Key Points
Human RUNX1a orthologs are only found in primates.
Alternative splicing of Runx1 involving exon 6 affects the pool size of hematopoietic stem cells.
Abstract
RUNX1 is an important transcription factor for hematopoiesis. There are multiple alternatively spliced isoforms of RUNX1. The best known isoforms are RUNX1a from use of exon 7A and RUNX1b and c from use of exon 7B. RUNX1a has unique functions due to its lack of C-terminal regions common to RUNX1b and c. Here, we report that the ortholog of human RUNX1a was only found in primates. Furthermore, we characterized 3 Runx1 isoforms generated by exon 6 alternative splicing. Runx1bEx6− (Runx1b without exon 6) and a unique mouse Runx1bEx6e showed higher colony-forming activity than the full-length Runx1b (Runx1bEx6+). They also facilitated the transactivation of Runx1bEx6+. To gain insight into in vivo functions, we analyzed a knock-in (KI) mouse model that lacks isoforms Runx1b/cEx6− and Runx1bEx6e. KI mice had significantly fewer lineage-Sca1+c-Kit+ cells, short-term hematopoietic stem cells (HSCs) and multipotent progenitors than controls. In vivo competitive repopulation assays demonstrated a sevenfold difference of functional HSCs between wild-type and KI mice. Together, our results show that Runx1 isoforms involving exon 6 support high self-renewal capacity in vitro, and their loss results in reduction of the HSC pool in vivo, which underscore the importance of fine-tuning RNA splicing in hematopoiesis.
Introduction
RUNX1, also known as AML1, PEBP2αB, and CBFα2, is a transcription factor crucial for hematopoietic cell development. Its gene was cloned from the breakpoint of t(8;21) translocation in acute myeloid leukemia,1 and its function has been best studied in the hematopoietic system.2 It binds to the promoters and enhancers of target genes, such as the M-CSF receptor, GM-CSF, and PU.1, to promote their expression.3-5 Conventional Runx1 knockout mice are embryonic lethal due to bleeding in the central nervous system and lack of definitive hematopoiesis.6 However, in Runx1 conditional knockout mice, adult hematopoiesis is maintained with defects in multiple lineages.7-9 Human RUNX1 is a frequent target of translocations or mutations in hematologic malignancies including acute myeloid leukemia, acute lymphoid leukemia, and myelodysplastic syndrome (MDS).2
Runx1 controls stem cell fate and proliferation in various organs, both in normal and malignant settings. In addition to its function in hematopoiesis, Runx1 is involved in development of blood vessels, muscles, neurons, and epithelial tissues including hair follicles, oral epithelia, and lacrimal glands.10-13
Runx1 is also expressed in the intestine and colon, although its role in these tissues should be validated in conditional knockout models.12 Human RUNX1 is overexpressed in cell lines and/or primary tissues of skin, breast, prostate, intestine, and ovarian cancers.12,14 A conditional knockout model showed that Runx1 is essential for the growth and survival of skin and oral squamous cell carcinoma and in part for ovarian carcinoma.12 RUNX1 single-nucleotide polymorphisms are associated with cancer and autoimmune disease.15 Thus, the importance of Runx1 is now recognized in a wide range of tissues and disease spectrum.
In vertebrates, Runx1 gene expression is under the control of 2 promoters, distal (P1) and proximal (P2) (Figure 1A).16 They generate transcripts that differ in 5′-untranslated regions and N-terminal coding sequences. The P1 transcript is predominant in hematopoietic stem cells (HSCs) in fetal liver, T cells and, to a lesser extent, B cells, whereas P2 is expressed predominantly or similarly to P1 in myeloid and other nonhematopoietic tissues.17,18 During blood cell development of the mouse embryo, P2 is first activated to specify hemogenic endothelium.19 When definitive hematopoietic cells emerge, both P1 and P2 promoters are active, with a skewing toward the P2.19,20 Similar results were observed in an in vitro model using human embryonic stem cells.21 P1 knockout mice did not exhibit any overt phenotypes.19
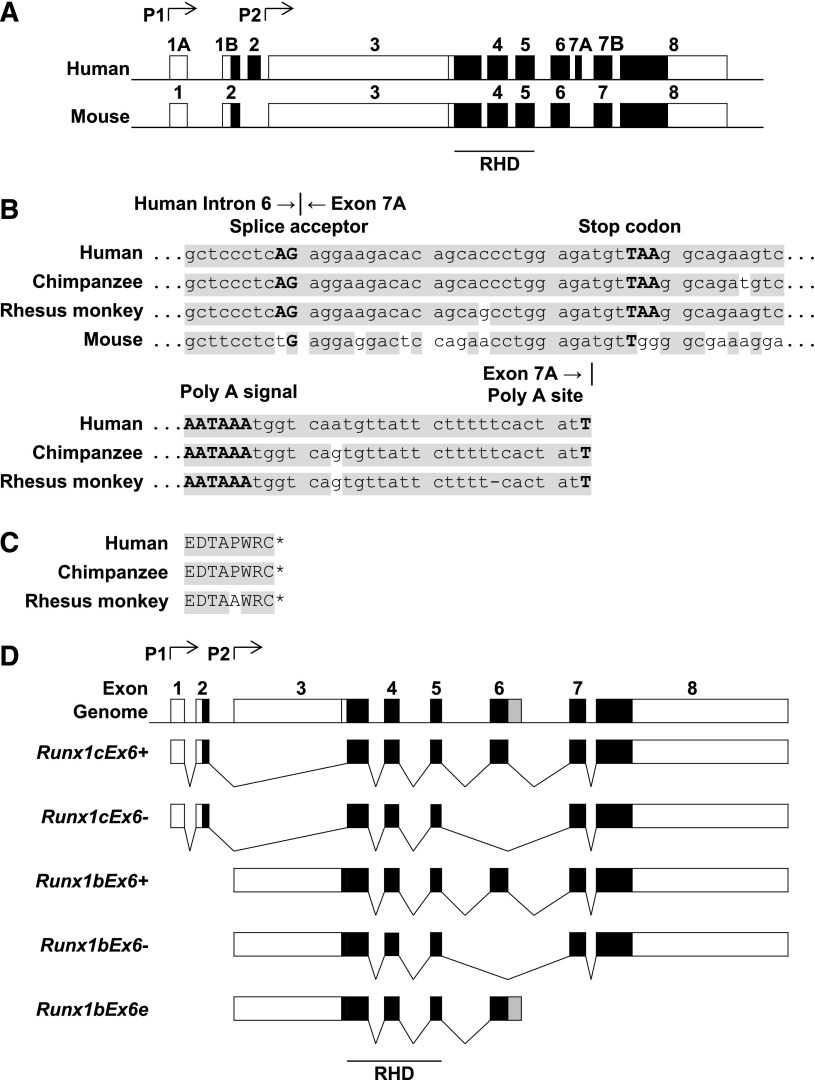
Figure 1.
Genome structure of Runx1 in various species. (A) Exon-intron structure of human RUNX1 and mouse Runx1. Genome sequences of Runx1 were collected from the GenBank database (NCBI). Boxes represent exons. Black boxes represent coding sequences. (B) Sequence homology of human RUNX1a intron 6 and exon 7A and their counterparts in other species. Top, Junction of human RUNX1a intron 6 and exon 7A. Bottom, Poly A signal and poly A site. Shaded areas show the common sequences. (C) Amino acid sequence encoded by human RUNX1a exon 7A and its counterparts in other species. Shaded area shows the common sequences. *, The end of the protein sequence. (D) Alternatively spliced isoforms of mouse Runx1. Boxes represent exons. Black boxes represent coding sequences. Gray box in Runx1bEx6e shows the part of extended exon 6 located in intron 6. P1, distal promoter; P2, proximal promoter.
In addition to promoter usage, alternative splicing results in multiple isoforms of Runx1. In human RUNX1, the P1 promoter encodes RUNX1c, and P2 encodes RUNX1a and RUNX1b.18 RUNX1a transcript stops at alternative exon 7A and therefore lacks exons 7B and 8 encoding C-terminal regulatory domains common in RUNX1b and RUNX1c. Several studies have described the functional differences among these 3 Runx1 isoforms.21,22 RUNX1a is specifically enriched in the immature fraction of cord blood cells.22 RUNX1a/b isoforms are expressed consistently throughout hematopoietic differentiation, whereas RUNX1c is only expressed at the emergence of definitive HSCs in an in vitro model using human embryonic stem cells.21 Overexpression of RUNX1a resulted in increased competitive engraftment of mouse bone marrow (BM) cells in vivo and increased proliferation of progenitors in vitro; overexpression of Runx1b showed opposite effects.22 RUNX1a works both in human and mouse systems to expand the HSC population in vitro and in vivo.22-24 Furthermore, RUNX1a promotes hematopoietic differentiation of human pluripotent stem cells,25 suggesting that it is a positive regulator of proliferation in immature cells. In B cells, RUNX1c, but not RUNX1b, inhibits proliferation due to its unique N-terminal domain.26 These results demonstrate that the tight regulation of RNA splicing is critical for controlling RUNX1 activity during hematopoiesis.
In addition to alternative usage of exon 7A and exon 7B to produce RUNX1a, b, and c, isoforms of RUNX1 that skip exon 6 have been reported in both human and mouse cells.18,27-31 Furthermore, RUNX1 without exon 6 in human ovarian cancer and atypical RUNX1-RUNX1T1 (RUNX1-ETO) fusion protein that includes exon 6 (the most common fusion includes exon 5 but not exon 6) have been reported.27,32 However, the RUNX1 isoforms generated by alternative splicing of exon 6 have not been thoroughly described and characterized.
Here, we examined human and murine Runx1 gene sequences and found that the RUNX1a ortholog is missing in mice due to the absence of a splice acceptor sequence. With further cross-species comparison, RUNX1a seemed to exist only in primates and not in other phylogenetic orders. Furthermore, we evaluated biological functions of the Runx1 isoforms involving exon 6 by overexpressing them in mouse BM cells and by using a knock-in (KI) mouse model. Our results show that exon 6 coding region affects Runx1 protein stability and transcription activity, which leads to different colony formation potential in the replating assay and maintenance of hematopoietic stem cell frequency in vivo. This is the first report about the significant roles of Runx1 exon 6–related alternative splicing in controlling clonogenicity of hematopoietic progenitors in vitro and pool size of hematopoietic stem cells in vivo.
Materials and methods
Genome structure analysis of Runx1
Genome sequences were collected from GenBank (National Center for Biotechnology Information [NCBI]). The following genome sequences were used: Homo sapiens (human) chromosome 21, NC_000021.8; Pan troglodytes (chimpanzee) chromosome 21, NC_006488.2; Macaca mulatta (rhesus monkey) chromosome 3, NC_007860.1; Bos taurus (cattle) chromosome 1, AC_000158.1; Sus scrofa (pig) chromosome 13, NC_010455.4; Rattus norvegicus (Norway rat) chromosome 11, NC_005110.3; Mus musculus (mouse) strain C57BL/6J chromosome 16, NC_000082.6; Takifugu rubripes (fish) chromosome 8, NC_018897.1. Sequences were aligned using CLC Sequence Viewer 6.
Mice
Wild-type (WT) C57Bl/6 (CD45.2) and syngeneic (CD45.1) mice were purchased from The Jackson Laboratory. Homozygous Runx1–internal ribosome entry site–green fluorescent protein (Runx1-IRES-GFP) KI mice were previously described.33 Age- and gender-matched mice (6-9 weeks old) were used for analysis. Procedures were approved by the institutional animal care and use committee. Details of transplantation experiments are described in supplemental Methods (see supplemental Data available on the Blood Web site).
Constructs
Full-length Runx1b (Runx1bEx6+) and Runx1bEx6e coding sequences were cloned from mouse total BM cells. Runx1b without exon 6 (Runx1bEx6−) was made by polymerase chain reaction (PCR) mutagenesis. All the inserts were N-terminally hemagglutinin (HA)–tagged by PCR and subcloned into murine stem cell virus (MSCV)–IRES-puro (MIP) or MigR1 vector.
Reverse transcription PCR
Total RNA was extracted from mouse whole BM cells with TRIzol (Life Technologies) and treated with DNaseI (Qiagen). Complementary DNA (cDNA) was synthesized using the Superscript III kit (Life Technologies). PCR was done using Red Taq DNA polymerase (BioPioneer). Primers used were as follows: forward: CGG CAG AAC TGA GAA ATG CT, R1: TCG GAG ATG GAC GGC AGA GTA GGG A, R2: GGG ACT CCA GCA AAG ACA GA; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward: GAG CTG AAC GGG AAG CTC ACT GG, reverse: CAA GAG AGT AGG GAG GGC TCC CTA G. Thermal cycle was 94°C for 2 minutes, 30 cycles of 94°C for 1 minute, 60°C (for R1) or 55°C (for R2) for 1 minute, 72°C for 1 minute, followed by final extension of 72°C for 5 minutes.
Cell culture and transfection
293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 10 mM glutamine, and penicillin (100 IU)/streptomycin (100 µg/mL). Mouse BM cells were cultured in Iscove's modified Dulbecco's medium supplemented with 20% FBS, 4% stem cell factor conditioned medium, 4% interleukin-3 (IL-3) conditioned medium, and penicillin/streptomycin.34 Transfection was done using polyethylenimine (Polysciences Inc).
Western blotting
Lineage-negative c-Kit+ adult BM cells (sorted by magnetic beads; Miltenyi Biotec) or 293T cells transfected with Runx1 constructs were directly denatured in preheated sample buffer, and western blotting was performed following a standard protocol. Primary rabbit anti-RUNX1 antibody that detects amino acids 3-17 IPVDASTSRRFTPPS and anti-α-Tubulin antibody (T9026; Sigma-Aldrich) were used. For the protein stability assay, 100 ng/mL cycloheximide (CHX; Sigma-Aldrich) or 10 µM MG132 (Peptide International Inc) dissolved in dimethyl sulfoxide (DMSO) was added to the culture medium and incubated for the indicated time. The final concentration of DMSO was 0.1%. Anti-phospho S276 and S303 antibodies were obtained from the laboratory of Dr Kraft (Medical University of South Carolina, Charleston, SC).35,36
Flow cytometry
Primary cells were treated with red blood cell lysis buffer (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM EDTA) and stained with the antibodies detailed in supplemental Methods. Cells were analyzed with FACSCanto or FACSAria (both from BD Biosciences). Data were interpreted with FACSDiva (BD Biosciences) or FlowJo (TreeStar). Sorting was done using FACSAria II (BD Biosciences).
Colony-forming assay
Lineage-negative cells infected with MIP-based retroviruses were selected for 1 week in methylcellulose (MethoCult M3134; STEMCELL Technologies) supplemented with 50 ng/mL mouse stem cell factor, 10 ng/mL mouse IL-3, 10 ng/mL human IL-6 (all from PeproTech), penicillin/streptomycin, and 1 µg/mL puromycin. Cells (1 × 105) were seeded into methylcellulose in duplicate. One week later, colony and cell numbers were counted. Colony cells were used for replating, or cytospin preparations with or without sorting followed by Wright-Giemsa staining (Sigma-Aldrich). Images were acquired at room temperature using an Olympus BX51 microscope equipped with a DP71 camera and DP controller/DP Manager software (Olympus).
Luciferase assay
293T cells were transfected with each Runx1 expression construct together with CBFβ expression construct in pCMV vector, M-CSFR reporter construct,37 and Renilla luciferase expression vector. Total DNA amount of Runx1 and/or MIP was adjusted to 2.5 μg using MIP vector DNA. Forty-eight hours later, cells were assayed for transcriptional activity by the Dual-Luciferase Reporter Assay System (Promega) and Monolight 3010 Luminometer (BD Biosciences).
Statistical analysis
Results are presented as mean ± standard deviation (SD) unless otherwise stated. Statistical significance was determined by Student t test. The differences were considered significant if P < .05 and marked with an asterisk in the figure images.
Results
Mice lack RUNX1a ortholog
Human RUNX1a has very unique functions. It enhances engraftment and promotes proliferation in HSCs.22 It also improves hematopoietic lineage development from human pluripotent stem cells.25 Its expression is due to the use of RUNX1 gene exon 7A (Figure 1A). It lacks several well-recognized regulatory domains in the C terminus.18,38 These facts prompted us to do a cross-species comparison to examine evolutionary conservation of this isoform. We found that the homologous mouse sequence of human exon 7A lacked sequences relating to splice acceptor AG and stop codon TAA, indicating that there is no ortholog of human RUNX1a in mice (Figure 1A-B). We further compared the entire Runx1 gene structure in vertebrate species that were available in the NCBI database (described in “Materials and methods”). The majority of the Runx1 genomic structure was well preserved among the various species (data not shown). The intron sequence 3′ to the runt homology domain (RHD; exons 3-5 in human) was examined and the chimpanzee and rhesus monkey genomes displayed highly homologous sequences to human RUNX1 exon 7A, including sequences relating to a polyadenylation signal and a polyadenylation site (Figure 1B-C). However, other species, such as cattle, pig, rat, and fish, did not have the same homologous DNA or amino acid sequence to exon 7A for human RUNX1a when translated in 3 open reading frames (data not shown). These results suggest that RUNX1a is unique to primates.
In the NCBI database, there are 4 major isoforms for the mouse Runx1 gene (Figure 1D). Runx1c with or without exon 6 (Runx1cEx6+ and Runx1cEx6−, respectively) is transcribed from the distal promoter (P1), whereas Runx1b with or without exon 6 (Runx1bEx6+ and Runx1bEx6−, respectively) is transcribed from the proximal promoter (P2). Runx1cEx6+ and Runx1bEx6+ are mouse orthologs of human RUNX1c and RUNX1b, respectively. Runx1cEx6− and Runx1bEx6− naturally omit exon 6, which encodes 64 amino acids. Although it is not in the NCBI database, a human RUNX1 gene product lacking exon 6 has been reported.18,27-30
Although no RUNX1a ortholog was found in mice, a truncated isoform Runx1bEx6e was identified in a mouse embryonic cDNA library.39 It was a transcript from P2, and had an extended exon 6 with an additional 63 amino acids in the C terminus. There are 13 conventional polyadenylation signals (“AATAAA”) 3′ to the stop codon in intron 6 (data not shown). Thus, including Runx1bEx6e, there are 5 Runx1 isoforms in mice (Figure 1D).
Runx1bEx6− and Runx1bEx6e have higher colony formation ability
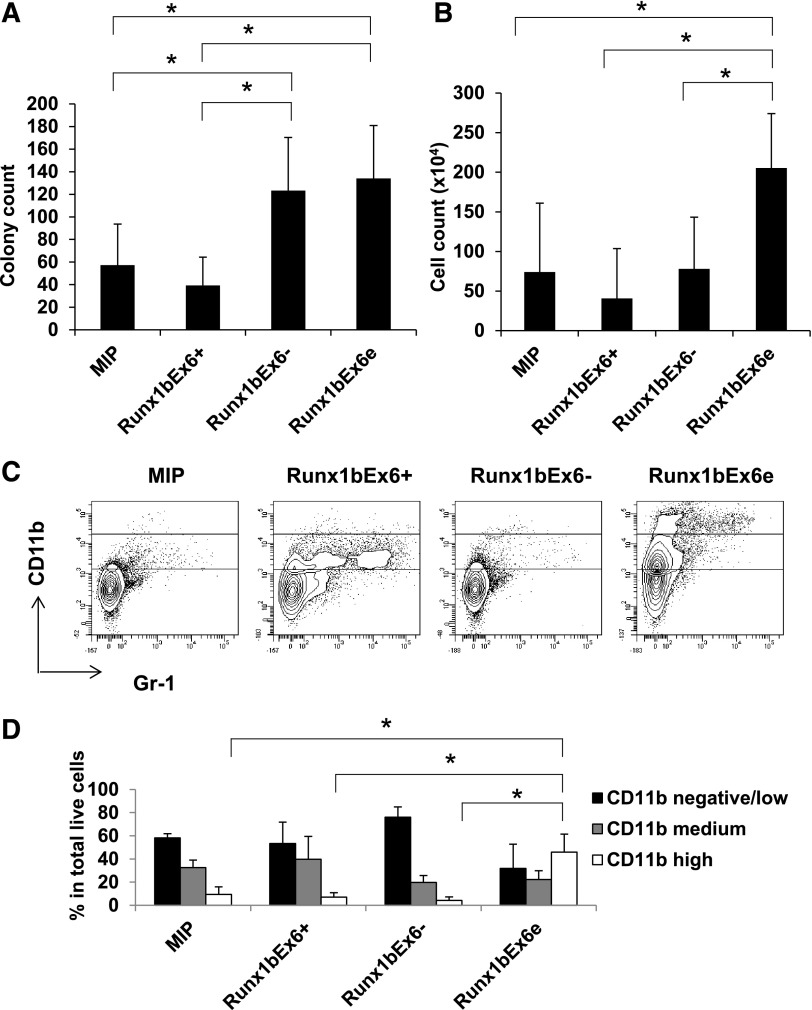
To gain insight into the functions of exon 6, we evaluated the biological effects of Runx1 isoforms Runx1bEx6+, Runx1bEx6−, and Runx1bEx6e in vitro. Protein expression of the constructs was confirmed in 293T cells and mouse total BM cells (supplemental Figures 1-2). Colony-forming assays using lineage-negative BM cells overexpressing Runx1 isoforms showed that Runx1bEx6e and Runx1bEx6− resulted in higher colony counts compared with empty vector (MIP) or Runx1bEx6+ (Figure 2A). Runx1bEx6e had the highest cell counts (Figure 2B). Similar results were also generated with sorted cells transduced with the MigR1 (GFP+) series of retrovirus expressing these Runx1 isoforms (data not shown). Only Runx1bEx6e cells could be replated at least 5 times; other groups could be replated up to 2 times (data not shown). Flow cytometry showed higher CD11b and reduced Gr-1 expression in Runx1bEx6e colony cells (Figure 2C-D). On the other hand, CD11b-negative cells were dominant in Runx1bEx6− cells. The sorted colony cells based on CD11b expression showed unique morphology in each population (supplemental Figure 3). The CD11b high population contained round, macrophage-like cells with multiple vacuoles. CD11b-medium cells were a mixture of neutrophils and monocytes. CD11b-negative/low cells had dense red granules like eosinophils. Differential counts of cytospins of whole colony cells correlated with fluorescence-activated cell sorter patterns, showing Runx1bEx6e colony cells had significantly higher macrophage-like cells than other groups (supplemental Figure 4). These results suggest that Runx1bEx6− and Runx1bEx6e have higher colony-forming ability, and Runx1bEx6e has a stronger effect on myeloid differentiation than the other isoforms.
Figure 2.
Runx1bEx6− and Runx1bEx6e have higher colony-forming ability than full-length Runx1bEx6+. Lineage-negative BM cells were infected with indicated retroviral constructs. After 1-week selection in 1 μg/mL puromycin, 1 × 105 cells were seeded into methylcellulose in duplicate. One week later, (A) colony counts and (B) cell counts per plate were evaluated. Mean ± SD of 5 independent experiments is shown. (C) Flow cytometric analysis of colony cells. The gating of CD11b-negative/low, -medium, and -high (from bottom to top) is shown in the plots. Representative result of 3 independent experiments is shown. (D) Quantification of the results in panel C. Mean ± SD of 3 independent experiments is shown.
Runx1bEx6− and Runx1bEx6e cooperate with Runx1bEx6+ in regulating gene promoter activities
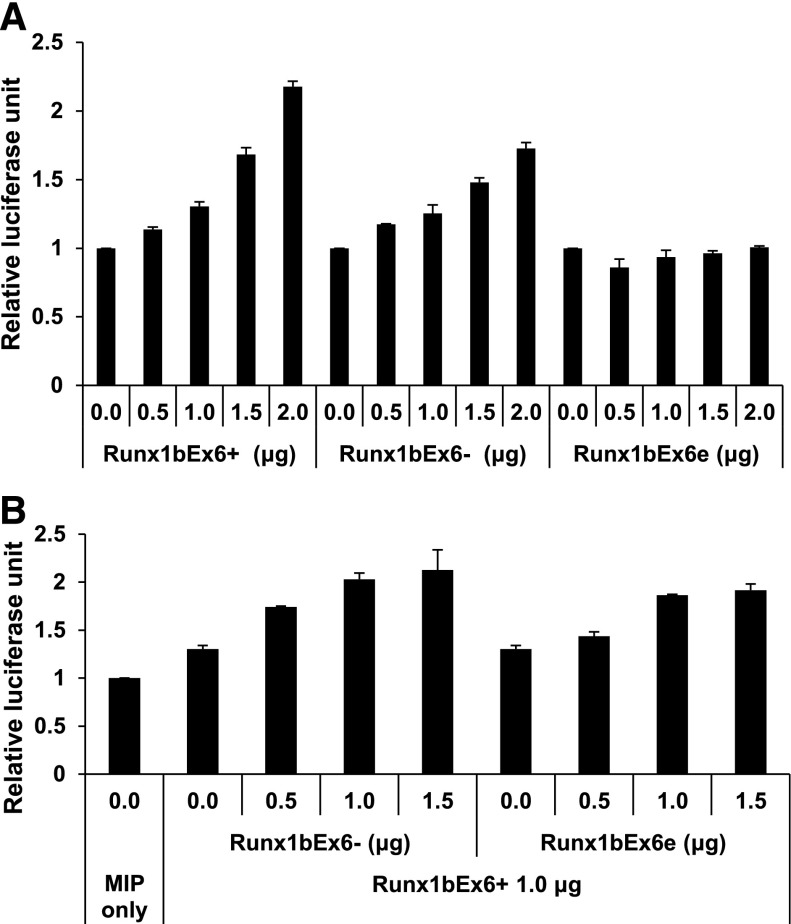
To study the functions of these isoforms in transcription, we conducted promoter-luciferase assay with the M-CSF receptor promoter-luciferase DNA construct.37 Runx1bEx6− showed weaker transcriptional activity compared with Runx1bEx6+, and Runx1bEx6e alone did not activate the promoter (Figure 3A). However, cotransfection of Runx1bEx6+ with Runx1bEx6− or Runx1bEx6e showed an additive or synergistic effect, respectively (Figure 3B). Thus, Runx1bEx6− and Runx1bEx6e enhance the transactivation ability of Runx1bEx6+.
Figure 3.
Runx1bEx6− and Runx1bEx6e have lower transactivation efficiency than full-length Runx1bEx6+. 293T cells were transfected in duplicate with the indicated construct together with CBFβ expression construct, M-CSF receptor promoter-firefly luciferase reporter construct and Renilla luciferase reporter construct as a transfection efficiency control. (A) Transfection with single Runx1 isoform. Runx1bEx6− shows lower transactivation than Runx1bEx6+. Runx1bEx6e has no transactivation. Values were normalized to Renilla luciferase signal, and promoter activity of MIP-transfected cells was set to 1. Representative result of 2 independent experiments is shown. (B) Cotransfection of MIP, Runx1bEx6−, or Runx1bEx6e with Runx1bEx6+. Runx1bEx6− and Runx1bEx6e show additive and synergistic effect, respectively. Values were normalized to Renilla signal, and MIP was set to 1. Representative result of 2 independent experiments is shown.
Runx1 isoforms have various protein stabilities
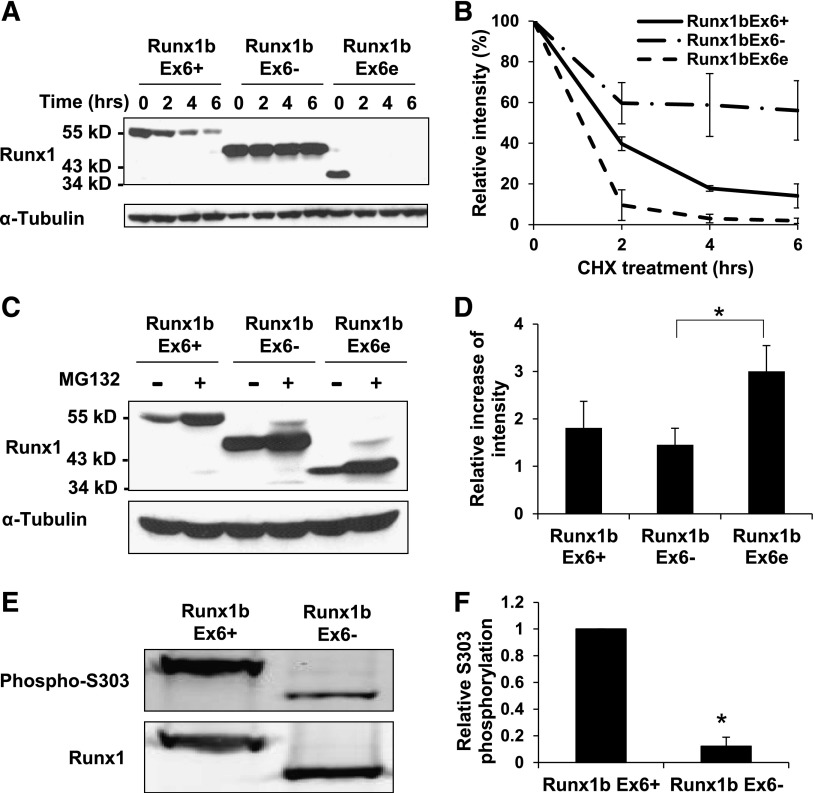
Expression of the 3 Runx1 isoforms in 293T cells showed a lower protein level of Runx1bEx6e than that of Runx1bEx6+ and Runx1bEx6− (supplemental Figure 1). This led us to compare the protein stability of these Runx1 isoforms. 293T cells expressing each Runx1 isoform were treated with CHX and the half-lives of the proteins were evaluated (Figure 4A-B). Runx1bEx6− was the most stable, degrading <50% over 6 hours. On the other hand, Runx1bEx6e was the least stable, with a half-life of 1 hour.
Figure 4.
Runx1 isoforms have various protein stability and susceptibility to proteasome-mediated degradation. (A-B) Protein stability of Runx1 isoforms. 293T cells were transfected with indicated construct. Forty-eight hours later, cells were treated with 100 ng/mL CHX for indicated time. α-Tubulin served as a loading control. (A) Representative western blotting result of 3 independent experiments. (B) Quantification of the results in panel A. Intensities at 0 h were set to 100%. Mean ± SD of 2 independent experiments is shown. (C-D) Inhibition of proteasome-mediated degradation pathway. 293T cells were transfected with the indicated construct. Forty-eight hours later, cells were treated with 10 µM MG132 for 24 hours. α-Tubulin served as a loading control. (C) Representative western blotting result of 3 independent experiments is shown. (D) Quantification of the results in panel C. Runx1 signals were normalized to those of tubulin, and vehicle (DMSO)–treated signals were set to 1. Mean ± SD of 3 independent experiments is shown. (E) Western blot of phosphorylation of “S303”. Replicate membranes were probed with anti-Runx1 and anti-phospho S303 antibodies, respectively. (F) Quantification of panel E. Mean ± SD of 3 independent experiments is shown.
Runx1 is known to be posttranscriptionally modified by several pathways which regulate protein stability.35,40,41 To study how much the proteasome-mediated degradation pathway is involved, the cells were treated with proteasome inhibitor MG132 (Figure 4C-D). As expected, Runx1bEx6e was the most affected isoform, followed by Runx1bEx6+ and Runx1bEx6−. Runx1bEx6+ has 9 lysine residues, all of which are located in exons 1-6. Runx1bEx6− has 2 fewer lysine residues due to the loss of exon 6. On the other hand, Runx1bEx6e has 4 “additional” lysine residues in its extended exon 6.39 These residues may give more potential targets for the ubiquitin-proteasome pathway, which result in a shorter half-life.40
We also examined phosphorylation of 2 serine residues which are important for ubiquitin-mediated RUNX1 degradation. They are S276 and S303 on exon 7b in human RUNX1c, which correspond to S249 and S276 in Runx1bEx6+, respectively.35,36 Runx1bEx6e lacks both residues and Runx1bEx6− has these residues. Western blotting showed significantly reduced phosphorylation of “S303” in Runx1bEx6− (Figure 4E-F). Phosphorylation of “S276” was not clearly detected in the current condition (data not shown). These results suggest that exon 6 is important for the phosphorylation of Runx1 “S303” residue, and that reduced phosphorylation is likely one of the causes of enhanced stability of Runx1bEx6−.
Three Runx1 alternative splicing isoforms are missing in Runx1-IRES-GFP KI mice
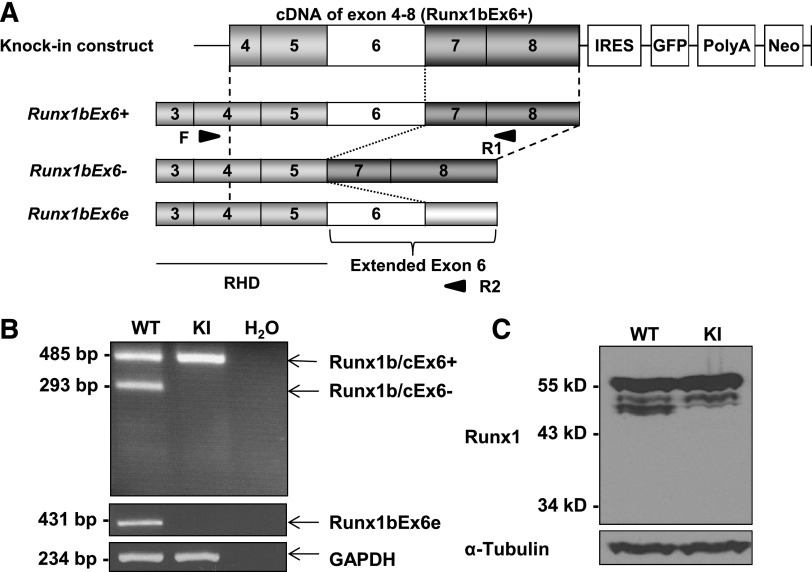
Next, we wanted to evaluate the roles of the Runx1 isoforms in vivo. In Runx1-IRES-GFP KI mice (hereafter KI mice), the cDNA sequence of Runx1 gene containing exons 4-8 is inserted into the endogenous exon 4 locus (Figure 5A).33 This affects the expression of alternatively spliced isoforms involving this region. Based on the genomic structure of Runx1, KI mice lack Runx1bEx6−, Runx1cEx6−, and Runx1bEx6e, all involving exon 6. We confirmed the loss of these isoforms by reverse transcription PCR (RT-PCR) (Figure 5B). Western blotting of lineage-negative c-Kit+ BM cells showed similar expression levels of the most dominant band in WT and KI cells (Figure 5C, supplemental Figure 5). On the other hand, several minor bands, one of which corresponds to Runx1bEx6− (47 kDa, supplemental Figure1), were missing in KI BM cells, consistent with the results from RT-PCR. However, there were no bands around 38 kDa, where Runx1bEx6e should be detected (supplemental Figure 1), even in the WT lane, possibly due to its instability (Figure 4A-B) or low expression level. These results show that KI mice lack 3 isoforms involving exon 6 without affecting total amount of Runx1.
Figure 5.
Runx1-IRES-GFP KI mice lack the expression of 3 Runx1 isoforms. (A) Structure of the Runx1-IRES-GFP KI construct. Boxes with numbers represent exons. Light gray boxes are RHD, and dark gray boxes are regulatory domains. Arrowheads show the positions of primers (F, R1, and R2) used for panel B. (B) Expression of Runx1 isoforms in total BM cells by RT-PCR. KI cells lack Runx1b/cEx6− and Runx1bEx6e. GAPDH serves as a loading control. Water lane is the negative control. Representative result of 3 independent experiments is shown. (C) Protein expression of Runx1 in lineage-depleted c-Kit–enriched BM cells. KI lane has the same level of the major band although it lacks smaller bands which correspond to Runx1b/cEx6−. Runx1bEx6e band is undetectable in either WT or KI lane. α-Tubulin serves as a loading control. Representative result of 2 independent experiments is shown. Neo, neomycin resistance gene; PolyA, polyadenylation signal.
HSC frequency is reduced in KI mice
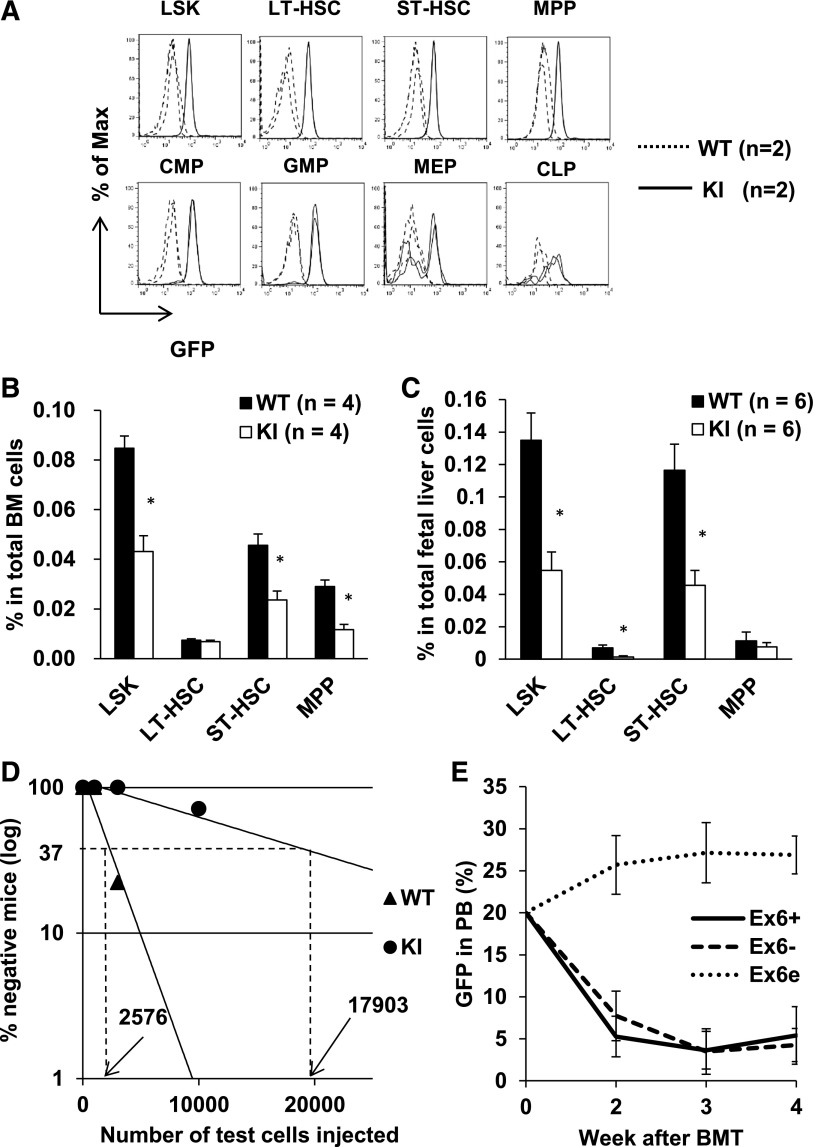
KI mice have normal appearance and blood indices.33 Homozygous KI mice were obtained at the expected frequency from heterozygous mice.33 We also examined embryonic day 14.5 (E14.5) KI embryos and did not observe any apparent abnormality (data not shown). GFP intensity correlates with Runx1 expression level, and shows lineage-specific changes during maturation. However, the behavior of hematopoietic stem/progenitor cells (HSPCs) was not examined in the previous report. Because the KI mice lose 3 isoforms of Runx1 and we detected unique characteristics of Runx1bEx6− and Runx1bEx6e, we hypothesized that these isoforms may play special roles during early hematopoiesis. We therefore examined the HSPCs in homozygous KI mice. To determine the expression pattern of Runx1 in HSPCs, we analyzed GFP expression in various HSPC populations of adult KI BM cells, including the lineage-Sca1+c-Kit+ (LSK), common myeloid progenitor (CMP), granulocyte/monocyte progenitor (GMP), megakaryocyte/erythroid progenitor (MEP), and common lymphoid progenitor (CLP) fractions (Figure 6A). A GFP signal can be detected in all of these cells, indicating that the Runx1 promoter is active in all of these cells. Next, we compared the frequency of HSPC populations in BM cells between WT and KI mice. The frequency of LSK cells was significantly lower in KI mice compared with controls (Figure 6B). In addition, subpopulations of the LSK fraction, including short-term HSCs (ST-HSCs) and multipotent progenitors (MPPs), were significantly decreased. E14.5 fetal liver cells also displayed a decrease in LSK fractions (Figure 6C). On the other hand, there was no difference in the frequency of CMPs or CLPs (data not shown). We performed a competitive repopulation assay to compare the frequency of functional HSCs in BM. BM cells from WT and KI mice were mixed 1:1, and combined with a fixed number of syngeneic supportive cells. Because KI cells have GFP fluorescence, we can successfully distinguish 3 origins of cells by the combination of CD45 (CD45.1 and CD45.2) and GFP expression. CD45.2−GFP− cells are from recipient/supportive cells, CD45.2+GFP− cells are from WT donors, and CD45.2+GFP+ are from KI donors. Mice were analyzed 4 months after transplantation. Calculated HSC frequency was sevenfold higher in WT than in KI mice (1 of 2576 vs 117 903) (Figure 6D, Table 1). Thus, deletion of the 3 Runx1 isoforms (Runx1bEx6−, Runx1cEx6−, and Runx1bEx6e) resulting from the alternative splicing of exon 6 reduces the HSC pool.
Figure 6.
Runx1-IRES-GFP KI mice have decreased HSC pool. (A) GFP expression in HSPCs. GFP fluorescence in KI cells indicates Runx1 promoter activity. GFP histograms are shown by gating the indicated population. Dashed and solid lines denote WT and KI cells, respectively (2 mice each). (B) Flow cytometric analysis of adult BM. (C) Flow cytometric analysis of E14.5 fetal liver cells. (D) Competitive repopulation unit assay. The detailed protocol is described in supplemental Methods. WT cells have 7 times higher frequency of stem cells than KI cells. This experiment was repeated twice with similar results. (E) BMT of Runx1 overexpressing cells. One million cells with 20% GFP+ cells were injected into recipient mice. GFP percentage in peripheral blood was followed up. Ex6+, n = 4. Ex6−, n = 3. Ex6e, n = 4. Ex6+, Ex6−, and Ex6e represent Runx1bEx6+, Runx1bEx6−, and Runx1bEx6e, respectively. LT-HSC, long-term HSC.
Table 1.
Limiting dilution competitive repopulation analysis
| Injected cell no. | WT | KI |
|---|---|---|
| 30 000 | 8/8 | 8/8 |
| 10 000 | 7/7 | 2/7 |
| 3000 | 7/9 | 0/9 |
| 1000 | 0/5 | 0/5 |
Mice positive for donor engraftment were defined as those with >1% CD45.2+GFP− or CD45.2+GFP+ cells (for WT or KI cells, respectively) in the peripheral blood in both the lymphoid and myeloid gates. The proportion of positive mice is given as (number of positive mice)/(number of analyzed mice). Representative data from 2 independent experiments are shown.
To see the effects of overexpression of these isoforms, BM transplantation using MigR1 constructs was performed (Figure 6E). Similar to the effect of human RUNX1b and RUNX1a, Runx1bEx6+ recipients showed the lowest engraftment of GFP+ cells, whereas the truncated isoform Runx1bEx6e showed a continuous increase of GFP+ cells in blood.22 Interestingly, Runx1bEx6− recipients showed a decrease of GFP percentage. This result showed that Runx1bEx6e, but not Runx1bEx6−, significantly enhances repopulation ability in vivo.
Discussion
We analyzed genome sequences of Runx1 in various species and revealed that the chimpanzee and rhesus monkey have highly homologous sequences pertaining to human RUNX1 exon 7A, but that this homology did not carry over to mice or other species (Figure 1A-C and data not shown). The exon 7A sequence that is critical for generating human RUNX1a was found only in primates in our analysis. Similarly, the mouse “extended exon 6” sequence was not found in humans or other species that we have studied. It is intriguing that primates and mice express C-terminal truncated isoforms. Although the molecular mechanism and amino acid sequences of the C-terminal ends of these short forms are different, both of them have similar biological function. The cross-species comparison implies that the fine-tuning of hematopoiesis emerged during evolution, and it also tempts us to go back into the early evolution of animals whether they have the homologs of those additional isoforms.
Runx1 exon 6 encodes the negative regulatory region of DNA binding which inhibits association of Runx1 with DNA.42 This region also binds to corepressor Sin3A,43 and interacts with cyclin D3, which decreases expression of downstream target PU.1.44 These studies suggest that exon 6 is an inhibitory domain of Runx1.2,38 Runx1 exon 6 is also 1 of the 2 interaction sites with Ets-1.45 In this report, we examined the biological function of Runx1 isoforms generated by exon 6 alternative splicing using colony formation and luciferase assays (Figures 2-3). Runx1bEx6− and Runx1bEx6e had higher colony-forming ability.31 Runx1bEx6e colony cells had higher CD11b expression than other types of cells, implying differential effects on differentiation. In the luciferase assay, Runx1bEx6− and Runx1bEx6e had lower transactivation activity compared with Runx1bEx6+. It has been reported that Runx1bEx6− has 20% to 45% of the transcription activity whereas DNA binding ability is 2 to 3 times stronger compared with Runx1bEx6+ when tested in the P19 mouse teratocarcinoma cell line, implying that this isoform might work as a dominant-negative form.31 Here, we showed that both Runx1bEx6e and Runx1bEx6− had synergistic or additive, not dominant-negative, effects over Runx1bEx6+ (Figure 3B). However, human RUNX1b without exon 6 showed a dominant-negative effect over full-length RUNX1b in a luciferase assay using the plasminogen activator inhibitor-1 promoter as a reporter and the immortalized ovarian surface epithelial cell T29 line.27 The difference may be due to the presence of different cooperating factors in different cell lineages and different cis-acting elements adjacent to the Runx1 binding sites in promoters.
We previously reported that overexpression of the Runt homology domain causes expansion of HSCs and ultimately results in MDS in a mouse transplantation model.34 We showed that overexpression of this truncation form not only caused effects similar to those caused by knocking out Runx1, but also had unique gene expression changes in microarray data and in vivo phenotypes. This mutant had modest dominant-negative effects over full-length RUNX1, and even worked as a transactivator under specific conditions.34 These facts as well as the results from the Runx1bEx6e luciferase assay suggest that Runx1 isoforms without C-terminal regulatory domains may be more than just dominant-negative regulators. They may have additional features, especially in vivo, possibly due to interactions with other molecules. In addition to the importance of Runx1 in hematopoiesis, Runx1 has been recently recognized as a major player in the normal and oncogenic development of nonhematopoietic cells.11-15 It is expected that Runx1-related gene regulation should be complex and interesting to pursue.
The protein stability of Runx1 is regulated by the ubiquitin-proteasome pathway and by phosphorylation.35,40 S276 and S303 in RUNX1c are phosphorylated by Cdk1/cyclin B and Cdk2/cyclin A, and they affect not only stability but also transcriptional activation.35 The 3 Runx1 isoforms that were tested in this study displayed various half-lives and were differentially affected by proteasome-mediated protein degradation (Figure 4A-D). Runx1bEx6− lacks 2 “destruction boxes (D boxes)” consensus sequences on exon 6 required for the interaction between Runx1 and Cdc20, a substrate-targeting subunit of the ubiquitin ligase complex APC.35 In addition, Runx1bEx6− had significantly lower phosphorylation compared with Runx1bEx6+ (Figure 4E-F). Cdc20 promotes the degradation of S303-phosphorylated Runx1 but not nonphosphorylated Runx1.35 Lower phosphorylation and lack of D boxes may be the cause of the extreme stability of Runx1bEx6−. Lower phosphorylation might also partially explain the lower transactivation of Runx1bEx6− in the luciferase assay (Figure 3A). The exact mechanism of low phosphorylation is not clear. These results will further help to reveal the significance of specific modifications in regard to their overall activity.
“Runx1-IRES-GFP KI mice” were originally made as a tool for tracking Runx1 expression in vivo.33 There was no obvious phenotype in mature cell populations in adult animals.33 Here, we analyzed early hematopoietic events in the KI mice as an in vivo model that lacks the 3 Runx1 isoforms involving exon 6 (Figure 5). The HSC pool was diminished in fetal liver and adult BM in KI mice (Figure 6B-D). Cell-cycle analysis of the sorted LSK of BM cells did not show the difference between WT and KI cells (data not shown). By western blotting, we confirmed that the lower molecular weight bands below the major bands were missing in KI cells, suggesting that they correspond to Runx1b/c Ex6− proteins (Figure 5C). We did not detect Runx1bEx6e protein in lineage-depleted c-Kit–enriched BM cells even in WT cells, possibly due to its rapid turnover, although a small Runx1 isoform similar to Runx1bEx6e was reported in Gr-1–positive BM cells in a previous report.46 On the other hand, we did not observe any difference in the 2 higher molecular weight bands of Runx1 protein between WT and KI samples, which should include Runx1bEx6+ and Runx1cEx6+. The expression of Runx1bEx6+ and Runx1cEx6+ should be theoretically increased in KI mice because Runx1 regulatory elements are only responsible for these variants. These results suggest that the 3 missing isoforms are only a small portion of total Runx1 in the tested cells. We cannot rule out the possibility that a very slight increase of Runx1b/cEx6+ caused the decrease in HSC pool because RUNX1b overexpression inhibits proliferation and stem cell activity.22 However, it is likely that the detected phenotypes in KI mice are attributable to lack of Runx1 isoforms Runx1b/cEx6− and Runx1bEx6e because these isoforms support the self-renewal of HSPCs in vitro (Figure 2) and enhance transactivation of Runx1bEx6+ (Figure 3). There is also a possibility that the expression of GFP might cause toxicity, leading to the observed phenotypes. To test this, it is ideal to examine a mouse strain with GFP replacing the entire Runx1 allele, which is not available so far. However, GFP protein is commonly used to study hematopoiesis, suggesting that the potential toxicity of GFP is negligible. Lastly, Figure 5C showed similar levels of total Runx1 between WT and KI, ruling out the possibility that the phenotypes were due to the effects of posttranscriptional regulation.47 Thus, missing the Runx1 isoforms resulted in the reduction of HSCs in vivo.
Human RUNX1 isoforms lacking exon 6 are also reported in healthy or diseased human cells.18,27-30 Changes of RUNX1 isoform ratios or novel isoforms are also found in various malignancies.24,27,29,48 In acute myeloid leukemia, t(8;21) is a common form of chromosome translocation. It generates fusion transcripts between exon 5 of human RUNX1 and exon 2 of RUNX1T1. Recently, t(8;21) was detected in a few cases of advanced chronic myeloid leukemia.32 Interestingly, fusion between RUNX1 exon 6 and RUNX1T1 exon 2 was detected in at least 2 patients with chronic myeloid leukemia, which correlated with RUNX1-RUNXT1 fusion protein with additional 64 amino acids. Currently, it is unknown whether the presence or absence of exon 6 of human RUNX1 contributes to the distinguished phenotypes of these categories of leukemia. Furthermore, it is not yet clear how the RNA splicing around exon 6 is regulated. In rat pheochromocytoma PC12 cells, alternative splicing to produce the Runx1Ex6− isoform requires heterogeneous nuclear ribonucleoprotein K to bind to target motifs upstream of Runx1 exon 6, which affects neuronal differentiation.49 In different cell types, Runx1 isoforms may have unique tissue-specific functions. More detailed mouse models (knocking out a specific isoform) will certainly be useful to address such questions. Recently, mutations of several splicing factors were detected in MDS and leukemia.50 It would be interesting to analyze whether these mutations affect alternative splicing of RUNX1.
In conclusion, we presented the effects of mouse Runx1 isoforms involving exon 6 in vitro and in vivo. Because Runx1 is a master regulator of stem cell function and development, and various RUNX1 aberrations are detected in malignancy, revealing the functions of splicing isoforms or regulation of Runx1 alternative splicing may lead to novel therapeutic treatment options.
Acknowledgments
This work was supported in part by the National Institutes of Health (grant P01 DK080665). Y.K. was supported partially by the Sumitomo Life Social Welfare Services Foundation and the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.K., M.Y., and D.-E.Z. designed the experiments; Y.K., M.Y., and Y.-J.H. performed the experiments; S.M., M.-C.L., and K.L. provided technical support; J.R.D. provided the RUNX1-IRES-GFP KI mice; Y.K. wrote the manuscript; D.G.T. participated in critical discussion; and D.-E.Z. supervised data analysis and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, Moores UCSD Cancer Center, University of California San Diego, 3855 Health Sciences Dr, La Jolla, CA 92093-0815; e-mail: d7zhang@ucsd.edu.
References
- 1.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark Ed) 2012;17:1120–1139. doi: 10.2741/3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang DE, Fujioka K, Hetherington CJ, et al. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol Cell Biol. 1994;14(12):8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi A, Satake M, Yamaguchi-Iwai Y, et al. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86(2):607–616. [PubMed] [Google Scholar]
- 5.Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40(1):51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 6.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 7.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 8.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniuchi I, Osato M, Egawa T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 10.Philipot O, Joliot V, Ait-Mohamed O, et al. The core binding factor CBF negatively regulates skeletal muscle terminal differentiation. PLoS ONE. 2010;5(2):e9425. doi: 10.1371/journal.pone.0009425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi A, Senzaki K, Ozaki S, Yoshikawa M, Shiga T. Runx1 promotes neuronal differentiation in dorsal root ganglion. Mol Cell Neurosci. 2012;49(1):23–31. doi: 10.1016/j.mcn.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31(21):4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voronov D, Gromova A, Liu D, et al. Transcription factors Runx1 to 3 are expressed in the lacrimal gland epithelium and are involved in regulation of gland morphogenesis and regeneration. Invest Ophthalmol Vis Sci. 2013;54(5):3115–3125. doi: 10.1167/iovs.13-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planagumà J, Liljeström M, Alameda F, et al. Matrix metalloproteinase-2 and matrix metalloproteinase-9 codistribute with transcription factors RUNX1/AML1 and ETV5/ERM at the invasive front of endometrial and ovarian carcinoma. Hum Pathol. 2011;42(1):57–67. doi: 10.1016/j.humpath.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Scheitz CJ, Tumbar T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J Cell Biochem. 2013;114(5):985–993. doi: 10.1002/jcb.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93(5):1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telfer JC, Rothenberg EV. Expression and function of a stem cell promoter for the murine CBFalpha2 gene: distinct roles and regulation in natural killer and T cell development. Dev Biol. 2001;229(2):363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi H, Ohira M, Shimizu K, et al. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23(14):2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sroczynska P, Lancrin C, Kouskoff V, Lacaud G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009;114(26):5279–5289. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- 20.Bee T, Swiers G, Muroi S, et al. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115(15):3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- 21.Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol. 2010;38(5):403–416. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto M, Enver T. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med. 2007;4(5):e172. doi: 10.1371/journal.pmed.0040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuzuki S, Seto M. Expansion of functionally defined mouse hematopoietic stem and progenitor cells by a short isoform of RUNX1/AML1. Blood. 2012;119(3):727–735. doi: 10.1182/blood-2011-06-362277. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Zhang Q, Zhang DE, et al. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia. 2009;23(4):739–745. doi: 10.1038/leu.2008.350. [DOI] [PubMed] [Google Scholar]
- 25.Ran D, Shia WJ, Lo MC, et al. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013;121(15):2882–2890. doi: 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brady G, Elgueta Karstegl C, Farrell PJ. Novel function of the unique N-terminal region of RUNX1c in B cell growth regulation. Nucleic Acids Res. 2013;41(3):1555–1568. doi: 10.1093/nar/gks1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanjundan M, Zhang F, Schmandt R, Smith-McCune K, Mills GB. Identification of a novel splice variant of AML1b in ovarian cancer patients conferring loss of wild-type tumor suppressive functions. Oncogene. 2007;26(18):2574–2584. doi: 10.1038/sj.onc.1210067. [DOI] [PubMed] [Google Scholar]
- 28.Levanon D, Glusman G, Bangsow T, et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262(1-2):23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- 29.Chimienti G, Alaibac M, Marzullo F, Carbone A, Pepe G. The expression pattern of the AML1 gene in non-Hodgkin’s B-cell lymphomas and normal B lymphocytes. Blood Cells Mol Dis. 2000;26(3):186–192. doi: 10.1006/bcmd.2000.0295. [DOI] [PubMed] [Google Scholar]
- 30.Montero-Ruíz O, Alcántara-Ortigoza MA, Betancourt M, Juárez-Velázquez R, González-Márquez H, Pérez-Vera P. Expression of RUNX1 isoforms and its target gene BLK in childhood acute lymphoblastic leukemia. Leuk Res. 2012;36(9):1105–1111. doi: 10.1016/j.leukres.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Bae SC, Ogawa E, Maruyama M, et al. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14(5):3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solari L, Bauer T, Dicker F, et al. A novel recurrent AML1-ETO fusion: tight in vivo association with BCR-ABL1. Leukemia. 2013;27(6):1397–1400. doi: 10.1038/leu.2013.53. [DOI] [PubMed] [Google Scholar]
- 33.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103(7):2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 34.Matsuura S, Komeno Y, Stevenson KE, et al. Expression of the runt homology domain of RUNX1 disrupts homeostasis of hematopoietic stem cells and induces progression to myelodysplastic syndrome. Blood. 2012;120(19):4028–4037. doi: 10.1182/blood-2012-01-404533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol. 2006;26(20):7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Zhang Y, Soosairajah J, Kraft AS. Regulation of RUNX1/AML1 during the G2/M transition. Leuk Res. 2007;31(6):839–851. doi: 10.1016/j.leukres.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14(1):373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang LS, Ito K, Ito Y. RUNX family: regulation and diversification of roles through interacting proteins. Int J Cancer. 2013;132(6):1260–1271. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- 39.Tsuji K, Noda M. Identification and expression of a novel 3′-exon of mouse Runx1/Pebp2alphaB/Cbfa2/AML1 gene. Biochem Biophys Res Commun. 2000;274(1):171–176. doi: 10.1006/bbrc.2000.3112. [DOI] [PubMed] [Google Scholar]
- 40.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20(4):723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang Y, Zhao X, Xu X, et al. CHIP functions an E3 ubiquitin ligase of Runx1. Biochem Biophys Res Commun. 2009;386(1):242–246. doi: 10.1016/j.bbrc.2009.06.043. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Woo AJ, Waldon Z, et al. A Src family kinase-Shp2 axis controls RUNX1 activity in megakaryocyte and T-lymphocyte differentiation. Genes Dev. 2012;26(14):1587–1601. doi: 10.1101/gad.192054.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson LF, Boyapati A, Ranganathan V, et al. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005;25(23):10205–10219. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim WY, Sieweke M, Ogawa E, et al. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18(6):1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corsetti MT, Calabi F. Lineage- and stage-specific expression of runt box polypeptides in primitive and definitive hematopoiesis. Blood. 1997;89(7):2359–2368. [PubMed] [Google Scholar]
- 47.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci USA. 2009;106(1):238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi SJ, Oba T, Callander NS, Jelinek DF, Roodman GD. AML-1A and AML-1B regulation of MIP-1alpha expression in multiple myeloma. Blood. 2003;101(10):3778–3783. doi: 10.1182/blood-2002-08-2641. [DOI] [PubMed] [Google Scholar]
- 49.Cao W, Razanau A, Feng D, Lobo VG, Xie J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res. 2012;40(16):8059–8071. doi: 10.1093/nar/gks504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makishima H, Visconte V, Sakaguchi H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]