Abstract
Background:
The response of ovarian cancer patients to carboplatin and paclitaxel is variable, necessitating identification of biomarkers that can reliably predict drug sensitivity and resistance. In this study, we sought to identify dynamically controlled genes and pathways associated with drug response and its time dependence.
Methods:
Gene expression was assessed for 14 days post-treatment with carboplatin or carboplatin–paclitaxel in xenografts from two ovarian cancer models: platinum-sensitive serous adenocarcinoma-derived OV1002 and a mixed clear cell/endometrioid carcinoma-derived HOX424 with reduced sensitivity to platinum.
Results:
Tumour volume reduction was observed in both xenografts, but more dominantly in OV1002. Upregulated genes in OV1002 were involved in DNA repair, cell cycle and apoptosis, whereas downregulated genes were involved in oxygen-consuming metabolic processes and apoptosis control. Carboplatin–paclitaxel triggered a more comprehensive response than carboplatin only in both xenografts. In HOX424, apoptosis and cell cycle were upregulated, whereas Wnt signalling was inhibited. Genes downregulated after day 7 from both xenografts were predictive of overall survival. Overrepresented pathways were also predictive of outcome.
Conclusions:
Late expressed genes are prognostic in ovarian tumours in a dynamic manner. This longitudinal gene expression study further elucidates chemotherapy response in two models, stressing the importance of delayed biomarker detection and guiding optimal timing of biopsies.
Keywords: ovarian cancer, carboplatin, paclitaxel, xenograft, prognosis
Current standard treatment for ovarian cancer consists of carboplatin with or without paclitaxel following surgery (Cannistra, 2004). Although the majority of patients initially respond to therapy, most eventually relapse whereas others are resistant to treatment at the outset (International Collaborative Ovarian Neoplasm Group, 2002). Thus, it is important to understand and define which patients are likely to be sensitive to treatment and who have resistant disease. In this study, we analysed gene expression changes after treatment in two ovarian cancer models with distinct chemotherapy response profiles over a period of 14 days. The OV1002 model is derived from high-grade serous adenocarcinoma and is platinum sensitive, whereas HOX424 is of clear cell/endometrioid carcinoma origin and has reduced responsiveness to platinum.
Our emphasis was on pathways rather than individual genes. Previous studies have evaluated the association of gene expression directly with outcome in clinical specimens (Helleman et al, 2006; Tothill et al, 2008; Crijns et al, 2009; Konstantinopoulos et al, 2011) and have predominantly sought to identify candidate genes that link with outcome. Other studies have evaluated the temporal impact of platinum on sensitive and resistant ovarian cancer cells in vitro with a view to identifying resistance mechanisms (Li et al, 2007); however, the cisplatin-treated isogenic cell lines were monitored for a limited period of 24 h. Several genes have been found differentially expressed in ovarian cancer cell lines treated with platinum drugs or taxanes (Sherman-Baust et al, 2011). Helleman et al (2010) have used a panel of nine gene sets associated with platinum resistance to perform pathway analysis. Knowledge of the molecular mechanisms involved in drug response is essential to improve therapy and possibly reverse drug resistance (Shahzad et al, 2009).
The use of human ovarian cancer xenografts allows the analysis of drug treatment on temporal changes of gene expression in cancer tissue after treatment, a task not easily undertaken in patients. Comparison of models that are either chemosensitive or chemoresistant should help identify gene expression changes that are associated with response, and some of these genes are likely to be helpful to predict outcome after treatment. In this study, we used models with distinct response patterns to carboplatin and assessed their gene expression profiles after treatment with drugs. We identified time-related gene clusters and significant pathways with predictive value within independent clinical data sets.
Materials and methods
Xenografts
The OV1002 and HOX424 models were established at the Institute of Genetics and Molecular Medicine, University of Edinburgh, as previously described (Faratian et al, 2011). Adult female nu/nu mice were implanted subcutaneously with ovarian tumour fragments in the flanks and allowed to grow to 4–6 mm in diameter (over a period of ∼2 months). Animals were at least 8 weeks old at the time of experimentation and were maintained in negative pressure isolators (La Calhene, Cambridge, UK). The mice were treated with carboplatin (50 mg kg−1), carboplatin (50 mg kg−1)+paclitaxel (10 mg kg−1) or were untreated. Drugs were given as a single injection via the intraperitoneal route in saline on day 0. Tumour size was measured twice weekly using calipers and the volume calculated according to the formula π/6 × length × width2. Relative tumour volumes (RTV) were then calculated for each individual tumour by dividing the tumour volume on day t (Vt) by the tumour volume on day 0 (V0). Treated xenografts were harvested on days 1, 2, 4, 7 and 14, and untreated controls on days 0, 1, 7 and 14. A total of 101 xenografts were used, with 3–4 biological replicates per time point (except carboplatin–paclitaxel-treated HOX424 on day 2 that had 2 replicates; Supplementary Table 1, column D). Agreement between their expression levels was good as measured by the correlation coefficients (r) among replicates of each condition (mean r 95% confidence interval 0.987–0.990, Supplementary Table 1, column E).
Gene expression profiling and analysis
Total RNA was prepared from 10 to 50 mg of frozen tissue on the days of treatment, preincubated with RNAlater-ICE (Ambion, Austin, TX, USA) using the miRNeasy Mini kit (Qiagen, Hilden, Germany) and TissueRuptor (Qiagen) following the manufacturers' instructions. The RNA quality was checked by the RNA 6000 Nanoassay on the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). All samples were divided into two identical aliquots for independent labelling and hybridisation. Total RNA for each sample (0.5 mg) was amplified and biotinylated using the Illumina TotalPrep RNA Amplification Kit (Ambion) according to manufacturers' standard procedure. This consists of reverse transcription with an oligo(dT) primer bearing a T7 promoter using a reverse transcriptase (RT). The quality and quantity of cRNA in the samples was checked with an Agilent Bioanalyzer 2100 (Agilent Technologies), samples were diluted to 150 ng ml−1 and hybridised to Illumina HT-12 BeadChips in duplicate (Illumina, San Diego, CA, USA). This Illumina platform was previously validated by qPCR in a breast cancer-derived xenograft study (Sims et al., 2012).
Analysis of microarray data
Illumina chips contained 47 323 probes. Expression sets were processed using Bioconductor's lumi package (Du et al, 2008). The data set has been deposited to Gene Expression Omnibus (GEO) with accession number GSE49577. Bioconductor package limma (Smyth, 2005) was used for differential expression calculations. Treated samples from days 1, 2, 4, 7 and 14 post-treatment were contrasted to pooled controls in each cell line. Significant genes had FDR adjusted P-values of <0.05. Differentially expressed genes were classified as early (days 1–4) and/or late (days 7 and 14), and as either transient (expressed significantly in one time point) or sustained (expressed significantly in at least two time points). Means of log-fold change values for each time point of differentially expressed genes were hierarchically clustered using Cluster 3.0 (Eisen et al, 1998) to identify genes with similar temporal profiles. Heatmaps were visualised using Bioconductor. Overrepresented KEGG (Kyoto encyclopedia of genes and genomes) pathways (Kanehisa et al, 2012) were identified using DAVID (Database for Annotation, Visualisation and Integrated Discovery) (Huang et al, 2009).
Hierarchical clustering and Kaplan–Meier survival analysis
Two clinical ovarian cancer gene expression data sets from GEO, GSE9891 (Tothill et al, 2008) and GSE3149 (Bild et al, 2006), were used to assess the prognostic capacity of dynamically changing genes in response to treatment (Supplementary Table 2). Clinical data, such as survival time and censoring information, were available for both data sets. Our hypothesis was that, if gene sets play crucial roles in tumour progression, then patients with different gene expression patterns of such gene sets may also have different overall survival (OS) and prognosis. For each cluster of differentially expressed genes from both treatments and models in our study, the corresponding genes were selected in the two clinical ovarian cancer data sets. Unsupervised hierarchical clustering was applied to expression values in the clinical data sets in order to separate tumour samples based on expression profiles, as described previously (Liu et al, 2008). The hierarchical clustering was divided at the highest level to produce two groups of tumour samples having the most broadly different gene expression profiles. Kaplan–Meier plots were graphed to visualise the difference in OS between these groups and, thus, the capacity of the gene set to discriminate favourable from poor prognosis patients. Log rank P-values were calculated to assess the statistical significance of the difference in survival probabilities between the two patient groups. Using the same methodology, sets of differentially expressed genes with distinct temporal expression profiles were tested, as well as genes in selected overrepresented pathways. To mitigate multiple testing issues, only significant pathways known to be involved in ovarian cancer were tested. Except where noted, all genes in an overrepresented pathway that were extant in the clinical data set (whether they had been differentially expressed in our data set or not) were used as the gene set.
Results
Treatment with carboplatin or carboplatin-paclitaxel causes a reduction in tumour growth of ovarian cancer model xenografts
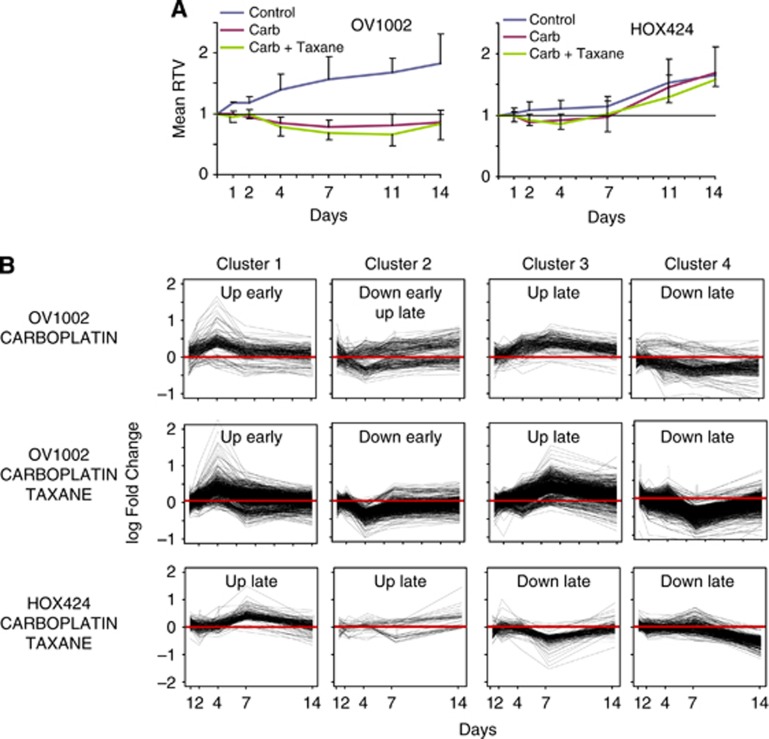
The effect of carboplatin and carboplatin–paclitaxel treatment on tumour volume growth was assessed 1, 2, 4, 7, 11 and 14 days post-treatment. Treated xenografts demonstrated an RTV reduction compared with control samples (Figure 1A). In OV1002 xenografts, both treatments resulted in a clear growth inhibition. Differences were significant at all time points (P<0.01; Student's t-test) with values diverging from day 1. The effect peaked at day 7 for carboplatin or day 11 for carboplatin–paclitaxel, where RTV dropped well below one in treated samples. By day 14, the growth inhibition effect showed signs of reversal. Reduction in RTV was steady and gradual in both groups, although carboplatin–paclitaxel had a larger effect than carboplatin at day 7 (P=0.04). Untreated HOX424 samples grew more slowly (Figure 1A). A small growth reduction effect due to both treatment types compared with control was observed, but this was significant only at days 2–7 (carboplatin, P<.03) or day 4 (carboplatin–paclitaxel, P<0.001).
Figure 1.
Relative tumour volume (RTV) growth with respect to dynamic gene expression patterns. (A) Post-treatment tumour volume relative to day of treatment (day 0) for the OV1002 and HOX424 xenografts. (B) Mean expression change post-treatment of genes with similar temporal profiles in carboplatin- and carboplatin–paclitaxel-treated OV1002 and in carboplatin–paclitaxel-treated HOX424 xenografts.
Carboplatin dynamically modulates DNA repair, cell cycle and apoptosis control genes in OV1002 xenografts, but has limited effect in gene expression of HOX424
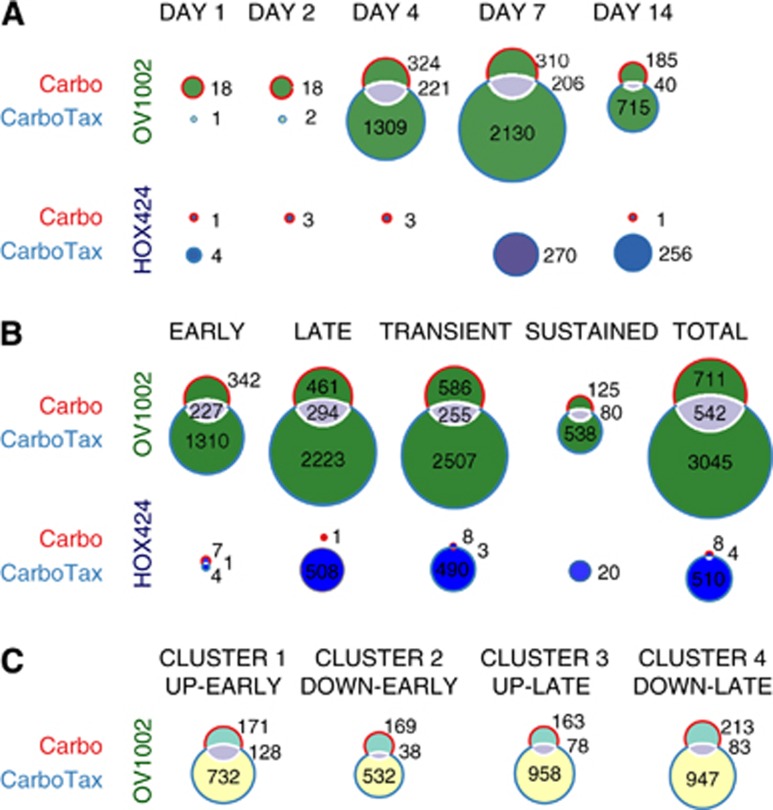
In OV1002 xenografts, a total of 711 genes were differentially expressed mainly late and transiently after carboplatin treatment (Figures 2A and B). A complete listing is shown in Supplementary Table 3, and genes with the largest expression change compared with control are shown in Supplementary Table 4. Several pathways involved in cell cycle control were overrepresented within the pool of differentially expressed genes from all time points, such as cell cycle, DNA replication and p53 signalling (Table 1A, complete pathway statistics in Supplementary Table 5). Hierarchical clustering separated the genes into four main clusters (Figure 1B).
Figure 2.
Differential expression analysis of treated OV1002 and HOX424 xenografts. Venn diagrams of differentially expressed gene counts in each time point (A) and according to their temporal characteristics (B) in carboplatin- and carboplatin–taxane-treated OV1002 and HOX424 cells. Gene expression was analysed in 5 time points post-treatment (days 1, 2, 4, 7 and 14). Early (late) genes were differentially expressed in days 1, 2 and 4 (days 7 and 14). Transient (sustained) were differentially expressed in a single time point (more than one time point). (C) Venn diagrams of gene overlaps in each corresponding cluster with similar temporal expression profiles in carboplatin- and carboplatin–paclitaxel-treated OV1002 cells.
Table 1. KEGG pathways overrepresented in dynamically changing genes upon treatment with carboplatin or carboplatin taxane in OV1002 and HOX424 xenografts.
| A | ||||
|---|---|---|---|---|
|
OV1002 carboplatin | ||||
|
All differentially expressed genes |
Cluster 1: up early |
Cluster 2: down/up early |
Cluster 3: up late |
Cluster 4: down late |
| hsa04110: cell cycle hsa00970: aminoacyl-tRNA biosynthesis hsa00240: pyrimidine metabolism hsa04115: p53 signalling pathway hsa04914: progesterone-mediated oocyte maturation hsa04114: oocyte meiosis hsa00230: purine metabolism hsa03030: replication hsa05130: pathogenic Escherichia coli infection | hsa00970: aminoacyl-tRNA biosynthesis hsa04110: cell cycle hsa00670: one carbon pool by folate hsa00240: pyrimidine metabolism hsa03030: DNA replication | hsa04612: antigen processing and presentation hsa00240: pyrimidine metabolism hsa04623: cytosolic DNA-sensing pathway | hsa04110: cell cycle hsa04114: oocyte meiosis hsa04914: progesterone-mediated oocyte maturation hsa04115: p53 signalling pathway hsa04540: Gap junction | hsa00051: fructose and mannose metabolism hsa00240: pyrimidine metabolism |
| B | ||||
|---|---|---|---|---|
|
OV1002 carboplatin–paclitaxel | ||||
|
All differentially expressed genes |
Cluster 1: up early |
Cluster 2: down early |
Cluster 3: up late |
Cluster 4: down late |
| hsa03010: ribosome hsa04110: cell cycle hsa03030: DNA replication hsa03420: nucleotide excision repair hsa04115: p53 signalling pathway hsa00970: aminoacyl-tRNA biosynthesis hsa00100: steroid biosynthesis hsa00240: pyrimidine metabolism hsa03410: base excision repair hsa03430: mismatch repair hsa00250: alanine, aspartate and glutamate metabolism hsa03050: proteasome hsa05130: pathogenic Escherichia coli infection hsa00230:Purine metabolism hsa00520: amino sugar and nucleotide sugar metabolism hsa03440: homologous recombination hsa00670: one carbon pool by folate hsa00270: cysteine and methionine metabolism hsa00051: fructose and mannose metabolism hsa00052: galactose metabolism hsa04612: antigen processing and presentation hsa00480: glutathione metabolism | hsa03030: DNA replication hsa04110: cell cycle hsa00970: aminoacyl-tRNA biosynthesis hsa00240: pyrimidine metabolism hsa03410: base excision repair hsa03420: nucleotide excision repair hsa03430: mismatch repair hsa00230: purine metabolism hsa00670: one carbon pool by folate hsa04115: p53 signalling pathway hsa03440: homologous recombination hsa00630: glyoxylate and dicarboxylate metabolism hsa00250: alanine, aspartate and glutamate metabolism hsa04350: TGF-β signalling pathway | hsa00020: citrate cycle (TCA cycle) hsa00190: oxidative phosphorylation hsa04142: lysosome hsa00280: valine, leucine and isoleucine degradation hsa05322: systemic lupus erythematosus hsa00100: steroid biosynthesis hsa04360: axon guidance | hsa03050: proteasome hsa04110: cell cycle hsa04114: oocyte meiosis hsa05130: pathogenic Escherichia coli infection hsa05416: viral myocarditis hsa05330: allograft rejection hsa04612: antigen processing and presentation hsa04914: progesterone-mediated oocyte maturation hsa04115: p53 signalling pathway hsa05332: graft-versus-host disease hsa04514: cell adhesion molecules (CAMs) hsa05320: autoimmune thyroid disease hsa04144: endocytosis hsa04940: type I diabetes mellitus hsa04810: regulation of actin cytoskeleton hsa04672: intestinal immune network for IgA production | hsa03010: ribosome hsa03420: nucleotide excision repair hsa00051: fructose and mannose metabolism hsa00280: valine, leucine and isoleucine degradation hsa00520: amino sugar and nucleotide sugar metabolism hsa00561: glycerolipid metabolism hsa04722: neurotrophin signalling pathway hsa00650: butanoate metabolism hsa03040: spliceosome hsa00410: β-alanine metabolism |
| C | ||||
|---|---|---|---|---|
|
HOX424 carboplatin–paclitaxel | ||||
|
All differentially expressed genes |
Cluster 1: up late (day 7) |
Cluster 2: up late (day 14) |
Cluster 3: down late (day 7) |
Cluster 4: down late (day 14) |
| hsa03010: ribosome hsa05130: pathogenic Escherichia coli infection hsa05210: colorectal cancer hsa03040: spliceosome hsa04210: apoptosis hsa04530: tight junction hsa04510: focal adhesion hsa04662: B-cell receptor signalling pathway hsa00450: selenoamino acid metabolism hsa00510: N-glycan biosynthesis hsa04310: Wnt signalling pathway hsa05416: viral myocarditis | hsa03010: ribosome hsa00450: selenoamino acid metabolism hsa04210: apoptosis hsa00460: cyanoamino acid metabolism hsa00430: taurine and hypotaurine metabolism hsa00590: arachidonic acid metabolism | — | hsa03010: ribosome hsa05130: pathogenic Escherichia coli infection hsa04510: focal adhesion hsa04310: Wnt signalling pathway hsa03050: proteasome | hsa03010: ribosome hsa03040: spliceosome hsa05210: colorectal cancer hsa05130: pathogenic Escherichia coli infection hsa05211: renal cell carcinoma hsa04670: leukocyte transendothelial migration |
Abbreviations: IgA=immunoglobulin A; KEGG=Kyoto encyclopedia of genes and genomes; TCA=tricarboxylic acid; TGFβ=transforming growth factor-β; tRNA=transfer RNA.
Pathways are shown for the set of all differentially expressed genes (left column) and for gene sets with similar temporal profiles. (A) Carboplatin-treated OV1002, (B) carboplatin–paclitaxel-treated OV1002 and (C) carboplatin–paclitaxel-treated HOX424.
Genes in cluster 1 were mainly upregulated early in treated samples. Many were implicated in cell cycle regulation and DNA repair. Cluster 2 contained genes implicated in apoptosis that were either upregulated or downregulated early and later upregulated. Genes in cluster 3 were mostly upregulated late in treated samples. It contained several genes implicated in the cell cycle and p53 signalling pathways, as well as HIF1α and its target MET. Cluster 4 genes were mainly downregulated with late expression. This group contained several HIF1α targets and genes with anti-apoptotic activity (Supplementary Table 6). The results suggest that in OV1002, cell cycle control, DNA repair and pro-apoptotic genes dominated the early response to carboplatin. From day 7, more cell cycle regulators and p53 signalling genes were detected.
In HOX424 xenografts, very few differentially expressed genes were observed upon treatment with carboplatin. Significant genes are shown in Supplementary Table 3 (complete list) and Supplementary Table 7 (largest changes).
Carboplatin–paclitaxel treatment has distinct time-related effects in each ovarian cancer model
In OV1002 xenografts, a total of 3045 genes were differentially expressed upon carboplatin–paclitaxel treatment. The majority of differential genes expressed late and transiently (Figures 2A and B). Genes with the largest expression change upon treatment are shown in Supplementary Table 8, and a complete list of differentially expressed genes is shown in Supplementary Table 3. Cell proliferation and DNA repair-related pathways such as cell cycle, DNA replication, p53 signalling, nucleotide excision repair, base excision repair, homologous recombination and mismatch repair were overrepresented within differentially expressed genes (Table 1B, complete pathway statistics in Supplementary Table 9). Glutathione metabolism was also significant. Hierarchical clustering separated the genes into four main clusters (Figure 1B).
Genes in cluster 1 were predominantly upregulated early and were implicated in cell cycle control, DNA repair, apoptosis, TGFβ and Wnt signalling pathways. Cluster 2 genes were generally downregulated early in treated samples. This cluster contained genes implicated in oxidative phosphorylation and citrate cycle, several HIF1α targets and STAT3, RBL2 and RAB2B. The third cluster contained genes that were mainly upregulated late in treated samples. It included several genes implicated in cell cycle control and p53 signalling and in immune system-related pathways. Other significant genes were members of MAPK and ERBB signalling pathways and PARP9 and PARP10. Genes in cluster 4 were mostly downregulated late and included HIF1α targets, some of which were implicated in mTOR signalling. Several heat shock proteins, including several hsp70 species, and Wnt signalling pathway targets were also detected. Other significant genes included DYRK1B, ESR1 and H3F3B (Supplementary Table 6).
In HOX424, there were 510 significant genes with exclusively late/transient expression patterns that were clustered in four groups (Figure 1B and Supplementary Table 3). Genes with the largest expression fold change are shown in Supplementary Table 10. Overrepresented KEGG pathways included apoptosis, Wnt signalling and ribosome pathways (Table 1C and Supplementary Table 9).
Upregulated clusters 1 and 2 were implicated in apoptosis, in cell cycle regulation and other genes with known cancer involvement. Downregulated genes in clusters 3 and 4 were involved in Wnt and MAPK signalling pathways. Other genes in this cluster included HIF1α, MSH2 and PMS1 (Supplementary Table 6).
These results suggest distinct modes of response to carboplatin–paclitaxel treatment in each ovarian cancer model. In OV1002, the early response was dominated by cell cycle control, DNA repair, pro-apoptotic genes, TGFβ and Wnt signalling pathways, whereas aerobic metabolic pathways were downregulated. From day 7, more cell cycle regulators, p53 signalling genes, immune system related, MAPK and ERBB pathway genes were detected. In HOX424, the cell cycle and DNA repair response was limited and late. Instead, several pro-apoptotic genes were induced, whereas Wnt and MAPK signalling pathways were downregulated.
The combined treatment instigated a more extensive response compared with carboplatin alone, judging from the larger count of differentially expressed genes in both xenografts. In OV1002, there was significant overlap of responsive genes between treatments, with 542 genes in common. There was significant overlap among corresponding upregulated groups, especially among the early expressed (Figure 2). Most of the differentially expressed genes in carboplatin-treated xenografts were significant in the combined treatment also, but EME1 (upregulated early), MYC (upregulated late), SIPA1, SIAH1, IGFBP3 and ERCC1 (downregulated) were specific for carboplatin. In carboplatin–paclitaxel-treated OV1002, there were more significant genes within pathways observed in carboplatin-treated cells, but also genes of new pathways altogether (Figure 3).
Figure 3.
Heatmaps of gene sets with significant increased (red) or decreased (green) expression in response to treatment relative to control samples.(A) Carboplatin-treated OV1002, (B) carboplatin–paclitaxel-treated OV1002 and (C) carboplatin–paclitaxel-treated HOX424 xenografts. Clusters are annotated with significant overrepresented functions.
Dynamic gene expression analysis reveals variation between the two ovarian models in their response to chemotherapeutic agents
When treated xenografts were compared with control samples, more differentially expressed genes were found in OV1002 than in HOX424 after either treatment. There were 206 genes in common between the two xenografts in the combination treatment, of which 143 had the same regulation. For instance, PARP2 and FANCD2 were positively whereas CASP6, MAPK9 and BNIP3L were negatively regulated in both cell lines. A total of 63 genes had opposite regulation (up/down compared with control). Some of these were upregulated in OV1002, such as ETS1, RAPH1, NEK2, MAD2L1, PSMD1, or in HOX424, for instance CAT, HSPA12A, FASTK and WDR6. Pro-apoptotic TNFRSF1A of the MAPK pathway and Wnt pathway modulator SENP2 were upregulated in OV1002, but downregulated in HOX424. Xenografts differed in the time patterns of gene expression, with the chemoresistant HOX424 responding 7 days post-treatment, whereas platinum-sensitive OV1002 demonstrated a more balanced response across time points.
Dynamically changing genes and overrepresented pathways are prognostic of outcome in independent data sets
The relationship to clinical outcome in the clinical ovarian cancer data sets (Supplementary Table 2) was assessed for sets of differentially expressed genes, both for all genes identified from a condition and genes clusters. Gene sets from downregulated late clusters in carboplatin–paclitaxel-treated xenografts were found prognostic of OS in both data sets (Table 2). Other gene sets, such as upregulated late and downregulated early clusters, were successful at differentiating prognosis in one data set.
Table 2. Prognostic capacity of gene sets with similar temporal expression profiles in three ovarian cancer data sets (log rank P-values).
| Data set | Bild et al (2006) | Tothill et al (2008) |
|---|---|---|
| Data set ID |
GSE3149 |
GSE9891 |
| Microarray platform |
Affymetrix U133A |
Affymetrix U133 Plus 2 |
| Samples |
133 |
285 |
| Platinum/taxane |
|
243/195 |
| Tumour grade (1/2/3/NA) | 3/4/125/1 | 19/97/164/5 |
|
End point |
|
OS |
OS |
PFS |
|---|---|---|---|---|
| Gene set | Gene set size | |||
| All OV1002 carboplatin sets |
711 |
0.0473 |
0.00292 |
0.000642 |
| All OV1002 carboplatin/taxane sets |
3045 |
0.128 |
0.00101 |
2.88E−05 |
| All HOX424 carboplatin/taxane sets |
510 |
0.00931 |
0.000136 |
0.000268 |
| OV1002 carboplatin set 1 up early |
171 |
0.421 |
0.315 |
0.87 |
| OV1002 carboplatin set 2 down/up early |
169 |
0.0406 |
0.062 |
0.00216 |
| OV1002 carboplatin set 3 up late |
163 |
0.577 |
0.112 |
0.059 |
| OV1002 carboplatin set 4 down late |
213 |
0.789 |
0.589 |
0.0935 |
| OV1002 carboplatin/taxane set 1 up early |
733 |
0.0936 |
0.152 |
0.463 |
| OV1002 carboplatin/taxane set 2 down early |
533 |
0.0257 |
0.809 |
0.0304 |
| OV1002 carboplatin/taxane set 3 up late |
958 |
0.263 |
0.008 |
6.48E−05 |
| OV1002 carboplatin/taxane set 4 down late |
947 |
0.0288 |
0.0001 |
4.71E−05 |
| HOX424 carboplatin/taxane set 1 up late |
148 |
0.0285 |
0.468 |
0.469 |
| HOX424 carboplatin/taxane set 2 up late |
27 |
0.869 |
0.065 |
0.532 |
| HOX424 carboplatin/taxane set 3 down late |
119 |
0.0398 |
0.0003 |
0.00126 |
| HOX424 carboplatin/taxane set 4 down late |
226 |
0.000463 |
0.00009 |
0.000102 |
| KEGG apoptosis |
88 |
0.00095 |
0.776 |
0.423 |
| KEGG oocyte meiosis |
114 |
0.647 |
0.00654 |
0.0203 |
| KEGG progesterone-mediated oocyte maturation |
87 |
0.886 |
0.0114 |
0.0847 |
| KEGG p53 signalling pathway |
69 |
0.372 |
0.0139 |
6.40E−05 |
| KEGG cell cycle |
128 |
0.0453 |
0.138 |
0.443 |
| OV1002 carboplatin/taxane cell cycle |
51 |
0.0438 |
0.00814 |
0.00301 |
| OV1002 carboplatin/taxane set 1 up early cell cycle |
25 |
0.0272 |
0.00665 |
0.00683 |
| KEGG glutathione metabolism |
50 |
0.0259 |
0.00895 |
0.726 |
| KEGG neurotrophin signalling pathway |
126 |
0.0282 |
0.00133 |
7.54E−06 |
| KEGG β-alanine metabolism | 22 | 0.0488 | 0.0414 | 0.000998 |
Abbreviations: KEGG=Kyoto encyclopedia of genes and genomes; NA=not assigned; OS=overall survival; PFS=progression-free survival. Significant P-values (P<0.05) are shown in bold.
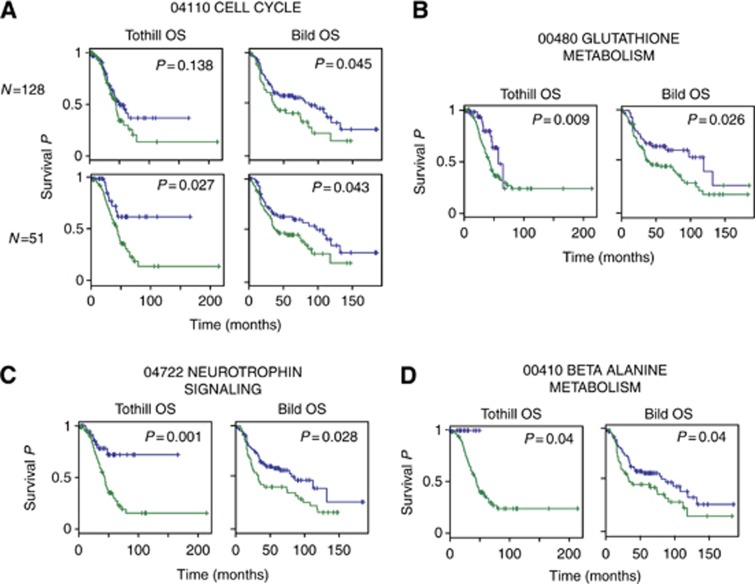
The relationship to clinical outcome was also assessed for overrepresented pathways. Several overrepresented pathways in the overall set of differentially expressed genes (leftmost columns in Table 1) like oocyte meiosis, apoptosis and cell cycle were prognostic of OS in at least one independent data set. Another seven pathways including Wnt signalling pathway, nucleotide excision repair and focal adhesion were significant predictors (P<0.05) of OS in one of the two independent data sets. Glutathione metabolism pathway was a significant predictor of OS in both independent data sets. Although all gene members of the cell cycle pathway were prognostic of OS in one data set, subsets of genes found significant in the early upregulated combined treated OV1002 xenografts were prognostic of OS in both data sets. Two pathways in late downregulated gene sets (rightmost columns in Table 1) were prognostic of OS (Table 2): neurotrophin signalling pathway and β-alanine metabolism (Figure 4).
Figure 4.
Kaplan–Meier distributions of patient groups clustered according to gene set expression. The KEGG pathway-derived gene sets were prognostic of overall survival in two independent clinical studies (Bild et al, 2006; Tothill et al, 2008). Genes from (A) cell cycle, (B) glutathione metabolism, (C) neurotrophin signalling pathway and (D) β-alanine metabolism were overrepresented in the carboplatin–paclitaxel-treated OV1002 xenografts. The complete cell cycle pathway (N=128) gene set was significant in one independent data set, whereas the subset of genes found significant in this study (N=51) was prognostic in both data sets.
Discussion
Despite the progress in identification of genes involved in the development of resistance to platinum- and taxane-based chemotherapeutic agents, the exact timing of the transcriptional response post-treatment warrants further research. Two xenograft models with distinct platinum response profiles were used to characterise drug-induced gene expression patterns. This longitudinal study spanned a 14-day period and allowed the analysis of gene expression in multiple time points. Such insight in dynamic expression changes can indicate not only markers associated with platinum sensitivity or resistance, but also the time they become detectable. Clinical snapshots cannot provide such a dynamic view of the transcriptome and may potentially miss informative changes in expression (Taylor et al, 2010).
The two models displayed distinct transcriptional profiles before and after treatment. Up to 4 biological replicates were used for each time point in this study, with high concordance within each condition. A significant RTV effect was observed in the platinum-responsive, high-grade OV1002 xenografts. Early upregulated genes were involved in DNA repair, cell cycle and apoptosis regulation, suggesting a driving effect on these pathways immediately after treatment administration. DNA damage-induced apoptosis seems to be responsible for the large RTV reduction in OV1002. In carboplatin–paclitaxel-treated cells, a concurrent downregulation of oxygen-consuming metabolic processes was observed, suggesting a reduction in the metabolic rate and a switch to hypoxic processes. The number of genes was increased in late time points and included downregulated apoptosis inhibitors and heat shock proteins. The RTV effect was gradually reduced in late time points after expression of anti-apoptotic regulators such as aurora kinases and possibly because of hypoxic adaptation. Treatment-based upregulation of HIF1α may also be responsible for the gradual RTV reduction observed after day 7.
Carboplatin uptake triggers activation of DNA repair pathways and, in case damage is beyond repair, cell cycle arrest and apoptosis (Galluzzi et al, 2012). In accordance with this, early response in OV1002 involved DNA repair, cell cycle control and apoptosis genes, which were upregulated in both treatments, with many more in the combined. Several gene functions essential for drug chemosensitivity were observed, providing plausible explanations for the response to treatment by OV1002 and the gradual change to chemoresistance. For instance, CHEK1, which activates branches of MAPK pathway, and CHEK2, shown to convey lethal signals in response to carboplatin (Damia et al, 2001; Pabla et al, 2008), were upregulated in treated xenografts. Several PARPs, including PARP2, PARP6, PARP9 and PARP10, were significant after day 4, potentially influencing the growth rate during late time points. Downregulation of ERCC1 has been correlated with survival and responsiveness to platinum treatment in various cancers, including ovarian (Dabholkar et al, 1992). This gene was downregulated in carboplatin-treated samples. Early expression of the tumour suppressor XAF1, which antagonises apoptosis inhibitors (Plenchette et al, 2007), was observed in carboplatin-treated xenografts, whereas pro-apoptotic FAS was upregulated in carboplatin–paclitaxel-treated samples. TRIB3, a pro-apoptotic, negative regulator of AKT1 and NFkB that is involved in hypoxia response (Wennemers et al, 2011), had the largest expression increase upon both treatments. Heat shock proteins protect against platinum drug activity, thus promoting resistance (Ren et al, 2008). Several heat shock proteins, including hsp70s, were found downregulated in carboplatin–paclitaxel-treated OV1002 xenografts in accordance to the chemoresponsiveness of this cell line.
In the slower growing HOX424, which displays reduced platinum sensitivity, the RTV effect was smaller. Transcriptional response to carboplatin was very limited, suggesting that carboplatin is not internalised in sufficient quantities possibly because of an effective transport mechanism potentially driven by P-type ATPases or ABC transporters. Alternatively, HOX424 may be capable of inactivating carboplatin in the cytoplasm by a glutathione pathway-based mechanism. The response to the combined treatment was exclusively late and contained apoptosis and cell cycle regulators. The modest RTV effect may be attributed to the activation of pro-apoptotic genes (Figure 3). Some, such as FOSB, were not observed in OV1002. The MSH2 downregulation may partly explain the lesser RTV effect in HOX424, as mismatch repair pathway underexpression has been associated with carboplatin resistance (Fink et al, 1998). The effect diminished after day 7, possibly because of the presence of efficient inherent oxidative stress adaptations and failure to trigger apoptosis.
Taxanes, such as paclitaxel, act by stabilising microtubules, thereby causing a G2–M arrest followed by apoptosis (Shah and Schwartz, 2001). Gene expression differences between treatments may be attributed to paclitaxel addition, yet this was not translated in a dramatic RTV effect. The RTV reduction in carboplatin–paclitaxel compared with carboplatin was significant only in OV1002 day 7. There was substantial overlap between genes and pathways targeted by carboplatin and paclitaxel; however, several known paclitaxel targets, such as TRAF3 (Blagosklonny et al, 1997), were found in OV1002 and many tubulin isotypes in both xenografts.
Both treatments triggered changes in cell cycle pathway at early and late time points, but the response was more comprehensive in carboplatin–paclitaxel treatment. A larger number of cell cycle control genes were expressed late including cyclins, cyclin-dependent kinases and CDCs, with more expressed after carboplatin–paclitaxel treatment. Many genes affecting G2–M checkpoint were upregulated late after carboplatin–paclitaxel treatment, consistent with the known paclitaxel effect on inhibition of microtubule spindle disassembly.
The two distinct treatment responses of the xenografts provide a measure of interpatient gene expression diversity over time. OV1002 demonstrated the largest RTV reduction and consistently responded to treatment in early time points, whereas HOX424 was growing slower and lacked a sustained RTV reduction effect. The marked variation in the two cell lines may be useful to assess patient responses to new treatment, to investigate mechanisms of drug resistance and possibly to find ways to circumvent it.
Late downregulated genes in both cell lines were associated with prognosis in two clinical data sets. Consistent with their functions, increased expression of SPRY1, CITED2, FZD6 and downregulation of PPFIBP1 were associated with the good prognosis patient clusters. Members of the SPRY family are MAPK inhibitors and RAS antagonists, usually acting as tumour suppressors (Fong et al, 2006; Schaaf et al, 2010). CITED2 interferes with the binding of the transcription factors HIF1α or STAT2 (Yanagie et al, 2009; Yoon et al, 2011), whereas FZD6 is a negative regulator of the canonical Wnt signalling pathway, acting as an inhibitor of oncogenic transformation and cell proliferation (Golan et al, 2004). PPFIBP1 interacts with S100A4, a calcium-binding protein related to tumour invasiveness (Kriajevska et al, 2002). Expression levels of ALDH1A1, ALDH5A1, ALG13, TMEM106B, SPAG16 and N4BP2L1 were also elevated in good prognosis clusters; however, their involvement in tumourigenesis is not established. Interestingly, elevated SPAG16 has been reported in various tumours (Siliņa et al, 2011), whereas N4BP2L1 maps in the region of BRCA2 (13q13.1). Downregulation of these genes coincides with the gradual loss of platinum sensitivity, as seen in the diminishing RTV reduction effect after day 7, and may be driving the switch from a responsive to a nonresponsive phenotype (Table 2). Heatmaps in Supplementary Figure 1 indicate the potential value of these genes as biomarkers of outcome.
Several overrepresented pathways identified from the xenografts were found associated with prognosis in the clinical data sets, including the neurotrophin signalling pathway overrepresented in late downregulated genes from OV1002 carboplatin–paclitaxel-treated xenografts. Neurotrophin factors 4/5 are expressed in breast cancer and have been targeted to inhibit breast tumour growth (Vanhecke et al, 2011). Neurotrophin plays a role in the early development of follicles acting via TRKB receptors (Dorfman et al, 2011), and it is plausible that related pathways become dysregulated during tumourigenesis and potentially have prognostic value.
Although the aim of this study is to characterise the dynamic response to platinum drugs and taxanes, rather than assembling a resistance or survival-associated gene signature, several genes from these clusters are promising leads that warrant further investigation. The discovery of gene sets and pathways derived from the late downregulated clusters that were associated with prognosis has been possible only through a dynamic analysis of gene expression over a period of several days. It would be interesting to explore their expression patterns before and after treatment in patients or in additional cell line xenografts.
In this study we demonstrate for the first time that genes responding in a time-related manner to carboplatin–paclitaxel can predict prognosis in primary tumours treated with these chemotherapeutic drugs. Further clinical assessment in pre- and post-treatment tumours from responding or resistant patients will help determine which of those have the potential to be used as predictors of response.
Acknowledgments
We thank Medical Research Scotland and the Scottish Funding Council for supporting this study. We thank the Wellcome Trust Clinical Research Facility in Edinburgh for processing the Illumina Bead chips. We are grateful to In Hwa Um and Charlene Kay for their help in processing the mRNA and protein samples. This work was supported by Medical Research Scotland (FRG353 to VAS), the FP7-Directorate-General for Research and Innovation of the European Commission (EU HEALTH-F4-2012-305033 to Coordinating Action Systems Medicine to DJH); the Chief Scientist Office of Scotland (to DJH) and the Scottish Funding Council (to DJH and SPL).
Author contributions
AK wrote the manuscript and performed all computational analyses; SPL performed all biological experiments; VAS and DJH conceived the study. All authors read and commented on the manuscript and agreed upon a final version together.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr, Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Giannakakou P, el-Deiry WS, Kingston DG, Higgs PI, Neckers L, Fojo T. Raf-1/bcl-2 phosphorylation: a step from microtubule damage to cell death. Cancer Res. 1997;57:130–135. [PubMed] [Google Scholar]
- Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- Crijns AP, Fehrmann RS, de Jong S, Gerbens F, Meersma GJ, Klip HG, Hollema H, Hofstra RM, te Meerman GJ, de Vries EG, van der Zee AG. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009;6:e24. doi: 10.1371/journal.pmed.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabholkar M, Bostick-Bruton F, Weber C, Bohr VA, Egwuagu C, Reed E. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl Cancer Inst. 1992;84:1512–1517. doi: 10.1093/jnci/84.19.1512. [DOI] [PubMed] [Google Scholar]
- Damia G, Filiberti L, Vikhanskaya F, Carrassa L, Taya Y, D'incalci M, Broggini M. Cisplatinum and taxol induce different patterns of p53 phosphorylation. Neoplasia. 2001;3:10–16. doi: 10.1038/sj.neo.7900122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman MD, Kerr B, Garcia-Rudaz C, Paredes AH, Dissen GA, Ojeda SR. Neurotrophins acting via TRKB receptors activate the JAGGED1-NOTCH2 cell-cell communication pathway to facilitate early ovarian development. Endocrinology. 2011;152:5005–5016. doi: 10.1210/en.2011-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faratian D, Zweemer AJ, Nagumo Y, Sims AH, Muir M, Dodds M, Mullen P, Um I, Kay C, Hasmann M, Harrison DJ, Langdon SP. Trastuzumab and pertuzumab produce changes in morphology and oestrogen receptor signalling in ovarian cancer xenografts revealing new treatment strategies. Clin Cancer Res. 2011;17:4451–4461. doi: 10.1158/1078-0432.CCR-10-2461. [DOI] [PubMed] [Google Scholar]
- Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- Fong CW, Zhua MS, McKie AB, Ling SH, Mason V, Li R, Yusoff P, Lo TL, Leung HY, So SK, Guy GR. Sprouty 2, an inhibitor of mitogen-activated protein kinase signaling, is down-regulated in hepatocellular carcinoma. Cancer Res. 2006;66:2048–2058. doi: 10.1158/0008-5472.CAN-05-1072. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- Golan T, Yaniv A, Bafico A, Liu G, Gazit A. The human Frizzled 6 (HFz6) acts as a negative regulator of the canonical Wnt·β-catenin signaling cascade. J Biol Chem. 2004;279:14879–14888. doi: 10.1074/jbc.M306421200. [DOI] [PubMed] [Google Scholar]
- Helleman J, Jansen MP, Span PN, van Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC, van der Burg ME, Stoter G, Nooter K, Berns EM. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2006;118:1963–1971. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- Helleman J, Smid M, Jansen MP, van der Burg ME, Berns EM. Pathway analysis of gene lists associated with platinum-based chemotherapy resistance in ovarian cancer: the big picture. Gynecol Oncol. 2010;117:170–176. doi: 10.1016/j.ygyno.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- International Collaborative Ovarian Neoplasm Group Paclitaxel plus carboplatin versus standard chemotherapy with either single-agent carboplatin or cyclophosphamide, doxorubicin, and cisplatin in women with ovarian cancer: the ICON3 randomised trial. Lancet. 2002;360:505–515. doi: 10.1016/S0140-6736(02)09738-6. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Cannistra SA, Fountzilas H, Culhane A, Pillay K, Rueda B, Cramer D, Seiden M, Birrer M, Coukos G, Zhang L, Quackenbush J, Spentzos D. Integrated analysis of multiple microarray datasets identifies a reproducible survival predictor in ovarian cancer. PLoS One. 2011;6:e18202. doi: 10.1371/journal.pone.0018202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriajevska M, Fischer-Larsen M, Moertz E, Vorm O, Tulchinsky E, Grigorian M, Ambartsumian N, Lukanidin E. Liprin β1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1) J Biol Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- Li J, Wood WH, 3rd, Becker KG, Weeraratna AT, Morin PJ. Gene expression response to cisplatin treatment in drug-sensitive and drug-resistant ovarian cancer cells. Oncogene. 2007;26:2860–2872. doi: 10.1038/sj.onc.1210086. [DOI] [PubMed] [Google Scholar]
- Liu J, Campen A, Huang S, Peng SB, Ye X, Palakal M, Dunker AK, Xia Y, Li S. Identification of a gene signature in cell cycle pathway for breast cancer prognosis using gene expression profiling data. BMC Med Genomics. 2008;1:39. doi: 10.1186/1755-8794-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–6583. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- Plenchette S, Cheung HH, Fong WG, LaCasse EC, Korneluk RG. The role of XAF1 in cancer. Curr Opin Investig Drugs. 2007;8:469–476. [PubMed] [Google Scholar]
- Ren A, Yan G, You B, Sun J. Down-regulation of mammalian sterile 20-like kinase 1 by heat shock protein 70 mediates cisplatin resistance in prostate cancer cells. Cancer Res. 2008;68:2266–2274. doi: 10.1158/0008-5472.CAN-07-6248. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Hamdi M, Zwijnenburg D, Lakeman A, Geerts D, Versteeg R, Kool M. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumours carrying a mutated RAS gene. Cancer Res. 2010;70:762–771. doi: 10.1158/0008-5472.CAN-09-2532. [DOI] [PubMed] [Google Scholar]
- Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin Cancer Res. 2001;7:2168–2181. [PubMed] [Google Scholar]
- Shahzad MM, Lopez-Berestein G, Sood AK. Novel strategies for reversing platinum resistance. Drug Resist Updat. 2009;12:148–152. doi: 10.1016/j.drup.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Baust CA, Becker KG, Wood Iii WH, Zhang Y, Morin PJ. Gene expression and pathway analysis of ovarian cancer cells selected for resistance to cisplatin, paclitaxel, or doxorubicin. J Ovarian Res. 2011;4:21. doi: 10.1186/1757-2215-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliņa K, Zayakin P, Kalniņa Z, Ivanova L, Meistere I, Endzeliņš E, Ābols A, Stengrēvics A, Leja M, Ducena K, Kozirovskis V, Linē A. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother. 2011;34:28–44. doi: 10.1097/CJI.0b013e3181fb64fa. [DOI] [PubMed] [Google Scholar]
- Sims AH, Zweemer AJM, Nagumo Y, Faratian D, Muir M, Dodd M, Um I, Kay C, Hasman M, Harrison DJ, Langdon SP. Defining the molecular response to trastuzumab, pertuzumab and combination therapy in ovarian cancer. Br J Cancer. 2012;106:1779–1789. doi: 10.1038/bjc.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK.2005Limma: linear models for microarray data Bioinformatics and Computational Biology Solutions Using R and BioconductorRRW Gentleman, V Carey, S Dudoit, R Irizarry, WRW Huber Huber (eds) pp397–420.Springer: New York [Google Scholar]
- Taylor KJ, Sims AH, Liang L, Faratian D, Muir M, Walker G, Kuske B, Dixon JM, Cameron DA, Harrison DJ, Langdon SP. Dynamic changes in gene expression in vivo predict prognosis of tamoxifen-treated patients with breast cancer. Breast Cancer Res. 2010;S12:R39. doi: 10.1186/bcr2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I, Australian Ovarian Cancer Study Group. Gertig D, DeFazio A, Bowtell DD. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Vanhecke E, Adriaenssens E, Verbeke S, Meignan S, Germain E, Berteaux N, Nurcombe V, Le Bourhis X, Hondermarck H. Brain-derived neurotrophic factor and neurotrophin-4/5 are expressed in breast cancer and can be targeted to inhibit tumour cell survival. Clin Cancer Res. 2011;17:1741–1752. doi: 10.1158/1078-0432.CCR-10-1890. [DOI] [PubMed] [Google Scholar]
- Wennemers M, Bussink J, Scheijen B, Nagtegaal ID, van Laarhoven HW, Raleigh JA, Varia MA, Heuvel JJ, Rouschop KM, Sweep FC, Span PN. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 2011;13:R82. doi: 10.1186/bcr2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagie H, Hisa T, Ogata A, Miyazaki A, Nonaka Y, Nishihira T, Osada I, Sairennji T, Sugiyama H, Furuya Y, Kidani Y, Takamoto S, Takahashi H, Eriguchi M. Improvement of sensitivity to platinum compound with siRNA knockdown of upregulated genes in platinum complex-resistant ovarian cancer cells in vitro. Biomed Pharmacother. 2009;63:553–560. doi: 10.1016/j.biopha.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Yoon H, Lim JH, Cho CH, Huang LE, Park JW. CITED2 controls the hypoxic signaling by snatching p300 from the two distinct activation domains of HIF-1α. Biochim Biophys Acta. 2011;1813:2008–2016. doi: 10.1016/j.bbamcr.2011.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.