Abstract
Regulation of symmetrical cell growth in the culm is important for proper culm development. So far, the involvement of gibberellin (GA) in this process has not yet been demonstrated in sorghum. Here, we show that GA deficiency resulting from any loss-of-function mutation in four genes (SbCPS1, SbKS1, SbKO1, SbKAO1) involved in the early steps of GA biosynthesis, not only results in severe dwarfism but also in abnormal culm bending. Histological analysis of the bent culm revealed that the intrinsic bending was due to an uneven cell proliferation between the lower and upper sides of culm internodes. GA treatment alleviated the bending and dwarfism in mutants, whereas the GA biosynthesis inhibitor, uniconazole, induced such phenotypes in wild-type plants— both in a concentration-dependent manner, indicating an important role of GA in controlling erectness of the sorghum culm. Finally, we propose that because of the tight relationship between GA deficiency-induced dwarfism and culm bending in sorghum, GA-related mutations have unlikely been selected in the history of sorghum breeding, as could be inferred from previous QTL and association studies on sorghum plant height that did not pinpoint GA-related genes.
Dwarfism in plants is brought about by an irregularity in one or more of the various growth-related mechanisms, and may involve physical defects in some cellular growth processes, or problems in the production and action of phytohormones. So far, aberrant cellular division or elongation has already been reported to cause dwarfism in some mutants such as the rice d61, bent uppermost internode 1 (bui1)2, jmj7033 and dwarf and gladius leaf (dgl-1)4, and the Arabidopsis lue15. However, the relationship between dwarfism and phytohormones has been more widely studied using various kinds of plant species. In the case of auxin, some of the reported dwarf mutants include the maize brachytic 2 (br2)6,7 and the sorghum dwarf 3 (dw3)6 which both have semi-dwarfism due to a defect in auxin transport; the rice small organ 1 (smos1), which has a defective transcription factor that disrupts auxin signaling8, and the auxin-deficient tdd19; and the Arabidopsis sax1 which shows high sensitivity to auxin10, and bud1 which has a defect in auxin metabolism11. Another growth-promoting hormone is brassinosteroid (BR). Mutants that have a defect in BR biosynthesis and signalling show severe dwarf phenotypes as in the case of the Arabidopsis BR-deficient de-etiolated 2 (det2)12 and seuss-1 (seu-1)13, and the brassinosteroid-insensitive bri114; the rice BR signalling mutants, d61-415 and erect leaf 1 (elf1-1)16, and the BR-deficient ebisu dwarf (d2)17; and the tomato br6ox (BR biosynthesis) mutant “Micro-Tom”, a commercial cultivar18. BR-related mutations can also give rise to semi-dwarfism, and among them, a barley mutant, uzu, was used for producing a lodging-resistant variety19. Recently, strigolactones (SLs), a group of newly identified plant hormones that control plant shoot branching, have also been implicated for dwarfism in high-tillering dwarf mutants as in the case of the SL-deficient d10 and the SL-insensitive d53 and d14 mutants of rice20,21.
Among the phytohormones, gibberellin (GA) is the most well known to be involved in controlling stem elongation, and a deficiency or insensitivity to GA could easily result in severe dwarfism as reported in many different kinds of plant species such as the rice mutants independently mutated in any of the six GA biosynthetic enzymes, ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA 20 oxidase (GA20ox), and GA 3-oxidase (GA3ox)22; the pea GA-deficient na-123; the Arabidopsis GA-insensitive short internodes (shi)24 and ga-insensitive (gai)25, and the GA-deficient ga1 – 3 mutants26; the barley GA-deficient grd2c mutant27, and the potato andigena (adg) mutant28. Across different species, GA-related mutants with severe dwarfism show several consensus phenotypes such as small dark-green leaves, delayed germination, defective flowering, reduced seed production and male sterility22,28,29,30,31,32,33,34, whereas, semi-dwarf mutants only show mild height reduction with no adverse effects on overall morphology or agronomical traits. This is the reason why GA-related mutants with semi-dwarfism were widely used in the history of crop production. Actually, the rice semi-dwarf1 (sd1) and wheat Reduced height-1 (Rht-1) mutants became the highlights of the Green Revolution, which avoided the imminent food shortage of the mid-20th century35. Later on, the SD1 gene was characterized as a GA 20-oxidase36,37, which catalyses the penultimate step of GA biosynthesis, while Rht-1 gene was found to encode a constitutively active repressor of GA signaling38. Further, another GA-deficient mutant defective in KO, Tan-ginbozu/d35, also contributed to rice crop productivity due to its lodging resistance39.
In contrast to the other plant species above, there are still no other reports of dwarfing mechanisms (including those relating to GA) in sorghum aside from the auxin-related dw3 mutant mentioned earlier. Until now, even the underlying mechanisms behind the major dwarfing QTL genes (dw1, dw2 and dw4) used in the history of sorghum breeding have remained unknown40,41,42. Aside from these classical dwarfing genes, increasing the number of available dwarfing mechanisms for sorghum should be important not only for breeding but also for basic research. Thus, in this study, we mutagenized sweet sorghum to create a mutant library, and searched it for plants with varying degrees of dwarfism. In the process, we found five severe dwarf mutants showing identical unusually bent culms which have never been reported in sorghum. We hereby attempted to isolate and analyse these mutants namely, bending dwarf 1 (bdw1), bdw2, bdw3-1, bdw3-2 and bdw4. We found that the mutants were mutated in four GA biosynthetic genes encoding enzymes catalysing the earlier steps of GA biosynthesis (Supplementary Fig. S1), and that their loss of function directly promotes culm bending. Thus, we reveal for the first time that GA-deficiency can pleiotropically induce culm bending or prostrate growth in sorghum. We also hypothesize that this could possibly be related to why GA-related mutations have not been utilized for sorghum breeding, unlike the case of rice and wheat.
Results
The bdw mutants show an abnormal bent culm
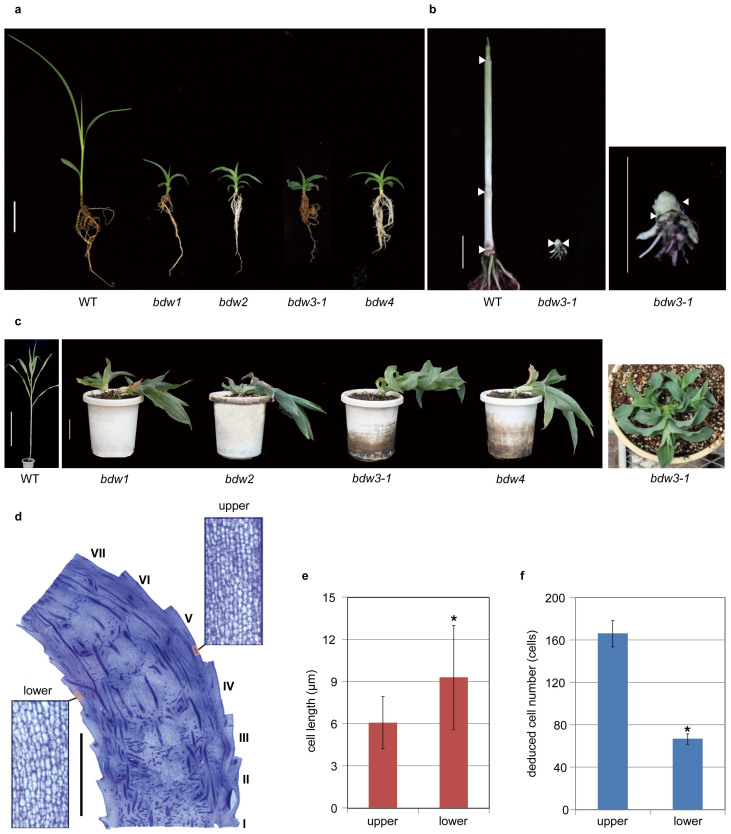
In order to increase the diversity of sorghum dwarf mutants and to further our knowledge of sorghum dwarfing mechanisms, we produced a mutant library through gamma-ray irradiation. During the process of screening for dwarf phenotypes, we isolated several unusual dwarf mutants, which developed bent culms. Succeeding analyses revealed that they were mutated in four different loci, thus we named them as bending dwarf (bdw)1 - bdw4, with bdw3 having two different alleles (bdw3-1 and -2). At the seedling stage, the bending culm phenotype was not evident in the mutants (Fig. 1a) but severe inhibition of internode elongation was observed (Fig. 1b). At about three and a half months after sowing, the bent culm phenotype was clearly observed (Fig. 1c). The mutants bent in a prostrate manner with leaves extending from the sides of the culm (rightmost in Fig. 1c).
Figure 1. Phenotypic analysis of the bdw mutants.
(a) The bdw and WT plants at the seedling stage (30 days after germination, DAS). Bar = 4 cm. (b) Culm structure of WT (left) and bdw3 (center and right) at the vegetative stage. Bars = 2 cm. Nodes are indicated by arrowheads. (c) WT and the bent bdw mutants at the vegetative stage (110 DAS). Top view of two-month old bdw3 is shown at the rightmost panel. Bars: 50 cm, WT; 4 cm, mutants. (d) Longitudinal section of bdw3 culm. Roman numerals at the right side indicate internode position from bottom to top. Bar = 5 mm. (e–f) The length and number of cells at the upper and lower sides of the fifth internode in the bent region shown in panel (d). Error bars for cell length represent the standard deviation calculated from approximately 100 cells. Error bars for cell number represent the standard deviation calculated from 5 cell files (see Methods). Asterisks indicate significant differences at 0.1% (P<0.001).
In order to study the underlying reason for the bending, we carried out a histological analysis of the bdw3-1 culm (Fig. 1d). The cell length at the upper side of the bent region was significantly shorter than that of the lower side (Fig. 1d, e), suggesting that there is a faster rate of cell proliferation at the upper side. Actually, the cell number at the upper side was more than two-folds higher than that at the lower side (Fig. 1f). These observations suggest that the bending was due to a difference in cell proliferation rates between the upper and lower sides of the bent culm.
The bdw mutants have reduced gravitropic response
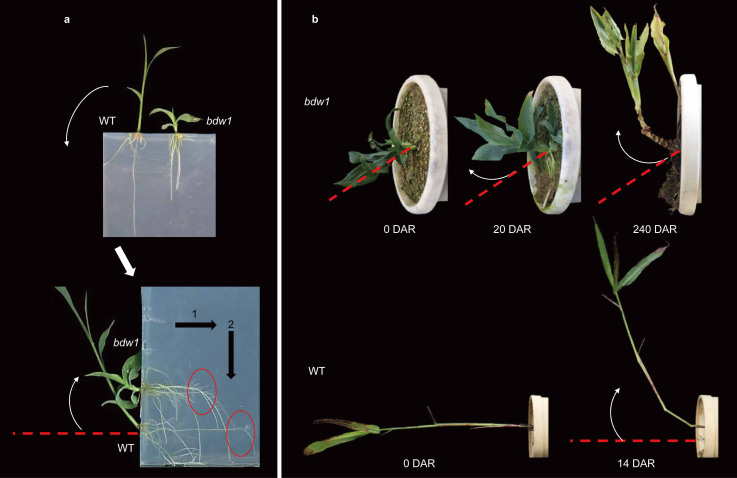
Since we suspected that the gravitropic perception of the mutants might be defective, we directly examined the response of the bdw1 culm to gravity at two stages, seedling and vegetative (Fig. 2). The aerial part of bdw1 at the seedling stage responded positively to gravistimulation, but with a much weaker response compared to the WT (Fig. 2a), possibly due to its naturally inhibited culm elongation. Such weak gravitropic response was also observed at the late vegetative stage (Fig. 2b). Aside from the culm, the roots of bdw1 also responded positively but much more slowly to gravistimulation at the seedling stage (Fig. 2a), as observed in another bdw mutant, bdw3-1 (Supplementary Fig. S2), strongly suggesting that the bdw mutants have diminished ability to respond to gravity.
Figure 2. Gravitropism test of bdw1.
(a) The gravitropic response at the seedling stage. Top, plants at 4 days after germination (DAG) in upright position before gravistimulation; bottom, 4-DAG plants were rotated 90 degrees and further grown for 4 days after rotation (DAR). The horizontal “1” and vertical “2” arrows indicate the gravity directions before and after rotation, respectively. (b) The gravitropic response of bdw1 (top) and WT (bottom) at the vegetative stage. Red dashed lines indicate the original direction of main culm before rotation. White arrows indicate the direction of culm movement.
A loss of function of GA biosynthetic genes causes the bending dwarf phenotype
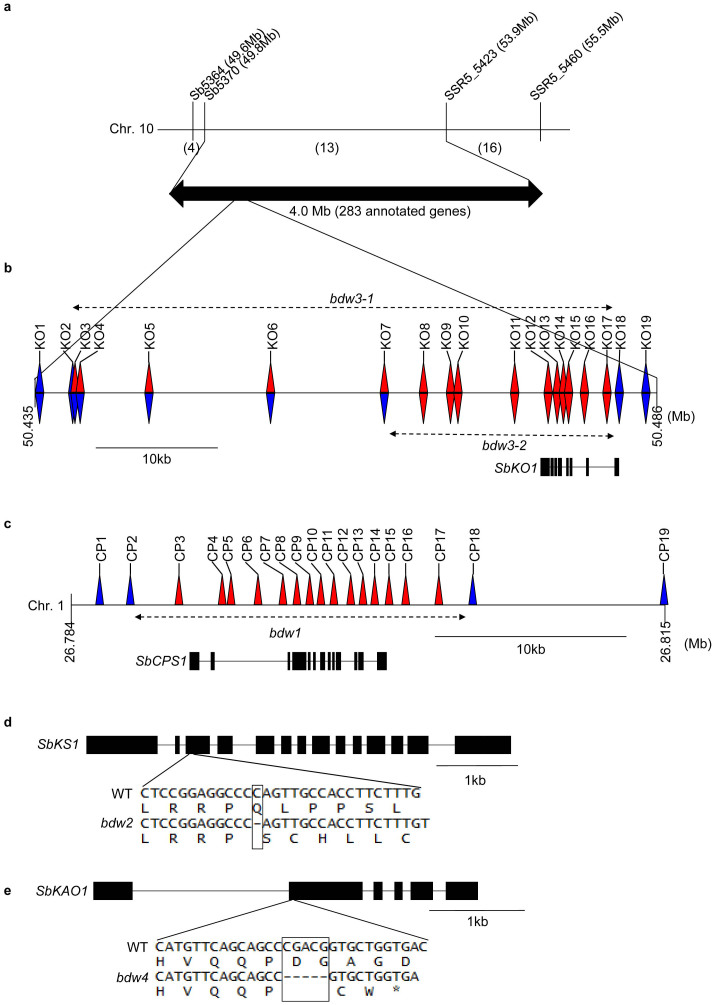
To further characterize the bdw mutants, we carried out positional cloning of the BDW3 gene since there was a pair of allelic mutants for this gene among the five bdw mutants. We used F2 plants derived from the cross between bdw3-1 and the cultivar bmr-6. These plants segregated into two phenotypic groups, tall and dwarf, with a segregation ratio of 3∶1 (P<0.05), respectively. By genotyping 249 F2 plants, we narrowed down the locus of BDW3 to approximately 4.0-Mb region between the markers Sb5370 and SSR5_5423 (Fig. 3a). This region contains 283 genes annotated in the sorghum genomic DNA sequence database (http://www.phytozome.net/), including Sobic.010G172700 (Supplementary Table S1), a gene which is homologous to the rice ent-kaurene oxidase (KO) that encodes a GA biosynthetic enzyme22. In rice and Arabidopsis, the loss of function of KO causes a severe dwarf phenotype with small dark-green leaves22,43, which was also evident in bdw3-1 except that it had an added bent culm trait (Fig. 1c). The deduced amino acid sequence of Sobic.010G172700 showed a high similarity to the entire sequence of rice KO (67%) and also to that of Arabidopsis KO (56%) (Supplementary Fig. S3c)22,43. We searched for mutations in the genomic DNA sequence encompassing the SbKO gene by PCR, and obtained no PCR products from the genome of bdw3-1 (upright red triangles, Fig. 3b), whereas the same primers produced PCR products from the WT genome at the same condition. By using primers covering the flanking sequences of the SbKO1 gene, we successfully located sequences that yielded PCR products (upright blue triangles KO1, KO2, KO18 and KO19, Fig. 3b) and predicted the deletion around the SbKO1 gene to be at about 45 kb from KO3 to KO17. We also performed the same experiment on another bdw3 mutant, bdw3-2, and found that it contained a shorter deletion (∼17 kb) involving all portions of its SbKO1 gene (inverted red or blue triangles in Fig. 3b). A genomic DNA blotting analysis using the promoter sequence of the SbKO1 gene as probe also confirmed the large deletion in the corresponding region of the bdw3-1 genome (Supplementary Fig. S4c, d).
Figure 3. Identification of the causal genes of the bdw mutants.
(a) A physical map of the candidate region of bdw3. The top line shows the position of markers around the target region of chromosome 10. The numbers in parentheses indicate the number of recombinant plants between the markers. The bdw3 mutation locates between Sb5370 and SSR5_5423. (b) Deletion in the bdw3-1 and bdw3-2 genome as detected by PCR analysis. Presence or absence of PCR products is indicated by blue and red triangles, respectively, whereas all PCR products were successfully obtained from the WT genome. Upright and inverted triangles correspond to the bdw3-1 and bdw3-2 genomes, respectively. Dashed arrows at the top and beneath the triangles show estimated deletions around the SbKO1 gene. The SbKO1 structure is shown at the bottom, where boxes and lines between them indicate exons and introns, respectively. (c) Deletion in the bdw1 genome as detected by PCR analysis. The presentation is the same as in panel (b). (d) One-nucleotide deletion in the SbKS1 gene of bdw2. The presentation is the same as in panel (b). (e) Five-nucleotide deletion in SbKAO1 gene of bdw4. The presentation is the same as in panel (b).
The above results suggest that mutation in the SbKO1 gene could be the reason for the severe dwarfism in the bdw3 mutants. To determine whether such loss of function of GA biosynthetic genes is behind the phenotype of all the bdw mutants, we examined the sequences of the other GA biosynthetic genes (CPS, KS, KAO, GA20ox, and GA3ox) in the remaining mutants. As expected, the genome of bdw1 contained an approximately 16-kb deletion involving the entire genomic region of SbCPS1 (Sobic.001G248600) (Fig. 3c; Supplementary Fig. S4a, b). That of bdw2 had a one-nucleotide deletion in the exon 3 of SbKS1 (Sobic.006G211500) (Fig. 3d), while bdw4 contained a five-nucleotide deletion in the exon 2 of SbKAO1 (Sobic.010G007700) (Fig. 3e). These demonstrate that all of the five bdw mutants have defects in GA biosynthetic genes.
To confirm that the concerned GA biosynthetic genes in sorghum really function in vivo, we used them to transform corresponding GA-deficient mutants of rice, namely, oscps1-1, osks1-1, osko2-1 and oskao-122. We introduced a 7,545-bp sorghum genomic DNA fragment containing the entire SbKO1 sequence into rice osko2-1 via Agrobacterium tumefaciens-mediated transformation and found that it completely rescued the severe dwarfism of the mutant (Fig. 4). Similarly, introduction of SbCPS1, SbKS1, and SbKAO1 genes rescued the dwarfism of corresponding rice mutants (Fig. 4). These observations confirmed that the isolated sorghum genes have biological activity in rice, and therefore the dwarf phenotype of the bdw mutants should be caused by the loss of function of these genes.
Figure 4. Introduction of sorghum GA biosynthetic genes rescued the corresponding mutants of rice, oscps1-1, osks1-1, osko2-1, and oskao-1.
Scale bar = 4 cm. Plants at the left and right in each set were transformed with the complementary WT genes from sorghum and the vector control, respectively.
GA rescues the bending culm phenotype, whereas, uniconazole induces bending in WT plants
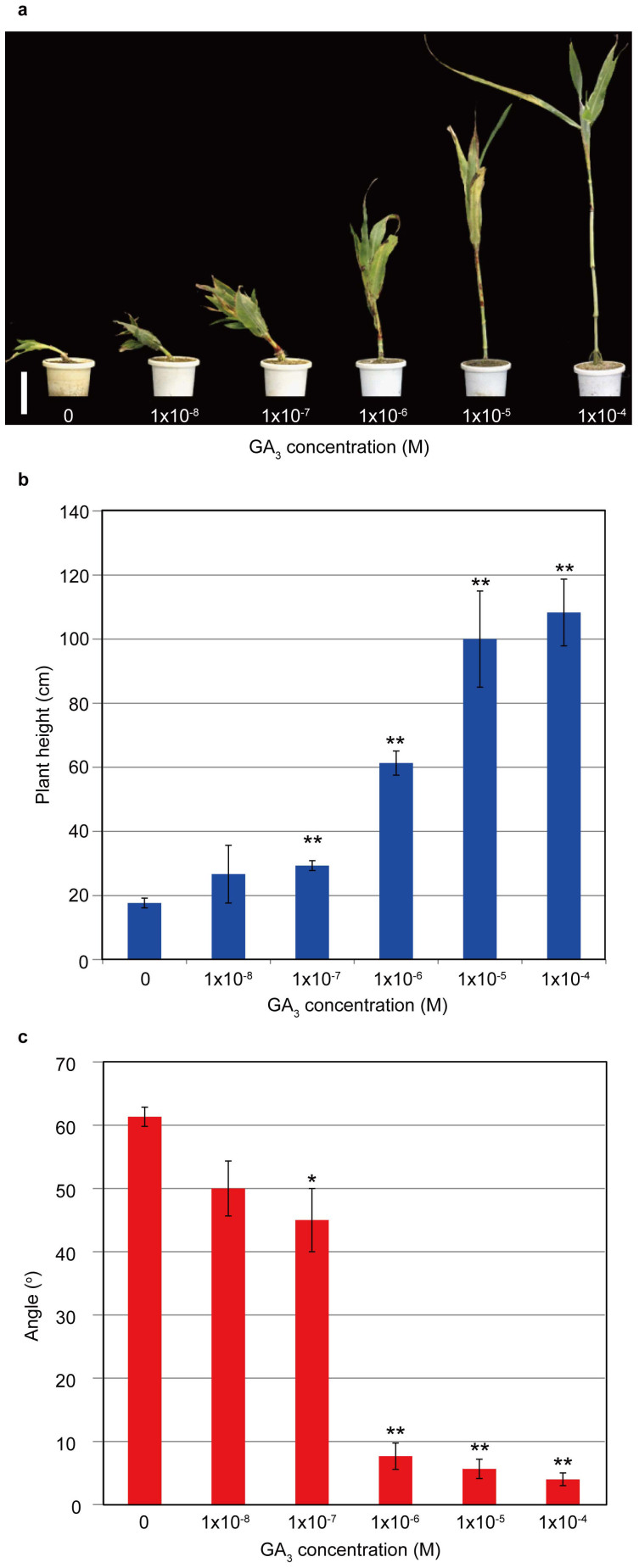
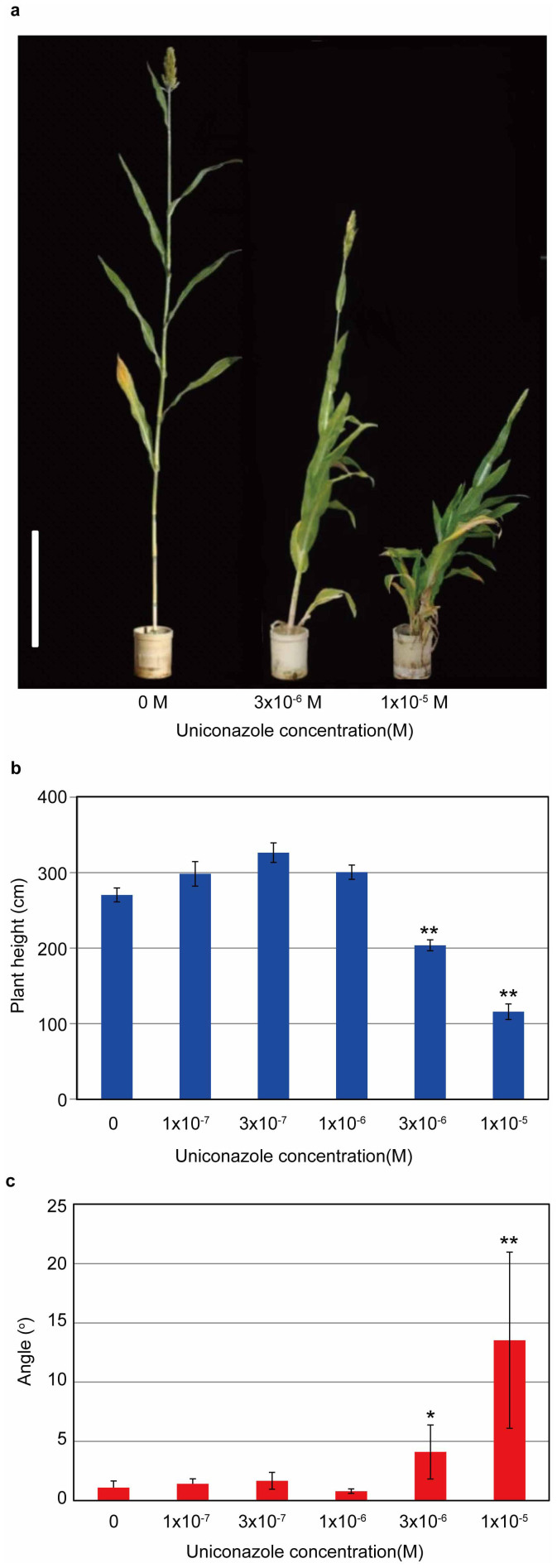
The results described above indicate that loss of function of GA biosynthetic genes causes dwarfism accompanied by unusual culm bending in the mutants. We further confirmed that GA deficiency causes the culm bending by using two approaches. First, we treated the bdw3-1 mutant with different concentrations of GA3 for 4 months. Both the dwarf phenotype and the bending culm phenotype were rescued in a dose-dependent manner by GA3, and were almost normal at 10−4 M GA3 (Fig. 5). At 10−6 M GA3, the dwarfism was partially rescued to about 60% but the culm remained at a significantly bent state (Fig. 5). Secondly, we treated WT with various concentrations of uniconazole (Fig. 6), an inhibitor of GA biosynthesis. As a result, treatment with 10−5 M uniconazole induced WT to bend in a similar manner observed in the mutants (Fig. 6a). At 3 × 10−6 M uniconazole, the plant height was reduced to about 25% while the bending was significantly induced from 0 to about 5 degrees against the vertical axis. These results show that the alleviation of the dwarf and bent phenotype of the mutants by GA was exactly opposite to the effect of uniconazole on WT plants, and that dwarfism and bending are tightly correlated (Fig. 5a, 6a).
Figure 5. Dose-dependent recovery of the mutant phenotypes by exogenous GA treatment.
(a) The bdw3-1 plants grown in different concentrations of GA3 for four months. Bar = 10 cm. (b) Dose-dependent recovery of plant height. (c) Dose-dependent recovery of the bending culm phenotype, which was determined by measuring the angle between the main culm and the vertical axis. Error bars represent the standard deviation calculated from 3 replicates. Single (*) and double asterisks (**) indicate significant difference with the mock (0 M GA3) at 0.5% (P<0.005) and 0.1% (P<0.001) levels of significance, respectively, as determined by t-test.
Figure 6. Effect of uniconazole on the plant height and culm bending of WT plants.
(a) Gross morphology of the uniconazole-treated plants. The plants at the left, center, and right were treated with 0, 3 × 10−6 and 1 × 10−5 M uniconazole, respectively. Bar = 50 cm. Dose-dependent inhibition of plant height (b) and induction of culm bending (c) in WT plants. Error bars represent the standard deviation calculated from 5 replicates. Single (*) and double asterisks (**) indicate significant difference between the mock and 3 × 10−6 and 1 × 10−5 M uniconazole at 5% (P<0.05) and 1% (P<0.01) levels of significance, respectively, as determined by t-test.
Discussion
In this study, we were able to isolate five dwarf mutants (bdw1, 2, 3-1, 3-2, and 4) with abnormal culm bending from a mutant population of sweet sorghum (Fig. 1). Each of the bdw mutants had a defect in a specific GA biosynthetic gene, namely, SbCPS1, SbKS1, SbKO1 and SbKAO1 (Fig. 3; Supplementary Fig. S1)— the genes involved in the early part of GA biosynthesis. Further analyses of these mutants revealed that both dwarfism and culm bending were caused by GA deficiency (Fig. 5, 6).
Although previous studies in maize, rice, and Arabidopsis have shown that loss-of-function mutations in GA biosynthetic genes result in the suppression of internode elongation and dwarfism22,43,44, there was no mention of culm bending or GA being involved in such a phenomenon. In barley, however, the loss of function of GA3ox1, which is involved in the last step of GA biosynthesis, resulted not only in dwarfism but also prostrate growth in the grd2c mutant27. Also in wheat, a strong allele of Rht-B1, Rht-B1c, induced severe dwarfism that correlated with an increase in tiller angle45. These indicate that culm bending (prostrate growth) is one of the pleiotropic effects of GA-related gene mutations and such event appears to be species-dependent. Our conclusion that GA deficiency causes culm bending in sorghum was supported by three experimental evidences. First, the independent loss-of-function mutants defective in four different GA biosynthetic genes were found to have bent culms (Fig. 1, 3). Second, the bent culm phenotype was reverted by exogenous GA treatment in a dose-dependent manner (Fig. 5). Lastly, uniconazole treatment of WT mimicked the bending of the mutants also in a dose-dependent manner (Fig. 6).
Although the exact molecular mechanism controlling bending in sorghum under GA deficiency is still unclear, there was a strong indication that the bending in the bdw3 mutants was due to the asymmetric growth of culm internodes (Fig. 1d, f), which could be partially due to the reduced gravitropic response of the mutants as compared to the WT (Fig. 2; Supplementary Fig. S2). A comparable situation was observed in the GA-deficient barley grd2c mutant27, which showed a slower pulvinar gravitropic response than the WT, and to a greater extent, to sln1c, a GA-hypersensitive mutant. The evidence indicated that gravity-induced auxin asymmetry leads to an asymmetry of GA distribution, and that GAs play a role in facilitating faster gravitropic response by inducing rapid cell elongation on the lower side of the pulvini, following an asymmetric localization of auxin which serves as an initial trigger for bending. Further, just like the bdw mutants, the barley grd2c mutant was also reported to have a prostrate growth habit and GA appears to be affecting the gravitropic set-point angle (GSA) of its lateral shoots (tillers)27. Recently, using pea and Arabidopsis, GSA values were found to be dynamically specified by auxin throughout development by regulating the magnitude of the anti-gravitropic offset component via TIR1/AFB-Aux/IAA-ARF-dependent auxin signalling within the gravity-sensing cells of the root and shoot46, suggesting that the bending in the bdw mutants could also be auxin-related. Our results show that GA deficiency triggers prostrate growth habit in sorghum in the presence of auxin, which is consistent with the case of barley grd2c27, strongly indicating that GA also has a crucial role in controlling the GSA of the culm, at least in these species.
Mutations involving partial GA-deficiency and semi-dominant GA-insensitivity have long been harnessed in breeding programs to induce the favourable semi-dwarf trait made famous by the Green Revolution rice (sd1) and wheat (Rht-B1b and Rht-D1b), respectively36,38. In such instances, semi-dwarfism led to increased lodging resistance and high yield. However, the present study strongly suggests that the use of GA-related genes for sorghum breeding remains to be a big challenge because of the bending side-effect linked with GA deficiency. Actually, results presented in Fig. 5 and 6 demonstrated that even semi-dwarf sorghum plants show a potentially destabilizing (e.g. lodging-prone) culm curvature under reduced GA levels. Our assumptions are also being upheld by the fact that there are still no reports of GA-related genes used for sorghum semi-dwarf breeding to date.
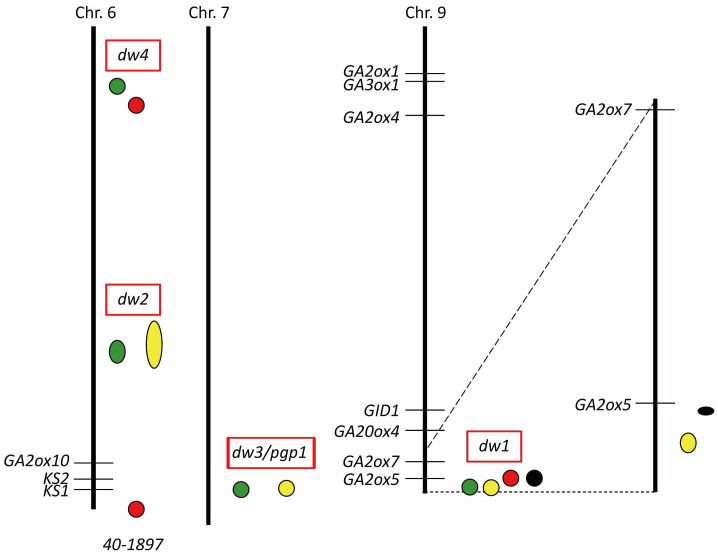
So far, it is widely known that four major classical dwarfing genes (dw1 - dw4) had been introgressed into elite varieties in the early history of sorghum breeding47,48,49,50,51. Among these dwarf genes, only the causal gene of dw3 had been identified, namely, a gene encoding a phosphoglycoprotein involved in auxin transport6. Because the location of dw1, dw2 and dw4 have also been identified40,41,42,52 (Supplementary Table S2), we compared them against the loci of GA-related genes (Fig. 7 and Supplementary Table S1) and found that the loci of dw2, dw3 and dw4 do not coincide with mapped positions of known GA-related genes. On the other hand, dw1 is in close proximity to a GA 2-oxidase-like gene (Fig. 7), Sobic.009G230800 (referred to as GA2ox5 in Supplementary Table S1), which was previously discussed by two independent groups to be the possible causal gene of dw140,41. However, based on our experiments, it is unlikely that a GA2ox is behind the phenotype of dw1 because in that case, bending would become inevitable. Since GA2ox is involved in GA catabolism, its effect on sorghum should be similar to that of a GA synthesis inhibitor, uniconazole. To investigate whether dw1 is the same as GA2ox5, we directly compared the DNA sequence and expression level of the gene in a tall (Dw1Dw1) and dwarf (dw1dw1) isogenic sorghum cultivar carrying gain-of-function and loss-of-function alleles of Dw1, respectively. As expected, there was no difference in the genome sequence and expression level of GA2ox5 between the two cultivars (Supplementary Fig. S5). This result supports the above idea that a GA2ox is unlikely to be the causal gene of dw1.
Figure 7. Comparison between the map positions of previously reported dwarfing QTL genes and the GA-related genes of sorghum.
The chromosomes carrying the dwarfing QTL genes previously reported are presented. Ovals and circles at the right side of each chromosome indicate the positions of respective previously reported dwarfing QTLs (red rectangles), where green, yellow, red and black shades represent the mapping work of Morris et al. (2013), Thurber et al. (2013), Wang et al. (2012) and Brown et al. (2008), respectively. The positions of GA-related genes are presented at the left side of each chromosome with their corresponding names as presented in Supplementary Table S1.
As mentioned earlier, in wheat, the use of a strong allele of Rht-B1 caused severe dwarfism and bending45, and such traits were further aggravated by overexpression of a GA-inactivating enzyme, GA2ox29,53. In contrast, the weak Rht-B1 mutation used in the Green Revolution only resulted in semi-dwarfism, with no obvious bending38. On the other hand, the Green Revolution rice and even the severe dwarf rice mutants carrying null alleles for GA biosynthetic or signalling genes do not show culm bending at all22,36,54. These observations suggest that, because of the exceptionally tight relationship between GA deficiency-induced dwarfism and bending in sorghum, the use of GA-related mutations to induce semi-dwarfism, without compromising the straightness and lodging resistance of the culm, may not be possible. Taken altogether, our results strongly suggest that the classical dwarfing genes that have been utilized for sorghum breeding in the past may not be related to GA. As in the case of dw36, the isolation and characterization of dw1, along with those of dw2 and dw4 will reveal novel mechanism(s) for semi-dwarfism in sorghum. Once identified, these genes can also be exploited in rice and other crops as additional or alternative options for lodging resistance breeding.
Methods
Plant materials and mutant library construction
We used Sorghum bicolor cv. SIL-05 (sweet sorghum) for constructing a mutant library. Mutagenesis was carried out by irradiating seeds with gamma-ray (175–250 Gray (Gy)). The resulting M1 seeds were sown in seedling trays and were grown inside the greenhouse at 25°C for more than 3 weeks before planting in the field. About 5,000 of such plants were self-pollinated to obtain M2 lines which were screened for dwarfism at the seedling (greenhouse) and vegetative (field) stages. Identified mutants and all plants for analysis were grown in pots inside the greenhouse under ambient lighting conditions.
Microscopic analysis
A culm longitudinal section of the bdw3-1 mutant was made with a cryotome. For staining, hematoxylin and eosin stains were used. Photographs were taken using an Olympus photomicroscope connected to a CCD camera. Resulting photographs were stitched to create a panoramic image (Photostitch, Canon). In order to understand the underlying cause of the bending in the bdw3-1 mutant, the length and number of the cells at both the upper side and lower side of the fifth internode, where the greatest degree of bending took place, was quantified. This was done by measuring the length and number of cells in 5 cell files within a 500-μm long region (orange rectangles, Fig. 1d) at both the upper and lower sides of the fifth internode and then estimating the total number of cells in the internode.
Gravitropism test
Seeds of the bdw1 and bdw3-1 mutants (as representatives of the phenotypically identical bdw mutants) and WT were disinfected with 0.5% benlate overnight at 4°C and sowed on MS medium with 0.9% agar in plastic rectangular culture plates. Plants were grown for 4 days after germination (DAG) in an upright position and then rotated 90 degrees and examined 4 days after rotation (DAR). The culture plates were imaged by using a flatbed digital scanner. Also, bent bdw1 plants at the vegetative stage were rotated 90 degrees following the direction of culm bending and compared their response with the WT.
Genome mapping of the bdw3 locus and sequence analysis
To carry out gene mapping, we crossed heterozygous siblings of the bdw3-1 mutant (sweet sorghum (SIL-05) background) with bmr-6 (grain sorghum cultivar) to produce F1 plants that were progeny-tested to obtain the desired F2 population for analysis. Segregation ratio of the tall and dwarf phenotypes was observed and analysed by chi-square test.
We screened about 1,500 Simple Sequence Repeats (SSR) markers55 and selected 162 polymorphic markers. For rough mapping of the candidate region, we used the SSR markers on 16 F2 seedlings with tall phenotype. For the fine mapping, we expanded the analysis to 249 F2 tall plants.
We used the amino acid sequences of known GA biosynthetic genes of rice as queries for the TBLASTN program (http://www.phytozome.net/search.php?show=blast&org=Org_Sbicolor_v2.1) to identify homologous sequences in the sorghum genome. PCR analysis of the identified genes, namely, SbCPS1 (Sobic.001G248600), SbKS1 (Sobic.006G211500), SbKO1 (Sobic.010G172700), SbKAO1 (Sobic.010G007700), SbGA20ox2 (Sobic.003G379500) and SbGA3ox2 (Sobic.003G045900) was done on all the bdw mutants and the WT. PCR primers used are listed in Supplementary Table S3. DNA sequencing was done with the ABI Prism 310-10 sequencer following the manufacturer's protocol.
Amino acid sequences of sorghum CPS1, KS1, KO1 and KAO1 were compared with those of other species by aligning with ClustalW (http://www.genome.jp/tools/clustalw/) using default settings (slow/accurate) and then manually adjusted to optimize alignments. The unrooted phylogenetic tree with branch length (N-J) tree file output was used to create a phylogenetic tree with FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) using default settings for the rectangular tree layout.
For DNA gel blot analysis of SbCPS1 and SbKO1, 1 µg genomic DNAs from bdw1, bdw3-1 and WT were analysed as previously described56. Primers used to amplify the probes are listed in Supplementary Table S3.
Complementation of rice GA-deficient mutants with WT GA biosynthetic genes from sorghum
To examine the bioactivities of the identified sorghum GA biosynthetic genes, we first cloned 11,287, 6,938, 7,628, and 5,909 bp of sorghum genomic DNA fragments containing entire SbCPS1, SbKS1, SbKO1 and SbKAO1 genes along with their native promoters into a pCAMBIA1300 binary vector, respectively. These chimeric plasmids were introduced into corresponding mutants of rice, oscps1-1, osks1-1, osko2-1 and oskao-1, through Agrobacterium-mediated transformation22,57. The transformants were screened on selection medium with hygromycin and the regenerated plants were documented.
Exogenous gibberellin treatment and uniconazole treatment
For GA treatment, two-week old bdw3 seedlings were transplanted into pots. A set of three pots (with holes in the bottom) were placed in a square plastic vat containing a designated concentration of GA3 (0, 1 × 10−8, 1 × 10−7, 1 × 10−6, 1 × 10−5 and 1 × 10−4 M) and were grown continuously in the solution for four months inside the greenhouse. For uniconazole treatment, a set of five 4-week-old WT plants (SIL-05) were grown continuously for 2 months in a vat containing specified concentrations of uniconazole (0, 1 × 10−7, 3 × 10−7, 1 × 10−6, 3 × 10−6 and 1 × 10−5 M). In both GA and uniconazole experiments, the vats were regularly checked and augmented with water whenever necessary to guard against evaporation and fluctuation in the concentration of the original solution until the completion of the experiment. Measurement of plant height was done with a rope by tracing the contour of the main culm from the base to the longest tip. Culm angle was taken by measuring the angle formed by the culm against the vertical axis.
DNA and RNA-seq analysis by Next Generation Sequencer (NGS)
Two isogenic varieties, tall white sooner milo (Dw1Dw1) and dwarf white milo (dw1dw1) carrying gain-of-function and loss-of-function alleles of Dw1, respectively, were utilized for genomic DNA sequence analysis using NGS. Sequencing was performed at Hokkaido System Science Co. Ltd. (Sapporo, Japan). Library for sequencing was prepared using TruSeq DNA/RNA Sample Prep Kit (Illumina, Inc.) according to the manufacturer's protocol. The resultant library was sequenced (2 × 101 cycles, paired-end) on HiSeq 2500/2000 instrument using TruSeq PE Cluster Kit and TruSeq SBS Kit (Illumina, Inc.) to obtain approximately 40 million raw sequence reads for analysis. Sequences were mapped to the BTx623 sorghum reference genome58 by using BWA version 0.6.1-r104 (http://bio-bwa.sourceforge.net/), and SNPs were called with the SAMtools version 0.1.18 (http://samtools.sourceforge.net/). The two varieties were also subjected to RNA-seq analysis by using elongating internode total RNAs isolated by a modified Trizol method59. Total RNAs were utilized for RNA-seq experiment as described previously60.
Author Contributions
R.L.O., Y.I., S.K., H.K., M.M. and T.S. designed the experiments and wrote the manuscript. R.L.O. conducted almost all of the molecular analyses and plant assays. T.S. and T.T. grew the M1 and M2 populations. A.H. and Y.I. conducted the uniconazole test and mutant phenotyping in the field. K.O.S. made the construct for SbKO1 and assisted in genetic transformation, while H.M. did the RNA-seq analysis. And H.M. did the RNA-seq analysis.
Supplementary Material
Compilation of Supp Figs and Tables
Acknowledgments
This work was partially supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation), QTL5501, 5505, Genomics-based Technology for Agricultural Improvement, IVG-1004; Core Research for Evolutional Science and Technology (CREST) Program “Novel techniques of tailor-made breeding for energy crop improvement using high-throughput genotyping” of Japan Science and Technology Agency (JST); and Grants-in-Aid from the NC-CARP project, the Ministry of Education, Science, Sports and Culture of Japan. We thank Prof. Y. Inukai for the gamma-ray treatment of sorghum seeds, A. Kondo-Watanabe and S. Araki-Nakamura for their technical assistance, and K. Aya, K. Yano and T. Hosaki for handling DNA- and RNA-seq results.
References
- Sato Y. et al. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23, 661–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. USA. 110, 1953–1958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komorisono M. et al. Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol. 138, 1982–1993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T., Mattsson O., Naested H., Foster R. & Mundy J. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801 (2003). [DOI] [PubMed] [Google Scholar]
- Multani D. S. et al. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302, 81–84 (2003). [DOI] [PubMed] [Google Scholar]
- Knoller A. S., Blakeslee J. J., Richards E. L., Peer W. A. & Murphy A. S. Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J. Exp. Bot. 61, 3689–3696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K. et al. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 55, 897–912 (2014). [DOI] [PubMed] [Google Scholar]
- Sazuka T. et al. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 60, 227–241 (2009). [DOI] [PubMed] [Google Scholar]
- Ephritikhine G., Fellner M., Vannini C., Lapous D. & Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18, 303–314 (1999). [DOI] [PubMed] [Google Scholar]
- Dai Y., Fu Z. & Li J. Isolation and characterization of an Arabidopsis bush and dwarf mutant. Acta Bot. Sin. 45, 621–625 (2003). [Google Scholar]
- Noguchi T. et al. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120, 833–839 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S., Rueschhoff E. E., Bhatti H. & Franks R. G. Synergistic disruptions in seuss cyp85A2 double mutants reveal a role for brassinolide synthesis during gynoecium and ovule development. BMC Plant Biol. 10, 198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D., Langford M. & McMorris T. C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A. et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140, 580–590 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kitano H. & Fujioka S. An E3 ubiquitin ligase, ERECT LEAF1, functions in brassinosteroid signaling of rice. Plant Signal. Behav. 8, e27117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z. et al. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E., Gisbert C., Bishop G. J., Dixon M. S. & Garcia-Martinez J. L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 57, 2037–2047 (2006). [DOI] [PubMed] [Google Scholar]
- Chono M. et al. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 133, 1209–1219 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T. et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 (2009). [DOI] [PubMed] [Google Scholar]
- Jiang L. et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T. et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S. E., Elliott R. C., Helliwell C. A., Poole A. T. & Reid J. B. The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol. 131, 335–344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I., Kuusk S., Moritz T. & Sundberg E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11, 1019–1031 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P. & Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang C. M., Davies N. W., Taylor S. A. & Ross J. J. Gravistimulation leads to asymmetry of both auxin and gibberellin levels in barley pulvini. Physiol. Plant. 131,140–148 (2007). [DOI] [PubMed] [Google Scholar]
- Bamberg J. B. & Hanneman R. E. Characterization of a new gibberellin related dwarfing locus in potato (Solanum tuberosum L.). Am. Potato J. 68, 45–52 (1991). [Google Scholar]
- Hedden P. & Phillips A. L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530 (2000). [DOI] [PubMed] [Google Scholar]
- Fleet C. M. & Sun T. P. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85 (2005). [DOI] [PubMed] [Google Scholar]
- Tanimoto E. Regulation of root growth by plant hormones–Roles for auxin and gibberellin. Crit. Rev. Plant Sci. 24, 249–265 (2005). [Google Scholar]
- Wang Y. & Li J. The plant architecture of rice (Oryza sativa). Plant Mol. Biol. 59, 75–84 (2005). [DOI] [PubMed] [Google Scholar]
- Chhun T. et al. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19, 3876–3888 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S. F. et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the Green Revolution. Trends in Genet. 19, 5–9 (2003). [DOI] [PubMed] [Google Scholar]
- Sasaki A. et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702 (2002). [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., Ellis M. H. & Chandler P. M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99, 9043–9048 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999). [DOI] [PubMed] [Google Scholar]
- Itoh H. et al. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54, 533–547 (2004). [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Bible P., Loganantharaj R. & Upadhyaya H. D. Identification of SSR markers associated with height using pool-based genome-wide association mapping in sorghum. Mol. Breed. 30, 281–292 (2012). [Google Scholar]
- Morris G. P. et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 110, 453–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber C. S., Ma J. M., Higgins R. H. & Brown P. J. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 14, R68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A. et al. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 95, 9019–9024 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S. et al. Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol. 88, 1367–1372 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiol. 157, 2120–2130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhry S., Del Bianco M., Kieffer M. & Kepinski S. Auxin controls gravitropic setpoint angle in higher plant lateral branches. Curr. Biol. 23, 1497–1504 (2013). [DOI] [PubMed] [Google Scholar]
- Quinby J. R. & Karper R. E. Inheritance of height in sorghum. Agron. J. 46, 211–216 (1954). [Google Scholar]
- Hadley H. H. An analysis of variation in height in sorghum. Agron. J. 49, 144–147 (1957). [Google Scholar]
- Quinby J. R. Sorghum Improvement and the Genetics of Growth (Texas A&M University Press, College Station, TX, 1974). [Google Scholar]
- Smith C. W. & Frederiksen R. A. Sorghum: Origin, history, technology, and production, 191–223 (John Wiley & Sons, New York, 2000). [Google Scholar]
- Klein R. R. et al. Effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Crop Sci. 48, S12–S26 (2008). [Google Scholar]
- Brown P. J., Rooney W. L., Franks C. & Kresovich S. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180, 629–637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleford N. E. et al. Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J. Exp. Bot. 58, 3213–3226 (2007). [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005). [DOI] [PubMed] [Google Scholar]
- Yonemaru J. et al. Development of genome-wide simple sequence repeat markers using whole-genome shotgun sequences of sorghum (Sorghum bicolor (L.) Moench). DNA Res. 16, 187–193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K. et al. A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. USA 106, 11218–11223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y. & Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834 (2008). [DOI] [PubMed] [Google Scholar]
- Paterson A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- Chomczynski P. & Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
- Mizuno H. et al. Global transcriptome analysis reveals distinct expression among duplicated genes during sorghum-Bipolaris sorghicola interaction. BMC Plant Biol. 12, 121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Compilation of Supp Figs and Tables