekly household visits and NS were collected during episodes of acute respiratory illness for identification of respiratory viruses.4, 5 For NS sample collection, one polyester swab was placed into each nostril sequentially and rotated beneath the turbinates, placed into a tube with 3 mL of Remel M4RT viral transport medium, and transported in envelopes with cold packs to the local research laboratory within 8 hours of collection. At the local research laboratory, two 800-μl aliquots were preserved as original samples and three 200-μl aliquots were preserved in lysis cryovials with 300 μl lysis buffer (MagNA Pure LC, Roche® and MagMAX, Ambion®). All vials were labeled and stored at − 70°C in the local research laboratory.
NP samples were collected monthly without regard to the presence of respiratory illness following WHO recommendations for identification of pneumococcal colonization.1, 2 NP samples were collected with a rayon wire-handled swab inserted deep into the nasopharynx and rotated, and were immediately placed in a tube with 1 mL of STGG. The specimens were transported in envelopes with cold packs to the local laboratory within 8 hours of collection. Specimens were labeled and preserved with the swab in the medium at −70°C. All NS and NP specimens were collected and processed by the same study workers in the same laboratory and were shipped on dry ice to Vanderbilt for testing. NP specimens had been previously thawed and refrozen once for bacterial culture and PCR.
We selected a random sample of 226 paired NS and NPs that were collected from the same child on the same date, during a scheduled household visit. Both NS and NP samples were analyzed at Vanderbilt for identification of influenza, human metapneumovirus (MPV), respiratory syncytial virus (RSV), human rhinovirus (HRV), parainfluenza virus 3 (PIV3), and adenovirus (ADV) using nucleic acid extraction and real-time RT-PCR methods previously described.5-8 To maximize the efficiency of the comparisons, NP samples were tested for the targets identified in the paired NS, and for completeness, a random sample of NP paired to NS that had tested negative for all study viruses were also tested for all targets. For both NS and NP, samples with a minimum volume of 200 μl were included, and the samples were considered positive if the RT-PCR CT value was <40. All specimens tested positive for human RNase P to ensure RNA integrity.
We calculated the percent agreement and the agreement beyond chance between NS and NP samples using the Kappa statistic.9 To further assess the agreement between NS and NP, we compared CT values between paired NP and NS samples plotting the mean CT by the CT differences using Bland-Altman plots, and calculated limits of agreement for each study virus.10 We also compared CT distributions for each virus between groups of NS and NP specimens using a 2-tailed Wilcoxon signed-rank test for matched-pairs. Although some viral co-infections were anticipated, for simplicity we considered each virus detection as independent and reported them individually. Statistical analyses were done in STATA 12.1.
RESULTS
The paired study samples included 133 NS with single detections: 49 HRV, 28 RSV, 24 PIV3, 12 MPV, 11 ADV and 9 influenza; 30 co-detections including two viruses (28 including HRV or ADV); 2 co-detections including three viruses, RSV, HRV and ADV; and, 61 NS that tested negative for all study viruses. Among the paired samples, the detection of common respiratory viruses ranged from 13%-50% among NS samples and from 16%-53% among NP samples. The percent agreement between NP and NS results was 89-99% (Table 1). The Kappa statistic indicated high agreement between the strategies and ranged from 0.71 for ADV to 0.97 for MPV. The Kappa for all viruses other than ADV was ≥86 (Table 1).
Table 1.
| Virus (number of paired NP-NS samples) |
Positive in NS |
Positive in NP |
% Agreement | Kappa |
|---|---|---|---|---|
| Influenza A (69) | 9 (13%) | 11 (16%) | 97.1% (67/69) | 0.88 |
| MPV (80) | 19 (24%) | 20 (25%) | 98.8% (79/80) | 0.97 |
| RSV (97) | 32 (33%) | 34 (35%) | 93.8% (91/97) | 0.87 |
| HRV (132) | 66 (50%) | 70 (53%) | 93.2% (123/132) | 0.86 |
| PIV3 (88) | 24 (27%) | 25 (28%) | 96.6% (85/88) | 0.92 |
| ADV (83) | 16 (19%) | 19 (23%) | 89.2% (74/83) | 0.71 |
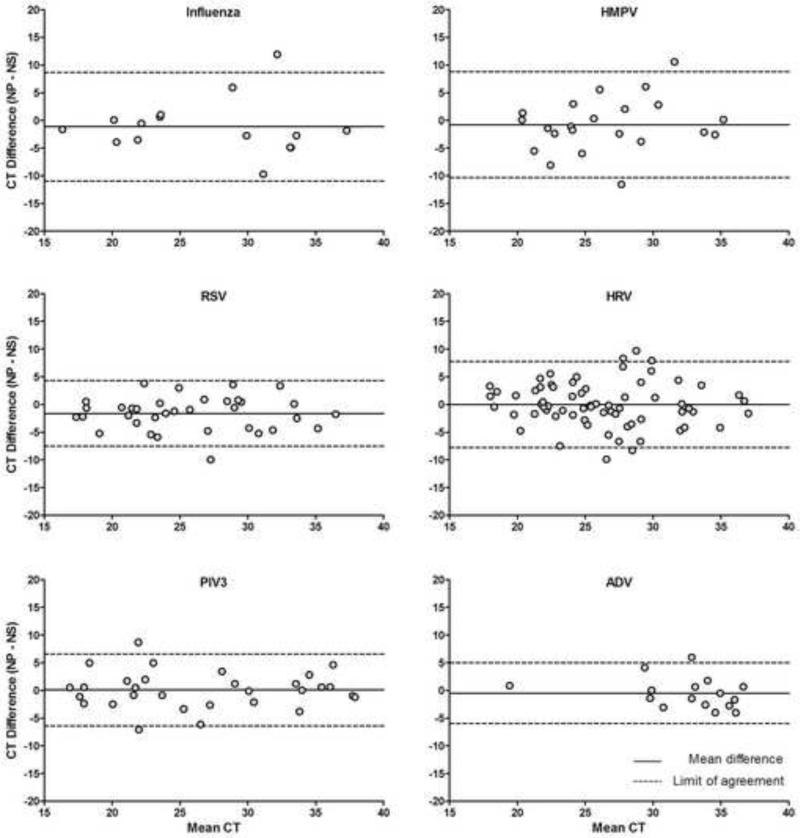
The CT differences between NP and NS measurements were small and close to 0 for most paired specimens (<10 cycles across all RT-PCR targets). CT values did not vary in a systematic way to suggest differential detection by NS versus NP methods (Figure 1). For ADV, agreement was lower and CTs higher than for other viruses. When the distributions of CT values for each virus were compared between paired NS and NP samples, CT for influenza and RSV were slightly lower in NP than in NS samples, but CT for all other viruses were similar in both samples (Table 2).
Figure 1.
Agreement between NS and NP measurements
Footnote: the limits of agreement are calculated as 2 standard deviations above/below the mean difference between strategies. The cycle threshold (CT) indicates the PCR cycle at which the sample was positive, and is a surrogate measure of sensitivity of detection. A positive CT difference (y-axis; NP-NS) indicates that the NS was more sensitive, while a negative CT difference indicates that the NP was more sensitive.
Table 2.
Comparison of CT distributions between NS and NP specimens by virus
| Virus | NS Median CT (IQR) | NP Median CT (IQR) | P |
|---|---|---|---|
| Influenza A | 23.7 (22.5 – 35.0) | 24.1 (20.1 - 28.5) | 0.015 |
| MPV | 26.3 (23.9 – 31.0) | 23.4 (21.5 - 31.8) | 0.212 |
| RSV | 25.9 (22.0 - 31.5) | 23.8 (20.3 - 29.1) | <0.001 |
| HRV | 25.7 (22.1 - 30.2) | 25.5 (22.6 - 30.7) | 0.982 |
| PIV3 | 26.2 (20.4 – 34.0) | 23.5 (19.8 - 32.9) | 0.511 |
| ADV | 33.4 (30.2 - 36.4) | 33.0 (30.7 - 34.8) | 0.438 |
The distributions of CT were compared between NS and NP using a 2-tailed Wilcoxon signed-ranks test for matched-pairs.
DISCUSSION
We demonstrated that the agreement in detection of common respiratory viruses was very high between NS and NP samples. Furthermore, the comparison of CT values indicates that the differences in detection are small, suggesting that similar quantities of viral material were present in both samples. While there were significant differences in CT for some viruses, the majority of the samples had CTs that fell within a detectable range, and thus this slight discordance would not be expected to cause false negative results.
A previous study used samples from Thailand-Myanmar and documented high agreement between nasopharyngeal aspirates preserved in viral transport medium and NP samples preserved in STGG for detection of influenza, RSV and ADV among children with pneumonia.11 Our study, which used samples from young Andean children with acute respiratory illness, complements the prior report and expands it to a larger number of samples including MPV, HRV and PIV3; NS samples, which are commonly obtained; and importantly, by comparing CT values between detection strategies. Notably, even though our NP samples had undergone a previous freeze-thaw cycle, the detection of viruses was remarkably similar between the 2 sampling strategies.
In conclusion, our observations support the use of STGG-NP samples, the current standard for pneumococcal colonization studies,1, 2 as a simple and reliable strategy for identification of both pneumococcus and respiratory viruses, facilitating the evaluation of the interactions between these pathogens. This strategy could greatly simplify prospective field epidemiologic studies of respiratory illness in resource-limited settings. Furthermore, existing stored NP specimens preserved in STGG could be retrospectively tested for respiratory viruses with confidence as to the reliability of the results.
Highlights.
Studies of respiratory viruses frequently use nasal swabs and viral transport medium (NS)
Pneumococcal colonization studies use nasopharyngeal swabs and STGG medium (NP)
We compared RT-PCR viral detections in paired NS and NP collected from the same child on the same date
We observed very high agreement in viral detections, and similar CT values in both strategies
NP samples preserved in STGG allow efficient study of both pneumococcus and respiratory viruses
Acknowledgements
On behalf of the study of Respiratory Infections in Andean Peruvian children (RESPIRA PERU): Vanderbilt University: Marie R. Griffin, John V. Williams, Kathryn M. Edwards, Philip J. Budge, Yuwei Zhu, Monika Johnson and Carlos G. Grijalva; Emory University: Jorge E. Vidal and Keith P. Klugman; Instituto de Investigacion Nutricional: Hector Verastegui, Ana I. Gil and Claudio F. Lanata. We are indebted to the communities of San Marcos, Cajamarca, Peru for their participation in this study. We also appreciate the approval and continuous support of the Cajamarca Health Region authorities. We are also indebted to the field workers and field supervisors whose efforts in difficult geographical areas and harsh weather conditions allowed this study to be conducted.
Funding
This study was supported by the Thrasher Research Fund (grant 02832-9), Vanderbilt University CTSA grant UL1 RR024975-01 from the US National Institutes of Health, and an investigator initiated research grant IIR WS1898786(0887X1-4492) from Pfizer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
CGG received research support for the study from Pfizer and has served as consultant for Glaxo-Smith-Kline and Pfizer. MRG receives grant support from MedImmune. JVW serves on the Scientific Advisory Board of Quidel. CFL is an advisor to Takeda Vaccines Division.
Ethical approval
The study was approved by the IRB of Vanderbilt University and the Instituto de Investigacion Nutricional (Lima, Peru).
REFERENCES
- 1.O’Brien KL, Nohynek H. Report from a WHO working group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J. 2003;22:133–40. doi: 10.1097/01.inf.0000048676.93549.d1. [DOI] [PubMed] [Google Scholar]
- 2.Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: Updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2013;32:165–79. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 3.Grijalva CG, Griffin MR, Edwards KM, Williams JV, Gil AI, Verastegui H, et al. The Study of Respiratory Pathogens in Andean Children. Int J Epidemiol. 2013 doi: 10.1093/ije/dyt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin MR, Walker FJ, Iwane MK, Weinberg GA, Staat MA, Erdman DD. Epidemiology of respiratory infections in young children: insights from the new vaccine surveillance network. PediatrInfectDisJ. 2004;23:S188–S92. doi: 10.1097/01.inf.0000144660.53024.64. [DOI] [PubMed] [Google Scholar]
- 5.Poehling KA, Edwards KM, Weinberg GA, Szilagyi P, Staat MA, Iwane MK, et al. The underrecognized burden of influenza in young children. The New England journal of medicine. 2006;355:31–40. doi: 10.1056/NEJMoa054869. [DOI] [PubMed] [Google Scholar]
- 6.Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–9. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. 2011;49:2175–82. doi: 10.1128/JCM.02270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klemenc J, Asad Ali S, Johnson M, Tollefson SJ, Talbot HK, Hartert TV, et al. Real-time reverse transcriptase PCR assay for improved detection of human metapneumovirus. J Clin Virol. 2012;54:371–5. doi: 10.1016/j.jcv.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 10.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- 11.Turner P, Po L, Turner C, Goldblatt D, Nosten F. Detection of respiratory viruses by PCR assay of nasopharyngeal swabs stored in skim milk-tryptone-glucose-glycerol transport medium. J Clin Microbiol. 2011;49:2311–3. doi: 10.1128/JCM.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]