Abstract
Background
MicroRNAs are small (18–22 nucleotides) noncoding RNAs involved in posttranscriptional modification of many target genes. One of these, microRNA-21 (miR-21), has been shown to play a role in multiple hematologic and solid organ malignancies. We sought to determine the expression pattern of miR-21 in pancreatic cancers and its impact on clinicopathologic characteristics.
Methods
Eighty resected pancreatic cancer specimens were microdissected and tissue microarrays (TMA) created in duplicate. TMAs were also created for benign pancreas (N=12) and chronic pancreatitis (N=45). In situ hybridization (ISH) was undertaken utilizing locked nucleic acid probes for miR-21. RNA U6 and scrambled RNA served as positive and negative control, respectively. ISH was scored as 0 (absent), 1+ (faint/focal expression), or 2+ (strong expression). Kaplan– Meier survival curves were constructed and compared by log-rank analysis.
Results
MiR-21 expression was demonstrated in 63 (79%) pancreatic cancers (1+ in 49, 2+ in 14) compared to one of 12 (8%, p<0.0001) benign pancreas and 12/45 (27%, p<0.0001) chronic pancreatitis. None of the benign tissues demonstrated strong miR-21 expression. Although miR-21 expression did not correlate with tumor size, differentiation, nodal status, or T stage, strong miR-21 expression was predictive of poorer outcome compared to absent or faint/focal miR-21 expression in patients with node-negative disease (median 27.7 months vs. 15.2, p=0.037). Nodal status was also predictive of survival (p=0.029).
Conclusions
MicroRNA-21 is significantly overexpressed in pancreatic cancers as detected by in situ hybridization. Its strong expression predicts limited survival in patients with node-negative disease and may be an important biologic marker for outcome.
Keywords: MicroRNA, MiRNA, MiR-21, Pancreatic cancer
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related death in the United States. It is nearly uniformly fatal with its yearly mortality approaching its incidence with over 33,000 people succumbing from the disease in 2007.1 Chemoradiation has been shown to modestly prolong survival; however, the prognosis remains extremely poor with the median survival less than 24 months.2 These factors have led to the research and need to discover unique molecular targets and biologic therapies for pancreatic cancer.
MicroRNAs (miRNAs or miRs) are small (~18–22 nucleotides) noncoding RNAs which have critical functions in various biological processes.3 Over 450 human miRNAs have been reported and a number of them have been shown to play normal physiologic roles in cell proliferation, apoptosis, and differentiation. These naturally occurring miRNAs function by binding to target mRNAs, resulting in their degradation or translational inhibition based upon the degree of complimentarity with their target mRNA.4 MiRNAs have been proposed to contribute to oncogenesis by promoting the expression of oncogenes or by inhibiting tumor suppressors.5 These dysregulated miRNAs are often referred to as oncomiRs.
One such oncomiR, miR-21 has been shown to be overexpressed in multiple malignancies including pancreatic cancer,6,7 esophageal cancer,8 lung cancer,9 and colon cancer.10 This miRNA has been linked to tumor aggression and carcinogenesis, in part, by preventing apoptosis and, thus, functioning as an oncogene.11,12 Previous studies have primarily used real time polymerase chain reaction (RTPCR) or miRNA microarray technology to evaluate miRNA expression.13,14 The small size of mature miRNA leads to a low melting temperature of the miR/cDNA complex that is difficult to detect using in situ hybridization.15 Thus, in situ hybridization had not been used extensively to evaluate miRNA expression previously. This problem has been addressed by modifying the nucleotide bases with locked nucleic acids which markedly increases the melting temperature of the miR/cDNA probe.14 While very sensitive for miRNA detection, RT-PCR and micro-array chip technologies are unable to differentiate between expressions from malignant cells vs. contamination from surrounding stroma. This is especially important to distinguish in pancreatic cancer where the surrounding stroma demonstrates such an intense inflammatory reaction.
Herein, we sought to utilize in situ hybridization to answer two important questions. First, in pancreatic cancers, is tumor miR-21 expression derived primarily from malignant ductal epithelial cells or surrounding stroma? Secondly, does miR-21 expression predict survival in patients undergoing curative resection for pancreatic cancer?
Materials and Methods
Tissue Microarrays
After approval from the institutional review board, 80 formalin-fixed, paraffin-embedded pancreatic cancer specimens, 12 benign pancreas, and 45 chronic pancreatitis samples were obtained from the Department of Pathology's archival files at Ohio State University. Samples were microdissected and tissue microarrays (TMAs) were created in duplicate. Our method of TMA creation has been described previously.16 Briefly, 2 mm cores were punched out of each paraffin block in duplicate and transferred to the recipient TMA blocks using a precision instrument (Beecher Instruments, Silver Springs, MD, USA). The paraffin-embedded tissues were then cut in 4 μm slices and placed on a positively charged slide. The slides were heated to 40°C for 30 min, then leveled off and cooled to 4°C for 15 min.
In Situ Hybridization
The in situ hybridization was carried out with probes for miR-21 as well as appropriate controls. The TMA slides were incubated at 60°C for 30 min, deparaffinized in xylene, and rehydrated with graded alcohol washes. Subsequently, the slides were fixed in 4% paraformalde-hyde at 4°C for 10 min and then washed three times in phosphate buffered saline (PBS). The slides were then incubated in Proteinase K solution at 37°C for 20 min. After rinsing, they were immersed in formaldehyde for 10 min. The slides were prehybridized in hybridization buffer (no probe) at 53°C for 1 h. Digoxigenin (DIG)-labeled mercury locked nucleic acid probes for miR-21, U6 (positive control), and scrambled RNA (negative control; Exiqon, Woburn, MA) were hybridized to the slides for 20 h at 53°C. Strigency washes were performed at 53°C and the slides were then placed in a blocking solution for 1 h at room temperature. Sections were then incubated for 2 h at room temperature with preincubated blocking solution with alkaline phosphatase conjugated anti-DIG Fab fragment. After washing in 0.1% Tween-20 followed by PBS, they were stored at 4°C until the following day. The slides were then blotted and layed flat in a humidified chamber and subsequently incubated for 10 h with RTU BM purple AP substrate (Roche, Basel, Switzerland) at room temperature. The slides were then placed in stop solution for 5 min and mounted. Only the slides that stained appropriately for the controls were analyzed. The slides were then scored by two pathologists independently as negative (−), weak or focally positive (1+), or strongly positive (2+). Both pathologists were blinded to the patient's clinical outcome.
Data Acquisition and Statistics
Patient demographics, clinical presentation, hospital course, and outcome were extracted from hospital records. Data collected included age, gender, presence of jaundice, tumor size, T stage, nodal status, differentiation, and postoperative complications. Survival data was obtained from hospital and clinic records and the Social Security Death Index (http://www.ssdi.rootsweb.ancestry.com) as of February 14, 2007. Kaplan–Meier survival curves were constructed and compared by log-rank analysis. Categorical data were compared by Fisher's exact test.
Results
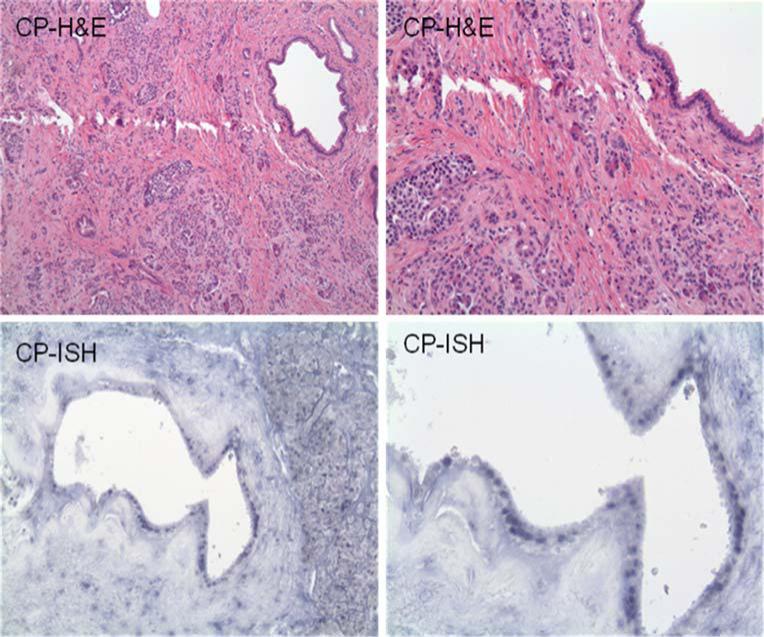
MiR-21 expression was demonstrated in 63 (79%) of the pancreatic cancers by in situ hybridization (Table 1). While most of the cancers demonstrated 1+ miR-21 expression, 14 had 2+ expression. MiR-21 expression was significantly less common in normal pancreas (8%, p<0.0001) and chronic pancreatitis (27%, p<0.0001; Fig. 1). In all cancer specimens, miR-21 expression was seen only in tumor cells and not in the surrounding stroma. None of the benign tissues (i.e., normal pancreas or chronic pancreatitis) demonstrated strong miR-21 expression. MiR-21 staining was seen predominately in the nuclei with some cytoplasmic stipling (Figs. 2 and 3).
Table 1.
MiR-21 Expression by In Situ Hybridization
| miR-21 expression | PCA (N=80) | CP (N=45) | NP (N=12) |
|---|---|---|---|
| 0 (negative) | 17 (21%) | 33 (73%) | 11 (92%) |
| 1+ (weak positive) | 49 (61%) | 12 (27%) | 1 (8%) |
| 2+ (strong positive) | 14 (18%) | 0 (0%) | 0 (0%) |
| All positive | 63/80 (79%) | 12/45 (27%)* | 1/12 (8%)* |
p<0.0001 vs. pancreatic cancer
Figure 1.
Hematoxylin and eosin stain (top panels) and in situ hybridization (bottom panels) for miR-21 in normal pancreas (NP) and chronic pancreatitis (CP) at ×40 magnification. No staining is seen for miR-21.
Figure 2.
Hematoxylin and eosin stain (top panels; ×40 magnification) and in situ hybridization (bottom panels; ×100 magnification) in chronic pancreatitis with 1+ expression of miR-21. Note darker nuclear staining in ductal epithelial cells.
Figure 3.
Hematoxylin and eosin stain (top panels; ×40 magnification) and in situ hybridization (bottom panels; ×100 magnification) of pancreatic adenocarcinoma (PA) with 2+ miR-21 expression. Note dark nuclear staining with cytoplasmic stipling, predominately in cancer cells with very little stromal staining.
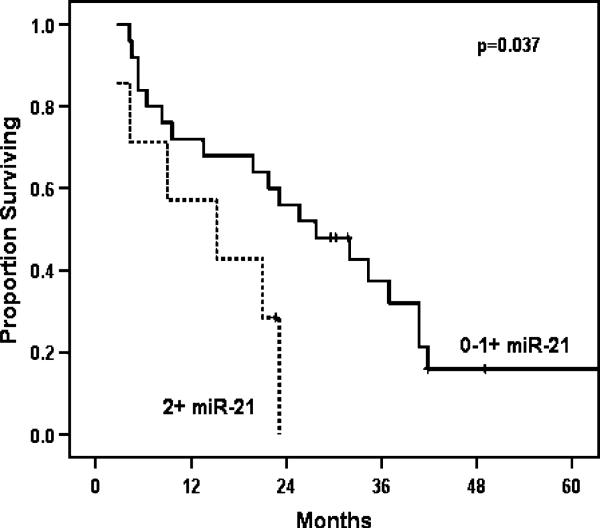
MiR-21 expression did not correlate with tumor size, differentiation, nodal status, or T stage. Of all variables tested, only T stage and nodal status were predictive of survival. A median survival of 14.3 months was seen in patients with positive nodes compared to 23.1 months for patients with node-negative disease (p=0.029; Fig. 4; Table 2). When miR-21 expression was considered in all pancreatic cancer patients, it was not a significant predictor of survival. However, in the subset of patients with node-negative disease, strong miR-21 expression was predictive of poorer outcome compared to absent or faint/focal miR-21 expression (Fig. 5). Those with node-negative disease and strong miR-21 expression had a median survival of 15.2 vs. 27.7 months for those who did not strongly overexpress miR-21 (p=0.037; Table 3).
Figure 4.
Overall survival curves for node-positive and node-negative pancreatic cancers.
Table 2.
Overall Survival Based Upon Nodal Status
| Median (months) | 1 year (%) | 5 year (%) | |
|---|---|---|---|
| Node negative | 23.1 | 71.8 | 17.2 |
| Node positive | 14.3 | 64.3 | 5.8 |
Figure 5.
Overall survival in node-negative patients strong vs. weak or no expression of miR-21.
Table 3.
Overall Survival in Node-Negative Patients with Strong vs. Weak or No Expression of miR-21
| miR-21 | Median (months) | 1 year (%) | 5 year (%) |
|---|---|---|---|
| 0–1+ | 27.7 | 72 | 16 |
| 2+ | 15.2 | 57.1 | 0 |
Discussion
MiRNA profiles have been established for many solid and hematologic malignancies. In particular, miR-21 has been reported to be important in many cancers but associations with clinical outcomes and survival are largely unknown. Similarly, few biological markers have been shown to predict survival in pancreatic cancer, likely due to a universally poor prognosis. In this study, we show that strong miR-21 expression in pancreatic cancer by in situ hybridization may be predictive of poor survival in a group of patients that would otherwise be considered as having a favorable pathology (i.e., nodes negative).
We have previously shown that miR-21 is significantly overexpressed in pancreatic cancer using miRNA micro-array technology.5 While microdissection was utilized to minimize contamination by surrounding stroma, the intense inflammatory reaction often associated with pancreatic cancer calls into question the cell of origin for miR-21 expression. In the present study using in situ hybridization, tumoral miR-21 expression was only seen in malignant cells and not in the surrounding stroma. Interestingly, when miR-21 was expressed, albeit weakly, in benign pancreas, it was only seen in ductal epithelial cells. Staining was predominately nuclear, suggesting binding to precursor miR-21 as well as the mature sequence seen in the cytoplasm. Given that a strong correlation between precursor and mature miRNA has been shown previously, such nuclear staining is not surprising.17
Not clear in this study still is whether miR-21 expression plays a role in oncogenesis in pancreatic cancer or is a late event, perhaps even being incited by reactive stromal cells. The low expression levels seen in the chronic pancreatitis specimens suggest that miR-21 expression is fairly specific to malignancy, however. Our previous microarray data demonstrated that miR-21 expression is able to discriminate, in part, between chronic pancreatitis and pancreatic cancer.6 Hence, miR-21 appears to play an important role in carcinogenesis.
MiR-21 expression did not correlate with tumor size, differentiation, nodal status, or T stage. As expected, the presence of metastatic disease in the lymph nodes decreased survival significantly. When all patients were considered, miR-21 expression did not have an impact on survival. However, in the patients who were expected to have the best survival (i.e., those with lymph node-negative disease), strong overexpressed of miR-21 was associated with a significantly decreased median, 1-, and 5-year overall survival. The subset of patients in this study with node-negative disease is quite small and, therefore, a larger study to confirm these data is necessary and underway. These findings could be helpful in determining which patients should receive the most aggressive treatments and serve as an important biological marker of outcome.
Footnotes
Presented at American Hepato-Pancreato-Biliary Association Annual Meeting in Ft. Lauderdale, FL, March 27, 2008.
Contributor Information
Mary Dillhoff, Department of Surgery, The Ohio State University, 410 W. 10th Ave., N924 Doan Hall, Columbus, OH 43210, USA.
James Liu, Department of Pathology, The Ohio State University, Columbus, OH, USA.
Wendy Frankel, Department of Pathology, The Ohio State University, Columbus, OH, USA.
Carlo Croce, Department of Molecular Virology, Immunology and Molecular Genetics, The Ohio State University, Columbus, OH, USA.
Mark Bloomston, Department of Surgery, The Ohio State University, 410 W. 10th Ave., N924 Doan Hall, Columbus, OH 43210, USA.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Yeo CJ, Abrams RA, Grochow LB, Sohn TA, Ord SE, Hruban RH, et al. Pancreaticoduodenectomy for pancreatic adenocarcinoma: postoperative adjuvant chemoradiation improves survival. A prospective, single-institution experience. Ann Surg. 1997;225(5):621–33. doi: 10.1097/00000658-199705000-00018. doi:10.1097/00000658-199705000-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNA's: genomics biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. doi:10.1016/S0092-8674(04) 00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Dillhoff M, Wojcik S, Bloomston M. MiRNAs in solid tumors. J Surg Res. 2008 doi: 10.1016/j.jss.2008.02.046. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. doi:10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. doi:10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 7.Lee E, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2006;120:1046–1054. doi: 10.1002/ijc.22394. doi:10.1002/ ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–260. doi: 10.1016/j.jtcvs.2007.08.055. doi:10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. doi:10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Schetter A, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. doi:10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. doi:10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 12.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24(29):4677–4684. doi: 10.1200/JCO.2005.05.5194. doi:10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33(17):5394–5403. doi: 10.1093/nar/gki863. doi:10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. doi:10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 15.Nuovo GJ. PCR In Situ Hybridization: Protocols and Applications. 3rd ed. Williams and Wilkins, Raven; Lippincott, New York: 1996. [Google Scholar]
- 16.Bloomston M, Bhardwaj A, Ellison EC, Frankel WL. Epidermal growth factor receptor expression in pancreatic carcinoma using tissue microarray technique. Dig Surg. 2006;23(1–2):74–79. doi: 10.1159/000093497. doi:10.1159/000093497. [DOI] [PubMed] [Google Scholar]
- 17.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cells lines and tumors. RNA. 2008;14(1):35–42. doi: 10.1261/rna.804508. doi:10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]