Abstract
Objective
Since community viral load (CVL) measurements are associated with incidence of new HIV-1 infections in a population, we hypothesized that similarly measured community drug resistance (CDR) could predict prevalence of transmitted drug resistance (TDR).
Methods
Between 2001 and 2011, the prevalence of HIV-1 drug resistance for patients with established infection receiving HIV care (i.e. CDR) and TDR in recently infected patients was determined in San Diego. At each position in HIV-1 reverse transcriptase (RT) and protease (pro), drug resistance was evaluated both as the overall prevalence of resistance associated mutations and by weighting each resistance position to concurrent viral load of the patient and its proportion to the total viral load of the clinic (CVL). The weighting was the proportion of the CVL associated with patients identified with resistance at each residue. Spearman ranked correlation coefficients were used to determine associations between CDR and TDR.
Results
We analyzed 1,088 resistance tests from 971 clinic patients and baseline resistance tests for 542 recently infected patients. CDR at positions 30, 46, and 88 in pro was associated with TDR between 2001 and 2011. When CDR was weighted by viral load of patients, CDR was associated with TDR at position 103 in RT. Each of these associations was corroborated at least once using shorter measurement intervals.
Conclusions
Despite evaluation of a limited percentage of chronically infected patients in San Diego, CDR correlated with TDR at key resistance positions and therefore may be a useful tool to predict the prevalence of TDR.
Keywords: HIV; Drug resistance, viral; Mutation; Antiretroviral therapy, highly active; Genotype; Viral load
Introduction
Mutations in the HIV-1 pol gene can confer decreased susceptibility to antiretroviral therapy (ART)(1). Failure of ART to fully suppress viral replication (i.e. treatment failure) can select for viral populations that harbor these mutations in the presence of ongoing therapy. These mutations can also be found in viral populations among people who have never taken ART, i.e. transmitted drug resistance (TDR)(2). The primary source of TDR is from patients on failing ART regimens with incomplete viral suppression in whom drug resistance mutations have been selected or from subjects with TDR. Since measures of HIV-1 viral load at the community level (i.e. community viral load) have recently been shown to be associated with the incidence of new HIV infections in general populations as well as specific high-risk groups(3, 4), we questioned if similar measures of community drug resistance (CDR) could be related to the prevalence of overall TDR. Specifically, we tested the hypothesis that CDR, calculated from a limited group of patients receiving care at a local HIV clinic with sufficient viral loads and resistance data available, could be associated with the prevalence of TDR in people recently infected with HIV in the same community over the same time period.
Methods
Study populations and testing
This project was approved by the local committee for the protection of human subjects. Chronic infection cohort: The UCSD Owen Clinic is a multidisciplinary clinic that provides comprehensive health care services to approximately 30% of the patients receiving care for HIV/AIDS in San Diego County, California. The total number of HIV-infected individuals living in San Diego in 2011, the last year of the study, was 7,221 (5). It is estimated that 85% of HIV-infected individuals in San Diego are aware of their diagnosis, 54% are linked to care, 32% receive regular care, and 31% have suppressed viral load (unpublished data from County of San Diego Health and Human Services, HIV, STD and Hepatitis Branch, HIV/AIDS Epidemiology Unit). Of those Owen Clinic participants who entered care since 2005, the median and mean lengths of follow-up are 651 and 813 days respectively, and the average attrition rate is 11.7% per year. Attrition was defined as patients not returning for a clinical visit within the periods of observation. Clinic data on attrition rates before 2005 were less complete and likely not as reliable. Available data between 2001 and 2011 included patient age, sex, race, ethnicity, HIV risk factor(s), CD4+ T-lymphocyte counts, HIV-1 viral loads, and results of resistance testing (either Genseq® HIV or PhenoSense GT; Monogram Biosciences, Inc., South San Francisco, California). Standard of care for San Diego County in the study period of observation was for viral loads and CD4 counts to be measured every three months, and over 80% of patients followed longitudinally between 2001 and 2011 met this standard(6). Resistance testing was performed in patients on ART with a viral load of ≥1,000 HIV RNA copies/mL throughout the study period, and baseline resistance testing on treatment naïve patients was implemented in the clinic in 2006. Since these data were obtained in a clinical cohort, it was not always clear if resistance testing occurred in the setting of treatment failure. All patients with an available resistance test result between 2001 and 2011 were included in CDR calculations for the main analysis. Given the longitudinal nature of this cohort, some patients had more than one resistance test included in the analysis, although only one resistance test per patient was included for a given calendar year. For patients with more than one resistance test for a given year, the last result was included. Approximately 14% of all patients who underwent viral load testing during this time period had an available resistance test result.
Primary infection cohort (PIC): Patients enrolled in the San Diego Primary Infection Program between 2001 and 2011 were included in this analysis. Patients were diagnosed with acute or early HIV infection based on results of HIV RNA testing, standard and detuned enzyme immunoassays, and Western blots(7). All resistance testing was performed using Genseq® HIV (Monogram Biosciences, Inc., South San Francisco, California) for plasma viral load ≥1,000 copies/mL and Viroseq v2.0 (Celera Diagnostics, Alameda, California) when plasma viral load was <1,000 copies/mL. Resistance testing was performed at baseline enrollment into the cohort. Only San Diego PIC participants with available baseline resistance tests were included in this analysis.
Drug resistance measurement and statistical analysis
CDR and TDR were measured as the prevalence of HIV-1 drug resistance at important positions within HIV-1 reverse transcriptase (RT) or protease (pro) according to the WHO 2009 surveillance TDR mutation list(8) for each calendar year. At each residue, CDR was evaluated as the overall prevalence of resistance among Owen Clinic patients receiving resistance testing during each calendar year. To account for possible differences in infectivity among these patients, prevalence estimates at each resistance position were also weighted by the proportion of the total viral load of patients with drug resistance (resistant viral load, RVL) to the calculated clinic community viral load (CVL). In other words, the RVL was the total viral load of all patients with resistance at a certain residue, a CDR weighted by the viral load. The theoretical underpinning of this weighting by viral load was that viral load is positively associated with HIV transmission (i.e. higher viral load equals higher infectiousness), so viral load could be a marker of likelihood of transmission of a particular resistance mutation. For each patient, the viral load that was obtained at or closest to the time of resistance testing was used to calculate RVL. CVL was the total viral load of all patients from the Owen Clinic during each year, including those with and without resistance testing. To avoid problems with repeated measurements for individual subjects, the average viral load was determined for patients who had multiple viral load measurements during a given year and was used in the calculation of CVL. Weighting was the proportion of the clinic CVL associated with patients identified with resistance at each interrogated residue (i.e. RVL). Since viral loads in the recipients, i.e. SD PIC participants, would not be relevant to infectiousness from potential sources, TDR was not weighted by viral loads in the participants and non-weighted resistance prevalence estimates were used to measure TDR. Spearman ranked correlation coefficients were calculated to determine the association between CDR and TDR.
Secondary analyses
In order to assess the reproducibility of our findings, as well as to evaluate different intervals for measuring CDR, we performed additional analyses by measuring CDR and TDR results at 3-, 4-, and 6-month intervals. CDR and TDR measurements were calculated as described above for each 3-, 4-, or 6-month period rather than for calendar year, and Spearman ranked correlation coefficients were calculated to determine the association between CDR and TDR. Only time intervals for which measurements were available for both CDR and TDR were included in these calculations.
Viral load analyses
Each individual position in HIV-1 RT and pro was evaluated for a significant difference in mean log-transformed viral load in subjects with and without resistance at that position using a two-tailed t-test. Further evaluation of the individual and combined effects of cooccurring resistance mutations, specifically at positions 82 in pro and 184 in RT, on viral load was performed using a two-way ANOVA, also using log-transformed viral loads. A similar analysis of the effects of co-occurrence of resistance at positions 184 and 190 in RT was performed using a two-way ANOVA.
Results
A total of 1,088 resistance test results from 971 HIV-infected patients receiving care at the Owen Clinic (i.e. chronically infected patients) were analyzed for the main analysis that was based on calendar years. The majority of the cohort was male (87.2%), with a median age of 40 years at the time of initial resistance testing, and about two thirds of the cohort were men who have sex with men (MSM). These demographics are consistent with the HIV epidemic in San Diego County(9). The median CD4 counts and viral loads were 253 cells/mm3 (range 0-1,344) and 4.35 log10 copies/mL (range <1.6-6.49 log10 copies/mL), respectively. At time of resistance testing, most patients were either ART naïve or off of ART (38.9%). Nearly one-third of all subjects were receiving protease inhibitor (PI) based therapy (32.2%), approximately 12.5% were on non-nucleoside RT inhibitor (NNRTI) based therapy, and 7.4% were on regimens containing both a PI and a NNRTI. Treatment data for remaining 9% of patients were incomplete. Viral loads were measured a mean of every three months for patients followed longitudinally, and results from 6,983 patients receiving care at the Owen Clinic, including those with and without resistance testing, were available for CVL estimations. This sampling represents approximately a third% of HIV-infected individuals living in San Diego during this time(5). Initial resistance test results were available for 542 recently infected patients, and 30% had at least one resistance associated mutation.
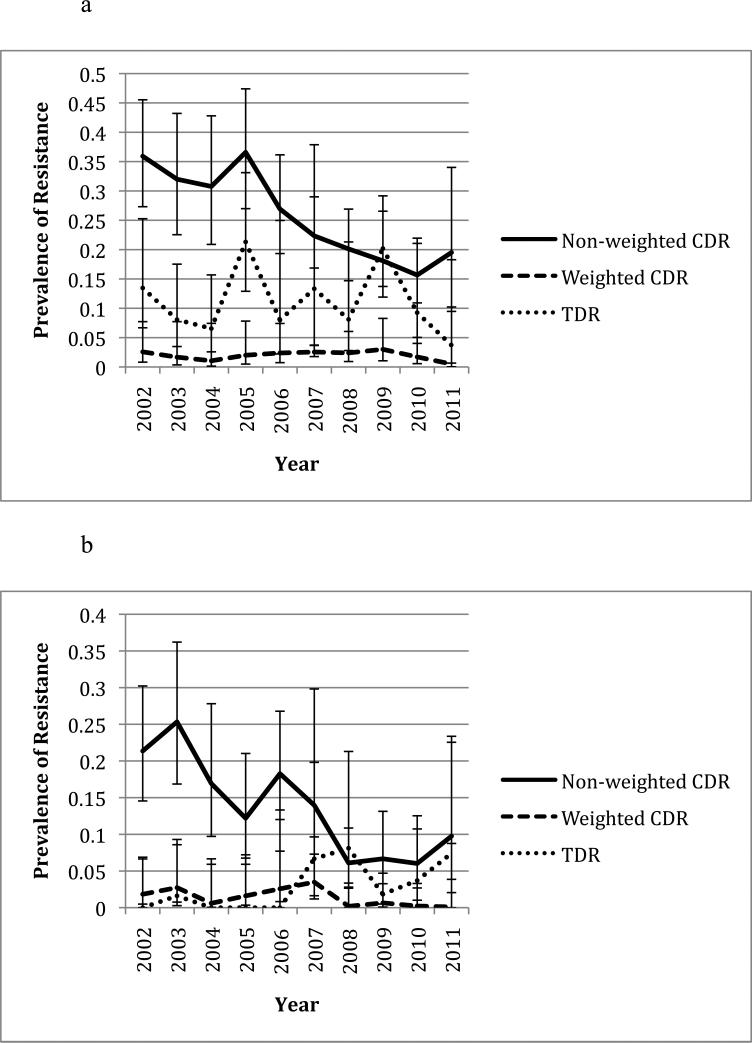
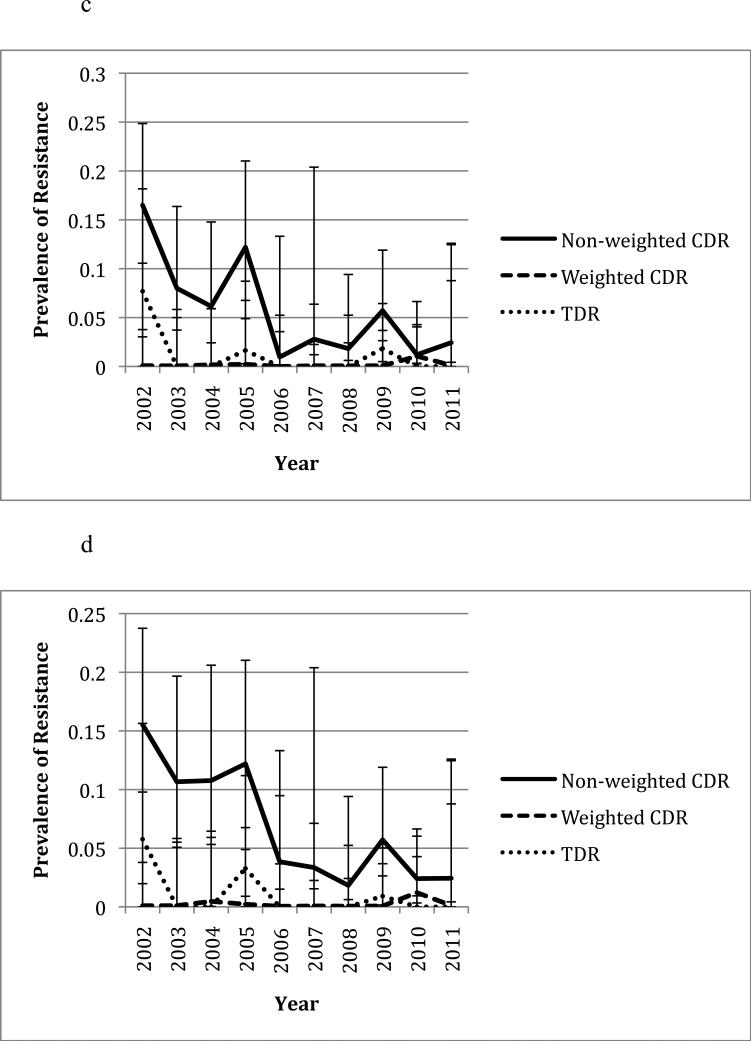
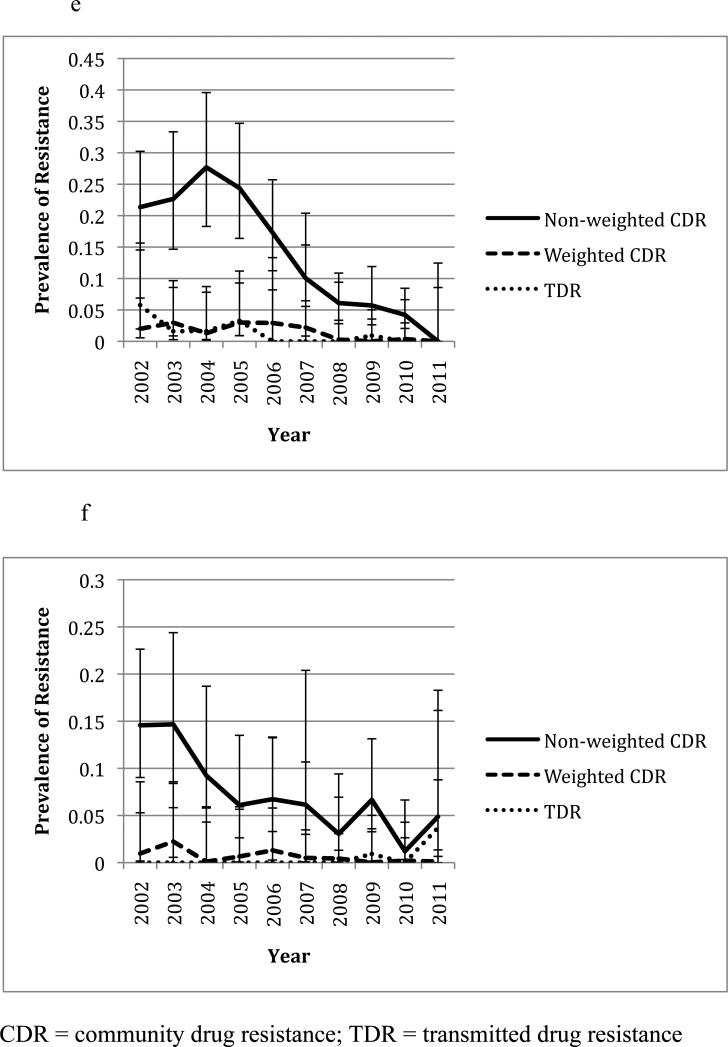
In the six common resistance associated positions (K103, V82, D30, N88, M46, and G190), CDR was lower during the second half of the study period (2006-2011) than in the first half (2002-2005) (Figure 1), which is consistent with previous observations(10, 11). The most common mutations in Owen Clinic and newly infected patients are shown in Table 1. When only considering the overall prevalence of drug resistance in the participants, i.e. without weighting by CVL, significant correlations between CDR and TDR were observed at positions 30 (rho=0.65, p=0.031), 46 (rho=0.74, p=0.008) and 88 (rho=0.68, p=0.021) in pro and position 75 in RT (rho=0.66, p=0.027, see Table 2). When CDR was weighted by CVL, CDR was associated with TDR at positions 103 (rho=0.76, p=0.009) and 190 (rho=-0.68, p=0.022, see Table 3). The yearly prevalence of CDR (i.e. non-weighted) and TDR at these positions ranged from 0 to 75.0% and 0 to 21.3% respectively. Figures 1a to 1f illustrate the changes in CDR and TDR over the course of the study period at various positions in HIV-1 RT and pro.
Figure 1.
Changes in prevalence of CDR and TDR over time at various resistance positions in HIV-1 RT and pro. Error bars represent 95% confidence intervals for each prevalence estimate. All years are included except for 2001 due to the low number of sampled individuals. The non-weighted CDR is the total prevalence of the resistance mutation in the Owen Clinic cohort, while the weighted CDR has been adjusted based on the patient's viral load. CDR during the years 2002-2005 was compared to that during the years 2006-2011 at each position using a t-test (see p-values in parentheses).
Table 1.
List of most common mutations among chronically infected Owen Clinic patients and newly infected Primary Infection Cohort patients. The average yearly prevalence of each mutation throughout the study period is shown in parentheses.
| Chronic Infection Cohort | Newly Infected Cohort | ||
|---|---|---|---|
| Mutation | Prevalence (%) | Mutation | Prevalence (%) |
| M184V, I | 39 | K103N, S | 11 |
| K103N, S | 30 | V179F | 7 |
| T215Y, F, I, S, C, D, V, E | 24 | M41L | 4 |
| M41L | 22 | T215Y, F, I, S, C, D, V, E | 4 |
| M46I, L | 17 | V82A, T, F, S, C, M, L | 3 |
| V82A, T, F, S, C, M, L | 17 | K101E, P | 3 |
| D67N, G, E | 16 | D67N, G, E | 2 |
| K70R, E | 16 | T69D, Ins | 2 |
| L90M | 16 | L90M | 2 |
| V179F | 15 | Y181C, I, V | 2 |
| I54V, L, M, A, T, S | 14 | M184V, I | 2 |
| T69D, Ins | 14 | K219Q, E, N, R | 2 |
Table 2.
Summary of mutations in HIV-1 pro and RT for which significant correlations were found between the overall prevalence of community drug resistance (CDR) (i.e. non-weighted CDR) and transmitted drug resistance (TDR) measurements; p values were calculated with Spearman ranked correlation coefficient.
| Mutation | Rho (p-value) | Measurement Interval |
|---|---|---|
| D30N (pro) | 0.65 (0.031) | Yearly |
| 0.61 (0.005) | Every 6 months | |
| 0.57 (0.00l) | Every 4 months | |
| 0.43 (0.008) | Every 3 months | |
| M46I,L (pro) | 0.74 (0.008) | Yearly |
| 0.43 (0.066) | Every 6 months | |
| 0.44 (0.016) | Every 4 months | |
| 0.22 (0.199) | Every 3 months | |
| N88D,S (pro) | 0.68 (0.021) | Yearly |
| 0.58 (0.009) | Every 6 months | |
| 0.59 (<0.001) | Every 4 months | |
| 0.45 (0.006) | Every 3 months | |
| L74V,I (RT) | 0.46 (0.151) | Yearly |
| 0.35 (0.137) | Every 6 months | |
| 0.40 (0.032) | Every 4 months | |
| 0.24 (0.160) | Every 3 months | |
| V75M,T,A,S (RT) | 0.66 (0.027) | Yearly |
| 0.39 (0.101) | Every 6 months | |
| 0.18 (0.34) | Every 4 months | |
| 0.24 (0.157) | Every 3 months | |
| T215Y,F,I,S,C,D,V,E (RT) | 0.32 (0.336) | Yearly |
| 0.56 (0.012) | Every 6 months | |
| 0.30 (0.110) | Every 4 months | |
| 0.44 (0.006) | Every 3 months | |
Table 3.
Summary of mutations in HIV-1 pro and RT for which significant correlations were found between community drug resistance (CDR) weighted for individual viral loads and transmitted drug resistance (TDR) measurements; p values were calculated with Spearman ranked correlation coefficient.
| Mutation | Rho (p-value) | Measurement Interval |
|---|---|---|
| I54V,L,M,A,T,S (pro) | −0.05 (0.87) | Yearly |
| 0.20 (0.40) | Every 6 months | |
| 0.38 (0.040) | Every 4 months | |
| 0.24 (0.154) | Every 3 months | |
| V82A,T,F,S,C,M,L (pro) | −0.46 (0.154) | Yearly |
| −0.56 (0.013) | Every 6 months | |
| −0.16 (0.41) | Every 4 months | |
| −0.26 (0.115) | Every 3 months | |
| D67N,G,E (RT) | −0.21 (0.54) | Yearly |
| 0.22 (0.37) | Every 6 months | |
| 0.40 (0.032) | Every 4 months | |
| 0.38 (0.019) | Every 3 months | |
| K103N,S (RT) | 0.76 (0.009) | Yearly |
| 0.40 (0.091) | Every 6 months | |
| 0.39 (0.037) | Every 4 months | |
| 0.19 (0.26) | Every 3 months | |
| G190A,S,E (RT) | −0.68 (0.022) | Yearly |
| −0.24 (0.32) | Every 6 months | |
| −0.08 (0.70) | Every 4 months | |
| 0.03 (0.87) | Every 3 months | |
For secondary analyses using measurements calculated at 3-,4-, and 6-month intervals rather than by calendar year, data from January 2002 to June 2011 were included in CDR and TDR measurements, accounting for 1084 resistance tests from Owen Clinic and 492 resistance tests from the primary infection cohort. Data from 2001 and the second half of 2011 were not included in these analyses, due to the low number of data points that would have been included in each shorter measurement interval. As with yearly measurements, significant correlations between the prevalence of drug resistance in the participants, i.e. non-weighted CDR, and TDR were observed at positions 30 and 88 in pro using measurement intervals of 3, 4 and 6 months, although the strength of the correlation decreased with shorter measurement intervals (see Table 2). Similarly, CDR and TDR remained significantly, albeit weakly, correlated at position 46 when measurements were taken every 4 months. When drug resistance was weighted by CVL, the significant correlation at position 103 in RT observed in the primary analysis was replicated with a measurement interval of 4 months but not at 3- and 6-month intervals (see Table 3). Shorter measurement intervals also resulted in positive correlations at some new positions using the prevalence of drug resistance in the patients only, i.e. non-weighted CDR, (positions 74 and 215 in RT) and viral load weighted CDR (position 67 in RT and position 54 in pro, see Tables 2 and 3).
At the majority of residues at which significant correlations between CDR and TDR were noted, a positive correlation was demonstrated. In other words increased CDR was associated with increased TDR, and decreased CDR was associated with decreased TDR.
Notable exceptions to this occurred at position 82 in pro and position 190 in RT, where higher CDR at these positions was associated with lower TDR in the same positions (see Table 3, Figures 1b and 1f). To evaluate if changes in replication capacity could explain the observations, further analyses of the viral loads of patients with and without resistance at these positions were conducted. There was no significant difference between the mean viral loads of patients with and without resistance at positions 82 and 190 (p=0.78 and 0.30 respectively).
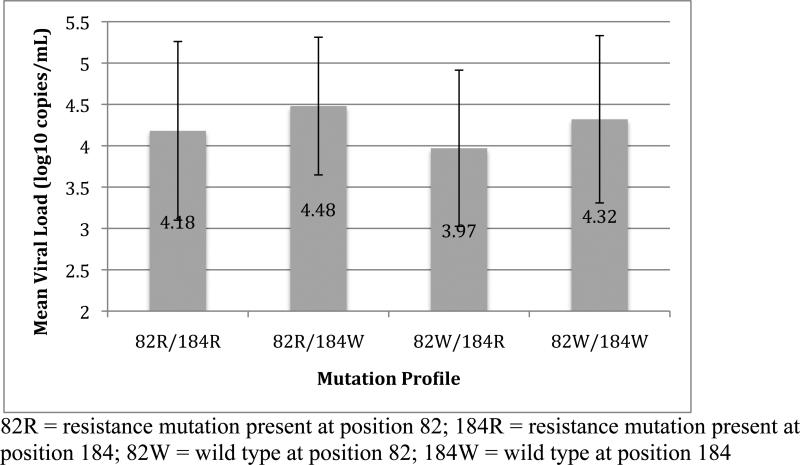
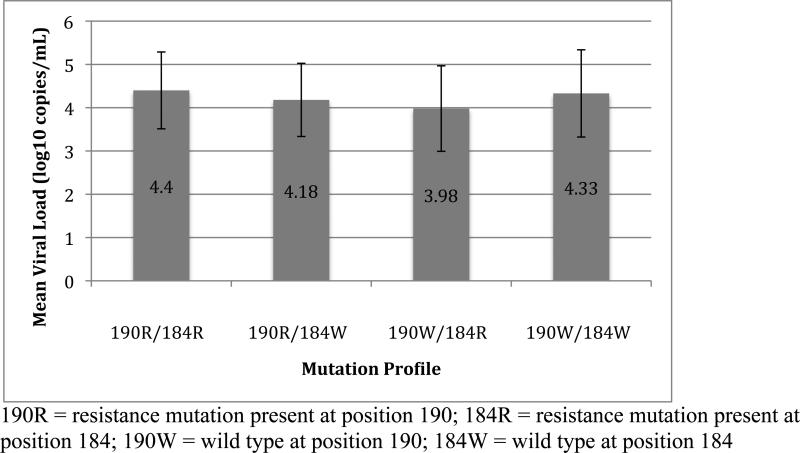
We then investigated if viral loads changed when resistance mutations at the positions co-occurred with the M184V,I mutations because of their overall common occurrence (34.7% of resistant genotypes) and association with decreased viral replication capacity(12). A two-way ANOVA performed to evaluate the individual and combined effects of resistance at positions 82 and 184 on viral load found that Owen Clinic patients with resistance at position 184 alone had the lowest mean viral load overall (See Figure 2), and this effect of resistance at position 184 was significant (F[1,1082] = 22.13, p<0.001). While the presence of resistance at position 82 did not have a significant effect on viral load (F[1,1082] = 0.35, p=0.55), the interaction of mutations at the two positions approached statistical significance (F[1,1085] = 3.79, p=0.052), with an apparent increase in mean viral load in Owen Clinic patients with mutations at both positions compared to those with M184V or I alone (See Figure 2). A two-way ANOVA performed to evaluate the effects of resistance at positions 184 and 190 in RT on viral load revealed a similar decrease in mean viral load of subjects with resistance at position 184 alone compared to subjects with other mutation profiles at these positions (See Figure 3). This effect of resistance at position 184 was significant (F[1,1082] = 22.2, p<0.001) and was abrogated when resistance at position 190 was also present. This was supported by a significant interaction between resistance at positions 184 and 190 (F[1,1085] = 7.03, p=0.008) on viral load. Resistance at position 190 alone did not have a significant effect on viral load (F[1,1082] = 0.73, p=0.39).
Figure 2.
Results of four-way comparison between patients with and without resistance at positions 82 in pro and 184 in RT. Mean viral loads and standard deviations for subjects with each mutation profile are shown.
Figure 3.
Results of four-way comparison between patients with and without resistance at positions 190 and 184 in RT. Mean viral loads and standard deviations for subjects with each mutation profile are shown.
Discussions
Since measurements of CVL have been associated with the incidence of new HIV infections among intravenous drug users(4) and MSM(3) over time, we evaluated if CDR could predict TDR. Despite representing approximately 30% of patients receiving HIV/AIDS care in San Diego and likely underutilization of resistance testing in patients with treatment failure, CDR significantly correlated with TDR at several important resistance positions in HIV-1 RT and pro. The strongest correlations were found in HIV-1 pro at positions 30, 46, and 88 and in HIV-1 RT at position 103 and likely reflect ART prescribing patterns for Owen Clinic patients.
Although ART data for the chronic infection cohort were limited, most of the patients on ART at the time of resistance testing were on PI-based therapy (52.6%), and about 14.9% of all patients were receiving either nelfinavir or atazanavir, particularly during the first half of the study period. Additionally, about 19.8% of all patients were receiving either efavirenz or nevirapine at the time of resistance testing. Further, we noticed an overall decrease in CDR during the second half of the study period compared to the first half.
This is likely due to the increased availability of more potent and well tolerated antiretroviral medications and subsequent changes in medications, and common use of resistance testing for ART-naïve patients. These changes in CDR were associated with similar patterns seen in TDR during this time period. Taking these prescribing patterns into account, our findings are biologically plausible, since a higher prevalence of resistance in the community and higher viral loads among these patients with resistance would be expected to increase the transmission potential of drug resistant virus.
Other factors, such as fitness costs associated with individual and combination mutations, likely also contribute to transmission potential and establishment of infection in a new host(13). For example, although M184V or I was present in over one third of the chronic infection group, it was only present in 2.6% of the TDR group (difference in cohorts chi-square, p<0.001), and there was no association between CDR and TDR at this position. This is likely due to the known fitness cost of M184V and I(14-16), as indicated by lower viral loads in clinic patients with isolated M184V or I compared to those without the mutations. However, for other mutations that are associated with fitness costs, such as K103N or S and G190A,S, or E(17, 18), this effect was not observed. In fact, CDR weighted by viral load at position 103 showed a strong positive correlation with TDR, while a negative correlation was noted at position 190. This indicates that some aspects of viral fitness that affect transmission potential and ability to establish infection in a new host may not always be reflected by blood viral load and that other viral and host factors, including combinations of mutations and frequency of risk behaviors, may also play a role in TDR.
Concerning the negative correlations observed in this study, resistance at RT position 184 did result in decreases in blood viral load, but this decrease in viral load seemed to be abrogated by resistance mutations at other sites, specifically at positions 82 and 190. These interactions do not fully explain the negative correlations observed between CDR and TDR at positions 82 and 190. They do indicate, however, that fitness costs and transmission potential likely are dependent in some part on the overall HIV-1 mutational profile of a patient. Therefore it is possible that, in some cases, the restriction of our analyses to individual positions in RT and pro was overly simplistic, since the overall resistance profile of a patient, including viral fitness costs and any compensatory mutations that may be selected in the presence of resistance and ongoing therapy, likely has an important impact on transmission potential. Nevertheless, the significant association of CDR and TDR at key positions in HIV-1 RT and pro that correspond to commonly prescribed NNRTIs and PIs among the clinic patients indicates that CDR is predictive of TDR.
Due to the nature of drug resistance monitoring at a community level, there are inherent limitations of any analysis of CDR. We based our hypothesis on the assumption that patients receiving HIV/AIDS care contribute significantly to new HIV transmissions in our community. While this is likely true and has been confirmed in many cases by phylogenetic clustering(19), other groups also contribute significantly to new HIV infections and therefore potentially to TDR. These include patients who are unaware that they are infected with HIV, including newly infected patients who have particularly high transmission potential, patients who are aware that they are infected but are not in care, and patients who were evaluated intermittently or lost to follow-up completely. This last group is especially pertinent in this study that demonstrated an 11.7% attrition rate. However, these populations are not accessible for surveillance purposes. While it would be interesting to compare clinical monitoring across patients with variable follow-up, it is beyond the scope of the current study. Patients who are failing therapy but do not receive a resistance test also contribute to potential HIV transmissions but could not be captured by our CDR measurements. Although most patients receiving ART with significant viremia presumably undergo resistance testing, this may not always be the case, and underutilization of resistance testing in patients with treatment failure has been demonstrated in similar cohorts(20). Further, drug resistance and viral load measurements do not capture frequency of risk behaviors such as sexual contact and needle use, which are also important determinants of HIV transmission potential. Finally, for each position in RT and pro we performed Spearman correlation tests to assess whether or not there was an association between the prevalence of CDR and TDR, and each residue was analyzed separately. Although mutation events at different positions may not be independent, correcting for multiple testing is not trivial. We tried to address this issue by weighting CDR estimates by contribution to the CVL, but this is an acknowledged limitation of these analyses. Despite these limitations, patients in care are still a major source of new HIV infections, and changes in CDR in these patients are likely to have an impact on TDR.
In conclusion, we observed significant associations between CDR measurements and TDR in San Diego County at several key resistance positions in HIV-1 RT and pro. These associations are likely reflective of the ART prescribing patterns at the time of resistance testing. Overall, CDR measurements represent a potentially useful tool to predict TDR using existing clinical data in conjunction with active surveillance measures.
Acknowledgements
This work was supported by grants from: the National Institutes of Health: DA034978, AI69432, AI007384, AI080193, AI096113, AI043638, MH62512, AI077304, AI36214, AI047745, AI74621, AI080353, AI100665, AI064086, AI080397, AI067039; the California HIV/AIDS Research Program RN07-SD-702; and the James B. Pendleton Charitable Trust.
M.W.T. assisted in study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report. G.O. assisted in the collection, analysis and interpretation of data and in the writing of the report. J.P-S. assisted in the analysis and interpretation of data and in the writing of the report. S.J.L assisted in study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report. D.D.R. assisted in study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report. W.C.M. assisted in study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report. R.H.H. assisted in study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report. D.M.S. conceived study design, assisted in the collection, analysis and interpretation of data and assisted in the writing of the report.
References
- 1.Richman DD. HIV drug resistance. AIDS Res Hum Retroviruses. 1992;8:1065–71. doi: 10.1089/aid.1992.8.1065. [DOI] [PubMed] [Google Scholar]
- 2.Erice A, Mayers DL, Strike DG, et al. Brief report: primary infection with zidovudine-resistant human immunodeficiency virus type 1. N Engl J Med. 1993;328:1163–5. doi: 10.1056/NEJM199304223281605. [DOI] [PubMed] [Google Scholar]
- 3.Das M, Chu PL, Santos GM, et al. Decreases in community viral load are accompanied by reductions in new HIV infections in San Francisco. PLoS One. 2010;5:e11068. doi: 10.1371/journal.pone.0011068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood E, Kerr T, Marshall BD, et al. Longitudinal community plasma HIV-1 RNA concentrations and incidence of HIV-1 among injecting drug users: prospective cohort study. BMJ. 2009;338:b1649. doi: 10.1136/bmj.b1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.HIV Reports and Statistics. San Diego County; 2001-2012. [Google Scholar]
- 6.Thompson MA, Aberg JA, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA. 2010;304:321–33. doi: 10.1001/jama.2010.1004. [DOI] [PubMed] [Google Scholar]
- 7.Le T, Wright EJ, Smith DM, et al. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N Engl J Med. 2013;368:218–30. doi: 10.1056/NEJMoa1110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009. 4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macchione N, Wooten W, Waters-Montijo K, Ginsberg M. HIV/AIDS Surveillance Program Epidemiology Report 2010. County of San Diego Health and Human Services Agency. 2010 [Google Scholar]
- 10.Aldous JL, Jain S, Mathews C, et al. Decreasing prevalence of drug resistance mutations over a 7 year period in the CFAR Network of Integrated Clinical Systems (CNICS) cohort.. 17th Conference on Retroviruses and Opportunistic Infections.; San Francisco, CA, USA. 2010. [Google Scholar]
- 11.Gill VS, Lima VD, Zhang W, et al. Improved virological outcomes in British Columbia concomitant with decreasing incidence of HIV type 1 drug resistance detection. Clin Infect Dis. 2010;50:98–105. doi: 10.1086/648729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nijhuis M, Deeks S, Boucher C. Implications of antiretroviral resistance on viral fitness. Curr Opin Infect Dis. 2001;14:23–8. doi: 10.1097/00001432-200102000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Leigh Brown AJ, Frost SD, Mathews WC, et al. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J Infect Dis. 2003;187:683–6. doi: 10.1086/367989. [DOI] [PubMed] [Google Scholar]
- 14.Cong ME, Heneine W, Garcia-Lerma JG. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J Virol. 2007;81:3037–41. doi: 10.1128/JVI.02712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paredes R, Sagar M, Marconi VC, et al. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J Virol. 2009;83:2038–43. doi: 10.1128/JVI.02154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain V, Sucupira MC, Bacchetti P, et al. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis. 2011;203:1174–81. doi: 10.1093/infdis/jiq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Bambara RA, Demeter LM, Dykes C. Reduced fitness in cell culture of HIV-1 with nonnucleoside reverse transcriptase inhibitor-resistant mutations correlates with relative levels of reverse transcriptase content and RNase H activity in virions. J Virol. 2010;84:9377–89. doi: 10.1128/JVI.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Dykes C, Domaoal RA, Koval CE, Bambara RA, Demeter LM. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology. 2006;348:462–74. doi: 10.1016/j.virol.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DM, May SJ, Tweeten S, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23:225–32. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham AG, Lau B, Deeks S, et al. Missing data on the estimation of the prevalence of accumulated human immunodeficiency virus drug resistance in patients treated with antiretroviral drugs in north america. Am J Epidemiol. 2011;174:727–35. doi: 10.1093/aje/kwr141. [DOI] [PMC free article] [PubMed] [Google Scholar]