Abstract
Background:
The miRNA deregulation is commonly observed in human malignancies, where they act as tumour suppressors or oncogenes. Despite the association of several miRNAs with bladder cancer, little is known about the miRNAs that contribute to bladder cancer progression from non-muscle invasive (NMI) to muscle-invasive (MI) disease.
Methods:
We first profiled the expression of miRNAs and mRNAs in a cohort of urothelial carcinomas and further characterised the role of miR-126 in invasion, as it emerged as the most downregulated miRNA between MI and NMI tumours.
Results:
We found that restoration of miR-126 levels attenuated the invasive potential of bladder cancer cells. Mechanistically, we identified the role of miR-126 in invasion through its ability to target ADAM9. Notably, a significant inverse correlation between miR-126 and ADAM9 expression was observed, where ADAM9 was upregulated in MI bladder cancer cells. While knockdown of ADAM9 attenuated the invasiveness of cells with low miR-126 levels, experimental upregulation of ADAM9 recapitulated the invasive phenotype. Furthermore, ADAM9 expression assessed by immunohistochemistry significantly correlated with poor prognosis in patients with urothelial carcinoma.
Conclusions:
In this study we describe the role of miR-126 in bladder cancer progression, identifying miR-126 and ADAM9 as potential clinical biomarkers of disease aggressiveness.
Keywords: bladder cancer, invasion, microRNA, miR-126, ADAM9
At tumour initiation, bladder cancer can be classified into two groups: low-grade papillary tumours and high-grade tumours (either papillary or non-papillary) (Cordon-Cardo, 2008). Chromosomal aberrations associated with the development of papillary tumours include deletion of chromosome 9 and activating mutations in oncogenes such as H-RAS, FGFR3, and PIK3CA (Cordon-Cardo, 2008). One frequently perturbed region is 9p21. This locus harbours CDKN2A (encodes p16 and p14ARF) and CDKN2B (encodes p15) genes (Goebell and Knowles, 2010). Homozygous deletion of CDKN2A is linked to papillary tumours of high grade and higher recurrence (Orlow et al, 1999; Chapman et al, 2005). Although it remains unclear whether loss of 9q precedes the loss of 9p, the loss of 9q phenotype is more frequent in non-muscle invasive (NMI) than muscle-invasive (MI) tumours (Simoneau et al, 2000).
In contrast, genetic alterations in tumour suppressors such as TP53, PTEN, and RB characterise initiation events in carcinoma in situ (CIS) lesions and MI tumours (Cairns et al, 1991, 1998; Cordon-Cardo, 2008). Additional chromosomal abnormalities that contribute to progression of high-grade tumours into MI UCs include deletion of 3p, 11p, and 18q (Presti et al, 1991; Li et al, 1996). Muscle invasion and metastasis in bladder cancer is a primary cause of patient mortality. Patients with unresectable metastatic bladder cancer have a median survival of 7–20 months, despite systemic chemotherapy, and the 5-year survival rate is only 5.5% (Cordon-Cardo, 2008; Howlader et al, 2009). Remarkably, little is known regarding the transition from superficial papillary tumours to MI tumours.
MicroRNAs (miRNAs) are noncoding RNA sequences that control gene expression by repressing the expression of specific mRNA targets (Schwarz et al, 2003). As regulators of gene expression, aberrant expression of miRNAs can be tumourigenic acting as tumour suppressors and oncogenes (He and Hannon, 2004; Esquela-Kerscher and Slack, 2006; Zhang et al, 2007). Several studies suggest that aberrant miRNA expression contribute to bladder cancer development and progression. Although the tumour suppressive roles of miR-1, miR-133a, and miR-145 in bladder cancer invasion have been characterised by a few groups (Chiyomaru et al, 2010; Yoshino et al, 2011; Chiyomaru et al, 2012; Hirata et al, 2012; Majid et al, 2012; Yamasaki et al, 2012; Yoshino et al, 2012; Noguchi et al, 2013), little is known about miRNAs that are misregulated in bladder cancer progression from NMI to MI disease.
In order to identify miRNAs with relevant roles in the progression of bladder cancers, we profiled the miRNA and mRNA expression of a repertoire of bladder UCs ranging from low-grade papillary NMI to high-grade MI tumours and we identified several miRNAs associated with tumour invasion. In particular, miR-126 was significantly downregulated in MI bladder cancer tissues compared with papillary NMI tumour tissues. Mechanistically, we found that miR-126 targets ADAM9, a member of the metzincin superfamily of matrix metalloproteinases implicated in the progression of tumours (Duffy et al, 2009). Specifically, ADAM9 has been shown to release and activate EGF and heparin-binding EGF and its expression has been correlated with progression of many human cancers (O'Shea et al, 2003). In this study, we demonstrated the role of the miR-126/ADAM9 axis on the invasion abilities of bladder tumours.
Materials and methods
Cell culture and human clinical samples
All cell lines were grown according to the manufacturer's protocols at 37 °C in a humidified air atmosphere at 5% CO2. The Human Urothelial Cell (HUC) line was purchased from ScienCell Research Laboratories (Carlsbad, CA, USA). EJ138 was a gift from Dr Sanchez-Carbayo (Tumor Markers Group, Madrid, Spain). MCF10A, TCCSUP, J82, and 293FT cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The following human bladder tumour tissue samples were obtained through institutional review board (IRB)-approved protocols: TaLG (n=3), TaHG (n=3), CIS (n=3), T1LG (n=3), T1HG (n=3), and T2HG (n=6). The CIS biopsies were dissected using laser capture microdissection as previously described (Jia et al, 2013) to isolate pure tumour cells from bladder stroma. The G1 tumours were designated as LG (low grade); G2 and G3 were designated as HG (high grade).
RNA extraction
The RNA was extracted from cell lines and tumour samples using the miRVANA Kit (Ambion, Carlsbad, MA, USA). The RNA from microdissected CIS samples was extracted using the RNAquesous-Micro Kit (Ambion). The quality of all RNA samples was verified using a RNA Pico Kit (Agilent, Santa Clara, CA, USA). Only sample qualities with RIN above 6.0 with clean 18S and 28S peaks were used, as previously described (Fleige and Pfaffl, 2006).
Microarray profiling and data analysis
Agilent Human miRNA Microarray (V3, based on Sanger miRbase release 12.0) was used for measuring miRNA expression in bladder samples. Samples used for the Agilent miRNA microarray were labelled as described by the manufacturer. Affymetrix Human U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA) were used for measuring gene expression in bladder samples. Samples used for Affymetrix microarray were amplified using the Ovation RNA Amplification System (NuGEN, San Carlos, CA, USA) and labelled using FL-Ovation cDNA Biotin Module V2 (NuGEN) following the manufacturer's protocol. Raw intensity miRNA data were normalised and median transformed using GeneSpring GX 12 (Agilent). The raw mRNA data were log transformed and analysed using Partek Genomics Suite 6.6 (Partek, Saint Louis, MO, USA). Only detected probe sets were used, and compromised or undetected probe sets were filtered out. One-way ANOVA and Tukey's honestly significant difference (HSD) post hoc test were performed across all samples to obtain miRNAs or mRNAs differentially expressed (P<0.05). Unsupervised hierarchical cluster analysis was performed on the list of differentially expressed probe sets with a fold change of ⩾2 (miRNAs) or a fold change of ⩾10 (mRNAs).
Plasmid generation
The RNA from MCF10A cells was used to PCR amplify pre-miRNA regions. The following sense and antisense oligonucleotides were used, respectively: miR-126, 5′-CATTCTCGAGTGGCTGTTAGGCAT-3′ and 5′-TGGGCGGCCGCCTCTGCACTTCTT-3′ Amplified pre-miR-126 was subcloned into an empty pLemiR vector (Open Biosystems, Pittsburgh, PA, USA) and verified by sequencing, hereby referred to as pLemiR-126.
Lentiviral plasmids PMB and CMV, retroviral plasmids VSVG and helper, and firefly luciferase plasmid (FUW-Luc) were gifts from Dr Jose Silva (Columbia University, New York, NY, USA). Renilla luciferase was subcloned into a modified pLPCX (Clontech Laboratories, Mountain View, CA, USA) plasmid that contains GFP, hereby referred to as pLPCX-Ren. The sense and antisense oligonucleotide sequences used to amplify Renilla luciferase from pRL-SV40 (Promega, San Luis Obispo, CA, USA) were 5′-AAAAACTCGAGCGCCACCATGACTTCGAAAGTTTATGATCC-3′ and 5′-AAAAAGAATTCTTATTGTTCATTTTTGAGAACTC-3′, respectively.
Human ADAM9 (accession no. BC126406.1) was expressed in vitro using the gateway vector pLenti6.2/V5-DEST (Invitrogen, Grand Island, NY, USA), hereby referred to as pLenti6.2-ADAM9. First, ADAM9 was amplified from pCR4-TOPO-ADAM9 (Thermo Scientific, Pittsburgh, PA, USA) using the following sense and antisense oligonucleotides: 5′-CACCGGCCGAGATGGGGTCT-3′ and 5′-AAATCTGTTTGCATATATAGGAACTTCTCTG-3′. Following the manufacturer's protocols, ADAM9 was then shuttled into the pENTR/D-TOPO vector (Invitrogen) and recombined into the pLenti6.2/V5-DEST (Invitrogen).
Transient transfection and infection
Transient knockdown of ADAM9 was conducted using the ON-TARGET plus SMARTpool siRNA against ADAM9 (Thermo Scientific) using Lipofectamine 2000 (Invitrogen). A final siRNA concentration of 25 nM was used.
For cell infections, 293FT cells were transfected with lentiviral or retroviral particles using Lipofectamine 2000 (Invitrogen). Media from 293FT at 48 h after transfection were collected to infect bladder tumour cell lines. Cell lines infected with pLPCX-Ren (designated J82_RL and EJ138_RL) were maintained using puromycin (0.5 μg ml−1). Cell lines infected with FUW-Luc (designated J82_FL and EJ138_FL) were subsequently infected with a pLemiR-126 (designated J82_FL_126 and EJ138_FL_126), and maintained using puromycin (0.5 μg ml−1) and geneticin (800 μg ml−1). J82_FL_126 and EJ138_FL_126 cell lines infected with pLenti6.2-ADAM9 (designated J82_FL_126_ADAM9 and EJ138_FL_126_ADAM9) were maintained using blasticidin (10 μg ml−1 for J82 cells and 20 μg ml−1 for EJ138 cells).
Quantitative real-time reverse transcription–PCR (qRT–PCR) expression profiling
TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used to convert miRNA to cDNA. Reverse transcription primers (44 nt) were designed so that the first 36 nt formed an internal stem loop and the last 8 nt were complementary to the mature miRNA sequence of interest. Qiagen Reverse Transcriptase System (Qiagen, Valencia, CA, USA) was used to produce cDNA from mRNA. Qiagen QuantiTect PCR (Qiagen) was used to measure quantitative expression of miRNA and mRNA. The PCR assays were performed as described by the manufacturer using a Stratagene MX3005P PCR system (Agilent). For normalisation, we used mir-92b or ACTIN for miRNA and mRNA, respectively. The miR-92b provided comparable expression levels across the bladder cell lines and has also been cited as a stable reference gene for miRNA qRT–PCR (Meyer et al, 2010).
Immunoblotting
For immunoblotting, cells were lysed in RIPA lysis buffer (Boston BioProducts, Ashland, MA, USA) and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA). Proteins were resolved by 4–20% SDS–PAGE and transferred to a nitrocellulose membrane. Blots were blocked with 5% non-fat dry milk and probed with primary antibodies as follows: ADAM9 rabbit monoclonal (2099; Cell Signaling, Danvers, MA, USA) and Actin rabbit monoclonal (A2066; Sigma-Aldrich, St Louis, MO, USA). Horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (GE Healthcare, Pittsburgh, PA, USA) was used. Reactive bands were visualised using ECL plus western blotting detection reagents (GE Healthcare).
TMAs of bladder cancer samples
To perform ADAM9 expression analysis in UC, we used three different tissue microarrays (TMAs) from two different institutions (Columbia University, New York, NY, USA, and Centro Nacional de Investigaciones Oncologicas (CNIO), Madrid, Spain). The TMAs were built following IRB-approved protocols. From each specimen, triplicate tissue cores with diameters of 0.6 mm were represented. Tissues were obtained from patients who underwent radical cystectomy or transurethral resection of a bladder tumour, including stages Ta to T4. None of the patients in our series received chemotherapy or radiation therapy before obtaining the sample analysed. Follow-up clinical information of the 103 patients included in these TMAs and their clinicopathological characteristics are summarised in Table 1. Follow-up data were available in 88 of the analysed cases.
Table 1. Stage and final evolution of analysed patient cohort.
| Samples (%) | |
|---|---|
|
Stage (n=103) | |
| TaLG | 21 (20.4%) |
| TaHG | 8 (7.8%) |
| T1HG | 25 (24.2%) |
| T2 | 22 (21.4%) |
| T3 | 20 (19.4%) |
| T4 |
7 (6.8%) |
|
Final evolution (n=88a) | |
| AWOD | 32 (36.4%) |
| AWD | 22 (25.0%) |
| DOD | 24 (27.2%) |
| DOOC |
10 (11.4%) |
| Follow-up (range) |
24.1 months (1.0–76.1) |
| ADAM9-positive phenotype (%) | 65 (63.1%) |
Abbreviations: ADAM9=ADAM metallopeptidase domain 9; AWD=alive with disease; AWOD=alive without disease; DOD=dead of disease; DOOC=dead of other causes.
Outcome available in 88 out of 103 patients.
Immunohistochemical analysis
Immunohistochemical analyses were performed on the three above-mentioned TMAs following the standard avidin–biotin procedure as previously described (Jia et al, 2013). To confirm the sensitivity and specificity of antibodies (ADAM9 rabbit monoclonal (2099; Cell Signaling)), we ran positive and negative controls in parallel with the TMAs. The expression of ADAM9 was observed in the cytoplasm of tumour cells at different intensities. Immunoreactivity was scored by assessing the percentage of cells that displayed a positive immunostaining profile (from undetectable (0%) to homogeneous expression (100%)), as well as the intensity of expression (from negative (0) to high intensity (2)). Average values of the representative cores from each arrayed sample were obtained and only cases that displayed a high intensity of expression in >10% of the tumour cells were considered positive for statistical purposes. Thus, a positive phenotype corresponded to a score of >20 (e.g., expression intensity of 2 in 10% of tumour cells).
Cell invasion assays
The in vitro invasion assays were carried out using 8-μm pore BD BioCoat Matrigel Invasion Chambers (BD Biosciences, San Jose, CA, USA) in 24-well and 6-well tissue culture plates according to the manufacturer's protocol (Supplementary Figure S1). Cells were placed in serum-free media 24 h before invasion assay set-up. Cells were seeded in a 50 : 50 mixture of firefly (FL) to Renilla luciferase (RL) lines to a total amount of 5 × 104 cells for 24-well and 1.5 × 106 cells for 6-well tissue culture plates. Because the RL cells were not infected with miR-126, these cells invade as would the native cell line; therefore, RL cells can normalise differences between individual transwell chambers and serve as an internal control. Both FL and RL expression were directly proportional to the cell amount in a linear manner (Supplementary Figure S2). Cells suspended in serum-free media were added to each chamber and allowed to invade towards the underside of the chamber for 20 h at 37 °C. After incubation, remaining cells inside the chamber were collected along with cells that have invaded the membrane. Firefly and Renilla luciferase activities were measured using the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase signals were normalised to that of Renilla luciferase. All quantifications of miR-126-induced invasion were calculated relative to invasion of the FL cell line.
An alternate method of visualising invasion was established by seeding only FL cells in 24-well tissue culture plates of 8-μm pore BD BioCoat Matrigel Invasion Chambers (BD Biosciences) at 5 × 104 cells per well. Cells were placed in serum-free media 24 h before invasion assay set-up. Cells were allowed to invade for 20 h at 37 °C. After incubation, a Q-tip was used to remove remaining cells in the chamber. Each chamber was fixed in cold 4% paraformaldehyde and washed in PBS. The membrane at the bottom of each chamber was excised and mounted onto glass coverslips using ProLong-Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen).
Cell proliferation and apoptosis assays
Cells were seeded at 1 × 104 cells per ml in a 96-well plate. Cell viability was determined at 24, 48, 72, 96, and 144 h by using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) kit (Promega) according to the manufacturer's protocol. Absorbance was measured at 490 nm. Data are presented as the mean for octuplicate wells.
Apoptosis assays were performed using the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer's protocol. The 4',6-diamidino-2-phenylindole (DAPI) was used as a vital dye. The fluorescence-activated cell sorting (FACS) analysis was performed to determine the apoptotic distribution of cells in each sample (BD FACSAria Cell Sorter, BD Biosciences). Data are presented as the mean for triplicates performed in two independent experiments.
Statistical analysis
The associations between protein expression values and pathological stage were assessed using the χ2-test. The association of ADAM9 expression with disease-specific survival and overall survival was assessed using the log-rank test. Survival curves were plotted using standard Kaplan–Meier methodology. A two-sided P-value of <0.05 was considered statistically significant. Statistical analyses were carried out with SPSS v19.0 (IBM, Armonk, NY, USA).
Results
The miRNA and mRNA microarrays cluster tumours based on tumour stage
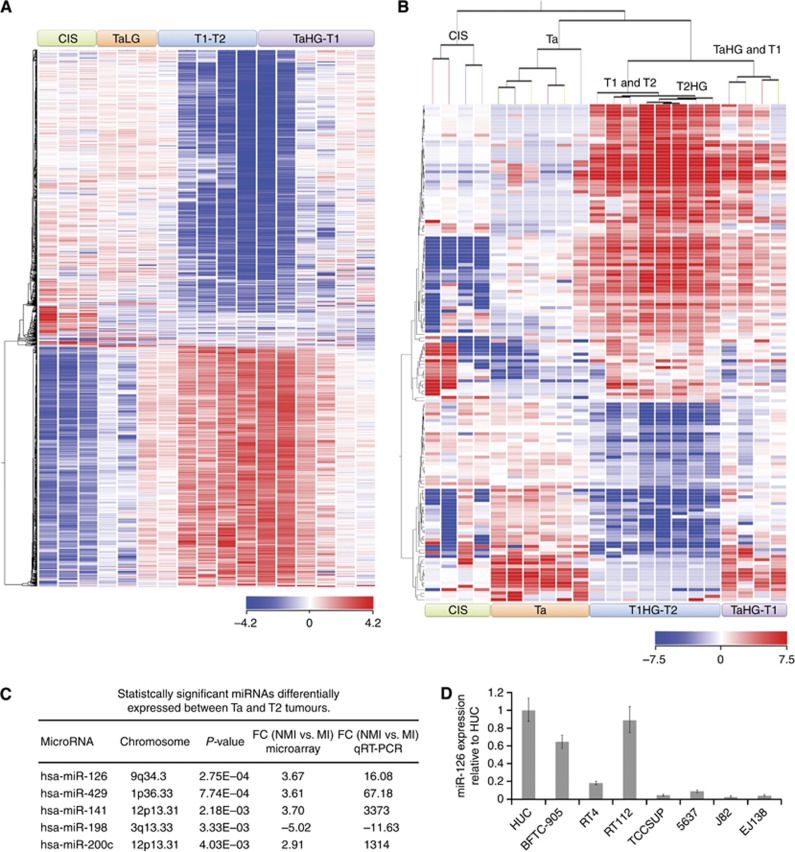
We first aimed to analyse the expression profiles of coding (mRNA) and noncoding (miRNAs) genes in a variety of bladder cancer tissues from LG papillary NMI tumours to HG MI cases (n=17 for mRNA analyses, n=22 for miRNA analyses). There were 1493 mRNAs (fold change ⩾10) differentially expressed across the 17 tumour samples that clustered tumours (unsupervised) depending on the pathological stage (Figure 1A). Similar to the mRNA profiles, 150 differentially expressed miRNAs (fold change ⩾2) also clustered tumours based on stage (Figure 1B). Interestingly, CIS tumours formed a separate branch from the rest instead of clustering with invasive tumours.
Figure 1.
The mRNA and miRNA expression profiles cluster bladder cancer by pathological stage. (A) Samples are in columns and mRNAs are in rows. Samples are clustered according to the expression signature of 1493 differentially expressed mRNAs (P⩽0.05, fold change (FC) ⩾10). (B) Samples are in columns and miRNAs are in rows. Samples are clustered according to the expression signature of 150 differentially expressed miRNAs (P⩽0.05, FC ⩾2). (C) List of statistically significant miRNAs differentially expressed between Ta (non-muscle invasive, NMI) and T2 (muscle-invasive, MI) bladder tumours and their respective FC according to the microarray. The qRT–PCR validation was performed on bladder cancer cell lines. (D) The expression of miR-126 in normal urothelium (HUC) and different-stage bladder cancer cell lines via qRT–PCR. Expression levels are calculated as log2 fold change.
In order to experimentally validate the role of miRNAs during bladder cancer progression, we transitioned our studies to tractable cell models. In all, 72 miRNAs were differentially expressed (fold change ⩾|2.5|) between Ta and T2 tumours (Supplementary Table S1); 31 miRNAs were upregulated in T2 tumours, whereas 41 miRNAs were upregulated in Ta tumours. We performed qRT–PCR studies to validate the expression of the 72 miRNAs in seven bladder cancer cell lines: HUC (normal primary urothelial cells), three NMI cell lines (BFTC-905, RT4, and RT112), and three MI cell lines (TCCSUP, J82, and EJ138). Of the 72 miRNAs, 5 had a fold change of >|10| between NMI and MI bladder cancer cells according to qRT–PCR (Figure 1C). These five miRNAs were: miR-200c, miR-141, miR-429, miR-126, and miR-198. With the exception of miR-198 that had the opposite trend, the four other miRNAs were expressed at higher levels in NMI cells than MI cells. As miR-126 was the most statistically significant differentially expressed miRNA (Figure 1D), we decided to further investigate the effect of miR-126 in bladder cancer cell function.
The miR-126 overexpression suppresses bladder cancer cell invasion
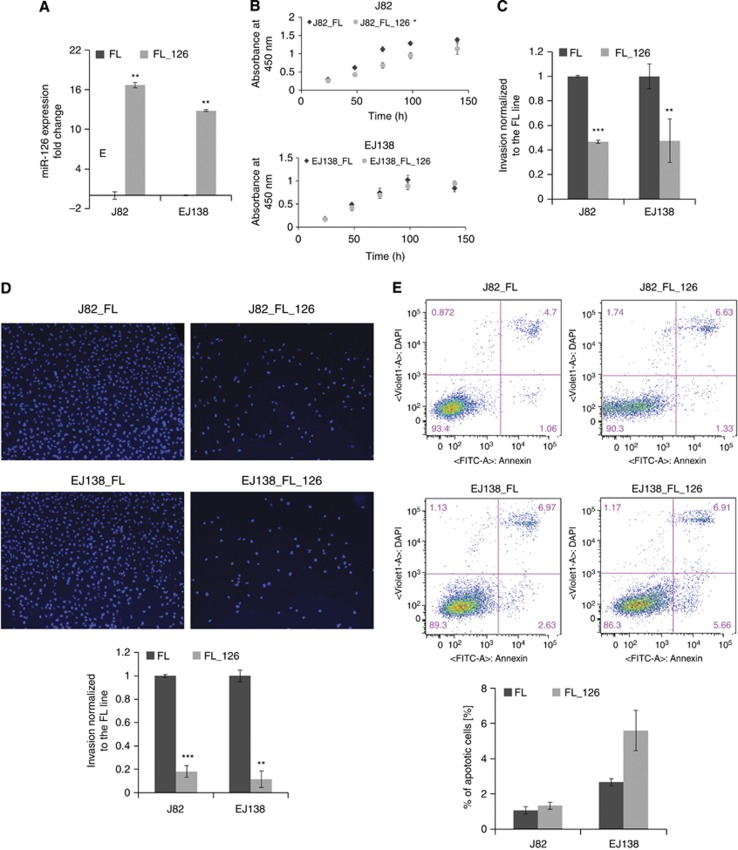
Quantitative RT–PCR confirmed that miR-126 was significantly overexpressed in both muscle-invasive cells lines: J82_FL_126 and EJ138_FL_126 (Figure 2A). To study the significance of miR-126 during invasion, we evaluated the ability of MI cells to migrate across a layer of matrigel deposited on a Boyden chamber when miR-126 was exogenously upregulated. Here we observed that miR-126 significantly inhibited invasion in both J82 and EJ138 by two-fold as demonstrated by luciferase assay (Figure 2C). Consistently, the same trend was observed after quantification of cells in the matrigel membrane after staining with DAPI (Figure 2D). In order to study whether this effect was mediated by changes in cell proliferation or apoptosis, we assessed cell viability at 24, 48, 72, 96, and 144 h through a colorimetric assay. Using Annexin-V-FITC and DAPI dye, FACS analysis for apoptosis was performed. Interestingly, we observed that miR-126 overexpression did indeed decrease cell proliferation in J82 cells, but these results were not consistent in EJ138, where no significant changes in cell proliferation were observed (Figure 2B). Finally, we observed that overexpression of miR-126 did not produce any effect on cellular apoptosis in any of the cell lines (Figure 2E), suggesting that the reduction in J82_FL_126 cell invasion may only in part be a consequence of inhibition of proliferation.
Figure 2.
Overexpression of miR-126 decreases invasion in J82 and EJ138 bladder cancer cell lines. (A) Validation of miR-126 overexpression in J82_FL and EJ138_FL cells stably infected with miR-126 by qRT–PCR. Effect of miR-126 overexpression in J82_FL and EJ138_FL on (B) cell proliferation (C and D) cell invasion and (E) cell apoptosis. Invasion of FL_126 cells is calibrated to the FL line (C). Invaded cells are stained with DAPI (D). Expression levels are calculated as log2 fold change. *P<0.05, **P<0.005, and ***P<0.0005 by Student's t-test.
The miR-126 directly targets ADAM9 in bladder cancer cells
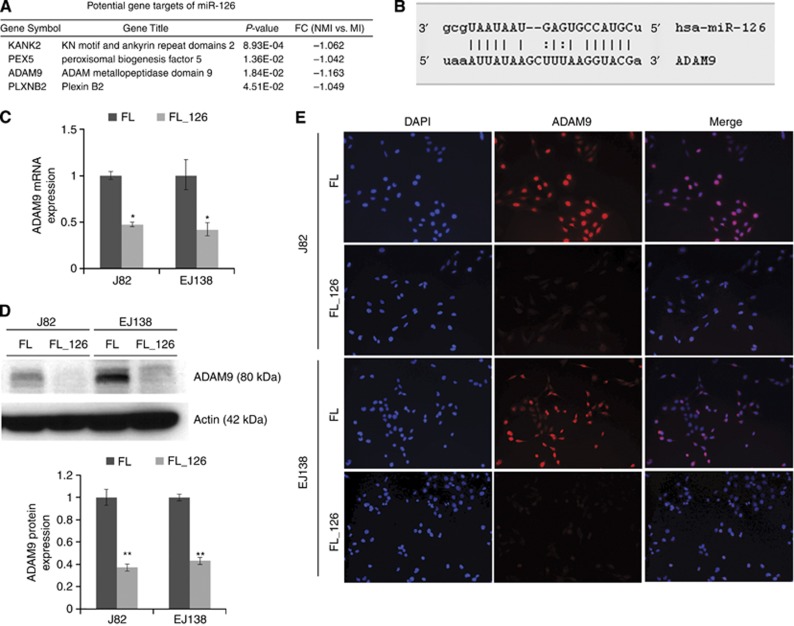
The in silico target prediction for miR-126 was performed using TargetScan (www.targetscan.org) on the list of 1493 differentially expressed mRNAs. The list of potential targets was modified to include only genes with expression patterns that inversely correlated with miR-126 expression. There were four unique gene targets predicted for miR-126 (Figure 3A). As predicted by TargetScan, miRanda, and miRWalk, miR-126 targets the 3′UTR of ADAM9 (Figure 3B). Of these four, ADAM9 displayed the largest magnitude of fold change and is of particular interest because of its implication in tumour progression (Duffy et al, 2009). To evaluate the impact of the miR-126 in the expression of ADAM9, we performed qRT–PCR, western blot, and immunofluorescence analyses in J82 and EJ138 cells stably infected with miR-126. Our data clearly showed a significant decrease in the expression of ADAM9 at both the mRNA level (Figure 3C) and protein level (Figure 3D and E). These results suggest that miR-126 targets ADAM9 directly by negatively regulating its expression.
Figure 3.
The miR-126 directly targets ADAM9 in bladder cancer cells. (A) Potential gene targets of miR-126 predicted by TargetScan using the mRNA microarray data. (B) Complementary miR-126 binding sequences in the ADAM9 3′UTR as predicted by miRanda, miRWalk, and TargetScan. (C) The ADAM9 mRNA expression in miR-126-infected cell lines. Results are expressed as mean±s.d. of three independent clones. (D) Representative western blot and densitometric analysis quantification of ADAM9 protein expression in five different clones of miR-126-infected cell lines, displayed as mean±s.d. (E) Representative immunofluorescence analyses of ADAM9 expression in miR-126-infected cell lines and their parental counterpart. *P<0.05 and **P<0.005 by Student's t-test.
The miR-126 regulates bladder cancer cell invasion by decreasing ADAM9 expression
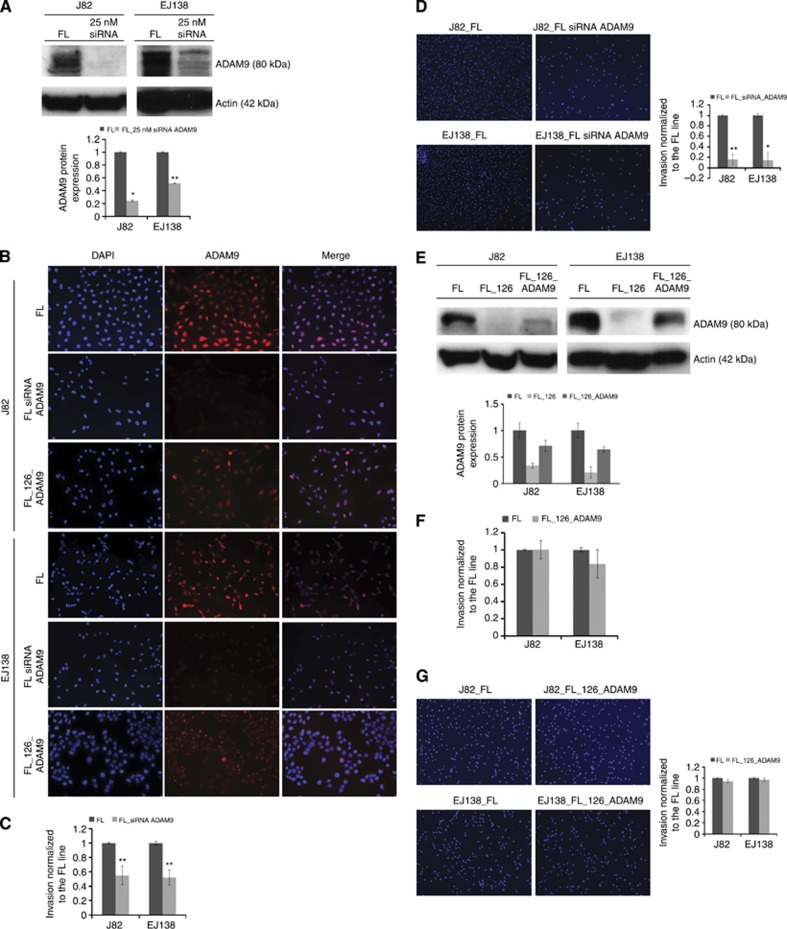
To study the contribution that regulation of ADAM9 by the miR-126 has on bladder tumour cell invasion, we first aimed to examine whether depletion of ADAM9 would affect cell invasion capabilities. Transient siRNA knockdown of ADAM9 was 80% in J82 and 50% in EJ138 (Figure 4A and B). As expected, transient knockdown of ADAM9 reduced cell invasion by two-fold (Figure 4C), and visualisation with DAPI staining confirmed the reduction in invasion at a level comparable to miR-126-induced inhibition (Figure 4D, compare with Figure 2D). These results demonstrate that overexpression of miR-126 and transient knockdown of ADAM9 suppressed cell invasion in a similar way. To further assess whether miR-126 prevents acquisition of invasive traits through suppression of ADAM9, we tested whether restoration of functional ADAM9 could reverse the effects of miR-126 and reestablish cell-invasive properties. To do so, a lentiviral ADAM9 construct, pLenti6.2-ADAM9, was introduced into J82_FL_126 and EJ138_FL_126 cells (Figure 4B and E). Noticeably, re-expression of ADAM9 in miR-126-overexpressing cells increased invasion back to parental cell levels in both the invasion luciferase assay (Figure 4F) and the invasion DAPI assay (Figure 4G). This suggests that miR-126 inhibits cell invasion by downregulation of ADAM9 expression.
Figure 4.
The knockdown of ADAM9 reduces cell invasion, and its overexpression in FL_126 cells reestablishes cell invasion potential. (A) Representative western blot and densitometric analysis quantification of ADAM9 protein expression at 48 h after transfection of 25 nM ADAM9 siRNA in five different clones of J82_FL and EJ138_FL. Expression levels are displayed as mean±s.d. (B) Representative immunofluorescence analysis of ADAM9 expression in FL cell lines transfected with ADAM9 siRNA, and FL_126 cell lines infected with pLenti6.2-ADAM9. (C and D) Effect of ADAM9 knockdown in J82_FL and EJ138_FL on cell invasion. (E) Representative western blot and densitometric analysis quantification of ADAM9 protein expression after infection with pLenti6.2-ADAM9 in five different clones of J82_FL_126 and EJ138_FL_126. Expression levels are displayed as mean± s.d. (F and G) Effect of ectopic expression of ADAM9 in J82_FL_126 and EJ138_FL_126 on cell invasion. Invasion of FL_126 and FL_126_ADAM9 cells is calibrated to the FL line (C and F). Invaded cells are stained with DAPI (D and G). *P<0.05 and **P<0.005 by Student's t-test.
ADAM9 is a marker of poor prognosis in bladder cancer
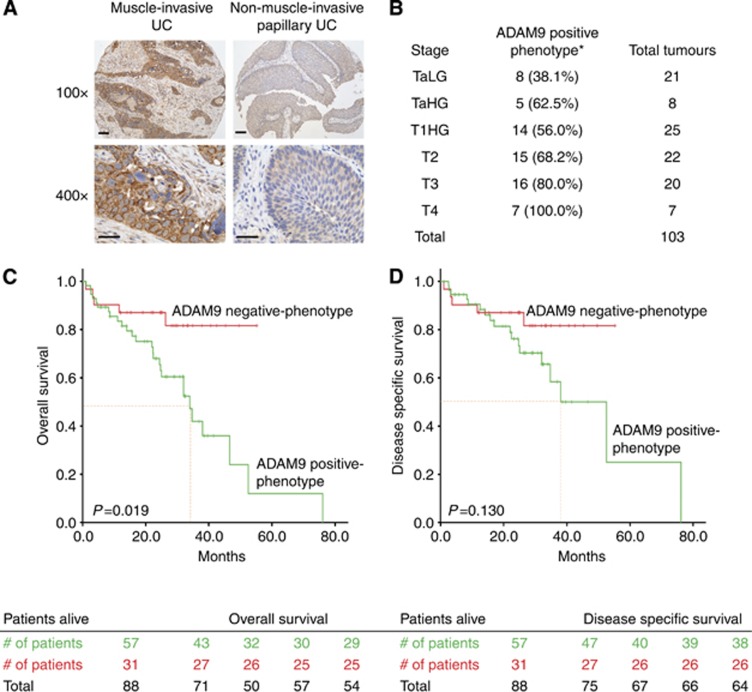
Finally, to examine the prevalence of ADAM9 in human bladder tumours and its significance in the clinical setting, we assayed the expression of ADAM9 in a panel of 103 UCs (Table 1). Notably, we observed that ADAM9 cytoplasmic expression in cancer cells significantly correlated with tumour stage (Figure 5A). Indeed, the percentage of tumours with an ADAM9-positive phenotype increased with higher stage, from 44.8% of Ta tumours to 100% of T4 tumours (P=0.022, Figure 5B). Furthermore, to determine whether ADAM9 has any prognostic significance, we performed Kaplan–Meier survival curve analyses in 88 of the patients with available follow-up. An ADAM9-positive phenotype (defined by an immunohistochemical score of >20) was associated with poor prognosis in patients, with both a shorter disease-specific survival (DSS) and overall survival (OS). More specifically, patients whose tumours displayed an ADAM9-positive phenotype had a significantly (P=0.019) lower median OS (34.0 months, 95% CI 24.9–43.2 months) compared with patients presenting with an ADAM9-negative phenotype tumour, for whom median OS was not reached at the current follow-up time (Figure 5C). Although not statistically significant, there is a clear trend that ADAM9 expression predicts a worse outcome for DSS, where an ADAM9-positive phenotype corresponded to a median DSS of 52.5 months (95% CI 33.1–71.9 months), whereas an ADAM9-negative phenotype did not reach median DSS (P<0.130) after a follow-up of 76.1 months (Figure 5D).
Figure 5.
Prognostic significance of ADAM9 in bladder UCs. (A) Immunohistochemical analyses of ADAM9 expression in representative cases of muscle-invasive UC (left panels) and non-muscle-invasive papillary UC (right panels). (B) Percentages of cases with an ADAM9-positive phenotype (defined by an immunohistochemical score of >20) distributed in the different tumour stages, *P =0.022 by χ2-distribution. (C and D) Kaplan–Meier analysis of ADAM9 expression for (C) overall survival (P =0.019) and (D) disease-specific survival (P <0.130). Median survival is indicated by dotted red line. For × 100, scale bar=200 μm; for × 400, scale bar=100 μm.
Discussion
To investigate miRNAs potentially involved in invasion, we profiled the expression of miRNAs and mRNAs in bladder tumours of various stages and grades. Both miRNA and mRNA expression levels clustered tumours based on stage. Of the 72 miRNAs differentially expressed between NMI (Ta) and MI (T2) tumours, 31 were upregulated and 41 were downregulated in MI tumours. From this list we identified two miRNAs, miR-198 and miR-126, previously not associated with bladder cancer.
In this study, we employed a novel method of assaying invasion (Supplementary Figure S1). The standard method to measure transwell matrigel invasion is by staining invasion chambers with crystal violet to visualise cells. However, it is a challenge to procure an accurate cell count. Because FL and RL levels increase linearly with cell amount, measurements of FL and RL expression could provide a more quantitative assessment of cell invasion. In our assay, errors that may arise because of physical variations between transwells and cell seeding irregularities are addressed by the use of RL-expressing cells that do not express miR-126 and a measure of cells that remain in the transwell (i.e., cells that did not invade).
Bladder cancer is a heterogeneous disease; once invasive and metastatic, it manifests an extremely poor prognosis and is frequently the primary cause of patient death (Howlader et al, 2009). Thus, the identification and characterisation of novel markers that can predict bladder cancer prognosis are critical to investigate new therapeutic approaches to improve patients' clinical outcome. Low miR-126 expression has been associated with an invasive phenotype in many tumours, such as ductal invasive tumours of the breast (Tavazoie et al, 2008), pancreatic adenocarcinoma (Hamada et al, 2012), and liver hepatocellular carcinoma (Chen et al, 2013). In addition, low serum levels of miR-126 in a three-miRNA plasma signature served as a significant prognostic biomarker for tumour progression in lung adenocarcinoma (Sanfiorenzo et al, 2013). Moreover, reduced miR-126 expression was a marker of tumour progression and nodal metastasis in oral squamous cell carcinoma (Sasahira et al, 2012). Our study confirms this trend in bladder cancer and shows for the first time the invasion suppressive activities of miR-126 in bladder cancer in vitro by direct downregulation of the metalloproteinase ADAM9. After analysing a cohort of 103 cases of bladder cancer tissue samples of different stages, we observed a significant decline in miR-126 in stage T2 (MI) tumours compared with stage Ta (NMI) tumours. The same trend was observed in bladder cancer cells, where miR-126 was highly expressed in the HUC line and NMI cell lines, but at barely discernible levels in MI cells. Four genes with increased expression in MI vs NMI bladder tumours were identified by TargetScan as potential miR-126 targets: KANK2, PEX5, ADAM9, and PLXNB2. Of these four, we decided to focus on ADAM9 as it is upregulated during tumour progression (Duffy et al, 2009). Elevated levels of ADAM9 has been correlated with breast cancer progression (O'Shea et al, 2003) and metastases to the liver (Le Pabic et al, 2003) and brain (Shintani et al, 2004) from colon and lung, respectively. A soluble form of ADAM9 secreted by hepatic stellate cells has been found to promote colon cancer invasion through interactions with α6β4 and α2β1 integrins (Mazzocca et al, 2005). Inhibition of ADAM9 in prostate cancer cell lines resulted in apoptotic cell death (Sung et al, 2006). Furthermore, loss of ADAM9 in a prostate cancer mouse model led to the development of well-differentiated prostate tumours as opposed to poorly differentiated tumours in control littermates (Peduto et al, 2005). To the best of our knowledge, functional studies of ADAM9 in the context of bladder cancer have not been reported.
Here we report that the introduction of miR-126 in invasive bladder cancer cells caused a significant reduction in ADAM9 at both the mRNA and protein levels. This is consistent with experiments in pancreatic cancer cells, where miR-126 was shown to directly target the 3′UTR of ADAM9 (Hamada et al, 2012). We were able to reproduce the same level of miR-126-induced suppression of cell invasion by a transient knockdown of ADAM9. Correspondingly, restoration of ADAM9 expression in miR-126-overexpressing cells reestablished cell-invasive properties. Although ADAM17 (tumour-necrosis factor α-converting enzyme) and ADAM10 (Kuzbanian) have been well studied, it is unclear what regulates ADAM9 expression (Blobel, 2005; Duffy et al, 2009). It is possible that reduced levels of miR-126 in invasive bladder cancer de-repress ADAM9 expression, leading to its accumulation. Although our data suggest that ADAM9 might be a prognostic marker with important clinical implications in bladder cancer patients, our cohort is too small to perform separate analyses in the different UC stages. Therefore, further studies in larger cohorts of both NMI and MI UCs should be performed to validate these findings.
In conclusion, we have provided evidence that miR-126 is downregulated in invasive bladder cancer. We also document for the first time overexpression of ADAM9 in invasive bladder cancer. Our study demonstrates that miR-126 exerts its tumour suppressive role by targeting ADAM9 to inhibit cell invasion. These results suggest that restoration of miR-126 may represent a potential therapeutic approach in the treatment of invasive bladder cancer.
Acknowledgments
We thank the members of the Dr Cordon-Cardo Laboratory for their support, and the staff at Mount Sinai Hospital and Herbert Irving Cancer Center, Columbia University. This work has partially been funded by the P01-CA-087497-12 NIH Grant (to CC-C and MC-M).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6 (1:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Cairns P, Evron E, Okami K, Halachmi N, Esteller M, Herman JG, Bose S, Wang SI, Parsons R, Sidransky D. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16 (24:3215–3218. doi: 10.1038/sj.onc.1201855. [DOI] [PubMed] [Google Scholar]
- Cairns P, Proctor AJ, Knowles MA. Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinoma. Oncogene. 1991;6 (12:2305–2309. [PubMed] [Google Scholar]
- Chapman EJ, Harnden P, Chambers P, Johnston C, Knowles MA. Comprehensive analysis of CDKN2A status in microdissected urothelial cell carcinoma reveals potential haploinsufficiency, a high frequency of homozygous co-deletion and associations with clinical phenotype. Clin Cancer Res. 2005;11 (16:5740–5747. doi: 10.1158/1078-0432.CCR-05-0411. [DOI] [PubMed] [Google Scholar]
- Chen H, Miao R, Fan J, Han Z, Wu J, Qiu G, Tang H, Peng Z. Decreased expression of miR-126 correlates with metastatic recurrence of hepatocellular carcinoma. Clin Exp Metastasis. 2013;30 (5:651–658. doi: 10.1007/s10585-013-9569-6. [DOI] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol. 2012;30 (4:434–443. doi: 10.1016/j.urolonc.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102 (5:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Cardo C. Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol Suppl. 2008;218:154–165. doi: 10.1080/03008880802291915. [DOI] [PubMed] [Google Scholar]
- Duffy MJ, McKiernan E, O'Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15 (4:1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6 (4:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27 (2–3:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Goebell PJ, Knowles MA. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010;28 (4:409–428. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10 (1:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5 (7:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7 (11:e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations) National Cancer Institute: Bethesda, MD; 2009. [Google Scholar]
- Jia AY, Castillo-Martin M, Domingo-Domenech J, Bonal DM, Sanchez-Carbayo M, Silva JM, Cordon-Cardo C. A common MicroRNA signature consisting of miR-133a, miR-139-3p, and miR-142-3p clusters bladder carcinoma in situ with normal umbrella cells. Am J Pathol. 2013;182 (4:1171–1179. doi: 10.1016/j.ajpath.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clement B, Theret N. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37 (5:1056–1066. doi: 10.1053/jhep.2003.50205. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang ZF, Reuter VE, Cordon-Cardo C. Chromosome 3 allelic losses and microsatellite alterations in transitional cell carcinoma of the urinary bladder. Am J Pathol. 1996;149 (1:229–235. [PMC free article] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS, Chang I, Yamamura S, Chiyomaru T, Fukuhara S, Tanaka Y, Deng G, Tabatabai ZL, Dahiya R. MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS One. 2012;7 (10:e46743. doi: 10.1371/journal.pone.0046743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzocca A, Coppari R, De Franco R, Cho JY, Libermann TA, Pinzani M, Toker A. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res. 2005;65 (11:4728–4738. doi: 10.1158/0008-5472.CAN-04-4449. [DOI] [PubMed] [Google Scholar]
- Meyer SU, Pfaffl MW, Ulbrich SE. Normalization strategies for microRNA profiling experiments: a ‘normal' way to a hidden layer of complexity. Biotechnol Lett. 2010;32 (12:1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S, Akao Y. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328 (2:353–361. doi: 10.1016/j.canlet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- O'Shea C, McKie N, Buggy Y, Duggan C, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Expression of ADAM-9 mRNA and protein in human breast cancer. Int J Cancer. 2003;105 (6:754–761. doi: 10.1002/ijc.11161. [DOI] [PubMed] [Google Scholar]
- Orlow I, LaRue H, Osman I, Lacombe L, Moore L, Rabbani F, Meyer F, Fradet Y, Cordon-Cardo C. Deletions of the INK4A gene in superficial bladder tumors. Association with recurrence. Am J Pathol. 1999;155 (1:105–113. doi: 10.1016/S0002-9440(10)65105-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduto L, Reuter VE, Shaffer DR, Scher HI, Blobel CP. Critical function for ADAM9 in mouse prostate cancer. Cancer Res. 2005;65 (20:9312–9319. doi: 10.1158/0008-5472.CAN-05-1063. [DOI] [PubMed] [Google Scholar]
- Presti JC, Jr., Reuter VE, Galan T, Fair WR, Cordon-Cardo C. Molecular genetic alterations in superficial and locally advanced human bladder cancer. Cancer Res. 1991;51 (19:5405–5409. [PubMed] [Google Scholar]
- Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8 (1:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, Kirita T, Kuniyasu H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107 (4:700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115 (2:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shintani Y, Higashiyama S, Ohta M, Hirabayashi H, Yamamoto S, Yoshimasu T, Matsuda H, Matsuura N. Overexpression of ADAM9 in non-small cell lung cancer correlates with brain metastasis. Cancer Res. 2004;64 (12:4190–4196. doi: 10.1158/0008-5472.CAN-03-3235. [DOI] [PubMed] [Google Scholar]
- Simoneau M, LaRue H, Aboulkassim TO, Meyer F, Moore L, Fradet Y. Chromosome 9 deletions and recurrence of superficial bladder cancer: identification of four regions of prognostic interest. Oncogene. 2000;19 (54:6317–6323. doi: 10.1038/sj.onc.1204022. [DOI] [PubMed] [Google Scholar]
- Sung SY, Kubo H, Shigemura K, Arnold RS, Logani S, Wang R, Konaka H, Nakagawa M, Mousses S, Amin M, Anderson C, Johnstone P, Petros JA, Marshall FF, Zhau HE, Chung LW. Oxidative stress induces ADAM9 protein expression in human prostate cancer cells. Cancer Res. 2006;66 (19:9519–9526. doi: 10.1158/0008-5472.CAN-05-4375. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451 (7175:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Yoshino H, Enokida H, Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Fuse M, Seki N, Nakagawa M. Novel molecular targets regulated by tumor suppressors microRNA-1 and microRNA-133a in bladder cancer. Int J Oncol. 2012;40 (6:1821–1830. doi: 10.3892/ijo.2012.1391. [DOI] [PubMed] [Google Scholar]
- Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104 (5:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino H, Enokida H, Chiyomaru T, Tatarano S, Hidaka H, Yamasaki T, Gotannda T, Tachiwada T, Nohata N, Yamane T, Seki N, Nakagawa M. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem Biophys Res Commun. 2012;417 (1:588–593. doi: 10.1016/j.bbrc.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171 (3:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.